Abstract
Optimization of primer sequences for polymerase chain reaction (PCR) and quantitative PCR (qPCR) and reaction conditions remains an experimental challenge. We have developed a resource, PrimerBank, which contains primers that can be used for PCR and qPCR under stringent and allele-invariant amplification conditions. A distinguishing feature of PrimerBank is the experimental validation of primer pairs covering most known mouse genes. Here, we describe a major update of PrimerBank that includes the design of new primers covering 17 076 and 18 086 genes for the human and mouse species, respectively. As a result of this update, PrimerBank contains 497 156 primers (an increase of 62% from the previous version) that cover 36 928 human and mouse genes, corresponding to around 94% of all known protein-coding gene sequences. An updated algorithm based on our previous approach was used to design new primers using current genomic information available from the National Center for Biotechnology Information (NCBI). PrimerBank primers work under uniform PCR conditions, and can be used for high-throughput or genome-wide qPCR. Because of their broader linear dynamic range and greater sensitivity, qPCR approaches are used to reanalyze changes in expression suggested by exploratory technologies such as microarrays and RNA-Seq. The primers and all experimental validation data can be freely accessed from the PrimerBank website, http://pga.mgh.harvard.edu/primerbank/.
INTRODUCTION
The quantitative polymerase chain reaction (qPCR) technique, also known as real-time PCR, was developed in the early 1990s and subsequently improved or modified by a large number of groups (1,2). Both probe-(allele-)specific as well as allele-non-specific technologies have been described. Real-time PCR is routinely employed for gene expression quantification, and has been widely used for the confirmation of results from high-throughput experiments such as DNA microarray expression profile studies or transcript abundance measurements by next-generation sequencing. Often the role of qPCR is to validate expression changes of selected genes showing profiles of interest in an initial genome-wide survey (3,4). However, with the appropriate design strategy, qPCR can also be performed in parallel for thousands of genes in a genome-wide fashion. Real-time PCR has several major advantages over DNA microarrays, the most important being a large dynamic range, high sensitivity and high specificity (5,6). Historically, a significant obstacle to genome-wide PCR had been the inability to identify robust oligonucleotide primers that could be used under identical conditions. Multiple design algorithms have been proposed (7–10), but few of them have been validated experimentally. To address the primer design challenge, we developed the PrimerBank database, which contains tens of thousands of computationally designed as well as experimentally validated primers. The primers can be applied either to small-scale experiments focusing on a few genes, or to high-throughput qPCR for thousands of genes. To allow for an efficient high-throughput qPCR process, PrimerBank primers have been designed and validated to perform at an invariant annealing temperature of 60°C. The high annealing temperature helps reduce non-specific amplification. The PrimerBank resource and its experimentally validated 26 855 primer pairs covering most known mouse genes have previously been described (11–13). The design of PrimerBank primers was founded in turn on an algorithm for the design of oligonucleotide probes for microarrays, with additional criteria that are specific to PCR (14). All PCR primers were designed to work with sequence-independent fluorescence detection methods such as SYBR Green-based qPCR (15,16). Thus, the design of target sequence-specific fluorescent probes has not been required, leading to significant savings.
In this work, we describe an updated primer design process for PrimerBank that reflects the maturation of our understanding of the murine and human genomes and reflects further refinements in the design strategy for primer selection. Current genomic annotations from the National Center for Biotechnology Information (NCBI) databases were consolidated as a consequence of the update. A total of 91 502 and 98 854 new primers for humans and mice, respectively, were designed and included in the database. Users can access all stored information (primer sequences and annotations, primer validation data, as well as gene annotations) from the PrimerBank website (http://pga.mgh.harvard.edu/primerbank/). The new primers cover most known human and mouse genes and have been selected from millions of primer candidates using an improved algorithm and current genomic information. Following this update, PrimerBank contains 497 156 primers, covering 94% of all known human and mouse protein-coding genes. All previously designed primers remain in the database, together with experimental validation data. Although several resources have been developed that contain collections of PCR primers for gene expression analysis, only a few thousand of the alternative primer pairs have been demonstrated experimentally (17,18). In addition, the primers have not been designed to work under the same temperature, so that parallel qPCR assays cannot be performed simultaneously. PrimerBank is designed to avoid these liabilities and has a strong predictive record reflected by its high resource utilization and high frequency of repeat users. PrimerBank is a BioDBcore compliant database (Supplementary Information).
UPDATE ON PRIMER DESIGN
Gene sequence selection
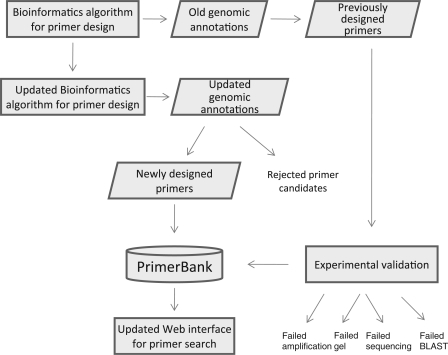
The workflow for the update process is presented in Figure 1. In our previous design, protein-coding sequences were extracted from GenBank and redundant sequences were removed by an automated bioinformatics pipeline prior to primer selection (14). The removal of sequence redundancy was necessary because each human or mouse gene was (and still is) typically represented by many sequences in GenBank. In the updated version, sequence redundancy has been removed by relying on the RefSeq database. In recent years, the NCBI RefSeq project has advanced to the point that RefSeq entries span nearly all the widely accepted human and mouse candidate genes (19). One major goal of the RefSeq database has been to manually curate known gene sequences by organizing them according to genomic locus. From the transcripts found at each locus, representative sequences were selected to uniquely represent the genes. In this way, sequence redundancy has been greatly reduced and confidence in the sequence assignment has been greatly improved. The RefSeq database is now widely accessed to identify known genes, especially those derived from human and murine sources.
Figure 1.
Workflow for the updating process of PrimerBank.
We have adopted RefSeq sequences as our template for the design of gene-specific PCR primers. The reliance on RefSeq only affects the internal primer design process, and has no effect on the user search interface at the PrimerBank website. In humans and mice, alternative splicing is a common phenomenon, leading to multiple distinct transcript isoforms for some genes. In the new design strategy, each primer pair was designed to cover all known isoforms from a given gene. As the first step, the shortest transcript from all isoforms for a gene was selected for primer design. The sequence of the selected transcript was scanned from the 5′- to 3′-end and sequence regions that were not present in all isoforms were excluded from further consideration. In this way, each primer pair was designed to quantify the expression of a gene by profiling all known isoforms.
Primer design process
With the template sequences available, the next step was to apply a stringent bioinformatic algorithm for primer selection. This new algorithm is similar to our previous algorithm (11), with some modifications. The sequence template selection strategy is guided by RefSeq as described above. In addition, the workflow of the design algorithm has been significantly improved, leading to faster performance. In the updated algorithm, the PCR amplicon size has been set to default to a narrower range than in the initial algorithm (100–250 versus 100–350 bp initially). A relatively short amplicon size is important for PCR amplification efficiency, especially if the RNA quality is low (e.g. extracted from archived clinical samples). On the other hand, if the amplicon size is well below 100 bp, it would be a challenge to differentiate specific PCR products from potential non-specific primer dimers when using gel electrophoresis or PCR melting curves for the specificity check. Most selected primer pairs (99.5%) were designed to generate amplicons in the default size range. In cases in which the size requirement could not be satisfied, alternative ranges were adopted for primer selection (60–99 and 251–550 bp).
One important design consideration is the homogeneity of the PCR assays. All the primers have Tm values in the range of 60–63°C as calculated by the nearest neighbor basepair stability method (20). The narrow range of Tm is helpful for homogeneous primer annealing, especially if multiple assays are to be performed in a single plate. In addition, the primer GC content is restricted to 35–65% to ensure uniform primer annealing. The design algorithm also avoids sequence regions with a high potential for secondary structure formation as site inaccessibility has been reported to lead to insufficient primer annealing (11).
The success of qPCR is dependent on the assay specificity, since non-specific PCR amplification results in inaccurate template quantitation. In the design presented here, multiple bioinformatics filters were implemented to select primers with high specificity. To avoid non-specific annealing resulting from low template sequence complexity, the DUST program was used and sequences of low complexity were identified and rejected (21). The 3′-end nucleotides of a primer contribute significantly to non-specific extension of primers, especially if the annealing of these nucleotides is favored (22). The design algorithm evaluates the free energy (ΔG) for the annealing of the last five bases in a primer, and primer candidates with stable 3′-ends (less than −9 kcal/mol) were rejected. For SYBR Green-based qPCR (or other similar quantification methods), one particular challenge is the formation of primer dimers. Previous studies indicate that DNA polymerase extension can be greatly reduced by a single mismatch in the 3′-end of a primer (23,24). To pass the 3′-end criterion, the last four bases in a primer are not allowed to anneal perfectly to itself or to any of the candidate primers in a proposed pair. One important specificity filter is the rejection of primer candidates with a stretch of contiguous bases matched perfectly to other unintended transcript templates. The screening for contiguous base match was done using an efficient computational algorithm described previously (14). To further search for potential primer cross-reactivity, BLAST search against all known sequences in the transcriptome was also performed to reject non-specific primers.
Primer design statistics
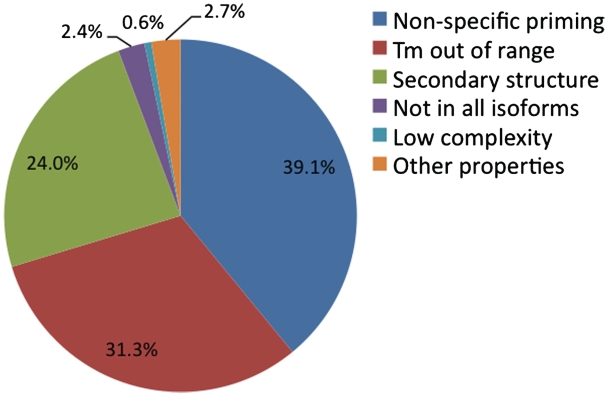
During the primer selection process, 99% of primer candidates were rejected for failing at least one screening filter. The distribution of rejected primer candidates for human genes is presented in Figure 2. The design statistics for mouse primer selection are similar to those for human primers. The specificity filters collectively rejected 39% of all primer candidates. About 31% of the primer candidates were then rejected because they were out of the required Tm range. Another major category of screening failures was generated by secondary structure (sequence complementarity) filters, which collectively rejected 24% of the primer candidates. In addition, other screening filters, such as the requirement to cover all gene isoforms, resulted in the elimination of a significant number of primer candidates.
Figure 2.
Distribution of all rejected human primer candidates by various bioinformatics screening filters. Combined together, 99% of all primer candidates were rejected by these screening filters.
Up to three new primer pairs were designed for each human and mouse gene. These new primers are designated as the version ‘b’ primers, and assigned PrimerBank IDs consisting of the numerical transcript ID followed by the suffix b1, b2 or b3 (see Figure 3 for an example). Among all the genes covered by the new ‘b’ primers, the vast majority (81%) have three primer pairs (Table 1); the rest have one or two ‘b’ primer pairs. In total, 190 356 new primers (including all ‘b1’, ‘b2’ and ‘b3’ primers) were designed and then imported into the PrimerBank database. Previously designed primers (version ‘a’ primers), from the previous version of the database, remain in the updated database with the PrimerBank IDs remain unchanged (each consisting of a numerical transcript ID followed by the suffix a1, a2 or a3). The statistics for all PrimerBank primers, including both newly designed ones and previous ones, are summarized in Table 2. An analysis of current genomic annotations with respect to the primer pairs that had been experimentally validated previously is presented in Table 3.
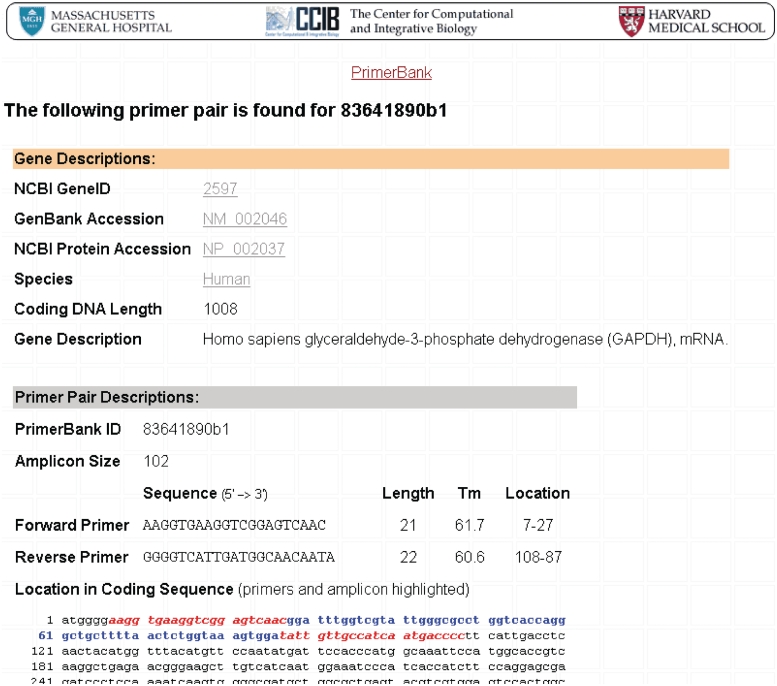
Figure 3.
A screenshot to demonstrate the PrimerBank search result. GAPDH is used here as an example.
Table 1.
Detailed statistics of newly designed human and mouse primers
| Number of new primer pairs | Number of human genes | Number of mouse genes | Number of genes from both species |
|---|---|---|---|
| 1 | 1854 | 1647 | 3501 |
| 2 | 1769 | 1537 | 3306 |
| 3 | 13 453 | 14 902 | 28 355 |
| Total | 17 076 | 18 086 | 35 162 |
Table 2.
Summary of primer statistics for the updated PrimerBank database
| Human | Mouse | Both species | |
|---|---|---|---|
| Number of newly designed primer pairs | 45 751 | 49 427 | 95 178 |
| Number of previous primer pairs | 83 941 | 69 459 | 1 53 400 |
| Total number of primer pairs designed | 1 29 692 | 1 18 886 | 2 48 578 |
| Number of currently annotated protein-coding genes | 18 932 | 20 305 | 39 237 |
| Number of genes represented by new primers | 17 076 | 18 086 | 35 162 |
| Number of genes represented by previous primers | 16 383 | 17 038 | 33 421 |
| Number of genes represented by all primers | 17 973 | 18 955 | 36 928 |
Table 3.
Experimentally validated mouse primers in the context of current genomic annotations
| Number of primer pairs representing current genes | |
|---|---|
| All tested primer pairs | 23 465 |
| Validated primer pairs based on all criteria | 15 473 (66%) |
| Validated Primer pairs based on gel electrophoresis | 19 561 (83%) |
| Validated primer pairs based on sequencing and BLAST | 17 242 (73%) |
| Primer pairs failed qPCR amplification | 1331 (5.7%) |
PRIMERBANK WEBSITE
The allowed database search criteria remain the same as previously, and include the following terms: GenBank accession number, NCBI protein accession number, NCBI gene ID, PrimerBank ID, NCBI gene symbol and gene description (keyword). In addition to the primer pair sequences and corresponding information, the cDNA and amplicon sequences have also been added. These can be viewed by clicking on the ‘cDNA and amplicon sequence’ link. A screenshot of a search result is presented in Figure 3.
All the newly designed PCR primers have been imported into PrimerBank and are accessible via the web search interface. In addition, previously designed version ‘a’ primers continue to be stored in the same database. Thousands of these previous primers have been experimentally validated (12). The validation data are an important consideration for users deciding which primer pairs to use. In addition, an unknown number of previous primers have been used and referenced in what we estimate are hundreds of peer-reviewed publications. Thus the previous primers need to be retained to preserve links that may have been made by prior research efforts presented in the literature. Given that multiple versions of primers are stored in PrimerBank, we have developed the following strategy to prioritize the primer pairs for web presentation: (i) All newly designed ‘b’ primers for the selected genes (up to three primer pairs per gene) are presented; (ii) all experimentally validated version ‘a’ primers for the selected genes, along with the validation data, are presented; and (iii) in most cases, other non-validated version ‘a’ primers are not presented. However, an exception is made when only version ‘a’ primers are available to represent the gene. In addition, another exception is made when searching by PrimerBank ID whereby all version ‘a’ or ‘b’ primers can readily be retrieved. In this way, researchers retain the ability to obtain the information for any primer from any design version when desired.
DISCUSSION
Technological advances in transcriptome analysis via transcriptional microarrays or next-generation sequencing has led to a significantly greater demand for experimental validation of gene expression levels. To this end, real-time PCR (qPCR) is widely considered the gold standard for validation of high-throughput expression data. One major challenge for qPCR is the design of robust PCR primers with high efficiency, specificity and homogeneity when multiple assays are simultaneously performed. To meet these requirements, we have developed a robust bioinformatics process for primer design. The algorithm has been used to design tens of thousands of PCR primers to cover most known human and mouse genes, all of which are freely accessible via the PrimerBank website.
High-throughput qPCR using PrimerBank primers has several advantages. The primers work under the same conditions and thousands have been experimentally validated. The expression profiles of thousands of genes can be determined at the same time, making the primers useful for high-throughput nanoliter-scale qPCR platforms, such as OpenArray from Life Technologies and BioMark from Fluidigm, in which thousands of qPCR assays can be performed in parallel (25,26).
PrimerBank is currently the largest public database for the retrieval of PCR and qPCR primers. In addition, PrimerBank contains thousands of experimentally validated primers, comprising the largest collection of its kind in the public domain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary File S1: List of the database core attributes in accordance with the BioDBcore standards.
FUNDING
National Institutes of Health (U01 HL066678 and R01 GM089784). Funding for open access charge: Center for Computational and Integrative Biology, Massachusetts General Hospital.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 2.Walker NJ. Tech.Sight. A technique whose time has come. Science. 2002;296:557–559. doi: 10.1126/science.296.5567.557. [DOI] [PubMed] [Google Scholar]
- 3.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 4.Arikawa E, Sun Y, Wang J, Zhou Q, Ning B, Dial SL, Guo L, Yang J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 6.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 7.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- 8.Podowski RM, Sonnhammer EL. MEDUSA: large scale automatic selection and visual assessment of PCR primer pairs. Bioinformatics. 2001;17:656–657. doi: 10.1093/bioinformatics/17.7.656. [DOI] [PubMed] [Google Scholar]
- 9.Kim N, Lee C. QPRIMER: a quick web-based application for designing conserved PCR primers from multigenome alignments. Bioinformatics. 2007;23:2331–2333. doi: 10.1093/bioinformatics/btm343. [DOI] [PubMed] [Google Scholar]
- 10.You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 15.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–958. 960, 962. [PubMed] [Google Scholar]
- 16.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–131. doi: 10.2144/97221bi01. 134–138. [DOI] [PubMed] [Google Scholar]
- 17.Lefever S, Vandesompele J, Speleman F, Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009;37:D942–D945. doi: 10.1093/nar/gkn777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput. Appl. Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 22.Rychlik W. Priming efficiency in PCR. Biotechniques. 1995;18:84–86. 88–90. [PubMed] [Google Scholar]
- 23.Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, Sninsky JJ. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang MM, Arnheim N, Goodman MF. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison T, Hurley J, Garcia J, Yoder K, Katz A, Roberts D, Cho J, Kanigan T, Ilyin SE, Horowitz D, et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006;34:e123. doi: 10.1093/nar/gkl639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]