Abstract
It has recently been found that among the 17 Arabidopsis myosins, six (XIC, XIE, XIK, XI-I, MYA1, and MYA2) have a major role in the motility of Golgi bodies and mitochondria in Nicotiana benthamiana and Nicotiana tabacum. Here, the same dominant negative tail fragments were also found to arrest the movement of Gogi bodies when transiently expressed in Arabidopsis plants. However, when a Golgi marker was transiently expressed in plants knocked out in these myosins, its movement was dramatically inhibited only in the xik mutant. In addition, a tail fragment of myosin XIK could inhibit the movement of several post-Golgi organelles, such as the trans-Golgi network, pre-vacuolar compartment, and endosomes, as well as total cytoplasmic streaming, suggesting that myosin XIK is a major player in cytoplasm kinetics. However, no co-localization of myosin tails with the arrested organelles was observed. Several deletion truncations of the myosin XIK tail were generated to corroborate function with localization. All deletion mutants possessing an inhibitory effect on organelle movement exhibited a diffuse cytoplasmic distribution. Point mutations in the tail of myosin XIK revealed that Arg1368 and Arg1443 are essential for its activity. These residues correspond to Lys1706 and Lys1779 from mouse myosin Va, which mediate the inhibitory head–tail interaction in this myosin. Therefore, such an interaction might underlie the dominant negative effect of truncated plant myosin tails and explain the mislocalization with target organelles.
Keywords: Arabidopsis thaliana, Golgi, myosin XIK, Nicotiana benthamiana
Introduction
In the past few years, progress has been made in understanding the roles of myosins in flowering plant cells. It has long been known that cytoplasmic streaming and organellar movement are mediated by the acto-myosin system (Shimmen and Yokota, 2004). Recently it was shown that myosins interact with the endoplasmic reticulum and are involved in its remodelling (Yokota et al., 2008, 2011; Sparkes et al., 2009; Ueda et al., 2010). Their role in cytoplasmic kinetics was further extended when it was shown that moving organelles led to hydrodynamic flow, which accelerates the distribution rate of soluble materials relative to the slow rate of diffusion (Esseling-Ozdoba et al., 2008). The total stiffness of the cytoplasm was shown to depend on the acto-myosin system, which had consequences on cytoplasm organization in plant cells (van der Honing et al., 2010). Plant myosins belong to two main groups of unconventional myosins: myosin XI and myosin VIII, both closely related to myosin V (Berg et al., 2001; Reddy and Day, 2001; Foth et al., 2006). The Arabidopsis myosin family contains 17 members: 13 myosin XI and four myosin VIII (Reddy and Day, 2001; Peremyslov et al., 2011). Recently, our view of the role of myosins in cytoplasmic dynamics was enhanced when the involvement of specific myosins in the movement of specific organelles was determined (Avisar et al., 2008, 2009; Sparkes et al., 2008). While no mutually exclusive connections between specific myosins and organelles were found, based on overexpression of a dominant negative tail construct, a group of six myosins (XIC, XIE, XIK, XI-I, MYA1, and MYA2) were found to be more important for the motility of Golgi bodies and mitochondria in Nicotiana benthamiana and Nicotiana tabacum (Avisar et al., 2008, 2009; Sparkes et al., 2008). Further, analysis of plants knocked out in these myosins revealed that the slowing of organelle movement was corroborated with phenotype. Plants knocked out in myosin xik and mya2 had shorter root hairs, and xik knockout plants showed trichome phenotypes (Ojangu et al., 2007; Peremyslov et al., 2008), suggesting a role for myosins in polar growth. Indeed, double, triple, and quadruple myosin knockout Arabidopsis plants were generated and it was found that the more myosin genes were knocked out, the shorter the plants became (Prokhnevsky et al., 2008; Peremyslov et al., 2010). In addition, knocking down the two myosin XIs in moss resulted in severe loss of cell polarity (Vidali et al., 2010).
Polar growth involves the preferential delivery and accumulation of wall and membrane materials to the correct location in cells via the secretory and endocytic pathways (Samaj et al., 2005). How myosins interact with their cargo organelles and vesicles in plant cells and what regulates this interaction are still not known. One hint emerged when RabD1 and RabC2a were found to interact with the tail of MYA2 in yeast two-hybrid and pull-down assays (Hashimoto et al., 2008). The localization of RabC2a to peroxisomes suggested that these small GTPases can mediate and regulate the interaction of MYA2 and its cargo organelles (Hashimoto et al., 2008). Other accumulated data regarding localization of plant myosins to organelles are confusing: green fluorescent protein (GFP) tail fusions of myosins MYA1, MYA2, XIC, XIE, XI-I, and XIK were able to arrest the movement of the Golgi bodies and mitochondria in tobacco plants (Avisar et al., 2008, 2009; Sparkes et al., 2008), suggesting their involvement in the movement of these organelles; however, no co-localization of these myosin tails and the arrested organelles was observed (Avisar et al., 2008, 2009; Sparkes et al., 2008). It was suggested that either negligible amounts of myosin were localized to the organelles, or the limited amounts of adaptor proteins that mediate the myosin–cargo interaction were sequestered by the overexpressed GFP–tail, which prevented both myosin–organelle co-localization and organellar movement (Avisar et al., 2009). However, in other reports, co-localization of myosin tails and different organelles was observed, but with no interference with movement (Li and Nebenführ, 2007; Reisen and Hanson, 2007). In addition, a fragment of MYA2 from N. tabacum containing amino acids 1007–1512, and a fragment of myosin XI-F from Arabidopsis thaliana containing amino acids 1272–1313 and the corresponding fragment of myosin XI-F from N. tabacum localized to chloroplasts (Natesan et al., 2009; Sattarzadeh et al., 2009). These contradictory observations led to the assumption that correct folding of different fragments of myosin tails is critical for their function and accurate localization in the cell, and that different fragments of the same myosin tail can fold differently and exhibit different performance in different cells.
Here, several of the above-mentioned issues were addressed. Tail fragments of myosins MYA1, MYA2, XIC, XIE, XI-I, and XIK were found to arrest Golgi body movement when transiently expressed in Arabidopsis plants. In addition, XIK tail fragment could inhibit the movement of several post-Golgi organelles such as the trans-Golgi network (TGN), pre-vacuolar compartment (PVC), and endosomes, as well as total cytoplasmic streaming, suggesting that myosin XIK is a major player in cytoplasm kinetics. However, no co-localization between myosin XIK and any of the organelles tested was observed. Several point mutations were introduced into the sequence of XIK based on that of XIG. It was found that two amino acids, Arg1368 and Arg1443, are essential for its activity.
Material and methods
Plants and transient expression
Nicotiana benthamiana plants were grown and infiltrated with Agrobacterium as previously described (Avisar et al., 2009). Arabidopsis thaliana ecotype Colombia, wild type (WT) and knockout lines SALK_019031 (At1g17580; myosin Mya-1), SALK_055785 (At5g43900; myosin Mya-2), GABI_262B03 (At1g08730; myosin XIC), SALK_072023 (At1g54560; myosin XIE), SALK_082443 (At4g33200; myosin XI-I), and SALK_067972 (At5g20490; myosin XIK), were as described (Peremyslov et al., 2008) and were grown accordingly. Transient expression in A. thaliana seedlings was as previously described (Marion et al., 2008).
Plasmids
Arabidopsis thaliana IQ tail constructs have been previously described (Avisar et al., 2009). Tail and short tails were amplified by PCR using an N. benthamiana XIK IQ tail template (Avisar et al., 2008) and the primers listed in Supplementary Table S1 available at JXB online. The amplified fragments were cloned directly into pART27 downstream of enhanced GFP (eGFP), with a 10-alanine linker between the restriction sites KpnI and XbaI. Point mutations in the N. benthamiana XIK tail were inserted by PCR using the primers listed in Supplementary Table S1 and cloned directly into pART27 as above.
The binary expression vectors were transformed into Agrobacterium tumefaciens strain GV3101 after sequence validation of each construct. Plasmids encoding GFP fusions of ARA6, ARA7, Syp21, Syp22, and Syp41 were kindly provided by Dr Takashi Ueda, RIKEN, Japan (Ueda et al., 2001, 2004; Uemura et al., 2004). The plasmid encoding GFP–FYVE was kindly provided by Dr Josef Samaj (Voigt et al., 2005). The plasmid encoding RabA4b was kindly provided by Dr Erik Nielsen from the University of Michigan, USA (Preuss et al., 2004). Golgi markers were from the TAIR collection (Nelson et al., 2007).
Microscopy
An Olympus IX81/FV500 laser-scanning microscope was used to observe fluorescently labelled cells with the following filter sets: for eGFP, 488 nm excitation, and BA505–525 were used; for monomeric red fluorescent prtoein (mRFP), 543 nm excitation and BA610 were used. The objective used was PlanApo 60×/1.00 WLSM /0.17. When eGFP and mRFP were detected in the same sample, dichroic mirror 488/543 was used. In all cases, where more than one colour was monitored, sequential acquisition was performed.
Organelle tracking
Time-lapse images were acquired every 1 s exclusively from cells expressing both myosin and the organelle-specific marker. Organelles were tracked using the Volocity 5.2 module of Improvision, and the velocity was calculated from hundreds of organelles in the presence of each myosin. Size, shape, and intensity parameters were specified so that most organelles would be defined by the software. Tracks combining 12–20 sequential images were calculated by the shortest path model, and mean velocity was calculated for each track. Statistical tests were performed with KaleidaGraph 4.03 (Synergy Software). General linear model analysis was carried out, followed by Scheffe's multiple comparison test. This method was selected due to the uneven number of replicates between the treatments. Statistically significant differences between the treatments were determined at P <0.05.
Western blot analysis
Equal amounts of infiltrated leaf pieces were ground to a fine powder in liquid nitrogen. The powder was boiled in Laemmli protein sample buffer (Laemmli, 1970) for 10 min and then centrifuged for 10 min at 14 000 rpm at room temperature. A 20 μl sample of the extract was separated by SDS–PAGE and blotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore). GFP fusion proteins were detected with anti-GFP antibody (Santa Cruz) and a secondary horseradish peroxidase (HRP)-conjugated antibody (Jacksom ImmunoResearch). For chemiluminescent reaction, the SuperSignal kit (Pierce) was used.
Results
Transient expression of Arabidopsis myosin tails in Arabidopsis leaves
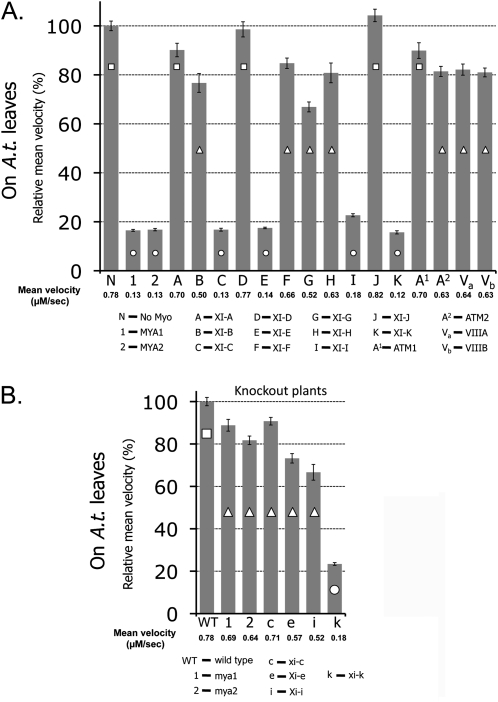
The involvement of all 17 Arabidopsis myosins in organelle movement was previously compared by expressing dominant negative fragments of their tails in leaves of tobacco. Tail fragments of myosins MYA1, MYA2, XIC, XIE, XI-I, and XIK were found to be able to arrest the motility of Golgi bodies and mitochondria in both N. benthamiana and N. tabacum (Avisar et al., 2009). To check whether these results were reproducible in Arabidopsis, a transient expression method was applied according to Marion et al. (2008). Each myosin tail fragment fused to GFP was transiently co-expressed in Arabidopsis seedlings, with an RFP marker for Golgi bodies. Several time-lapse movies were acquired and the velocity of a few hundred Golgi bodies in the presence of each myosin tail fragment was calculated using Volocity software. Fig 1A and Movie S1 at JXB online show that the same group of myosin tails—MYA1, MYA2, XIC, XIE, XI-I, and XIK—were major inhibitors of Golgi body movement in Arabidopsis leaves, allowing only ∼20% of the Golgi body's velocity compared with control cells (see Supplementary Table S2 for statistical analysis). The velocity of Golgi bodies was then checked in plants knocked out for mya1, mya2, xic, xie, xi-i, and xik (Peremyslov et al., 2008). When a Golgi marker was transiently expressed in these plants, its velocity was only dramatically reduced (∼20% of control) in the xik mutant, whereas in the other knockout plants, although significantly different (Supplementary Table S2), the Golgi's velocity remained at 70–85% of control WT plants (Fig. 1B; Supplementary Movie S2). Of note, Golgi movement was tested here in the mya2 SALK_055785 mutant, that exhibits a slightly weaker root hair phenotype compared with the SAIL_632_D12 mutant. This might explain the higher speed of Golgi body movement in the SALK_055785 mutant found here compared with their speed in the SAIL_632_D12 mutant previously described (Peremyslov et al., 2008). Taken together, it may be concluded that myosin XIK is a major player in the movement of Golgi bodies in plant cells.
Fig. 1.
Relative mean velocity of Golgi bodies in Arabidopsis epidermal cells in the presence of tail fragments of all Arabidopsis myosins and in Arabidopsis knockout plants. Actual mean velocity is shown at the bottom of the chart. Time-lapse movies were acquired from at least 10 cotyledon epidermal pavement cells from three different plants, and Golgi velocity was calculated using Volocity software. The graph represents velocity as a percentage of control (velocity of Golgi alone in A or Golgi in WT plants in B). Columns overlaid with different shapes are statistically different at P <0.05 as tested by Scheffe analysis. Standard error bars are shown. (A) GFP fusions of myosin tail fragments were transiently expressed together with an RFP–Golgi marker in Arabidopsis seedlings. (B) An RFP–Golgi marker was transiently expressed in Arabidopsis seedlings of WT or myosin knockout lines.
Myosin XIK is involved in the movement of vesicles from the post-Golgi network
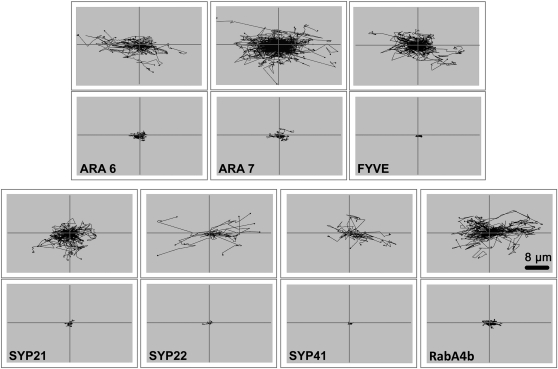
Involvement of the different myosin family members in organelle movement, using the dominant negative tail fragments, had been focused on the Golgi bodies (Avisar et al., 2008, 2009; Prokhnevsky et al., 2008; Sparkes et al., 2008), mitochondria (Avisar et al., 2008, 2009; Prokhnevsky et al., 2008; Sparkes et al., 2008), and peroxisomes (Avisar et al., 2008; Prokhnevsky et al., 2008; Sparkes et al., 2008). Here, the question of whether myosin XIK is also involved in the movement of post-Golgi vesicles, such as the TGN, endosomes, and PVCs, was addressed. The TGN was labelled with Syp41 (Uemura et al., 2004), the PVC with Syp21 and Syp22 (Uemura et al., 2004), endosomes with either ARA6, ARA7 (Ueda et al., 2004), or FYVE (Voigt et al., 2005), and exocytic vesicles with RabA4b (Preuss et al., 2004; Kang et al., 2011). Each marker, fused to GFP or yellow fluorescent protein (YFP), was expressed in leaves of N. benthamiana alone or together with the tail of myosin XIK from N. benthamiana (Avisar et al., 2008). Figure 2, and Supplementary Movie S3 at JXB online summarize the results. In the presence of myosin XIK tail fragment, the motility of each of the tested organelles was significantly reduced. In a recent elegant demonstration, the hydrodynamic flow of the cytoplasm was blocked by 2,3-butanedione monoxime (BDM), a myosin ATPase inhibitor (Esseling-Ozdoba et al., 2008). To check whether the tail of myosin XIK can arrest total cytoplasmic rearrangement, soluble GFP was used to stain the cytoplasm and follow the kinetics of cytoplasm patterns in control plants and in the presence of the dominant negative tail constructs of myosin XIK or MYA2 fused to RFP. To demonstrate cytoplasmic kinetics, time-lapse movies were acquired (represented in Supplementary Movie S4 at JXB online), from which three sequential frames of GFP are shown in Fig. 3 taken at time intervals of 20 s (0, 20, and 40 s) and coloured green, magenta, or blue, respectively: the more severe the inhibition of cytoplasmic dynamics, the more white pixels are expected (where magenta, green, and blue colours co-localize). Whereas in the control cells many coloured (magenta, green, or blue) pixels were observed and only a small number were white, in MYA2-expressing cells, fewer pixels were coloured and more were white. In XIK-expressing cells, almost all of the pixels were white. This supports the suggestion that myosin XIK is a major player in intracellular dynamics.
Fig. 2.
A tail fragment of myosin XIK arrests the motility of post-Golgi vesicles. Markers for endosomes (ARA6, ARA7, and FYVE), PVC (Syp21 and Syp22), TGN (Syp41), and exocytic vesicles/TGN (RabA4b) were transiently expressed in N. benthamiana leaf epidermal cells together with a tail fragment of N. benthamiana myosin XIK. Time-lapse movies were acquired, and velocity and tracks were analysed by Volocity. Black lines (which were artificially centralized) show the total tracks in a representative time-lapse movie. Scale bar=8 μm.
Fig. 3.
Dynamics of cytoplasm stained with soluble GFP in the presence of dominant negative myosin tail constructs fused to RFP. Nicotiana benthamiana leaves were infiltrated with a construct encoding GFP alone or GFP with an RFP fusion of coiled-coil tail domains from MYA2 or XIK (shown in magenta). First, one image was acquired including the two channels to ensure that both colours are present in the same cell (left and middle images) and then a time-lapse movie was acquired using only the GFP channel. Images were acquired every 2 s for 30 frames. Frames 0, 10, and 20 are coloured green, magenta, and blue, respectively and projected on each other (right images). White represents co-localization of the three colours in the same pixel. Scale bar=10 μm.
No co-localization of myosin XIK deletion mutants with specific organelles
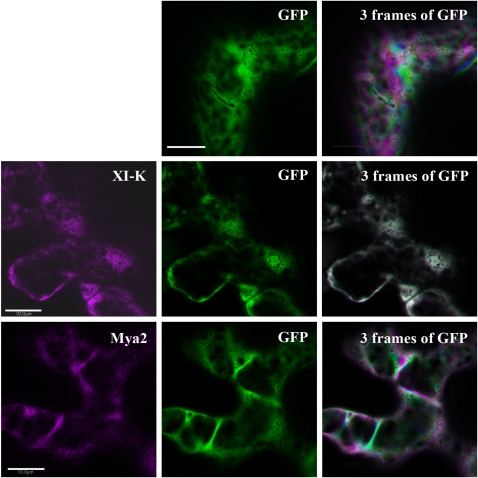
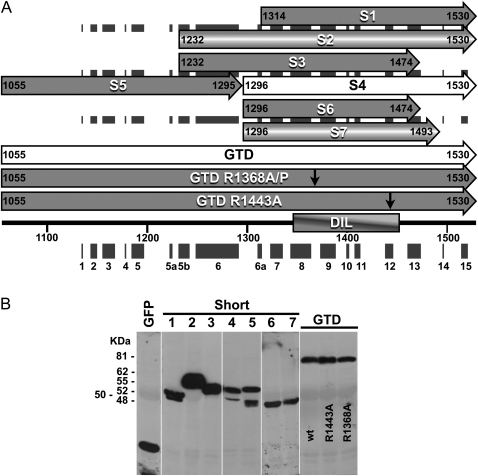
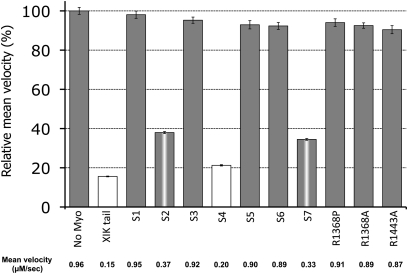
The strong interference by myosin XIK tail fragments with the kinetics of every intracellular marker tested again raised the question of how these fragments function without co-localizing with any of the vesicle or organelle markers. Since various fragments of myosin tails have been reported to localize to different organelles (Li and Nebenführ, 2007; Reisen and Hanson, 2007; Natesan et al., 2009; Sattarzadeh et al., 2009), a series of deletion fragments of the myosin XIK tail were created and localization was correlated with activity. To create a comprehensive series of deletions, the sequence of N. benthamiana myosin XIK was compared with that of yeast myosin V Myo2p, including its calculated 3D construction using the SWISS-MODEL website (http://swissmodel.expast.org) (Arnold et al., 2006) (Fig. 4). The deletions were planned according to a predicted helix conformation (Li and Nebenführ, 2007) in order to increase the chances of correct folding. Figure 4A shows the deletions. The full length of the globular tail domain (GTD) contained amino acids 1055–1530, including helixes 1–15. The following deletions were made: S1 (short1), amino acids 1314–1530, spanning helices 7–15; S2, amino acids 1232–1530, helices 5b–15; S3, amino acids 1232–1474, helices 5b–13; S4, amino acids 1296–1530, helices 6a–15; S5, amino acids 1055–1295, helices 1–6; S6, amino acids 1296–1474, helices 6a–13; and S7, amino acids 1296–1493, helices 6a–13. Each deletion was fused to GFP and its expression was checked by western blot analysis (Fig. 4B). The deletion fragments were transiently expressed in N. benthamiana leaves together with an RFP marker of Golgi bodies. Both the pattern of GFP distribution and the ability to arrest Golgi movement were followed (Fig. 5, Supplementary Movie S5 at JXB online). The shortest fragment that still inhibited Golgi movement was S4, which contained helix 6a–15, corresponding to SDII from yeast Myo2p (Pashkova et al., 2005) and GT2 from MYA1 (Li and Nebenführ, 2007). Its GFP pattern was diffuse in the cytoplasm. All other deletion tail fragments, regardless of their ability to interfere with the Golgi's movement, also exhibited a diffuse cytoplasmic pattern (Supplementary Movie S5). Taken together, the strong inhibitory effect of the tail of myosin XIK and its diffuse distribution in the cytoplasm again raised the question of how it works.
Fig. 4.
Serial deletions and point mutations in the globular tail domain (GTD) of N. benthamiana myosin XIK. (A) Scheme of the deletion mutants (S1–S7). Black boxes show the predicted helixes. Numbers indicate the first and last amino acid in each deletion. The two point mutations (R1368A/P and R1443A) shown by arrows were introduced into fragments spanning the whole GTD. (B) Western blot analysis of all deletion mutants. Proteins were detected using anti-GFP antibodies.
Fig. 5.
Relative mean velocity of Golgi bodies in the presence of different tail fragments and point mutations in the N. benthamiana myosin XIK tail. An RFP marker of the Golgi was transiently expressed in N. benthamiana leaves together with GFP fusions of the different deletions of myosin XIK from N. benthamiana. Time-lapse movies were acquired, and velocity was determined by Volocity. The graph shows relative velocity compared with control (Golgi marker alone). The actual mean velocity is shown at the bottom of the chart. Columns with different colours are significantly different as tested by Scheffe analysis (P <0.05). Standard error bars are shown.
Two amino acids from myosin XIK's tail are essential for its activity
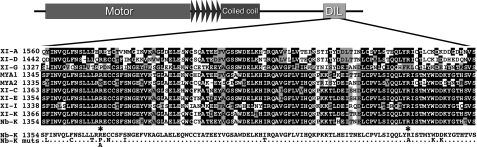
To elucidate further what is unique to the tail of myosin XIK, the amino acid sequences of the tail of myosin XIK from N. benthamiana (Avisar et al., 2008) and of all the inhibitory myosins from Arabidopsis: XIC, E, I, K, MYA1, and MYA2 (Avisar et al., 2009), were compared with those of myosins XI-A, D, and G (Fig. 6). The latter served as representatives of myosins with no effect on organelle movement and were thus termed ‘non-inhibitory’. Several conserved amino acids in the inhibitory myosins were found to be different from the non-inhibitory myosins (marked in Fig. 6). According to these differences, single or multiple mutations were introduced in the tail of myosin XIK from N. benthamiana as described in Fig. 6 and in Supplementary Fig. S1 at JXB online. Tail fragments containing these mutations were expressed in N. benthamiana and their ability to arrest Golgi body motility was tested. After the determination of all mutants shown in Fig. 6 and in Supplementary Fig. S1 it was concluded that a single change of Arg1386 to proline (as in XI-G) or a change of Arg1386 and Arg1443 to alanine could abolish myosin XIK's ability to arrest Golgi movement (Fig. 5, Supplementary Movie S6 at JXB online). Sequence alignment with mouse myosin Va and a further literature survey indicated that the corresponding amino acids (Lys1706 and Lys1779) from myosin Va are those that mediate the tail and head interaction in this myosin (Li et al., 2008). It has been suggested that tail folding and interaction with the head is a regulatory mechanism inhibiting myosin ATPase activity and the function of myosin Va (Li et al., 2008). This suggests that myosin XIK can fold and form a head–tail interaction. Of note, this option was previously proposed for MYA1 (Li and Nebenführ, 2007).
Fig. 6.
Sequence alignment of ‘inhibitory myosins’ and ‘non-inhibitory myosins’. A scheme of the differences in sequence that are conserved among the ‘non-inhibitory myosins’ (XI-A, D, G) relative to the ‘inhibitory myosins’ (MYA1, MYA2, XI-C, E, I, K, and XIK from N. benthamiana). Amino acids that were mutated in the sequence of the N. benthamiana myosin XIK GTD are shown below. Arg1368 and Arg1443, the mutation of which abolished the inhibitory ability of the tail of myosin XIK, are marked by asterisks.
Discussion
Herein, evidence is provided that myosin XIK is somehow different from the other 16 plant myosins in terms of its major role in powering the movements inside plant cells. It is shown that although dominant negative tail fragments of six myosins, MYA1, MYA2, XIC, XIE, XI-I, and XIK, can similarly interfere with organelle movement, when their null mutants are compared, only in xik plants is organelle movement inhibited to the same extent as in the presence of its truncated tail (20% of mean velocity in control WT plants). In all other null plants of mya1, mya2, xi-i, xic, and xie, organelle velocity was 70–90% of that in control WT plants. This suggests that the tail fragments of MYA1, MYA2, XIC, XIE, and XI-I can interfere with the function of myosin XIK; however, this is still to be determined. In addition, it is shown that unlike in the presence of the MYA2 tail, cytoplasmic rearrangement and movement of unknown vesicles freeze in the presence of the tail of myosin XIK, an effect that is similar to that of the actin-disrupting reagent latrunculin B (not shown). The observed superior function of myosin XIK is in agreement with various recent reports. It has been shown that endoplasmic reticulum streaming is only suppressed in myosin xik single mutants, whereas in single mutants of mya1 and mya2 no significant suppression is observed (Ueda et al., 2010). A triple mutant, mya1/mya2/xik, which exhibited smaller rosette leaves and shorter bolts than WT plants, fully recovered upon introduction of myosin XIK (Peremyslov et al., 2010).
Serial deletions of the tail of myosin XIK revealed that the smallest fragment which retains inhibition of Golgi movement is the fragment containing helixes 6a–15. The two subdomains, helixes 1–6 and 6a–15, termed GT1 and GT2, respectively, have been previously described (Li and Nebenführ, 2007), and have been defined by the crystal structure of myosin V Myo2p from yeast (Pashkova et al., 2006) which was compared with that of myosin XI (Li and Nebenführ, 2007). These two subdomains in Myo2p, termed SDI and SDII, were shown to have distinct binding domains: whereas SDI contains a specific domain for binding to the vacuole, SDII binds specifically to secretory vesicles and peroxisomes, and both are functional only when tightly bound to each other (Pashkova et al., 2005; Fagarasanu et al., 2009). Like SDI and SDII, GT1 and GT2 of MYA1 and MYA2 have been shown to interact with each other, although they are able to target organelles separately as shown by fluorescence co-localization (Li and Nebenführ, 2007). In agreement with the latter, fragment S4 of myosin XIK, which corresponds to GT2, was able to arrest Golgi movement by itself, but without fluorescence co-localization. Fragment S5, which corresponds to GT1, was inert to Golgi bodies and its GFP did not specifically localize to detectable organelles. In the present system, tail fragments of myosin XIK from N. benthamiana were transiently expressed in N. benthamiana abaxial epidermal cells. In the study of Li and Nebenführ (2007), the MYA1 and MYA2 tail fragments, GT1 and GT2, were of Arabidopsis origin and were expressed transiently in Arabidopsis leaves. Differential behaviour of GFP fusions of myosin fragments in different systems might result from misfolding. The inhibitory effect of GFP–myosin tail domain fusions on organelle movement without being physically bound to them might be explained by the existence of tail to head binding. The activity of mouse myosin Va has been reported to be regulated by the interaction of its tail and head, preventing it from being constantly active in the cells (Li et al., 2006, 2008). A similar regulatory mechanism has been reported for myosin VIIa (Umeki et al., 2009). Electron microscopy has revealed that myosin V has an extended conformation in the presence of high Ca2+ concentrations, whereas it forms a folded shape in the presence of EGTA, in which the tail domain is folded back toward the head (Wang et al., 2004). In that model, myosin V in the inhibited state is in a folded conformation such that the tail domain interacts with the head domain, inhibiting its ATPase and actin-binding activities. Cargo binding, high Ca2+, and/or phosphorylation may reduce the interaction between the head and tail domains, thus restoring its activity (Li et al., 2006). Lys1706 and Lys1779 were found to be the tail amino acids mediating this interaction in myosin Va (Li et al., 2008). The two amino acids from myosin XIK which were found to be essential for its ability to inhibit Golgi motility, Arg1386 and Arg1443, correspond to amino acids Lys1706 and Lys1779 from myosin Va (Li et al., 2008). Therefore, tail fragments of myosin XIK which inhibit organelle movement might do so by binding to endogenous head domains, leading to inhibition of their ATPase activity and actin binding. This might explain the diffuse cytoplasmic pattern of GFP–tail fusions, instead of being localized to the arrested organelles and vesicles. Whether the myosin head–tail interaction actually occurs in plant cells, and whether it occurs only in myosin XIK or with other plant myosins as well, and whether reciprocal interactions and Ca2+ regulation might exist, all remain to be determined. Curiously, myosin XI-A and XI-D tails, which were found to be inert to organelle movement, have arginine in positions corresponding to myosin XIK's Arg1368 and Arg1443. This might indicate that the context of the flanking amino acids is of functional importance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Golgi movement in the presence of the tail of myosin XIK with point mutations. (A) Relative mean velocity of Golgi bodies in the presence of the different mutations. (B) Schematic presentation of the mutations introduced. Mutants A, B, and C contain the mutations underlined. P1368R/mutA is mutant A in which only the proline was replaced by the original arginine.
Table S1. Primers used in this study.
Table S2. Scheffe statistical analysis of Golgi movement in the presence of myosin tail fragments.
Movie S1. Golgi movement in Arabidopsis epidermal cells in the presence of A. thaliana myosin tail fragments.
Movie S2. Golgi movement in epidermal cells of Arabidopsis mutant plants.
Movie S3. Movement of post-Golgi organelles in the presence of the myosin XIK tail in N. benthamiana leaf abaxial epidermal cells.
Movie S4. Cytoplasm dynamics (stained with soluble GFP) in the presence of tail fragments of MYA2 or myosin XIK fused to mCherry (RFP). Images acquired from N. benthamiana leaf abaxial epidermal cells.
Movie S5. Movement of the Golgi in the presence of tail deletions of myosin XIK in N. benthamiana leaf abaxial epidermal cells. Shown is a Golgi on the background of diffused GFP–tail distribution or Golgi alone.
Movie S6. Movement of the Golgi in the presence of a point mutation-containing tail of myosin XIK in N. benthamiana leaf abaxial epidermal cells. Shown is a Golgi on the background of diffused GFP–tail distribution or Golgi alone.
Acknowledgments
This work was supported by a research grant from the Israeli Science Foundation (ISF) 401/09 and research grant no. IS-4038-07 from BARD, the United States–Israel Binational Agricultural Research and Development Fund. The authors wish to thank V. Dolja for the myosin mutants.
References
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiology. 2009;150:700–709. doi: 10.1104/pp.109.136853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiology. 2008;146:1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Molecular Biology of the Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling-Ozdoba A, Houtman D, van Lammeren AAM, Eiser E, Emons AM. Hydrodynamic flow in the cytoplasm of plant cells. Journal of Microscopy. 2008;231:274–283. doi: 10.1111/j.1365-2818.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Mast FD, Knoblach B, et al. Myosin-driven peroxisome partitioning in S. cerevisiae. Journal of Cell Biology. 2009;186:541–554. doi: 10.1083/jcb.200904050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proceedings of the National Academy of Sciences, USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Igarashi H, Mano S, Takenaka C, Shiina T, Yamaguchi M, Demura T, Nishimura M, Shimmen T, Yokota E. An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. Journal of Experimental Botany. 2008;59:3523–3531. doi: 10.1093/jxb/ern202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA. Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans Golgi network compartments in Arabidopsis. Traffic. 2011;12:313–329. doi: 10.1111/j.1600-0854.2010.01146.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A. Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. Journal of Biological Chemistry. 2007;282:20593–20602. doi: 10.1074/jbc.M700645200. [DOI] [PubMed] [Google Scholar]
- Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. Journal of Biological Chemistry. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- Li XD, Jung HS, Wang Q, Ikebe R, Craig R, Ikebe M. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proceedings of the National Academy of Sciences, USA. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. The Plant Journal. 2008;56:169–179. doi: 10.1111/j.1365-313X.2008.03596.x. [DOI] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. Myosin XI is required for actin-associated movement of plastid stromules. Molecular Plant. 2009;2:1262–1272. doi: 10.1093/mp/ssp078. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Jarve K, Paves H, Truve E. Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma. 2007;230:193–202. doi: 10.1007/s00709-006-0233-8. [DOI] [PubMed] [Google Scholar]
- Pashkova N, Catlett NL, Novak JL, Wu G, Lu R, Cohen RE, Weisman LS. Myosin V attachment to cargo requires the tight association of two functional subdomains. Journal of Cell Biology. 2005;168:359–364. doi: 10.1083/jcb.200407146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO Journal. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Mockler TC, Filichkin SA, Fox SE, Jaiswal P, Makarova KS, Koonin EV, Dolja VV. Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiology. 2011;155:1191–1204. doi: 10.1104/pp.110.170720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiology. 2008;146:1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Dolja VV. Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. The Plant Cell. 2010;22:1881–1897. doi: 10.1105/tpc.110.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E. The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. The Plant Cell. 2004;16:1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proceedings of the National Academy of Sciences, USA. 2008;105:19744–19749. doi: 10.1073/pnas.0810730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biology. 2001;2:1–17. doi: 10.1186/gb-2001-2-7-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen D, Hanson MR. Association of six YFP–myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biology. 2007;7:6. doi: 10.1186/1471-2229-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Read ND, Volkmann D, Menzel D, Baluska F. The endocytic network in plants. Trends in Cell Biology. 2005;15:425–433. doi: 10.1016/j.tcb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Krahmer J, Germain AD, Hanson MR. A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Molecular Plant. 2009;2:1351–1358. doi: 10.1093/mp/ssp094. [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yokota E. Cytoplasmic streaming in plants. Current Opinion in Cell Biology. 2004;16:68–72. doi: 10.1016/j.ceb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L. Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell. 2009;21:3937–3949. doi: 10.1105/tpc.109.072249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Teanby NA, Hawes C. Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. Journal of Experimental Botany. 2008;59:2499–2512. doi: 10.1093/jxb/ern114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proceedings of the National Academy of Sciences, USA. 2010;107:6894–6899. doi: 10.1073/pnas.0911482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A. Functional differentiation of endosomes in Arabidopsis cells. The Plant Journal. 2004;40:783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO Journal. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Structure and Function. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- Umeki N, Jung HS, Watanabe S, Sakai T, Li XD, Ikebe R, Craig R, Ikebe M. The tail binds to the head–neck domain, inhibiting ATPase activity of myosin VIIA. Proceedings of the National Academy of Sciences, USA. 2009;106:8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing HS, de Ruijter NC, Emons AM, Ketelaar T. Actin and myosin regulate cytoplasm stiffness in plant cells: a study using optical tweezers. New Phytologist. 2010;185:90–102. doi: 10.1111/j.1469-8137.2009.03017.x. [DOI] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tuzel E, Bezanilla M. Myosin XI is essential for tip growth in Physcomitrella patens. The Plant Cell. 2010;22:1868–1882. doi: 10.1105/tpc.109.073288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Samaj J, et al. Actin-based motility of endosomes is linked to the polar tip growth of root hairs. European Journal of Cell Biology. 2005;84:609–621. doi: 10.1016/j.ejcb.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Wang F, Thirumurugan K, Stafford WF, Hammer JA, 3rd, Knight PJ, Sellers JR. Regulated conformation of myosin V. Journal of Biological Chemistry. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- Yokota E, Ueda H, Hashimoto K, Orii H, Shimada T, Hara-Nishimura I, Shimmen T. Myosin XI-dependent formation of tubular structures from endoplasmic reticulum isolated from tobacco cultured BY-2 cells. Plant Physiology. 2011;156:129–143. doi: 10.1104/pp.111.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E, Ueda S, Tamura K, Orii H, Uchi S, Sonobe S, Hara-Nishimura I, Shimmen T. An isoform of myosin XI is responsible for the translocation of endoplasmic reticulum in tobacco cultured BY-2 cells. Journal of Experimental Botany. 2008;60:197–212. doi: 10.1093/jxb/ern280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.