Abstract
DNA glycosylases specialized for the repair of alkylation damage must identify, with fine specificity, a diverse array of subtle modifications within DNA. The current mechanism involves damage sensing through interrogation of the DNA duplex, followed by more specific recognition of the target base inside the active site pocket. To better understand the physical basis for alkylpurine detection, we determined the crystal structure of Schizosaccharomyces pombe Mag1 (spMag1) in complex with DNA and performed a mutational analysis of spMag1 and the close homologue from Saccharomyces cerevisiae (scMag). Despite strong homology, spMag1 and scMag differ in substrate specificity and cellular alkylation sensitivity, although the enzymological basis for their functional differences is unknown. We show that Mag preference for 1,N6-ethenoadenine (εA) is influenced by a minor groove-interrogating residue more than the composition of the nucleobase-binding pocket. Exchanging this residue between Mag proteins swapped their εA activities, providing evidence that residues outside the extrahelical base-binding pocket have a role in identification of a particular modification in addition to sensing damage.
Keywords: base excision repair, DNA glycosylase, ethenoadenine, 3-methyladenine, protein–DNA interactions
Introduction
DNA is susceptible to alkylation damage from environmental toxins and from endogenous lipid peroxidation products and methyl donors (Friedberg et al, 2006). These agents produce a chemically diverse array of detrimental alkylated nucleobases that threaten genome integrity by causing mutations, DNA replication arrest and single- and double-strand breaks (Barnes & Lindahl, 2004). The toxic effects of alkylating agents are the rationale for their use in cancer chemotherapy, whereas the mutagenic potential of DNA alkylation damage leads to genomic instability and increases cancer risk. Alkylated DNA bases account for ∼23% of nucleobase damage in the genome (Friedberg et al, 2006), and have been detected in humans and rats after exposure to various carcinogens (Shuker et al, 1987; Holt et al, 1998).
A large number of toxic and mutagenic alkylpurines, including 3N-methyladenine (3mA), N7-methylguanine (7mG) and 1,N6-ethenoadenine (εA; Fig 1), are eliminated by DNA glycosylases, which initiate the base excision repair pathway by locating the modified bases and catalysing the hydrolysis of the N-glycosidic bond. DNA glycosylases specialized for alkylpurine lesions are found in all organisms and show an exceptionally broad substrate range. For example, in Escherichia coli, the constitutively active Tag (3-methyladenine DNA glycosylase I) enzyme is highly specific for cytotoxic 3mA lesions, whereas alkylation damage-inducible AlkA (E. coli 3-methyladenine DNA glycosylase II) recognizes a wide range of mutagenic substrates, including εA (reviewed in Rubinson et al, 2010a). Similarly, the human alkyladenine DNA glycosylase enzyme is a functional counterpart to AlkA and shows a robust activity towards etheno and oxidized DNA adducts (Saparbaev et al, 1995). Structural studies have illustrated how these enzymes use a common base-flipping mechanism to gain access to the lesion inside an active-site pocket on the surface of the enzyme (Rubinson et al, 2010a). In all cases, the extrahelical DNA conformation is stabilized by surface residues that intercalate into the DNA base stack and plug the gap left by the flipped base.
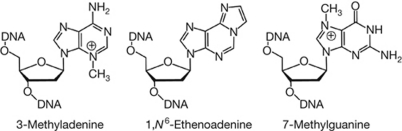
Figure 1.
Structures of alkylated bases relevant to this study.
The molecular basis for alkylpurine discrimination remains poorly understood, but is believed to be a consequence of shape and chemical complementarity between the extrahelical nucleobase substrate and the active-site pocket (Lau et al, 2000; Eichman et al, 2003; Metz et al, 2007). Recent work, however, has shown that some DNA glycosylases use the DNA plug residues as damage sensors by interrogating undamaged DNA before base flipping (Banerjee et al, 2006; Qi et al, 2009), suggesting that these interrogating residues might also be important for selection of a particular substrate. In addition, the specific catalytic mechanism of base excision and the thermodynamic stability of the lesion have been shown to influence the choice of substrates (Parikh et al, 2000; Stivers, 2004; Rubinson et al, 2010a).
In an attempt to understand the molecular basis for the selection of alkylation damage in particular, we carried out a structure–function analysis of two closely related yeast alkylpurine DNA glycosylases. Despite their extensive sequence homology, scMag (Mag from Saccharomyces cerevisiae) and spMag1 (Mag1 from Schizosaccharomyces pombe) have different DNA repair phenotypes and substrate preferences analogous to AlkA and Tag, respectively. Similarly to AlkA, scMag is induced by exposure to DNA-damaging agents, shows a strong mutator phenotype and excises a broad spectrum of alkylpurines, including εA (Chen et al, 1990; Chen & Samson, 1991; Saparbaev et al, 1995; Lingaraju et al, 2008). SpMag1, on the other hand, is constitutively expressed, has a much weaker mutator phenotype and has a restricted substrate preference (Memisoglu & Samson, 1996, 2000b). Specifically, spMag1 has been reported to lack εA excision activity (Alseth et al, 2005).
Here, we report the crystal structure of spMag1 bound to DNA, together with a mutational analysis of εA and 7mG excision, which enabled identification of the residues responsible for the yeast Mag substrate specificity differences. SpMag1 contains a unique histidine that contacts the minor groove outside the nucleobase-binding pocket. Substitution of this histidine with the corresponding serine residue in other Mag homologues resulted in an exchange of their relative εA activities, while not severely affecting 7mG activity. Surprisingly, mutation of residues in the extrahelical nucleobase-binding pockets had no effect on substrate specificity, challenging the previous notion that substrate recognition is based solely on steric exclusion of a lesion from the active-site pocket. These data provide evidence for how DNA glycosylases discriminate among different types of damage outside the active site, and suggest that alkylpurine selection might begin before base flipping.
Results and Discussion
Structure of the Mag1–DNA complex
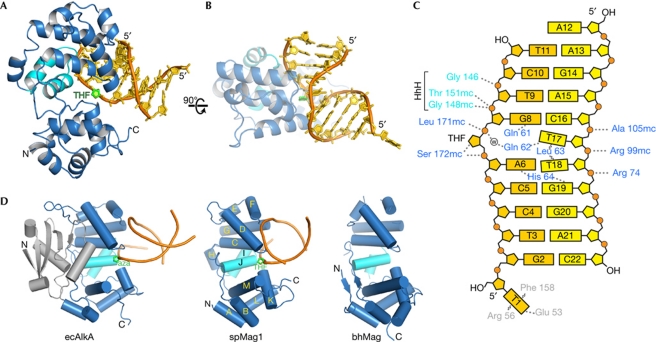
S. pombe Mag1 was crystallized in complex with DNA containing a tetrahydrofuran (THF) abasic site analogue, and the structure determined by single-wavelength anomalous dispersion (SAD) from selenomethionine (SeMet)-substituted spMag1–DNA crystals (supplementary Fig S1 online). The resulting crystallographic model consisting of two spMag1–DNA complexes in the asymmetric unit was refined against 2.2 Å native diffraction data (supplementary Table S1 online) to a crystallographic residual of 18.5% (Rfree=22.5%). The overall structure and DNA-binding mode of spMag1 is consistent with the helix–hairpin–helix (HhH) superfamily of DNA glycosylases (Huffman et al, 2005; Rubinson et al, 2010a). Two α-helical subdomains combine to form the extrahelical nucleobase-binding cleft at their interface (Fig 2). The DNA is anchored to the protein from the minor groove side primarily through electrostatic interactions between the HhH domain (helices αC–αJ) and the phosphate backbones of both strands (Fig 2A,B). The HhH motif (helices αI–αJ) binds to the phosphate backbone of the damaged strand immediately downstream from the THF abasic site (supplementary Fig S2 online), whereas helices αF–αG engage the strand opposite to the lesion (Fig 2C). The damaged strand is buried in the cleft between the two domains with the THF abasic site fully rotated 180° around the backbone into the extrahelical base-binding pocket. Importantly, the αC–αD loop intercalates into the duplex at the damage site, resulting in a 70° kink in the DNA. The arms of the duplex are primarily B-form DNA and are swung away from the protein towards the major groove (Fig 2B).
Figure 2.
The spMag1–DNA complex crystal structure. (A,B) Orthogonal views of spMag1 (blue ribbons) bound to DNA (gold) containing a tetrahydrofuran (THF) abasic site analogue (green). The helix–hairpin–helix (HhH) motif is light blue. (C) Schematic of spMag1–DNA interactions, with protein residues in blue, THF–DNA strand in gold, undamaged DNA strand in yellow and phosphates depicted as orange circles. Dotted and wavy lines represent hydrogen bonds and van der Waals interactions, respectively. (D) Structural alignment of 3-methyladenine DNA glycosylases: E. coli AlkA bound to 1-azaribose (aza) DNA (ecAlkA, PDB ID 1DIZ), S. pombe Mag1 and B. halodurans Mag (bhMag, PDB ID 2H56).
The DNA-binding mode of spMag1 is similar to that observed in the structure of AlkA bound to DNA containing a 1-azaribose transition state analogue (Fig 2D; supplementary Information online; Hollis et al, 2000). Despite the lack of the N-terminal β-sheet extension present in AlkA, the spMag1 structure is highly similar to AlkA residues 89–282 with an r.m.s.d. of 1.45 Å for main-chain atoms (supplementary Figs S3 and S4 online). Superposition of the two proteins results in a remarkable agreement in positions of the damaged DNA strands and the 1-azaribose and THF moieties. The only noticeable difference in the two DNA complexes is the trajectory of the duplex arms (supplementary Fig S4 online). With the structures of spMag1 and AlkA in hand, we were able to pinpoint putative active site and DNA-binding residues in both spMag1 and scMag for the purpose of explaining substrate specificity differences in the yeast proteins.
Base excision activity
A DALI search against the Protein Data Bank showed spMag1 to be most similar to an unpublished structure of a putative Mag orthologue from Bacillus halodurans (bhMag; PDB ID 2H56). SpMag1 and bhMag superimpose with an r.m.s.d. of 1.42 Å for main-chain atoms and share 27% sequence identity and 65% overall similarity. The high sequence and structural similarity among spMag1, scMag and bhMag (supplementary Table S2 online) prompted us to compare their base excision activities to understand Mag functional differences.
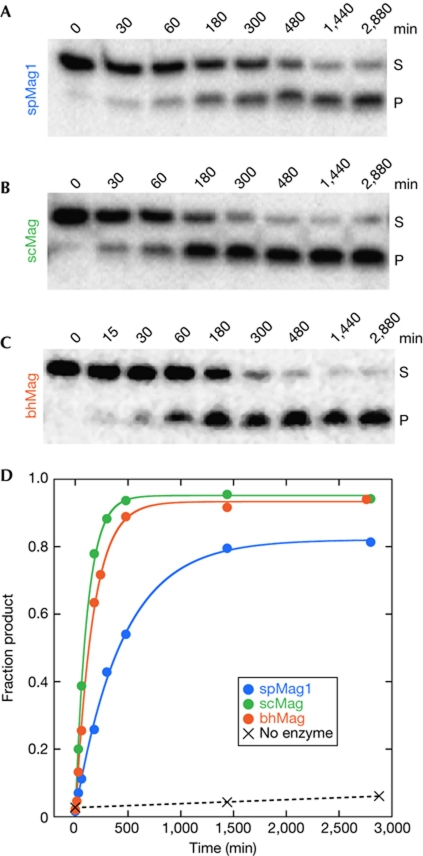
We measured the single-turnover kinetics of εA and 7mG excision from oligonucleotides containing a single lesion. Under the conditions of our assay, all three enzymes removed 7mG at equal rates (supplementary Table S3 online), whereas their activities towards εA differed (Fig 3). Contrary to a previous report (Alseth et al, 2005), we found that spMag1 can indeed excise εA at a low level of activity (Fig 3A). This discrepancy is most probably due to the specific conditions or DNA sequence used to test activity. Under the same conditions, scMag removed εA at a rate two- to threefold greater than spMag1 (Fig 3B)—a modest but significant difference (supplementary Table S3 online). The rate constant of (12.8±1.7) × 10−5 s−1 for the scMag/εA–DNA reaction is consistent with values previously reported for scMag and AlkA (O’Brien & Ellenberger, 2004; Lingaraju et al, 2008). Surprisingly, bhMag excised εA at a rate comparable to scMag despite its stronger similarity to spMag1 (Fig 3C,D; supplementary Table S2 online).
Figure 3.
1,N6-ethenoadenine (εA) excision activity of Mag orthologues. (A–C) Denaturing polyacrylamide gels showing the disappearance of εA-containing DNA substrate (S) and appearance of alkaline-cleaved abasic-DNA product (P) as a function of time after addition of spMag1 (A), scMag (B) and bhMag (C). (D) Quantification of the panels shown in A–C. Blue, spMag1; green, scMag; orange, bhMag. Rate constants calculated from the single-exponential fits to the data are shown in supplementary Table S3 online.
The nucleobase-binding pocket
To reconcile the low εA excision activity of spMag1 relative to its close homologues, we first compared the structural details of the base-binding pockets as preference for a particular substrate is determined in large part by its fit within the active site (Eichman et al, 2003). Consistent with our biochemical results, the spMag1-binding pocket is comparable in size to that of AlkA and can easily accommodate an εA base (Fig 4A,D). In fact, although we crystallized Mag1 in complex with an abasic site, we observed a nucleobase in this pocket from insertion of the 5′-terminal thymine base from an adjacent DNA molecule in the crystal through the large opening at the rear of the active site (supplementary Fig S5 online). This is a fortuitous lattice contact probably irrelevant to spMag1 function, given that spMag1 does not have a binding preference for 5′ overhangs (supplementary Table S4 online) and that this opening is normally occluded by the β-sheet domain in AlkA and presumably scMag. Nevertheless, it illustrates that the low activity towards εA is not a result of steric exclusion from the spMag1 active site.
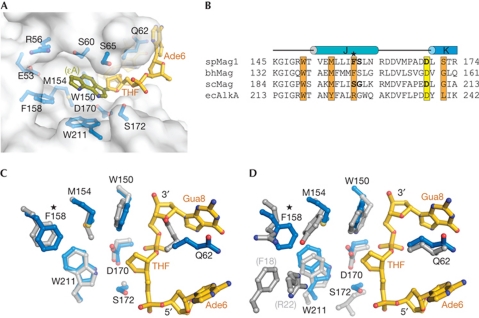
Figure 4.
SpMag1 nucleobase-binding pocket. (A) The base-binding pocket is shown as a transparent van der Waals surface. Protein side chains likely to contact an extrahelical base are shown in blue and the tetrahydrofuran (THF) abasic site and the two flanking nucleotides are shown in gold. The 1,N6-ethenoadenine (εA) base (olive) is modelled within the pocket from coordinates of εA–DNA bound to human alkyladenine DNA glycosylase (AAG; PDB ID 1F4R). (B) Sequence alignment of spMag1, bhMag and ecAlkA structures with scMag sequence. Residues predicted to contact the substrate base are highlighted orange, and the conserved helix–hairpin–helix (HhH) aspartic acid is yellow. The nonconserved Phe 158 is marked with a star. (C,D) Superposition of spMag1 (blue side chains, gold DNA) onto bhMag (C) and ecAlkA (D, grey).
There is remarkable agreement between the base-binding residues in the Mag enzymes and AlkA (Fig 4). Phe 158 in spMag1 is the only nonconserved residue predicted to contact the extrahelical nucleobase, and is the most significant difference between spMag1 and scMag sequences (Fig 4B) and the AlkA active site (Fig 4D). We therefore tested the contribution of Phe 158 and the spatially adjacent Ser 159 to εA excision activity by swapping the corresponding residues between spMag1 and scMag. Neither spMag1 F158S,S159G nor scMag S197F,G198S double mutant affected the εA or 7mG excision activity relative to wild type (supplementary Table S3 online; Fig 4D). Thus, the substrate specificity differences between spMag1 and scMag cannot be explained by the differences in residues contacting the extrahelical base.
In addition to steric exclusion, greater specificity towards 3mA and 7mG lesions might be influenced by the relatively weak catalytic power of some alkylpurine DNA glycosylases, as these bases have destabilized N-glycosidic bonds as a result of their formal positive charges, and therefore require minimal rate enhancement for removal over their spontaneous rate of depurination (Stivers & Jiang, 2003; Rubinson et al, 2010b). Most monofunctional DNA glycosylases contain a catalytically essential, conserved aspartate at the mouth of the nucleobase-binding pocket (Labahn et al, 1996). Whereas replacement of scMag Asp 209 with asparagine reduced activity towards both substrates to less than 1% of the wild-type enzyme, spMag1 D170N retained 20% and 5% activity towards εA and 7mG, respectively (supplementary Table S3 online). Interestingly, this residual activity in the spMag1 aspartate mutant was also observed in 3mA-specific MagIII (Eichman et al, 2003). The weaker catalytic potential of Asp 170 in spMag1 could potentially be influenced by a polar interaction with Ser 172, which is a glycine in scMag and bhMag (Fig 4B,C). Substitution of spMag1 Ser 172 to glycine decreased εA activity tenfold, whereas the corresponding Gly → Ser substitution in scMag did not have a significant effect (supplementary Table S3 online). Taken together, these data indicate that spMag1 and scMag might have subtle mechanistic differences in catalytic potential that could affect their ability to excise more stable alkylpurines.
Minor groove interactions influence substrate specificity
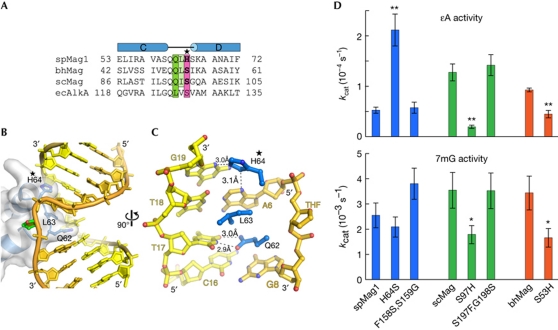
Outside the base-binding pocket, side chains that intercalate into the DNA base stack serve two functions—to interrogate the DNA duplex before base flipping and to stabilize the extrahelical base after flipping—and are thus essential for glycosylase activity (Stivers, 2004; Banerjee et al, 2006; Qi et al, 2009). In the HhH glycosylases, these interrogating residues are located at the tip of the αC–αD loop (Fig 5A,B). In spMag1, the extruded damaged strand and large kink in the duplex is stabilized by Gln 62, which plugs the gap in the damaged strand, and Leu 63, which wedges itself between the bases opposite the lesion (Fig 5C). Both of these residues are conserved in scMag and bhMag (Fig 5A). Replacement of spMag1 Gln 62 and Leu 63 with alanine resulted in an 80–90% and <99% decrease, respectively, in base excision activity for both εA and 7mG (supplementary Table S3 online), consistent with their importance to base excision.
Figure 5.
DNA interrogation by spMag1. (A) Structure-based sequence alignment of the αC–αD loops from Mag and AlkA enzymes. Plug/wedge residues are highlighted green/light green, and the new His 64 contact in spMag1 is magenta. His64 is labeled with a star in panels A, B, and C. (B) The spMag1 αC–αD loop is shown in blue within a transparent molecular surface and intercalating residues are rendered as sticks. The damaged DNA strand is coloured gold and tetrahydrofuran (THF) green. (C) Close-up of spMag1–DNA contacts at the lesion. (D) Single-turnover rates (kcat) of 1,N6-ethenoadenine (εA; top) and 7mG (N7-methylguanine; bottom) excision are plotted for wild-type and mutant spMag1 (blue), scMag (green) and bhMag (orange). Values are shown in supplementary Table S3 online and raw data are shown in supplementary Fig S6 online. Error bars represent the standard deviation (n=3). Asterisks denote P<0.05 (*) and P<0.002 (**).
SpMag1 contains an extra minor groove interaction adjacent to the plug and wedge residues that is not observed in other alkylpurine DNA glycosylases. The imidazole ring of His 64 is positioned to form a hydrogen bond with either the N3 nitrogen of the adenine immediately 5′ to the lesion or the N3 nitrogen of Gua 19 on the opposite strand, depending on the histidine conformer (Fig 5C). This residue is not conserved among the other Mag enzymes or AlkA, which all have a serine in the same position (Fig 5A). In an attempt to alter Mag specificity for εA, we changed spMag1 His 64 to serine, and scMag Ser 97 and bhMag Ser 53 to histidine and measured their activities towards both εA and 7mG (supplementary Table S3 and Fig S6 online). Interestingly, the spMag1 H64S mutation increased the εA excision activity threefold relative to the wild type, bringing the activity up to a level similar to scMag, but did not affect 7mG activity (Fig 5D). In contrast, scMag S97H and bhMag S53H decreased their εA excision activities down to spMag1 levels (Fig 5D), and had only marginal effect (less than twofold) on 7mG excision activity (supplementary Table S3 online). We therefore conclude that the minor groove interaction at this position in spMag1 has a significant role in defining the substrate preference among the Mag enzymes.
Mutation of the minor groove-intercalating residues in other glycosylases has been shown to abrogate base excision activity in several glycosylases (Jiang et al, 2001; Vallur et al, 2002; Eichman et al, 2003; Maiti et al, 2009). Recent work by Verdine and colleagues has illustrated that these residues in MutM and AlkA make intimate contacts with undamaged DNA and thus probably function as sensors to distinguish damaged versus undamaged DNA (Banerjee et al, 2006; Qi et al, 2009; Bowman et al, 2010). The effect of spMag1 His 64 extends these results by demonstrating that probe residues are also capable of discriminating between particular types of damage. One possible mechanism for this is that the side chain at this position senses a local perturbation in the duplex before base flipping. N3-substituted purines would be identified directly in the minor groove, whereas etheno adducts and N7 substitutions could be sensed by a perturbation in base pair structure or stability. The general loss of activity from the S → H substitutions in scMag and bhMag, but not spMag1, suggest that there might be subtly different modes of detection of 7mG and εA lesions. Our crystal structure, which represents the product of the reaction, does not rule out the possibility that the enzyme–substrate complexes might differ between spMag1 and scMag or AlkA, or that His 64 might function as an inhibitor by reducing the scanning rate as a result of the hydrogen bonds with the minor groove. Nevertheless, the structure and supporting biochemistry of base excision highlights the importance of residues outside the base-binding pocket in the lesion-recognition process.
The enzymological differences between the yeast Mag enzymes certainly have an important role in alkylation resistance in cells as protein expression has been shown to complement the alkylation sensitivity of a tag alkA E. coli strain with different levels of effectiveness (Chen et al, 1990; Chen & Samson, 1991; Saparbaev et al, 1995; Memisoglu & Samson, 1996, 2000b; Alseth et al, 2005; Lingaraju et al, 2008). The reduced dependence of spMag1 on alkylation repair in cells can also be partially explained by specific cellular responses to alkylation damage in S. pombe and S. cerevisiae apart from glycosylase activity (Memisoglu & Samson, 2000a). In addition to BER, nucleotide excision and recombination repair have significant roles in safeguarding S. pombe from alkylation damage (Memisoglu & Samson, 2000b; Alseth et al, 2005; Kanamitsu et al, 2007). Furthermore, S. pombe encodes a second Mag orthologue, Mag2, that lacks detectable glycosylase activity (S.A. and B.F.E., unpublished data; and Alseth et al, 2005) despite strong sequence similarity to Mag1, even in the functionally important residues described here. Thus, although the present work provides some biochemical insight into alkylation repair, the various other ways in which Mag proteins contribute to the alkylation response in yeast remain to be determined.
Methods
Protein purification. His6-tagged spMag1, scMag and bhMag proteins were overexpressed in E. coli and isolated by Ni-NTA (Qiagen) affinity chromatography, followed by cleavage of the His6 tag and purification by heparin and gel filtration chromatography. Mutant and SeMet proteins were purified in a manner similar to that of wild-type proteins, with minor adjustments to the buffers used, and their structural integrity was verified using circular dichroism spectroscopy (supplementary Fig S6 online).
X-ray crystallography. SpMag1 and 11-mer DNA (d(TGTCCA(THF)GTCT)/d(AAGACTTGGAC) were preincubated at a 1:1.2 (protein/DNA) ratio and crystallized by vapour diffusion against a reservoir solution containing 100 mM MES (pH 6.5), 20% (w/v) polyethylene glycol (PEG) 8K and 2.4% (v/v) glycerol. X-ray diffraction data (supplementary Table S1 online) were collected on flash-frozen crystals at the Advanced Photon Source beamlines 21-ID (native) and 22-BM (SeMet). SAD data were collected at the selenium absorption peak, and phases were determined from positions of ten Se atoms. A crystallographic model corresponding to amino acids 16–221 and nucleotides 1–22 for each of two protein/DNA complexes in the asymmetric unit was built into 2.8 Å Se-SAD electron density. The crystallographic model was refined against 2.2 Å native diffraction data using a maximum likelihood target. Improvements to the model were made by manual inspection of 2Fo−Fc and Fo−Fc electron density. The final model was validated using PROCHECK and deposited in the Protein Data Bank under the accession number 3S6I.
Enzymatic activity. Base excision activities were measured by following the alkaline cleavage of the abasic DNA product of alkylbase excision from a 25-mer oligonucleotide duplex [5′-32P-d(GACCACTACACCXTTTCCTAACAAC) annealed to 5′-d(GTTGTTAGGAAACGGTGTAGTGGTC)] containing a centrally positioned εA·C or 7mG·C base pair (X) as previously described (Rubinson et al, 2008). The 7mG-DNA was prepared enzymatically as previously described (Rubinson et al, 2008). Reactions were carried out at 25°C and contained 10 μM enzyme (saturating), 100 nM radiolabelled DNA duplex, 100 mM KCl, 2 mM DTT, and either 50 mM sodium acetate (pH 6.0) for εA–DNA or HEPES (pH 7.5) for 7mG-DNA. Reactions were initiated by addition of enzyme and were quenched by addition of 0.2 M NaOH and heated at 70°C for 2 min. The cleaved 12-mer product and unreacted 25-mer substrate oligonucleotides were separated by 15% polyacrylamide/7 M urea gel electrophoresis and quantified by autoradiography.
Detailed Methods are available in supplementary Information online.
Data deposition. PDB ID code 3S6I.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Tom Hollis for providing spMag1 and scMag plasmids, and the Advanced Photon Source (APS) LS-CAT and SER-CAT beamline staff. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). This work was funded by the National Institutes of Health (R01 ES019625) and the American Cancer Society (RSG-07-063-01-GMC).
Author contributions: B.F.E. planned the experiments; S.A. performed the research; S.A. and B.F.E. analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alseth I, Osman F, Korvald H, Tsaneva I, Whitby MC, Seeberg E, Bjoras M (2005) Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res 33: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL (2006) Structure of a DNA glycosylase searching for lesions. Science 311: 1153–1157 [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445–476 [DOI] [PubMed] [Google Scholar]
- Bowman BR, Lee S, Wang S, Verdine GL (2010) Structure of Escherichia coli AlkA in complex with undamaged DNA. J Biol Chem 285: 35783–35791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Derfler B, Samson L (1990) Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J 9: 4569–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Samson L (1991) Induction of S. cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res 19: 6427–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T (2003) Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J 22: 4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA Repair and Mutagenesis, 2nd edn. Washimgton, DC: ASM Press [Google Scholar]
- Hollis T, Ichikawa Y, Ellenberger T (2000) DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J 19: 758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S, Yen TY, Sangaiah R, Swenberg JA (1998) Detection of 1,N6-ethenoadenine in rat urine after chloroethylene oxide exposure. Carcinogenesis 19: 1763–1769 [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA (2005) DNA base damage recognition and removal: new twists and grooves. Mut Res 577: 55–76 [DOI] [PubMed] [Google Scholar]
- Jiang YL, Kwon K, Stivers JT (2001) Turning on uracil-DNA glycosylase using a pyrene nucleotide switch. J Biol Chem 276: 42347–42354 [DOI] [PubMed] [Google Scholar]
- Kanamitsu K, Tanihigashi H, Tanita Y, Inatani S, Ikeda S (2007) Involvement of 3-methyladenine DNA glycosylases Mag1p and Mag2p in base excision repair of methyl methanesulfonate-damaged DNA in the fission yeast Schizosaccharomyces pombe. Genes Genet Syst 82: 489–494 [DOI] [PubMed] [Google Scholar]
- Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, Ellenberger TE (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell 86: 321–329 [DOI] [PubMed] [Google Scholar]
- Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T (2000) Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA 97: 13573–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingaraju GM, Kartalou M, Meira LB, Samson LD (2008) Substrate specificity and sequence-dependent activity of the Saccharomyces cerevisiae 3-methyladenine DNA glycosylase (Mag). DNA Repair 7: 970–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Morgan MT, Drohat AC (2009) Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J Biol Chem 284: 36680–36688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu A, Samson L (1996) Cloning and characterization of a cDNA encoding a 3-methyladenine DNA glycosylase from the fission yeast Schizosaccharomyces pombe. Gene 177: 229–235 [DOI] [PubMed] [Google Scholar]
- Memisoglu A, Samson L (2000a) Base excision repair in yeast and mammals. Mut Res 451: 39–51 [DOI] [PubMed] [Google Scholar]
- Memisoglu A, Samson L (2000b) Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J Bacteriol 182: 2104–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AH, Hollis T, Eichman BF (2007) DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG). EMBO J 26: 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien PJ, Ellenberger T (2004) The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem 279: 26876–26884 [DOI] [PubMed] [Google Scholar]
- Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci USA 97: 5083–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL (2009) Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson EH, Adhikary S, Eichman BF (2010a) Structural studies of alkylpurine DNA glycosylases. In ACS Symposium Series: Structural Biology of DNA Damage and Repair, Stone MP (ed), Vol. 1041, Ch. 3, pp 29–45. Washington, DC: American Chemical Society [Google Scholar]
- Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF (2010b) An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature 468: 406–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson EH, Metz AH, O’Quin J, Eichman BF (2008) A new protein architecture for processing alkylation damaged DNA: the crystal structure of DNA glycosylase AlkD. J Mol Biol 381: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M, Kleibl K, Laval J (1995) Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res 23: 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker DE, Bailey E, Parry A, Lamb J, Farmer PB (1987) The determination of urinary 3-methyladenine in humans as a potential monitor of exposure to methylating agents. Carcinogenesis 8: 959–962 [DOI] [PubMed] [Google Scholar]
- Stivers JT (2004) Site-specific DNA damage recognition by enzyme-induced base flipping. Prog Nucleic Acid Res Mol Biol 77: 37–65 [DOI] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL (2003) A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev 103: 2729–2759 [DOI] [PubMed] [Google Scholar]
- Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB (2002) Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J Biol Chem 277: 31673–31678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.