Abstract
NF-κB is involved in immune responses, inflammation, oncogenesis, cell proliferation and apoptosis. Even though NF-κB can be activated by DNA damage via Ataxia telangiectasia-mutated (ATM) signalling, little was known about an involvement in DNA repair. In this work, we dissected distinct DNA double-strand break (DSB) repair mechanisms revealing a stimulatory role of NF-κB in homologous recombination (HR). This effect was independent of chromatin context, cell cycle distribution or cross-talk with p53. It was not mediated by the transcriptional NF-κB targets Bcl2, BAX or Ku70, known for their dual roles in apoptosis and DSB repair. A contribution by Bcl-xL was abrogated when caspases were inhibited. Notably, HR induction by NF-κB required the targets ATM and BRCA2. Additionally, we provide evidence that NF-κB interacts with CtIP–BRCA1 complexes and promotes BRCA1 stabilization, and thereby contributes to HR induction. Immunofluorescence analysis revealed accelerated formation of replication protein A (RPA) and Rad51 foci upon NF-κB activation indicating HR stimulation through DSB resection by the interacting CtIP–BRCA1 complex and Rad51 filament formation. Taken together, these results define multiple NF-κB-dependent mechanisms regulating HR induction, and thereby providing a novel intriguing explanation for both NF-κB-mediated resistance to chemo- and radiotherapies as well as for the sensitization by pharmaceutical intervention of NF-κB activation.
INTRODUCTION
The transcription factor family NF-κB regulates the expression of more than 150 genes and encompasses five members, namely RelA (p65), RelB, c-Rel, p50/p105 (NF-κB1) and p52/p100 (NF-κB2), which form various homo- and hetero-dimers (1). In the majority of cell types, these dimers are retained in the cytoplasm due to association with the inhibitory IκB-family proteins such as IκBα, IκBβ and IκBε. Different stimuli lead to NF-κB activation and almost all of them funnel in the activation of IκB-kinase (IKK) complex. The IKK complex consists of IKKα and IKKβ, which form the catalytic subunit, and the regulatory subunit IKKγ (NEMO). In the canonical pathway, activation of the IKK complex results in phosphorylation of IκBα on Ser32 and Ser36, which marks it for ubiquitination and proteasomal 26S degradation. It has been well-established that cytotoxic drugs and ionizing radiation activate NF-κB (1,2), which results in the induction of anti-apoptotic genes contributing to chemo- and radio-resistance of tumour cells. The pathway triggered by DNA damage merges with the canonical NF-κB pathway involving p65/p50 heterodimers and IκBα. Recent evidence has shown that the DNA damage sensor poly-(ADP-ribose)-polymerase-1 (PARP1) assembles Ataxia telangiectasia mutated (ATM) and SUMO-1 ligase PIASy resulting in phosphorylation and SUMOylation of IKKγ and thereby IKK activation (3).
Double-strand breaks (DSBs) in mammalian cells can be repaired by non-homologous end joining (NHEJ), single-strand annealing (SSA) and homologous recombination (HR) (4). NHEJ occurs throughout the cell cycle, while HR is limited to the S/G2-phase of the cell cycle. So far, only little is known about the contribution of NF-κB in DNA repair. Studies with murine fibroblasts showed that loss of p65 compromises DNA repair and genome stability during cellular senescence (5). Different observations were made in glioblastoma cells, where NF-κB promoted accumulation of ssDNA breaks and apoptosis (6). One report described that TNFα induces the interaction between p65 and the HR protein BRCA1 leading to enhanced NF-κB transcriptional activity (7). According to another report, this interaction was required for efficient NF-κB activation after camptothecin treatment (8). In the present work, we define the role of NF-κB in DSB repair, its downstream targets and the mechanism of action in different cell types.
MATERIALS AND METHODS
Cell lines and cultivation
K562(Δ/3′) cells originating from the human myeloid leukaemia cell line K562, carrying stably integrated Δ-EGFP/3′EGFP repair substrate (9) and parental K562 cells were cultivated in RPMI 1640 (PAA Laboratories, Pasching, Germany) supplemented with 10% FCS (PAA Laboratories) and 1% l-glutamine (Biochrom, Berlin, Germany). U87MG glioblastoma cell derivatives, stably expressing IκBα-SR (U87MG-SR), were generated as described in Karl et al. (6). HeLa knockdown cells (Control, Mre11, ATM, BRCA2, BRCA1) HeLa SilenciX were purchased from tebu-bio and maintained in DMEM High Glucose-Medium (PAA Laboratories), 10% FCS (PAA Laboratories) and 125 μg/ml Hygromycin B (Roche, Mannheim, Germany). The human primary osteogenic sarcoma cell line SaOS was cultivated in McCoys (Invitrogene Gibco, Karlsruhe, Germany) with 10% FCS (PAA Laboratories) and 1% l-glutamine. Human glioblastoma cells U87MG were maintained in DMEM High Glucose-Medium (PAA Laboratories), 10% FCS (PAA Laboratories) and 1% l -glutamine.
Transfections, DSB repair, drug treatment and gene silencing
DSB repair was examined by the use of the EGFP-based test system as described in Akyüz et al. (9). To determine DSB repair frequencies, K562(Δ/3′) cells were transfected via electroporation (Bio-Rad Laboratories, Hercules, CA, USA) with 10 μg pCMV-I-SceI, 10 μg pBS/wtEGFP plasmid (see below), 40 μg of pcDNA3.0-IκBα-SR expression plasmid/empty vector and/or 40 μg pcDNA3.0-p65 expression plasmid/empty vector. Cells were split into two aliquots and cultivated for 24 h when 10 ng/ml TNFα (Active Bioscience, Hamburg, Germany), 50 μM etoposide (Sigma-Aldrich, München, Germany) or 300 nM camptothecin (Sigma-Aldrich) were added. Transfected cells were cultivated for another 24 or 48 h when they were analysed flow cytometrically for EGFP positivity (Supplementary Figure S3). When studying different DSB repair pathways, the following repair substrates were used: EJ-EGFP, HR-EGFP/5′EGFP and 5′EGFP/HR-EGFP to evaluate NHEJ, HR and SSA, respectively (Figure 4A). K562 cells were electroporated with 10 μg pCMV-I-SceI, 10 μg pBS/wtEGFP plasmid, 10 μg of repair substrate, 30 μg of pcDNA3.0-IκBα-SR expression plasmid/empty vector and/or 30 μg of pcDNA3.0-p65 expression plasmid/empty vector. HeLa cells were transfected with FuGENE reagent (Roche) and a mixture of 4 μg DNA consisting of pCMV-I-SceI, pBS/wtEGFP plasmid, repair substrate to assess HR (HR-EGFP/5′EGFP) and pcDNA3.0-p65 expression plasmid/empty vector. To down-regulate expression of specific proteins during repair measurements in K562(Δ/3′) and K562 cells, cells were transfected with a mixture of two or three different knockdown plasmids generating target-specific shRNAs. To silence Bcl2, 40 μg of pRSBcl2-4 (5′-GCGACTTCGCCGAGATGTCCAGCCAGCTG-3′) and pRSBcl2-6 (5′-TGATTTCTCCTGGCTGTCTCTGAAGACTC-3′) or pRS control plasmid were added to the DNA mixture; 30 μg of pRSBcl-xL-4 (5′-TTAGTGATGTGGAAGAGAACAGGACTGAG-3′), pRSBcl-xL-6 (5′-CTCACTCTTCAGTCGGAAATGACCAGACA-3′) and pRSBcl-xL-7 (5′-CACCACATCCTCCGTCCAGCCGCCATTGC-3′) were used to knockdown Bcl-xL; 40 μg of pRSBAX-3 (5′-GACGAACTGGACAGTAACATGGAGCTGCA-3′), pRSBAX-5 (5′-TTTCTACTTTGCCAGCAAACTGGTGCTCA-3′) and pRSBAX-6 (5′-GTGCCTCAGGATGCGTCCACCAAGAAGCT-3′) were used to down-regulate BAX expression; 40 μg of pRSCtIP-6 (5′-TCGGTTAAGAGCAGGCTTATGTGATCGCT-3′) and pRSCtIP-8 (5′-GGTGAACAGAATAGGACTGAGTACGGTAA-3′) were used to silence CtIP; 40 μg of pRSBRCA1-4 (5′-AGGACCTGCGAAATCCAGAACAAAGCACA-3′) and pRSBRCA1-6 (5′-AAATGCCAGTCAGGCACAGCAGAAACCTA-3′) were utilized to knockdown BRCA1. All above-mentioned knockdown plasmids were purchased from Origene, Rockville, MD, USA, and specific targeted sequences are as indicated. To down-regulate expression of Ku70, knockdown plasmid as described in Uhl et al. (10) was applied.
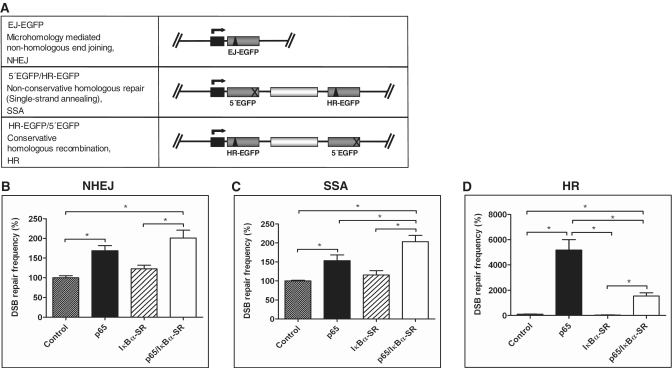
Figure 4.
NF-κB-dependent DSB repair stimulation affects mostly HR. (A) Substrate design for the assessment of different DSB repair pathways. In plasmid substrate EJ-EGFP, the I-SceI site (triangle) is flanked by 5 bp microhomologies for detection of microhomology-mediated NHEJ. Substrate HR-EGFP/5′EGFP enables the analysis of conservative HR (mostly gene conversion) and 5′EGFP/HR-EGFP assessment of non-conservative homologous repair (mostly SSA after I-SceI-mediated cleavage). Mutated EGFP genes: grey boxes; spacer sequence: white box; deleted EGFP sequence: cross; (B–D) K562 cells were co-transfected with pCMV-I-SceI plus pcDNA3.0-p65/pcDNA3.0 and pcDNA3.0-IκBα-SR/pcDNA3.0 plus repair substrates to assess NHEJ (B), SSA (C) or HR (D). DSB repair was measured 48 h after transfection. Mean values and SEMs from n = 6 (*P < 0.05), controls were defined as 100% each (absolute mean value for NHEJ: 2.7 × 10-3, SSA: 2.7 × 10−2, HR: 0.4 × 10−3).
Following cultivation for 48 or 72 h [K562(Δ/3′), K562] or 24 h (HeLa), 50 000–200 000 living cells were examined to determine EGFP-positive and EGFP-negative cells by the diagonal gating method in the FL1/FL2 dot plot (FACS Calibur® FACScan, BD Biosciences, Heidelberg, Germany) (9). Each transfected sample was accompanied by a sample transfected with the same DNA mixture except that the filler plasmid pBS was substituted by wtEGFP expression plasmid to determine the transfection efficiency. Each quantification of green fluorescent cells in repair assays (with pBS) was normalized by use of the individually determined transfection efficiency (with wtEGFP plasmid) to calculate the DSB repair frequency (absolute value). In this way, we corrected for potential differences in transfection, transcription, translation, proliferation and lethality. The statistical significance of differences was determined using Wilcoxon-matched pairs test, two tailed (P < 0.05), with the software GraphPad Prism version 5.01. In Figures and Supplementary Figures graphically presenting data, all statistically significant changes, i.e. after comparing each single mean value with each other in the graph, are indicated by brackets and 1–3 stars (*P < 0.05; **P < 0.01; ***P < 0.001).
Cell cycle, apoptosis and survival
Cell cycle and apoptosis data were obtained by FACS analysis on ethanol/acetone fixed and propidium iodide stained cells (10), which were processed under the conditions of the DSB repair assay.
For survival analysis K562(Δ/3′) cells were transfected with p65 expression/empty control vector as for DSB repair measurements. Twenty-four hours later the cells were seeded on 96-well plates at a density of 5000 cells per well and treated with 16–1000 µM of the PARP inhibitor IQD (Calbiochem). Cells were cultivated for 8 days during which fresh IQD was applied every 3 days, and MTT assay was performed.
Western blotting and antibodies
Cellular lysates were prepared by incubating cell samples in lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 2 mM EGTA; 2 mM EDTA; 25 mM NaF; 25 mM β-glycerophosphate; 0.1 mM NaV; proteinase inhibitor, Roche). To examine the phosphorylation status of proteins, phosphatase inhibitor was added into the lysis buffer (PhosSTOP Phosphatase Inhibitor Cocktail Tablets, Roche). Protein concentrations were determined by the BCA™ Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) and 60–80 μg of protein were loaded onto 8–15% SDS–PAGE gels. Electrophoresed protein samples were blotted onto Hybond™-C Extra or Hybond™-P (GE Healthcare, München, Germany) membranes and detected by use of the following antibodies: ATM mAb 5C2 (Abcam, Cambridge, UK), BAX mAb 2D2 (Calbiochem, Darmstadt, Germany), Bcl2 mAb 100 (Santa Cruz, Heidelberg, Germany), Bcl-xL mAb 2H12 (Pharmigen, BD Biosciences, Heidelberg, Germany), BRCA1 mAb Ab-1 Clone MS110 (Calbiochem), BRCA2 mAb Ab-1 Clone 2B (Calbiochem), CtIP anti goat T-16 (Santa Cruz), DNA-PKcs Cocktail Ab-4, Clone 18-12, 25-4, 42 psc (Neomarkers), IκBα rabbit serum C-21 (Santa Cruz), phospho-IκBα mAb 5A5 (Cell Signaling, Boston, MA, USA), Ku70 mAb S5C11 (Abcam), Mre11 rabbit serum (Novus Biologicals, Littleton, CO, USA), p65 rabbit C-20 (Santa Cruz), GAPDH mouse monoclonal antibody (Abcam) and tubulin mAb DM1A (Abcam). Western blot signals were visualised by Super Signal® West Pico Chemiluminescent Substrate or Super Signal® West Dura Extended Duration Substrate (Thermo Scientific). Densitometric quantification of band intensities on autoradiographs was performed using a ChemImager 5500 and corresponding software enabling background subtraction (Alpha Imunotech Corporation, San Leonardo, CA, USA). Values of band intensities for the protein of interest were corrected with the values obtained for the corresponding loading control each. Relative differences in band intensities mentioned in the text were calculated from the band intensities stemming from the representative western blot depicted in the corresponding figure each (from at least two western blots per protein). Please note that for quantification autoradiographs were scanned separately, which may explain different sensitivities in band detection regarding low protein levels.
Immunofluorescence analysis
SaOS cells were cultured on cover slips and exposed to bleomycin (7.5–100 mU, Sigma-Aldrich) alone or in combination with TNFα (10 ng/ml). For p65 expression, SaOs cells were transfected by nucleofection according to the amaxa protocol (Lonza, Cologne, Germany) with a mixture of 5 μg pBS and pcDNA3.0-p65 expression plasmid/empty vector DNA. After 24 h, cells were treated with 30 mU bleomycin. At the indicated times cells were fixed with 3.7% formaldehyde and permeabilized with 0.5% triton. To quantify replication protein A (RPA) or Rad51 foci cells were additionally pre-extracted with 0.2–0.5% triton solution. Cover slips with fixed cells were washed with PBS, 0.05% tween and blocked with 7% goat serum and immunostained with primary antibodies in 7% goat serum PBS solution. Subsequently, the cells were incubated with secondary antibody, stained with DAPI and the cover slips were mounted in a mixture of Mowiol and Dapco (3:1). Fluorescence micrographs were collected at RT on an Olympus BX51 epifluorescence microscope equipped with UPLAN FI objective lenses (40 × /0.75 and 100 × /1.3 from Olympus) fitted with a thermoelectronically cooled, charge-coupled device camera (model colour view 12 from Soft Imaging System). Grayscale source images were captured separately after fluorescence excitation with the following Olympus filter module: U-MNG2 (Alexa Fluor® 555), or U-MNU2 (DAPI). AnalySIS software including the mFIP module (version Auto 3.2, Soft Imaging System) was used for image acquisition, overlays and analysis. For the automated identification and visualization of nuclear foci with high fluorescence intensity versus diffuse staining, a colour intensity threshold was set and maintained for all images. Antibodies used for immunofluorescence analysis were: 53BP1 rabbit (Novus), RPA mAb clone 34-19 (Calbiochem), Rad51 rabbit H-92 (sc-8349, Santa Cruz), Alexa Fluor® 555 conjugated secondary antibodies (Invitrogen).
RESULTS
TNFα activates DSB repair
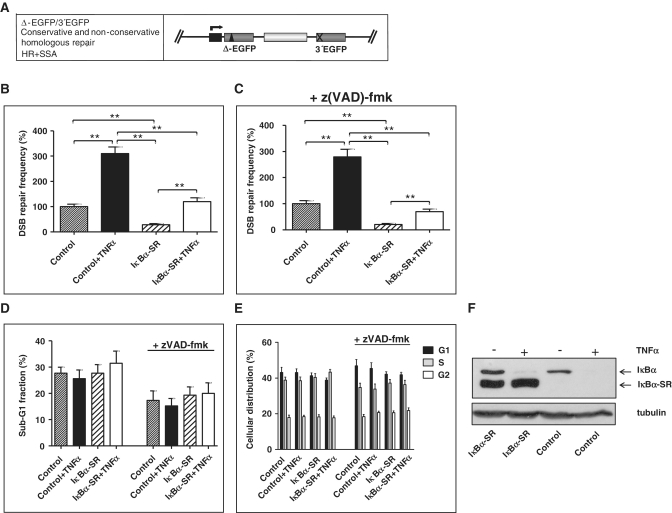
In order to analyse the potential role of NF-κB in DSB repair, we used an EGFP-based model system, where DSBs are introduced via meganuclease I-SceI-targeted cleavage in mutated EGFP variants (9). K562(Δ/3′) cells, which carry a stably integrated repair substrate supporting homologous DSB repair via HR and SSA (Figure 1A), were co-transfected with I-SceI- and IκBα-SR expression or control vectors. Expression of IκBα-SR is a well-known tool to block NF-κB-activation by conferring resistance to TNF-induced degradation (Figure 1F), (6). Cells were exposed to the NF-κB-activating stimulus TNFα 24 h post-transfection and FACS-analysed for EGFP positivity after another incubation period of 24 h. The results revealed 3.1-fold increased DSB repair in cells treated with TNFα (P = 0.0039). In the cells, additionally, expressing IκBα-SR both basal (3.5-fold) and TNFα-induced (2.7-fold) DSB repair was down-regulated (P < 0.01; Figure 1B). Since a TNFα-mediated induction of DSB repair could still be detected in cells expressing IκBα-SR, a NF-κB-independent effect of TNFα action is conceivable. However, we cannot exclude incomplete blockage of NF-κB activation by IκBα-SR (Supplementary Figure S1C).
Figure 1.
Effect of TNFα on DSB repair. (A) Design of substrate Δ-EGFP/3′EGFP to monitor conservative and non-conservative homologous DSB repair. The fluorescence-based DSB repair assay monitors reconstitution of wild-type (wt) EGFP after I-SceI meganuclease-mediated cleavage of the mutated EGFP gene Δ-EGFP, which is under the control of a CMV promoter (black box and kinked arrow), via determination of the fraction of green fluorescent cells within the total population (9). Each measurement was accompanied by the analysis of split samples after transfection of the cells with the same DNA mixture as in DSB repair measurements, but replacing filler pBS plasmid by wtEGFP-expressing plasmid of the DSB repair substrate design. Fractions of green fluorescent cells obtained were thus normalized with the individual transfection efficiency for each sample to give DSB repair frequencies, thereby excluding differences in transfection, transcription, translation, proliferation and lethality. Substrate Δ-EGFP/3′EGFP enables the analysis of conservative HR plus non-conservative SSA repair processes. Mutated EGFP genes: grey boxes; spacer sequence (hygromycin resistance gene cassette): white box; I-SceI site: triangle; deleted EGFP sequence: cross. (B) Influence of TNFα on DSB repair was analysed in K562(Δ/3′) cells carrying stably integrated homologous DSB repair substrate Δ-EGFP/3′EGFP. Cells were electroporated with pCMV-I-SceI for targeted substrate cleavage together with expression plasmid for the NF-κB antagonist IκBα-SR (pcDNA3.0-IκBα-SR) or with pcDNA3.0 in controls. After 24 h cells were treated with 10 ng/ml TNFα and cultivated for another 24 h, when DSB repair frequencies were determined. DSB repair frequencies in H2O-treated controls were defined as 100% each (absolute mean value 1 × 10−3). Mean values and SEMs from n = 9 (**P < 0.01). (C) In parallel, cells were treated with 25 μM caspase inhibitor z(VAD)-fmk or DMSO and results were evaluated as in (B). (D and E) Apoptosis (D) and cellular distribution (E) were determined by flow cytometry with propidium iodide-stained cells at the time point of repair measurements. Sub-G1 fractions were determined and viable cells were divided into G1, S and G2 cell cycle phases (presented in percentages). Mean values and SEMs from n = 6. (F) Expression levels of endogenous IκBα and exogenous IκBα-SR were examined by western blotting.
To prove that the NF-κB-signalling pathway is functional under these conditions, we analysed the phosphorylation status of endogenous IκBα, performed NF-κB DNA-binding studies and NF-κB-dependent reporter gene assays (Supplementary Figure S1). TNFα exposure led to marked IκBα phosphorylation, strong DNA binding as well as 4-fold (P = 0.0039) increased NF-κB-dependent GFP transcription. IκBα-SR expression compromised DNA binding and GFP expression in TNFα-treated samples. Since TNFα also activates caspase 8 and the pro-apoptotic JNK pathway (1), which might lead to degradation of repair proteins, the role of apoptosis was addressed in this scenario. For this purpose, cells were pre-treated with the broad range caspase inhibitor z(VAD)-fmk. Under these conditions, TNFα-induced DSB repair was found to be unchanged (Figure 1C and B). Furthermore, the suppressive effect of IκBα-SR on DSB repair (−/+TNFα) matched the effect in cells without z(VAD)-fmk suggesting that apoptosis-dependent processes are not involved in TNFα-mediated DSB repair activation. Because stable overexpression of IκBα-SR was reported to induce G1-arrest via p21, we also considered potential cell cycle-dependent effects of IκBα-SR on DSB repair (11). When analysing propidium iodide stained cells, we neither detected significant changes between TNFα- and mock treated nor between IκBα-SR expressing and control samples regarding sub-G1, G1-, S- or G2-phase fractions (Figure 1D and E).
Role of NF-κB in DSB repair triggered by DNA damage-inducing chemotherapeutic treatment
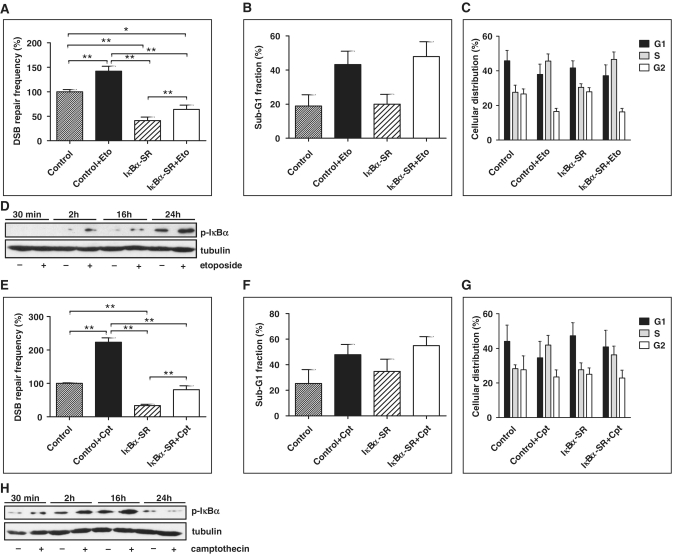
DNA damage, especially a DSB, represents one of the NF-κB-activating stimuli (2). To test whether exposure to DNA-damaging agents leads to NF-κB-dependent stimulation of DSB repair, K562(Δ/3′) cells were subjected to DSB repair analysis −/+ IκBα-SR expression and exposed to etoposide, camptothecin or doxorubicin. Treatment with 50 µM etoposide led to a 1.4-fold increase in DSB repair, which was 2.2-fold down-regulated by IκBα-SR (P < 0.01; Figure 2A). Camptothecin at a concentration of 300 nM stimulated repair 2.2-fold, which was 2.7 times down-regulated by IκBα-SR (P < 0.01; Figure 2E). When comparing the effect of 50 µM etoposide or 300 nM camptothecin on DSB repair without versus with IκBα-SR, we noticed similar fold induction (etoposide: 1.4- versus 1.5-fold, camptothecin: 2.2- versus 2.3-fold, P > 0.05). Applying low concentrations of these drugs still induced DSB repair, namely 1.3- (1 µM etoposide) and 1.2-fold (30 nM camptothecin), which was down-regulated 4.8- (etoposide) and 3.7-fold (camptothecin) by IκBα-SR (P < 0.01; Supplementary Figure S2A and B). However, under these low-dose treatment conditions drug-induced DSB repair was no longer detectable after IκBα-SR expression (P > 0.05). Similarly, when treating the cells with 0.3 µg/ml doxorubicin, DSB repair was increased (1.7-fold, P = 0.0313) in cells without but no longer in cells with IκBα-SR (Supplementary Figure S2C). When comparing the fold change of DSB repair after drug treatment between cells without and with IκBα-SR, a statistically significant difference was reached for 1 µM etoposide (1.3- versus 1.0-fold, P = 0.0195). These data suggested either that IκBα-SR cannot completely inhibit NF-κB after strong activation by high topoisomerase inhibitor concentrations or that NF-κB is only partially required for DSB repair induction as compared with other DNA damage signalling pathways triggered by the formation of DNA breaks.
Figure 2.
DSB repair stimulation by chemotherapeutics as a function of NF-κB activity. (A) K562(Δ/3′) cells were co-transfected with pCMV-I-SceI and pcDNA3.0-IκBα-SR/pcDNA3.0 as indicated, cultivated for 24 h and then treated with 50 μM etoposide (Eto). Cells were further cultivated for 48 h, when HR assays were performed. DSB repair frequencies in DMSO-treated controls were defined as 100% (absolute mean value 2 × 10−3). Mean values and SEMs from n = 9 (*P < 0.05; **P < 0.01). (B and C) The percentages of cells with sub-G1 DNA content (B) in G1, S or G2 cell cycle phases (C) were determined with propidium iodide-stained cells at the time point of the repair measurements. Columns, mean values of n = 4; bars SEMs. (D) IκBα phosphorylation in cells exposed to 50 μM etoposide was analysed by western blotting (p- IκBα). (E–H) K562(Δ/3′) cells were co-transfected with pCMV-I-SceI and pcDNA3.0-IκBα-SR/pcDNA3.0, treated with 300 nM camptothecin (Cpt) and evaluated as in (A–D). Note that IκBα degradation interfered with p-IκBα detection 24 h after camptothecin addition in (H).
Evaluation of the sub-G1 cellular fraction confirmed effectiveness of the cytostatics, since cell death was increased (Figure 2B and F). The same data sets excluded a significant influence of apoptosis on IκBα-SR-modulated DSB repair, since sub-G1 fractions in control and IκBα-SR-expressing samples treated with chemotherapeutics did not significantly change. Cell cycle analysis revealed that cells exposed to etoposide and camptothecin accumulated in S-phase (Figure 2C and G), notably, in a similar manner in IκBα-SR-negative and -positive samples. Increased phosphorylation of IκBα 2–24 h after etoposide and 0.5–16 h after camptothecin exposure verified NF-κB activation by chemotherapeutics (Figure 2D and H).
The NF-κB subunit p65 stimulates DSB repair independently of apoptosis
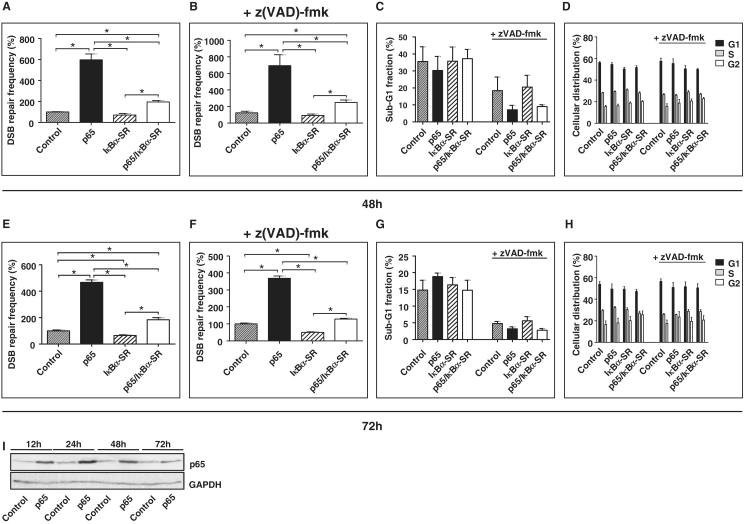
To ascertain the involvement of NF-κB in DSB repair, p65 was expressed in K562(Δ/3′) cells (Figure 3I), which were subjected to I-SceI-triggered DSB repair −/+ IκBα-SR expression. p65 expression stimulated DSB repair 5.9-fold, which was down-regulated 3.0-fold by IκBα-SR at 48 h and similarly at the 72 h time point with and without z(VAD)-fmk (P < 0.05; Figure 3A, B, E and F; primary flow cytometric data in Supplementary Figure S3). The fold change in cells with p65 was reduced by IκBα-SR expression (48 h: 5.9- versus 2.7-fold, P = 0.0625; 72 h: 4.6- versus 2.8-fold, P = 0.0313). Sub-G1 fractions of cells −/+ IκBα-SR did not differ, strengthening the data obtained with z(VAD)-fmk showing that DSB repair changes are independent of differences in apoptosis induction (Figure 3C and G). p65- or IκBα-SR expression also had no significant influence on the cell cycle distribution (Figure 3D and H). To further exclude potential NF-κB-dependent differences in DNA substrate cutting, we performed PCR analysis of the chromosomally integrated Δ-EGFP gene, which is cleaved by I-SceI. The results showed that Δ-EGFP was equally cleaved in cells with or without concomitant p65 expression, as indicated by the disappearance of the PCR product and quantitative real-time PCR data (Supplementary Figure S4A and B). Equivalent cutting was also observed for mock- and TNFα treatment.
Figure 3.
Expression of the NF-κB subunit p65 enhances DSB repair. (A and E) Homologous DSB repair was analysed in K562(Δ/3′) cells 48 h (A) and 72 h (E) after cells had been electroporated with pCMV-I-SceI, pcDNA3.0-p65/pcDNA3.0 and pcDNA3.0-IκBα-SR/pcDNA3.0 as indicated. Mean values from controls were defined as 100% (absolute mean values: 0.4 × 10−3 after 48 h, 0.9 × 10−3 after 72 h); Mean values and SEMs from n = 6; *P < 0.05. (B and F) In parallel experiments cells were treated with 25 μM z(VAD)-fmk. (C, D, G and H) Apoptosis (C and G) and cell cycle distribution (D and H) were analysed under the same conditions as in (A) and (E). Mean values and SEMs from n = 4. (I) Western blot analysis verifying p65 expression.
NF-κB stimulates DSB repair via the HR pathway
To clarify whether NF-κB activation enhances DSB repair in general or via specific repair pathways, K562 cells were transiently transfected with different EGFP-based repair substrates, namely with EJ-EGFP, 5′EGFP/HR-EGFP and HR-EGFP/5′EGFP (Figure 4A) together with I-SceI expression plasmid to assess NHEJ, non-conservative homologous DSB repair (mostly SSA) and conservative HR (mostly gene conversion). After p65 expression, we observed a 1.7- and 1.5-fold stimulation of NHEJ and SSA, respectively (P < 0.05; Figure 4B and C), indicating minor NF-κB-dependent effects. In contrast, HR was strongly stimulated by p65 expression (51.6-fold) and IκBα-SR-reduced p65-stimulated HR frequencies 3.3 times (P < 0.05; Figure 4D). When we examined DNA exchange without I-SceI, we again monitored a massive p65-mediated HR stimulation with HR-EGFP/5′EGFP (Supplementary Figure S4C). Fluorescent signals were absent in assays with EJ-EGFP lacking I-SceI, which is consistent with DSB dependence of NHEJ-mediated reconstitution of the wtEGFP reading frame (9). The substrate 5′EGFP/HR-EGFP is mostly repaired by SSA after I-SceI-mediated cleavage, but crossover events are still detectable without cleavage. Expression of p65 did not have a major effect on DNA exchange with 5′EGFP/HR-EGFP (Supplementary Figure S4C). These data excluded an indirect influence from I-SceI and confirmed that NF-κB modulates HR. We noted differences in the degree of HR stimulation by p65 with intrachromosomal (Figure 3A) as compared with extrachromosomal (Figure 4D) substrate (6- versus 52-fold), which might be explained by a better accessibility of extrachromosomal substrates to the end processing and strand-exchange machinery.
To better understand DSB repair pathway usage within chromosomally integrated substrate (ΔEGFP/3′EGFP in K562(Δ/3′) cells), we expressed I-SceI with or without p65 for 72 h, sorted green fluorescent and white, non-fluorescent cells flow cytometrically and performed genomic PCR analysis (Supplementary Figure S5) (9,10). Amplification of a fragment encompassing the ΔEGFP gene (Supplementary Figure S5A) was followed by restriction analysis using I-SceI and the control enzyme XhoI. In white cells, I-SceI cleavage failure was the result of error-prone NHEJ, in green cells the result of conservative HR (mostly gene conversion) in one of the chromosomally integrated ΔEGFP/3′EGFP copies (Supplementary Figure S5B). When comparing error-prone NHEJ-specific bands (corrected for loading), we quantified a 1.3-fold increase for cells expressing p65. When comparing HR-specific bands (corrected for loading), we quantified a 1.8-fold increase. When performing genomic PCR analysis specific for DSB repair events leading to spacer sequence deletion such as via SSA (9,10), we were unable to detect the corresponding PCR products both in control and p65 expressing cells (data not shown). To examine chromosomal DSB repair in another cell type, we correspondingly analysed WTK1(HR/3′) cells (Supplementary Figure S5C), which carry substrate HR-EGFP/3′EGFP (12). PCR fragments specific for error-prone NHEJ were hardly detectable, and quantification of band intensities did not indicate an increase for cells expressing p65. However, HR-specific bands (corrected for loading) revealed a 3.9-fold increase upon p65 expression. These data support the notion that NF-κB promotes the HR pathway in the extra- and intrachromosomal context.
To prove cell type-independency, we applied the EGFP-based repair assay on U87MG glioblastoma cell derivatives, stably expressing IκBα-SR (U87MG-SR). In accordance with the data obtained in K562 cells, HR was several-fold down-regulated in U87MG-SR (Supplementary Figure S6A and B). In the presence of the caspase inhibitor z(VAD)-fmk this HR frequency change became even more pronounced [without z(VAD)-fmk: 2.6-fold; with z(VAD)-fmk: 6.8-fold; P values < 0.01]. Moreover, when we analysed HR in response to TNFα-treatment, we measured a 9.3-fold (P = 0.0117) HR increase which was abrogated in the presence of IκBα-SR (P > 0.05) (Supplementary Figure S6C). These data indicated that NF-κB is necessary also for TNFα-mediated activation of the HR pathway.
NF-κB-dependent DSB repair induction is not mediated by Bcl2-family members
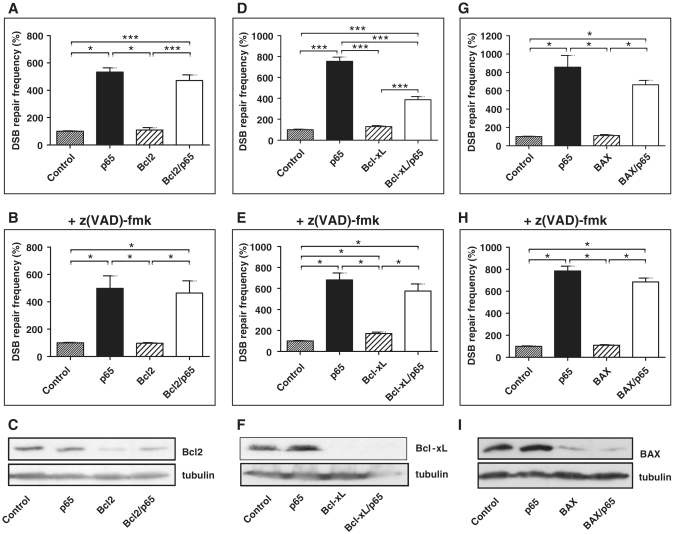
Proteins from the Bcl2-family represent classical NF-κB target genes that are well-known anti- and pro-apoptotic factors and have been implicated in HR (13–15). Consequently, we investigated whether these molecules are responsible for NF-κB-dependent DSB repair stimulation. Bcl2, Bcl-xL or BAX were silenced in K562(Δ/3′) cells via transfection with specific shRNA plasmids. Reduction of Bcl2 levels by 57–70% (Figure 5C) and BAX levels by 96% (Figure 5I) did not significantly change p65-stimulated DSB repair without or with caspase inhibition (Figure 5A, B, G and H). Differently, Bcl-xL depletion by 72% (Figure 5F) led to a 1.9-fold (P = 0.0005) down-regulation of p65-mediated DSB repair, but this effect was lost when apoptosis was blocked by z(VAD)-fmk (Figure 5D and E). Interestingly, when evaluating the effect of Bcl2, Bcl-xL and BAX on DSB repair without p65 expression, we noticed a 1.7-fold increase (P = 0.0313) of repair frequencies after Bcl-xL silencing, however, only after z(VAD)-fmk treatment. Taken together, we observed that contrary to the differential Bcl-xL effects, Bcl2 and BAX knockdown did not influence DSB repair. Consistently, immunoblotting confirmed Bcl-xL (2-fold up-regulation) but not Bcl2 or BAX as NF-κB-induced target gene.
Figure 5.
Role of Bcl2-family proteins in NF-κB-mediated DSB repair regulation. (A, D and G) K562(Δ/3′) cells were co-transfected with pCMV-I-SceI, pcDNA3.0-p65 or empty vector plus a mixture of two different shRNA expression plasmids directed against Bcl2, Bcl-xL or BAX or empty vector. DSB repair frequencies were measured 72 h post-transfection; mean values of controls were taken as 100% each (absolute mean value 1 × 10−3). Columns, mean values of n = 6–12; bars, SEM; *P < 0.05; ***P < 0.001. (B, E and H) Cells were treated with 25 μM z(VAD)-fmk and homologous DSB repair was assayed as in (A, D and G). (C, F and I) Immunoblot analysis showing Bcl2, Bcl-xL and BAX knockdown.
NF-κB-dependent enhancement of DSB repair requires up-regulated HR proteins
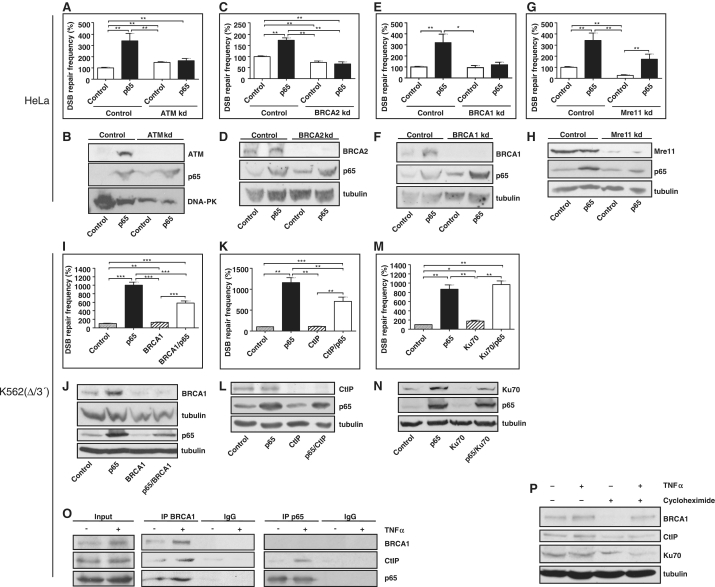
Since we found HR to be the DSB repair pathway stimulated by p65, we examined the NF-κB target gene products and HR proteins ATM and BRCA2 (16,17). HR analysis with transiently transfected HR-EGFP/5′EGFP was performed in HeLa derivatives, in which ATM or BRCA2 were stably silenced. In the control HeLa line, we observed on average 2.9-fold enhancement of HR by p65 (P = 0.0022). Both in the ATM and BRCA2 knockdown lines, this effect was completely abolished (Figure 6A and C) implying that ATM and BRCA2 are downstream effectors of p65 in DSB repair enhancement. ATM and BRCA2 protein levels were 11- and 1.8-fold up-regulated upon p65 expression, which was lost in the corresponding knockdown cells (Figure 6B and D). Interestingly, we also noticed a 7.3-fold BRCA1 accumulation, whereas Mre11 levels remained unaffected (Figure 6F and H). In BRCA1 knockdown cells HR stimulation by p65 was again abrogated (Figure 6E). In Mre11 knockdown cells, HR was reduced both in cells with and without p65 expression so that p65-dependent HR enhancement (6.4-fold, P = 0.0043) was still observable (Figure 6G).
Figure 6.
p65-dependent DSB repair involves the classical homologous DSB repair proteins ATM, BRCA2, BRCA1 and CtIP. (A, C, E and G) HeLa control and ATM, BRCA2, BRCA1 or Mre11 HeLa knockdown cell lines (ATM kd, BRCA2 kd, BRCA1 kd, Mre11 kd) were transfected with pCMV-I-SceI plus pcDNA3.0-p65/pcDNA3.0 and repair substrate to assess HR. After cultivation for 48 h, cells were FACS analysed for green fluorescence. Mean DSB repair frequencies with controls were set to 100% each (absolute mean value 0.5 × 10−3). Columns, mean values of n = 6; bars, SEM; *P < 0.05; **P < 0.01. (B, D, F and H) Western blots confirming ATM, BRCA2, BRCA1 or Mre11 knockdown and p65 expression. (I, K and M) Homologous DSB repair in K562(Δ/3′) cells was analysed after p65 expression with or without transient BRCA1, CtIP, or Ku70 knockdown by use of shRNA plasmids. Columns, mean values of n = 9–12; bars, SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (J, L and N) Western blots demonstrating BRCA1, CtIP or Ku70 knockdown and p65 expression. (O) K562(Δ/3′) cells were transfected with pCMV-I-SceI and 24 h later treated with 10 ng/ml of TNFα for 4 h, when immunoprecipitation was performed with antibodies directed against BRCA1 or p65 followed by immunoblotting with antibodies to detect BRCA1, CtIP and p65. (P) K562(Δ/3′) cells were transfected with pCMV-I-SceI, cultivated for 24 h, then, treated with 7 μM cycloheximide for 45 min and after that exposed to TNFα for 24 h (10 ng/ml). Cells were further cultivated for 24 h followed by protein extraction and immunoblotting analysis with antibodies against BRCA1, CtIP, Ku70 and tubulin.
Since a BRCA1–p65 interaction had been reported (7), we tested the involvement of BRCA1 in p65-regulated DSB repair also in another cellular system. In K562(Δ/3′) BRCA1 protein was up-regulated 2.4-fold by p65 (Figure 6J). Transient BRCA1 silencing by 51–74% reduced p65-dependent DSB repair stimulation 2.5-fold (P = 0.0005 for difference between fold induction) (Figure 6I and J). Because BRCA1 and CtIP form a complex which is important for ssDNA-end formation as a prerequisite for HR (18,19), we performed DSB repair analysis in K562(Δ/3′) with transient CtIP knockdown. Interestingly, silencing CtIP by 61–77% reduced p65-dependent DSB repair stimulation 1.8-fold (P = 0.0005 for difference between fold induction) (Figure 6K and L). Equivalent results were obtained with z(VAD)-fmk (data not shown). Finally, we also examined the effect of p65 expression on HR in lymphoblastoid cell lines derived from patients with mutations in BRCA2 or BRCA1 (20). As displayed in Supplementary Figure S7, expression of p65 stimulated HR 2.4-fold (P < 0.0001) in control lymphoblastoid cells. In accordance with the BRCA2/BRCA1-mutated status, HR was severely reduced in both patient cell lines, and p65 expression had no stimulatory effect on HR.
Because previous publications revealed competition between HR and NHEJ (4,21–23), we also tested the NF-κB target gene and NHEJ component Ku70 (24). Here, we noticed a 3.1-fold increase of Ku70 levels after p65 expression in K562(Δ/3′) cells and a 3.6-fold decrease following transient knockdown (Figure 6N). However, we did not observe a reduction of p65-dependent DSB repair stimulation upon Ku70 knockdown arguing against Ku70 involvement (Figure 6M).
Activation of NF-κB stimulates interactions with CtIP–BRCA1 complexes and accelerates the formation of RPA foci formation
To test whether BRCA1 interacts with NF-κB and whether this interaction is modulated by NF-κB activation in K562(Δ/3′), DSBs were induced by I-SceI expression followed by treatment with TNFα and subsequent immunoprecipitation with antibodies directed against BRCA1 and p65. Pull down of BRCA1 resulted in the co-precipitation of p65 and CtIP, pull down of p65 led to co-precipitation of CtIP, but not BRCA1 (Figure 6O), arguing for a direct and probably more stable contact of p65 with CtIP. In both cases, more CtIP protein was co-precipitated after TNF-α exposure, even though the total amount of p65 protein remained the same. For BRCA1, we had observed a p65-dependent increase of the protein level in different cell types (Figure 6F and J). To test whether NF-κB influences BRCA1 stability, cells were pre-treated with cycloheximide to block translation followed by TNF-α exposure. After TNF-α treatment, a 2-fold BRCA1 increase was observed (Figure 6O and P). Cycloheximide exposure resulted in complete disappearance of BRCA1 in untreated cells, while TNFα treatment allowed accumulation of BRCA1 even in cycloheximide-treated cells, suggesting that NF-κB activation stabilizes BRCA1 (Figure 6P). For comparison, the protein levels of CtIP or Ku70 were only marginally affected by TNFα.
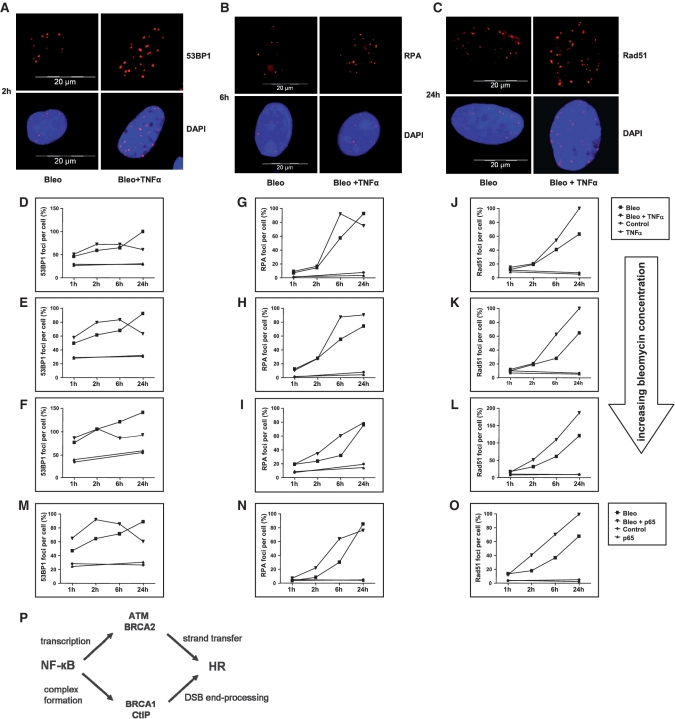
CtIP is known to promote DSB resection to generate ssDNA ends for HR (18) and to collaborate with BRCA1 in this process (25,26). Therefore, we investigated a potential role of NF-κB in the generation of resected ssDNA after introduction of DSBs into the cellular genome. Thus, we exposed SaOS cells, which also showed HR stimulation in response to NF-κB activation (data not shown), to increasing concentrations of the radiomimetic drug bleomycin alone or in combination with TNFα and monitored 53BP1, RPA and Rad51 foci assembly as markers of DSB, ssDNA and Rad51 filament formation, respectively. As expected, nuclear 53BP1 foci continuously accumulated in response to bleomycin treatment over a time period of 24 h (Figure 7A and D–F). After combined bleomycin- and TNFα treatment, we counted similar or even higher numbers of 53BP1 foci per cell within the first 2–6 h of treatment. At later time points (6–24 h), we noticed a decrease in 53BP1 foci numbers rather than a further increase, which is consistent with an enhanced homologous DSB repair in response to NF-κB activation. When analysing RPA, we observed accelerated foci formation in bleomycin and TNFα co-treated samples compared to bleomycin-treated cells as it became evident from the RPA foci number increase 6 h after drug exposure for different bleomycin concentrations (Figure 7B and G–I). Twenty-four hours after drug exposure the increase in RPA foci formation in TNFα co-treated samples disappeared, most likely due to continuation of DNA repair. Rad51 foci numbers were higher in TNFα co-treated samples 6–24 h after drug exposure (Figure 7C, J–L). Following transfection of cells with p65 expression or control plasmid and subsequent bleomycin treatment, 53BP1, RPA and Rad51 foci number changes were comparable to the ones observed in cells with or without TNFα co-treatment (Figure 7M–O). These data suggested that NF-κB activation stimulates DSB resection, which is mediated by CtIP–BRCA1 complexes, i.e. interaction partners of p65, and filament formation of Rad51, which binds the NF-κB target gene product BRCA2.
Figure 7.
Influence of TNFα on 53BP1, RPA and Rad51 foci assembly. SaOS cells were exposed to bleomycin (7.5–100 mU) alone or in combination with TNFα 10 ng/ml (A–L). In Figure 7M–O cells were transfected with pcDNA3.0 or pcDNA3.0-p65, cultivated for another 24 h and then treated with bleomycin. (A–C) At the indicated incubation times after treatment with 15 mU bleomycin, cells were processed for immunolabeling of 53BP1 (A), RPA (B), or Rad51 (C) to visualize foci assembly. (D–O) Immunolabeled foci from two independent experiments were scored by automated quantification in 50 nuclei from two separate slides for each time point. Bleomycin concentrations: 7.5 mU (D, G and J), 15 mU (E, H and K), 30–100 mU (F, I, L and M–O). Maximum scores were set to 100%. (P) Model of NF-κB-dependent DSB repair regulation.
DISCUSSION
A specific role of NF-κB in DSB repair
Activation of NF-κB by the DSB-induced kinase ATM provides an anti-apoptotic signal in balance with apoptosis induction by p53 (2). Despite the identification of NF-κB target genes, which encode DSB repair factors (16,17,24), very little was known about a functional role of NF-κB in DSB repair. We show that NF-κB strongly stimulates the removal of DSBs by enhancement of HR. Intriguingly, this again balances an antagonistic action of p53 in HR (27). The effectiveness of TNFα exposure, p65 expression and IκBα-SR-mediated suppression of NF-κB activation implies an involvement of the protoypical NF-κB heterodimer p50/p65, which is quickly activated following IκBα phosphorylation and degradation, thereby assuring a timely response for the removal of the most detrimental DNA lesion, namely a DSB.
Recent data identified the DNA single-strand break sensor PARP1 as critical NF-κB regulator (3). PARP1 senses stalled replication forks and promotes recombinative replication restart (28). NF-κB is most probably also activated by single-stranded breaks, because UV-induced NF-κB activation is abolished in XPC/D deficient primary fibroblasts, which are blocked before lesion excision creates ssDNA breaks (29). Experiments with hydroxyurea and aphidicolin identified also DNA replication arrest as NF-κB inducer (30). In our study, HR stimulation by NF-κB was not dependent on I-SceI-induced DSBs. To obtain first insight into the impact of NF-κB on replication stress sensitivity, we assessed cell viability following treatment of K562(Δ/3′) cells with the PARP inhibitor IQD, and noticed an ~50% increase of the IC50 value upon expression of p65. Thus, NF-κB triggers HR that removes any breaks arising spontaneously or in response to genotoxic stress.
Involvement of NF-κB downstream signalling
NF-κB is known best for its role in transcriptionally regulating a myriad of target genes regulating diverse biological processes (http://people.bu.edu/gilmore/nf-kb/). To avoid functional interactions with the target gene product p53 (31), which is also modulated by IκBα-SR (11), we chose p53-negative (K562, SaOS) or -depleted (HeLa) cells in this work. Nevertheless HR stimulation by NF-κB was not limited to p53-mutated cells, as it was also detected in wtp53-positive U87MG glioblastoma cells. The apoptosis-regulatory target genes Bcl-2, Bcl-xL and BAX were proposed to act downstream of NF-κB in DSB repair regulation, since the corresponding gene products had been implicated in HR (13–15). We did not observe an influence of Bcl-2 or BAX on NF-κB-mediated DSB repair. Regarding Bcl-xL, a partial contribution was seen but lost in cells treated with z(VAD)-fmk, suggesting that apoptosis suppression by Bcl-xL ameliorates NF-κB-mediated DSB repair stimulation. Interestingly, when NF-κB was not activated, our knockdown experiments revealed a moderate antagonistic effect of Bcl-xL. Our results may reconcile earlier discrepancies on the involvement of Bcl-xL in HR. Thus, HR stimulation by Bcl-xL observed by Kronenberg and co-workers (14) might have been influenced by an increased propensity of human lymphoblastoid TK6 cells to undergo apoptosis due to wtp53 expression in combination with active NF-κB in these cells.
The DNA-PK subunits Ku70 and Ku80 were shown to become up-regulated by NF-κB (24). In agreement with the fact that DNA-PK initiates canonical NHEJ processes rather than HR, we did not observe any contribution of Ku70 to NF-κB-mediated DSB repair. NF-κB was also proposed to transcriptionally induce polymerase β, which stimulates HR and causes genetic instabilities in tumour cells upon overexpression (32,33). Pathogenesis of Werner syndrome was also suggested to involve NF-κB signalling. Werner syndrome is caused by mutations in WRN, which has a role in the resolution step of homology-directed repair (34). We neither saw an effect of polymerase β nor WRN knockdown on NF-κB-mediated DSB repair (data not shown). To the contrary, when we analysed the involvement of the downstream gene products BRCA2 and ATM (16,17), we completely lost NF-κB-dependent HR enhancement. BRCA2 facilitates Rad51 filament assembly in HR (35) and ATM is the key DSB-signalling kinase that directly phosphorylates proteins, which are critically involved in HR. Moreover, it transmits the DNA damage signal to BRCA2 via Chk2 and to Rad51 via c-Abl (36,37). Knockdown of the initial HR component Mre11 compromised HR with or without NF-κB activation. These observations supported the notion that BRCA2- and ATM induction by NF-κB stimulate HR, whereas Mre11 complexes are equally necessary to exert basal and NF-κB-dependent HR processes.
Identification of a direct link between NF-κB and the DNA end processing CtIP–BRCA1 complex
Our data further provide evidence for a crucial role of CtIP–BRCA1 complexes in NF-κB-mediated HR. We hypothesize that direct interactions between p65 and CtIP–BRCA1 complexes stabilize BRCA1 protein, providing a novel mechanism for NF-κB-mediated downstream signalling in addition to its role as transcription factor. Indeed, protein stabilization represents a major mechanism for BRCA1 regulation (38). We further propose that these interactions have functional consequences, namely accelerated processing of DSBs. CtIP exonucleolytically generates extensive 3′ ssDNA overhangs at DSBs (18,39). CtIP does not influence canonical NHEJ but promotes HR, SSA and error-prone NHEJ, which may explain the modest stimulatory effects of NF-κB on microhomology-mediated NHEJ and SSA that we observed in this study (4). Recruitment of CtIP by BRCA1 is necessary for HR but not error-prone NHEJ shifting the balance of DSB repair to HR (26). Notably, Souto-Carneiro et al. (40) found that NF-κB is involved in controlling the exonucleolytic activity on the coding ends during V(D)J recombination, whereby the authors excluded an influence of NF-κB on terminal deoxynucleotide transferase, Artemis and DNA-PKcs.
Altogether, NF-κB utilizes multiple mechanisms to activate HR, as is outlined in the model in Figure 7P: In a rapid step, IkBα released NF-κB dimers associate with and stimulate the activity of CtIP–BRCA1 complexes to trigger DNA end processing, the initial step of homologous DSB repair. In a subsequent process, NF-κB transcriptionally up-regulates BRCA2 and ATM. BRCA2 channels homologous DSB repair into the HR pathway (35). Increased ATM kinase levels assure fully efficient HR through signalling to Mre11, Rad50, Nibrin, CtIP, BRCA2 and BRCA1 (36,37) and contributes to the manifestation of the pathway choice, since phosphorylation of BRCA1 by the ATM downstream kinase Chk2 promotes HR versus error-prone NHEJ (41). Our model does not take into account that many of the downstream components have also been described to act upstream of NF-κB: BRCA1 was shown to augment transcription by NF-κB (7) and ATM phosphorylates IKKγ in the nucleus (1,2). Activation of multiple molecular targets by NF-κB and signal amplification by feed forward loops might ensure strong and rapid activation of HR.
Implications
In high-risk MDS and AML ATM becomes activated during the transformation process resulting in constitutive NF-κB activation (42). NF-κB is also activated in the mammary progenitor population before formation of mammary luminal-type epithelial neoplasias (43). Of note, recombination-mediated bypass of stalled replication forks is necessary for replication restart and cellular survival during oncogene-induced replication stress in preneoplastic lesions (28). Here, we observed a requirement of NF-κB for induction of the DSB repair pathway HR in particular (Figure 4; Supplementary Figures S4C and S5). HR enhancement in response to constitutive NF-κB activation will permit accelerated proliferation of these preneoplastic cells. Thus, even though reduced HR activities are known to increase cancer proneness (35), uncontrolled excess recombination, in our study as a result of NF-κB activation, equally causes genetic instabilities and promotes carcinogenesis (33). In this context, it is of interest that TNFα, for which we demonstrated NF-κB-dependent HR induction, was shown to induce chromosomal instabilities in apoptosis-deficient leukaemia cells (44). TNFα may additionally induce homologous DSB repair pathways independently of NF-κB such as via JNK signalling (45).
Accumulating evidence indicates that NF-κB mediates resistance to anti-cancer therapies involving genotoxic treatment (46,47). In the light of our findings indicating that NF-κB may mediate DSB repair induction in response to topoisomerase inhibition at least partially or after low-dose treatment, it is of interest that CtIP and BRCA1 collaborate in the elimination of covalently bound polypeptides from DSBs after camptothecin- or etoposide treatment (25). NF-κB-mediated signalling to CtIP-BRCA1 may therefore underlie resistance to topoisomerase-trapping drugs and explain enhanced cytotoxicities when NF-κB inhibitors are administered in conjunction with cytostatic drugs. It will be of high interest to analyse whether accelerated removal of complex DSBs contributes to NF-κB-mediated adaptive resistance to radiotherapy (46). Our data further suggest that the NF-κB status may be an important parameter regarding the design of individual conditioning regimens during bone marrow transplantation therapies. In particular, immunodeficiency patients with mutations in the NF-κB pathway are predicted to show increased sensitivities to genotoxic therapies (48). Altogether, our findings on the function of NF-κB activation in regulating DSB repair may impact on the design of individualized treatment protocols for immunodeficiency and cancer patients.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG Klinische Forschergruppe 167: ‘Apoptoseregulation und ihre Störungen bei Krankheiten’, grants WI 3099/7-1 und -2).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 2.Habraken Y, Piette J. NF-kappaB activation by double-strand breaks. Biochem. Pharmacol. 2006;72:1132–1141. doi: 10.1016/j.bcp.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-Ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Mol. Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 4.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Jacob NK, Ladner KJ, Beg A, Perko JD, Tanner SM, Liyanarachchi S, Fishel R, Guttridge DC. RelA/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep. 2009;10:1272–1278. doi: 10.1038/embor.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karl S, Pritschow Y, Volcic M, Häcker S, Baumann B, Wiesmüller L, Debatin KM, Fulda S. Identification of a novel pro-apoptotic function of NF-κB in the DNA damage response. J. Cell. Mol. Med. 2009;13:4239–4256. doi: 10.1111/j.1582-4934.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benezra M, Chevallier N, Morrison DJ, MacLachlan TK, El-Deiry WS, Licht JD. BRCA1 augments transcription by the NF-κB transcription factor by binding to the Rel domain of the p65/RelA subunit. J. Biol. Chem. 2003;278:26333–26341. doi: 10.1074/jbc.M303076200. [DOI] [PubMed] [Google Scholar]
- 8.Habraken Y, Jolois O, Piette J. Differential involvement of the hMRE11/hRAD50/NBS1 complex, BRCA1 and MLH1 in NF-κB activation by camptothecin and X-ray. Oncogene. 2003;22:6090–6099. doi: 10.1038/sj.onc.1206893. [DOI] [PubMed] [Google Scholar]
- 9.Akyüz N, Boehden GS, Süsse S, Rimek A, Preuss U, Scheidtmann KH, Wiesmüller L. DNA substrate-dependence of p53-mediated regulation of double-strand-break-repair. Mol. Cell. Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhl M, Csernok A, Aydin S, Kreinberg R, Wiesmüller L, Gatz SA. Role of SIRT1 in homologous recombination. DNA Repair. 2010;9:383–393. doi: 10.1016/j.dnarep.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Kaltschmidt B, Kaltschmidt C, Hehner SP, Dröge W, Schmitz ML. Repression of NF-κB impairs HeLa cell proliferation by functional interference with cell cycle checkpoint regulators. Oncogene. 1999;18:3213–3225. doi: 10.1038/sj.onc.1202657. [DOI] [PubMed] [Google Scholar]
- 12.Restle A, Färber M, Baumann C, Böhringer M, Scheidtmann KH, Müller-Tidow C, Wiesmüller L. Dissecting the role of p53 phosphorylation in homologous recombination provides new clues for gain-of-function mutants. Nucleic Acids Res. 2008;36:5362–5375. doi: 10.1093/nar/gkn503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saintigny Y, Dumay A, Lambert S, Lopez BS. A novel role for the Bcl-2 protein family: specific suppression of the RAD51 recombination pathway. EMBO J. 2001;20:2596–2607. doi: 10.1093/emboj/20.10.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiese C, Pierce AJ, Gauny SS, Jasin M, Kronenberg A. Gene conversion is strongly induced in human cells by double-strand breaks and is modulated by the expression of BCL-x(L) Cancer Res. 2002;62:1279–1283. [PubMed] [Google Scholar]
- 15.Dumay A, Laulier C, Bertrand P, Saintigny Y, Lebrun F, Vayssière JL, Lopez BS. Bax and bid, two proapoptotic Bcl-2 family members, inhibit homologous recombination, independently of apoptosis regulation. Oncogene. 2006;25:3196–3205. doi: 10.1038/sj.onc.1209344. [DOI] [PubMed] [Google Scholar]
- 16.Wu K, Jiang SW, Thangaraju M, Wu G, Couch FJ. Induction of the BRCA2 promoter by nuclear factor-kappa B. J. Biol. Chem. 2000;275:35548–35556. doi: 10.1074/jbc.M004390200. [DOI] [PubMed] [Google Scholar]
- 17.De Siervi A, De Luca P, Moiola C, Gueron G, Tongbai R, Chandramouli GV, Haggerty C, Dzekunova I, Petersen D, Kawasaki E, et al. Identification of new Rel/NF-kappaB regulatory networks by focused genome location analysis. Cell Cycle. 2009;8:2093–2100. doi: 10.4161/cc.8.13.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Nievera CJ, Lee AYL, Wu X. Cell cycle-dependent complex formation of BRCA1-CtIP-MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 19.Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keimling M, Volcic M, Csernok A, Wieland B, Dörk T, Wiesmüller L. Functional characterization of clinically relevant mutations indicates dual roles of the ATM protein in regulating DNA-double strand break repair signalling pathways. FASEB J. 2011 doi: 10.1096/fj.11-185546. 21 July 2011 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 21.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delacôte F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl Acad. Sci. USA. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim JW, Kim H, Kim KH. Expression of Ku70 and Ku80 mediated by NF-κB and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J. Biol. Chem. 2002;277:46093–46100. doi: 10.1074/jbc.M206603200. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–464. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertrand P, Saintigny Y, Lopez BS. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muotri AR, Bottero V, Tergaonkar V, Correa RG. UV-mediated NF-kappaB activation is abolished in deficient XPC/D primary fibroblasts. Cell Cycle. 2006;5:1085–1089. doi: 10.4161/cc.5.10.2780. [DOI] [PubMed] [Google Scholar]
- 30.Wu ZH, Miyamoto S. Induction of a pro-apoptotic ATM–NF-κB pathway and its repression by ATR in response to replication stress. EMBO J. 2008;27:1963–1973. doi: 10.1038/emboj.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jänicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- 32.Faumont N, Le Clorennec C, Teira P, Goormachtigh G, Coll J, Canitrot Y, Cazaux C, Hoffmann JS, Brousset P, Delsol G, et al. Regulation of DNA polymerase beta by the LMP1 oncoprotein of EBV through the nuclear factor-kappaB pathway. Cancer Res. 2009;69:5177–5185. doi: 10.1158/0008-5472.CAN-08-2866. [DOI] [PubMed] [Google Scholar]
- 33.Canitrot Y, Capp JP, Puget N, Bieth A, Lopez B, Hoffmann JS, Cazaux C. DNA polymerase β overexpression stimulates the Rad51-dependent homologous recombination in mammalian cells. Nucleic Acids Res. 2004;32:5104–5112. doi: 10.1093/nar/gkh848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salminen A, Suuronen T, Huuskonen J, Kaarniranta K. NEMO shuttle: a link between DNA damage and NF-kappaB activation in progeroid syndromes? Biochem. Biophys. Res. Commun. 2008;367:715–718. doi: 10.1016/j.bbrc.2007.11.189. [DOI] [PubMed] [Google Scholar]
- 35.Jasin M. Homologous repair of DNA damage and tumourigenesis: the BRCA connection. Oncogene. 2002;2:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 36.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell. Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 37.Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008;27:3977–3985. doi: 10.1038/onc.2008.17. [DOI] [PubMed] [Google Scholar]
- 38.Blagosklonny MV, An WG, Melillo G, Nguyen P, Trepel JB, Neckers LM. Regulation of BRCA1 by protein degradation. Oncogene. 1999;18:6460–6468. doi: 10.1038/sj.onc.1203068. [DOI] [PubMed] [Google Scholar]
- 39.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souto-Carneiro MM, Fritsch R, Sepulveda N, Lagareiro MJ, Morgado N, Longo NS, Lipsky PE. The NF-kappaB canonical pathway is involved in the control of the exonucleolytic processing of coding ends during V(D)J recombination. J. Immunol. 2008;180:1040–1049. doi: 10.4049/jimmunol.180.2.1040. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosjean-Raillard J, Tailler M, Ades L, Perfettini JL, Fabre C, Braun T, De Botton S, Fenaux P, Kroemer G. ATM mediates constitutive NF-κB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2009;28:1099–1109. doi: 10.1038/onc.2008.457. [DOI] [PubMed] [Google Scholar]
- 43.Pratt MAC, Tibbo E, Robertson SJ, Jansson D, Hurst K, Perez-Iratxeta C, Lau R, Niu MY. The canonical NF-κB pathway is required for formation of luminal mammary neoplasias and is activated in the mammary progenitor population. Oncogene. 2009;28:2710–2722. doi: 10.1038/onc.2009.131. [DOI] [PubMed] [Google Scholar]
- 44.Beyne-Rauzy O, Recher C, Dastugue N, Demur C, Pottier G, Laurent G, Sabatier L, Mansat-De Mas V. Tumor necrosis factor alpha induces senescence and chromosomal instability in human leukemic cells. Oncogene. 2004;23:7507–7516. doi: 10.1038/sj.onc.1208024. [DOI] [PubMed] [Google Scholar]
- 45.Golding SE, Rosenberg E, Neill S. Extracellular signal-related kinase positively regulates Ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res. 2007;67:1046–1053. doi: 10.1158/0008-5472.CAN-06-2371. [DOI] [PubMed] [Google Scholar]
- 46.Ahmed KM, Li JJ. NF-kappa B-mediated adaptive resistance to ionizing radiation. Free Radic. Biol. Med. 2008;44:1–13. doi: 10.1016/j.freeradbiomed.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strair RK, Gharibo M, Schaar D, Rubin A, Harrison J, Aisner J, Lin HC, Lin Y, Goodell L, Anand M, et al. Nuclear factor-κB modulation in patients undergoing induction chemotherapy for acute myelogenous leukemia. Clin. Cancer Res. 2008;14:7564–7568. doi: 10.1158/1078-0432.CCR-08-1390. [DOI] [PubMed] [Google Scholar]
- 48.Courtois G, Smahi A. NF-κB-related genetic diseases. Cell Death Differ. 2006;13:843–851. doi: 10.1038/sj.cdd.4401841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.