Abstract
This review presents detailed information about the structure of triplet repeat RNA and addresses the simple sequence repeats of normal and expanded lengths in the context of the physiological and pathogenic roles played in human cells. First, we discuss the occurrence and frequency of various trinucleotide repeats in transcripts and classify them according to the propensity to form RNA structures of different architectures and stabilities. We show that repeats capable of forming hairpin structures are overrepresented in exons, which implies that they may have important functions. We further describe long triplet repeat RNA as a pathogenic agent by presenting human neurological diseases caused by triplet repeat expansions in which mutant RNA gains a toxic function. Prominent examples of these diseases include myotonic dystrophy type 1 and fragile X-associated tremor ataxia syndrome, which are triggered by mutant CUG and CGG repeats, respectively. In addition, we discuss RNA-mediated pathogenesis in polyglutamine disorders such as Huntington's disease and spinocerebellar ataxia type 3, in which expanded CAG repeats may act as an auxiliary toxic agent. Finally, triplet repeat RNA is presented as a therapeutic target. We describe various concepts and approaches aimed at the selective inhibition of mutant transcript activity in experimental therapies developed for repeat-associated diseases.
INTRODUCTION
In the early 1990s, the identification of a new class of disease-causing mutations caused considerable excitement in the community of human molecular geneticists. The mutations were inherited trinucleotide repeat (TNR) expansions, and the associated disorders became known as Trinucleotide Repeat Expansion Diseases (TREDs) (1). Over 20 neurological diseases have now been assigned to this group. Each disease is associated with a single defective gene, which triggers the process of pathogenesis through aberrant expression or toxic properties of mutant transcripts or proteins [reviewed in (2–4)]. Although researchers have been making efforts to develop treatments for TREDs for nearly two decades, they remain incurable.
TREDs include spinal and bulbar muscular atrophy (SBMA) (5), fragile X syndrome (FXS) (6), myotonic dystrophy type 1 (DM1) (7), Huntington's disease (HD) (8) and a number of spinocerebellar ataxias (SCA) (9,10). The first years of research on pathogenic mechanisms in TREDs resulted in clear mechanistic separation among different groups of the disorders. However, recent studies have begun to reveal that mutant RNA and mutant protein can act in parallel and exert their toxicities independently in some TREDs (11–13). Mutant transcripts may contribute to the pathogenesis of diseases driven by mutant proteins (11,12), and mutant proteins may contribute to the pathogenesis of disorders known as driven by toxic RNA (13). Thus, the long-standing borders between distinct pathomechanisms in TREDs are beginning to be crossed, and this crossing occurs in both directions.
Much of the recent excitement brought to the field of TREDs may be attributed to the rapid progress of research on various approaches to treat these diseases (14–16). All the approaches discussed here are aimed at targeting triplet repeat RNA sequences with the goal of disrupting their pathogenic interaction with sequestered proteins, inhibiting translation from the mutant allele or destroying mutant transcripts. In some of these approaches, detailed information on the structure of the target RNA is essential for the rational design of potent reagents that may become useful therapeutic tools in the future.
In this review, we summarize the results of detailed structural studies of triplet repeats present in transcripts of TRED genes, in either non-coding or protein coding regions. Relevant structural information is given to illustrate involvement of RNA structure in the mechanism of pathogenesis triggered by expanded repeats. Important recent findings are also presented in the context of TNR genomics. The genomic and transcriptomic perspectives are shown to better understand the abundance of various triplet repeats, i.e. their presence in the cells in which pathology develops and where selective targeting by various reagents must occur. The characteristics of interactions between TRED transcripts and specific proteins are also presented, as these interactions determine the downstream adverse effects of TNR mutations.
TRIPLET REPEATS ARE FREQUENT MOTIFS IN HUMAN TRANSCRIPTS
TNRs belong to simple sequence repeats (SSRs), also known as short tandem repeats or microsatellites, and are common motifs in the genomes of humans and many other species (17). The repeats mutate at a very high rate, are often polymorphic in length and functions proposed for the repeats are related to their variable length (18). They are copious not only in genomes but also in transcriptomes, and their abundance may be higher than originally thought due to the presence of bidirectional transcription across the majority of human genes and intergenic regions (19,20). Importantly, in translated sequences, TNRs are selected preferentially over dinucleotide or tetranucleotide repeats, because the length variation of TNRs does not change the reading frame (21).
Twenty different TNR motifs may potentially occur in RNAs if homotrinucleotide motifs are excluded and different phases of individual motifs are combined. The great abundance of some TNRs in cells raises questions about what roles these sequences might play in transcripts (22). TNRs differ in length, and the expression levels of their host transcripts vary greatly. The structures formed by TNRs in transcripts depend not only on the repeat motif, and the number of its units, but also on the presence of interruptions breaking the homogeneity of the repeat tract (23). TNRs that have beneficial structural features and functional properties are positively selected during evolution, and TNRs with deleterious features are selected against (24).
A number of studies have been performed and many resources developed to characterize the frequency and location of SSRs (including TNRs) in the human genome (17,25–27) and exome (28–33). The main questions that have been addressed are the following: how many human mRNAs contain TNRs? At what frequency do certain types of TNRs occur? What is the length distribution of various TNRs in transcripts? What is the preferred location of TNRs in mRNA? And what are the known and putative functions of these sequences?
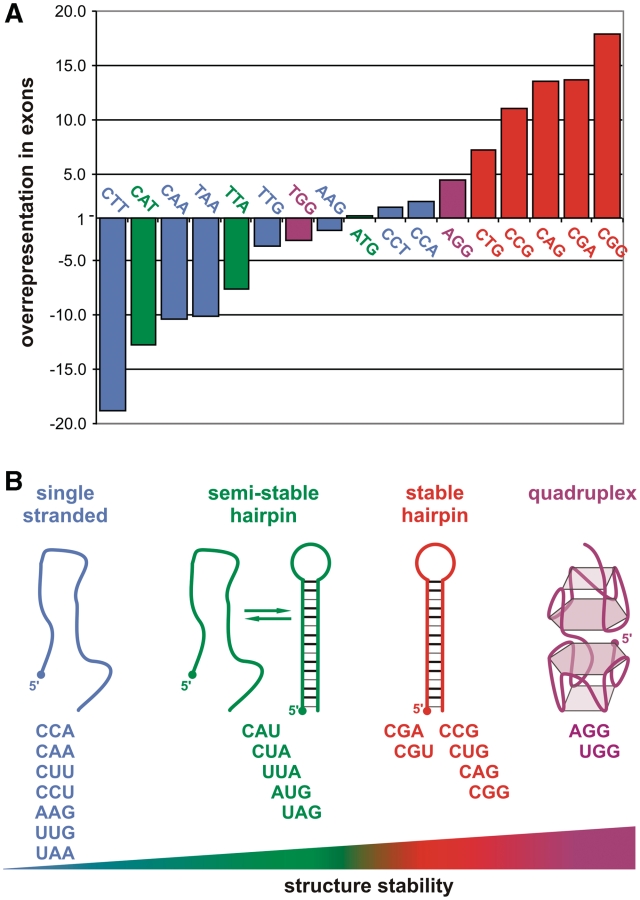
Three independent studies have provided the answers to these questions by identifying TNRs in the human genome reference sequence (27,29,34). In the most recent study, 32 448 tracts of uninterrupted TNRs composed of six or more repeated units were identified using the BLASTn algorithm (29). The relative frequencies of different TNR types were similar to those reported in earlier genome-wide surveys that used different repeat length and purity thresholds (25–27,34). As many as 1030 TNRs were identified in the exonic sequences of 878 genes. The TNRs that are strongly overrepresented in exons are CNG (where N is any nucleotide), CGA and AGG, whereas CTT, CAT, CAA, TAA and TTA are robustly underrepresented (Figure 1A). The shortest tracts are most prevalent, and the frequency of TNRs decreases roughly exponentially with their length. For the majority of TNR types, the longest tracts are <20 repeated units. However, for some TNRs such as AAG, several tracts >30 units have been identified (29). Of the 1030 exonic TNRs, 59% are located in the ORF, 28% are in the 5′-UTR and 13% are in the 3′-UTR. The CCA, CAG, CTG, CCT, AGG, AAG and ATG TNRs occur most frequently in the open reading frame (ORF) (~80%); AT-rich TNRs are more frequent in the 3′-UTR, whereas CCG and CGG repeats are most frequent in the 5’-UTR (52% and 62%, respectively) (29).
Figure 1.
The occurrence of various triplet repeats in the human transcriptome and their RNA structures. (A) Representation of TNRs composed of at least six repeat units in RefSeq mRNA sequences compared with the whole human genome sequence (17 out of 20 triplets are shown due to lack of CGT, CTA, TAG repeats in exons). Bar colors correspond to the four classes of RNA structure as shown in (B); (B) 20 different triplet repeat RNAs belong to four structural classes. On the diagram, they are ordered from left to right according to the increasing thermodynamic stability of their secondary structure: seven TNRs are single stranded even at low temperature (4°C), five AU-rich repeats form semi-stable hairpins in physiological conditions, the next six GC-rich TNRs form stable hairpin structures and two TNRs form the most stable quadruplexes.
To better characterize TNR sequences, their occurrence and genetic polymorphism have been investigated (35–38). Detailed information about triplet repeat length distribution in specific genes has been gathered experimentally for CAG and CTG repeats (36). A population genotyping study was conducted on 100 human genes selected to contain the longest runs of these repeats. The results demonstrated that very long and highly polymorphic repeat tracts are rare in genes not known to be associated with TREDs, which is in agreement with the results of a previous bioinformatics survey (23).
Functional association studies have been performed to gain some insight into the roles played by TNR-containing genes. It was found that genes coding for (i) proteins with transcription-related functions, (ii) proteins that interact with nucleic acids and (iii) proteins with nuclear localization are generally overrepresented among TNR-containing genes (27,29,37). These results as well as other lines of evidence suggest that the functions of TNRs can be expressed not only through proteins, but also at the DNA (24,35,39,40) and RNA (29,41–43) levels. Furthermore, TNR functions in RNA may strongly depend on the structures adopted by these sequences.
FOUR STRUCTURAL CLASSES OF TRIPLET REPEAT RNAs
To provide a basis for a functional analysis of triplet repeats in RNAs, the solution structures of oligoribonucleotides (ORNs) composed of the reiterations of specific triplets have been investigated under different experimental conditions using various methods. The ORNs that were first analyzed were four CNG motifs (N = A, C, G or U) reiterated 17 times (44). All these ORNs were found to form hairpin structures as demonstrated by chemical and enzymatic structure probing. The stem of the CNG repeat hairpin was shown to be composed of periodically occurring C–G and G–C base pairs and single N–N base mismatches. The hairpin loop was formed by either four or seven nucleotides. With the exception of the CGG repeat hairpin, the other three repeated CNG motifs form ‘slippery’ hairpins (i.e. tend to form alternative alignments unless they are fixed in one conformation by a G–C clamp) (44). Recently, the structures of all 20 different triplet motifs repeated either 17 or 20 times were subjected to a comparative analysis using biochemical structure probing as well as gel mobility analysis, and these structures were assigned to four classes (45). As shown in Figure 1B, AGG and UGG repeats form the most stable G-quadruplex structures; CGA, CGU and all CNG repeats form hairpins that are more stable than those of UAG, AUG, UUA, CUA and CAU repeats, whereas CAA, UUG, AAG, CUU, CCU, CAA and UAA repeats do not form any higher order structure. Further analysis of the hairpin and G-quadruplex structure-forming repeats was pursued using biophysical methods. UV-monitored structure melting revealed the following order of stability for CNG repeat hairpins: CGG > CAG > CUG > CCG; the stabilities of the AGG and UGG G-quadruplexes are roughly similar. CD spectra have shown that both G-quadruplexes are formed by parallel RNA strands (45). A shorter version of the G-quadruplex forming AGG RNA repeats (GGA)4 was also analyzed by NMR and CD (46,47). The intramolecular G-quadruplex was shown to be formed by a G:G:G:G tetrad plane and a G(:A):G:G(:A):G hexad plane (47). Other studies of triplet repeat ORN structures have included UV melting and/or CD studies of all CNG repeats or selected sequences of this group (48,49), gel mobility analysis of CGG repeats (50,51) and NMR studies of CGG repeats (52).
Crystal structures have been determined for short CUG (53,54), CAG (55) and CGG (56) repeats, which form intermolecular duplexes. The X-ray structures revealed details of the molecular architecture of these duplexes that are considered representative for stem portions of the CUG, CAG and CGG repeat hairpins. From a structural biology perspective, the nature and consequences of the periodic U:U, A:A and G:G mismatches were the most interesting findings. In the CUG repeat crystal structure, the U:U mismatches form stretched wobble interactions having only one hydrogen bond between the carbonyl O4 atom of one uracil residue and the N3 imino group of the opposite U residue (53). Similarly, in the CAG repeats, only one hydrogen bond is formed between the opposing adenine residues. This is an unusual and weak C2-H2•N1 bond. All the adenine residues are in the anti-conformation and serve as both hydrogen bond donors and acceptors (55). In the non-canonical G:G pairs found in CGG repeat duplexes, one guanosine residue is always in syn and the other is in anti conformation, and they form two hydrogen bonds, O6•N1H and N7•N2H. The helical structures of CGG repeats are more stable than those formed by CAG and CUG repeats (56).
Finally, an interesting correlation was found on comparing the occurrence of different triplet repeats in exons (described in a previous section) with their structures. As presented in Figure 1, TNRs that are strongly overrepresented in exons belong to the hairpin forming repeats, whereas underrepresented is the majority of repeat types that do not form any stable structure. The positive selection of repeats capable of forming hairpin structures in transcripts may suggest their importance in the regulation of gene expression, but the selection may also be acting at the level of amino acid repeats in proteins.
TRIPLET REPEAT EXPANSION DISEASES AND MECHANISMS OF PATHOGENESIS
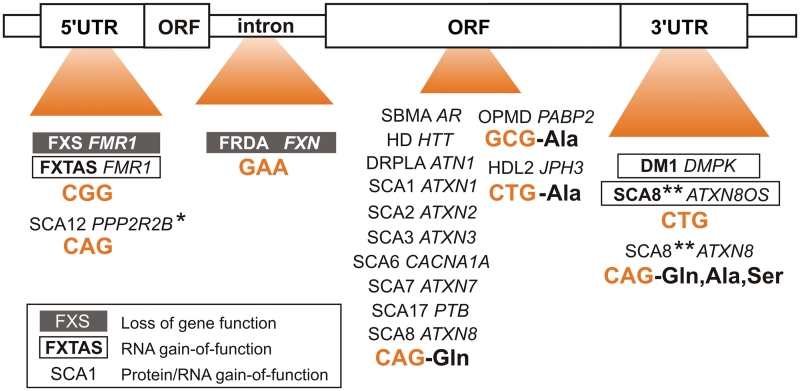
Over 20 different genes containing unstable TNRs have been implicated in the pathogenesis of human neurological diseases collectively named TREDs [reviewed in (1)]. Expanded CTG, CGG, GAA and CAG repeats are sources of degenerative changes leading to symptoms associated with DM1, FXTAS, Friedreich's ataxia (FRDA) as well as HD and a series of SCAs (Figure 2). These are typically late-onset inherited disorders and their causative repeat mutations are located in different parts of genes that primarily determine the number of potential toxic entities. The adverse effect of non-coding mutations in DM1 and FXTAS is principally determined by the expression of a mutant transcript (57–61), whereas the toxicity of coding mutations is pronounced by the presence of both RNA and protein, which harbor abnormally lengthened repeats (11,62). In two other non-coding repeat expansion disorders, FRDA and FXS, it is the diminished expression of specific proteins which triggers pathogenesis as a result of inhibited or abortive transcription across, respectively, expanded GAA and CGG repeats (63–67).
Figure 2.
Expanded triplet repeats are located in different parts of transcripts. Depending on the location of expandable repeats, neurological disease pathomechanisms can be divided into three groups. Repeats located in 5′-UTR, 3′-UTR or in introns cause diseases mainly via either loss of gene function or RNA gain-of-function, whereas repeats present in open reading frames trigger degeneration via gain-of-protein function in conjunction with RNA gain-of-function [in the figure legend, disease symbols (e.g. FXS), names of affected genes (e.g. FMR1), expanded repeat types and amino acids encoded are indicated]. In SCA12 CAG expansion in 5′-UTR also results in the elevated expression of the ppp2r2b protein (denoted by asterisk); In SCA8 bidirectional transcription also results in toxic proteins (denoted by double asterisks).
Among well-studied mechanisms underlying TREDs are: (i) toxic RNA gain-of-function caused by transcripts harboring expanded CUG, CAG or CGG repeats (2,61,68–70); (ii) toxic protein gain-of-function through expression of polyglutamine (polyQ) tract encoded by mutant CAG repeats (71,72); and (iii) aberrant loss-of-transcript and loss-of-protein function caused by GAA and CGG expansions (63–67). However, considering the results of the most recent reports one can speculate that the mechanistic complexity of pathogenesis in TREDs is higher and more variable. The presence of non-ATG initiated translation was recently reported by Ranum and colleagues (13,73), and bidirectional transcription through repeat regions was shown for several genes of TREDs (74–76). The bidirectional transcription is not only a source of sense and antisense transcripts, but also leads to the generation of triplet repeat-derived siRNAs targeting transcripts containing complementary repeats as shown by the groups of Bonini (77) and Richards (78). These results indicate the existence of novel toxic entities that may give rise to new potential pathomechanisms. Further studies will evaluate their importance to the pathogenesis of specific TREDs.
STRUCTURES OF TRIPLET REPEATS IN TRED TRANSCRIPTS
The involvement of mutant transcripts in the pathogenesis of DM1, and its possible contribution to the pathogenesis of FXTAS and polyQ diseases prompted researchers in the past decade to take on the detailed structure examination of triplet repeat regions in numerous TRED transcripts. A further argument for undertaking that effort was the conviction that the treatments of TREDs aimed at the allele-specific inhibition of mutant transcript or its destruction by direct repeat targeting will benefit from having a deeper insight into the structure of the target. The structural information gathered for ORNs (44,45) and described earlier in this review was insufficient for this purpose, as it did not provide answers to questions such as the following: what is the effect of repeat length on structures formed by repeats? What is the contribution of sequences flanking repeats to the structure of the repeat region? And what are the structural roles of various repeat interruptions?
The first study aimed at answering these questions was performed on the DMPK transcript implicated in DM1 pathogenesis (79). The study design included: (i) a selection of representative normal alleles of TRED genes based on population genotyping results; (ii) size selection of the repeat region based on RNA structure prediction; and (iii) PCR synthesis of DNA templates for in vitro transcription, RNA synthesis, end labelling and structure probing with the use of nucleases (80) and lead ions (81,82). Normal length transcripts containing 5, 11 and 21 CUG repeats revealed the conversion of a single-stranded repeat region into semi-stable slippery hairpins upon increasing the repeat length, whereas stable hairpins were formed by expanded 49 CUG repeats (79). The finding that double-stranded-like structures are formed by CUG repeats in expanded transcripts was instrumental to the later discovery that muscleblind-like 1 (MBNL1) protein is sequestered by mutant DMPK transcripts in DM1 patient cells (83).
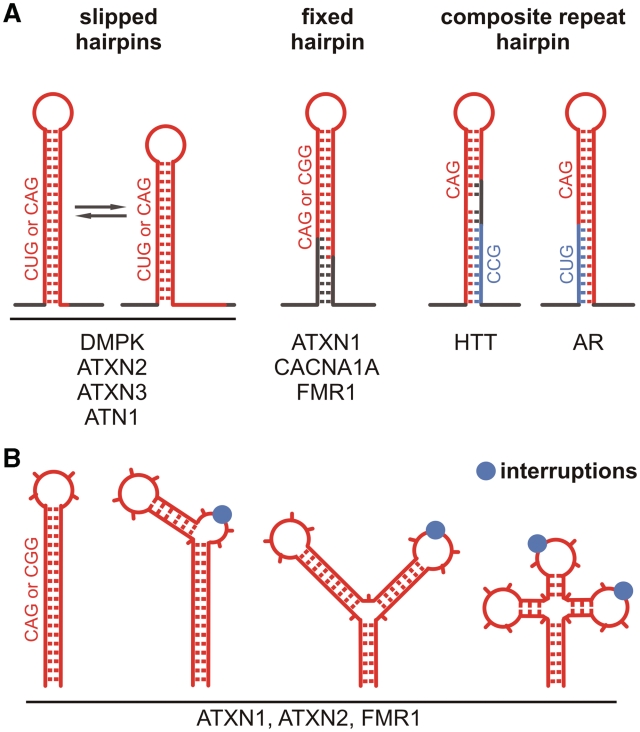
Similar in vitro structural analysis was further conducted for the triplet repeat regions of the majority of TRED transcripts and revealed their structural diversity. The contribution of sequences flanking the repeats to repeat hairpin stabilization was shown for ATXN1 (84), CACNA1A (85) and FMR1 (86) transcripts (Figure 3A). In contrast, flanking sequences did not influence the structures formed by repeats in DMPK (79), ATXN2 (87), ATXN3 or ATN1 transcripts (85). In HTT and AR transcripts, neighbouring repeats CCG and CUG, respectively, interact with CAG repeat tracts to form hairpins that have an unique composite architecture (88). It should be recognized, however, that the structures determined for triplet repeats in vitro may not fully recapitulate folding that occurs inside cells in the context of full-length RNA and various RNA binding proteins. The intracellular RNA structures and interactions, which were out of reach for a long time, are now amenable for investigation also for triplet repeats and on a transcriptome-wide scale. Such methods as global RACE (89), transcriptome-wide RNA structure probing (90) and HITS-CLIP (HIgh-Throughput Sequencing of CrossLinking and ImmunoPrecipitation products) (91) allow detecting products of RNA cleavages by endogenous nucleases or exogenous reagents, and determining high-resolution maps of RNA–protein interaction in vivo.
Figure 3.
Influence of sequences flanking TNRs and repeat interruptions on structures formed by TNRs in different TRED transcripts. (A) In some transcripts, the repeats themselves (red line) form autonomous hairpin structures that show slippage tendencies (two left panels). In several other transcripts, sequences flanking the repeat (dark gray line) influence the stability of the structure leading to the fixation of a single conformation (middle panel). In HTT and AR transcripts, two consecutive repeat sequences (red and blue lines) together with flanking sequences form a single composite hairpin structure (two right panels); (B) Sequences interrupting the triplet repeat region in the ATXN1, ATXN2 and FMR1 transcripts strongly influence structure formation. The interruptions can either break hairpin regularity or induce the formation of branched structures in which interruptions are predominantly present in terminal and internal hairpin loops.
In four TRED-related genes i.e. FMR1, ATXN1, ATXN2 and TBP, the majority of the normal alleles contain specific interruptions located within the repeat tracts. These are AGG triplets disrupting CGG repeats in the FMR1 gene, CAT triplets within CAG repeats in the ATXN1 gene and CAA interruptions within CAG repeats in both the ATXN2 and the TBP genes (87). Such repeat interruptions in DNA have been shown to function as protective elements, preventing pathogenic repeat expansion (92). But what could be their functions in transcripts? RNA structure probing revealed that AGG interruptions within the CGG repeat of the FMR1 transcript prevent single hairpin structure formation by the repeats (Figure 3B). Instead, branched hairpins are formed that have the substituted base either in the side loop or in an enlarged terminal loop, depending on the location of the interruption (86). Similar structural roles were demonstrated for CAU and CAA interspersions in the ATXN1 (84) and ATXN2 (87) transcripts (Figure 3B) and were predicted for the TBP transcript (87). It was hypothesized that the AGG interruptions may protect some premutation carriers from being prone to FXTAS by shortening the length of the hairpin composed of pure CGG repeats (86). In the cases of SCA1 and SCA2, rare carriers of expanded interrupted repeats have not developed any disease, and the RNA structure of the repeat region was found to be better correlated with pathogenesis than the length of the polyQ tract (84,87). These correlations suggested that RNA hairpin structure plays a more general role in the pathogenesis of TREDs.
TRIPLET REPEAT RNA INTERACTION WITH PROTEINS
The protein binding properties of TNR sequences have been mostly studied in relevance to the toxic features of mutant transcripts rather than in the context of the putative normal functions of TNR RNA (83,93,94). These studies took advantage of various methods to identify proteins that bind repeats and to characterize these interactions structurally. Most of this research has dealt with CUG repeats. First, the CUGBP1 protein was identified on the basis of its specific binding to single-stranded (CUG)8 incubated in HeLa cell nuclear extract (95). CUGBP1 is a member of the CELF (CUGBP and ETR-3-like factors) protein family, which regulate a number of post-transcriptional RNA processing steps including alternative splicing (96,97). The electron microscopy examination of the in vitro binding of recombinant CUGBP1 to transcripts containing expanded CUG repeats revealed that the protein localizes only to the base portion of the CUG repeat hairpin and its binding is not proportional to CUG repeat length (98). Studies have shown that CUGBP1 does not co-localize with mutant transcripts in DM1 cells (99,100).
Swanson and colleagues succeeded in identifying the RNA-binding protein, homologous to the Drosophila mbl proteins which binds to CUG repeats in a length-dependent manner and regulates alternative splicing (83). This protein was later shown to co-localize with mutant DMPK transcripts in a variety of DM1 patient cells and model organisms (99,101,102). Using the yeast two hybrid system, MBNL1 was shown to bind not only to CUG repeats but also to other types of repeated sequences, including CAG repeats (93). A filter-binding assay revealed a very similar in vitro binding affinity of MBNL1 to mutant CUG and CAG repeats (103). Moreover, the analysis of fluorescence recovery after photobleaching (FRAP) indicated that the affinity of the GFP-MBNL1 fusion protein to long CUG and CAG repeats is very similar in transfected cells (104). Most recently, HeLa and neuroblastoma cells expressing 5, 30, 70 or 200 exogenous CUG or CAG repeats were analyzed for colocalization repeat-containing transcripts with endogenous MBNL1 (12). Mutant transcripts with 70 or 200 CUG or CAG repeats were found to form nuclear inclusions that overlap with MBNL1. Other studies investigated the ability of CUG and CAG repeats to activate RNA-dependent protein kinase (PKR), the known cellular sensor of long dsRNA (105). CUG repeats of lengths with pathological consequences showed some activation of the kinase in vitro, and mutant CAG repeats of the HTT transcript were shown to activate PKR in human HD brain tissues (106).
The architecture of a CUG repeat hairpin of mutant length (54 repeats) in complex with MBNL1 was investigated using chemical and enzymatic structure probing and electron microscopy (103). MBNL1 multimers were shown to form ring-like structures that bind to the stem portion of the CUG repeat hairpin. The structures of very short oligomers containing CUG motifs in complex with MBNL1 were determined by crystallography, and the results suggested that MBNL1 may efficiently bind single-stranded CUG repeats (107). Very recently, MBNL1 was shown to bind (CUG)17 and (CAG)17 ORNs with similar affinity, whereas non-hairpin forming repeats of the same lengths composed of AUG or UUA repeats did not bind MBNL1 under the same assay conditions (12).
Interactions between CGG repeats and proteins have been recently investigated in cellular systems. Various cell lines were transfected with plasmids expressing 20, 40, 60 or 100 CGG repeats, and only the expression of long repeats supported the formation of nuclear aggregates in some but not all of the cell lines tested (108). These aggregates were shown to recruit the RNA-binding proteins SAM68, hnRNP-G and MBNL1. Earlier studies identified a number of other proteins that co-localize with long CGG repeats. Hagerman and colleagues used fluorescence-activated flow sorting, mass spectroscopy and immunohistochemistry to analyze the protein composition of RNA-containing intranuclear inclusions formed in astrocytes and neurons of FXTAS patients (109). These authors identified >20 proteins, including Lamin A/C, vimentin, hnRNP-A2/B1 and MBNL1. Furthermore, Pur alpha, hnRNP A2/B1 and CUGBP1 were shown to bind CGG repeats in the Drosophila model of FXTAS (94,110). The results of the protein binding studies suggest that MBNL1 sequestration by expanded CUG and CAG repeat transcripts is likely caused by direct RNA–protein interactions (103), and that SAM68 sequestration by expanded CGG repeats depends rather on indirect interaction (108).
RNA-MEDIATED PATHOGENESIS TRIGGERED BY TRIPLET REPEAT EXPANSION
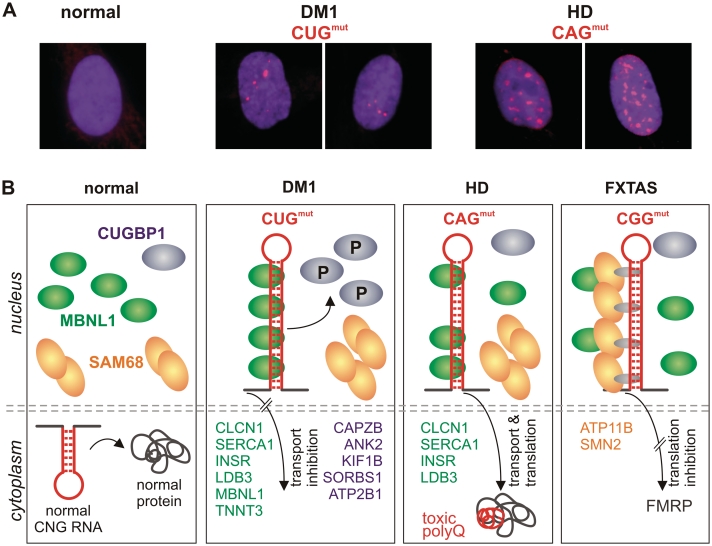
The ability of mutant CNG repeats to form long hairpin structures (44,79,98) is a significant determinant of toxic RNA-dependent pathogenesis in a number of TREDs. TNRs of mutant length gain a toxic function by binding essential proteins and consequently diminishing their functional cellular levels (Figure 4). A characteristic feature of the RNA–protein interactions in mutant TNR-expressing cells is the formation of nuclear foci [reviewed in (111)] in which sequestered proteins, such as MBNL1-3 and SAM68, become immobilized. These aggregates cause cells to develop degenerative changes that are manifested through misregulated alternative splicing and embryonic splicing patterns in adult tissues (108,112–114). In the following sections, we characterize the toxic effects of nuclear aggregates in cells expressing mutant CUG, CGG and CAG repeat RNA focusing on their adverse effect on alternative splicing.
Figure 4.
Dominant effects of toxic RNA repeats in different TREDs. (A) Mutant CUG and CAG repeat RNAs form nuclear foci (red) in DM1 and HD human fibroblasts. RNA FISH was performed using fluorescently labelled repeat probes: CAG in DM1 cells and CTG in HD cells, normal cells were treated with either probe; (B) In normal cells, transcripts with short CNG repeats are exported from the nucleus and translated into functional proteins (first panel). Thus, the availability of nuclear splicing factors CUGBP1 (blue), MBNL1 (green) and SAM68 (orange) is not compromised. In DM1 cells (second panel), the CUGmut transcript is retained in the nucleus, where it sequesters MBNL1 and causes the abnormal phosphorylation of CUGBP1, upregulating its nuclear levels. As a result, the incorrect splicing of several MBNL1- and CUGBP1-dependent transcripts occurs (examples of misspliced transcripts are specified as green and blue, respectively). In HD cells (third panel), expanded CAG repeat RNA partially sequesters MBNL1, but does not change the level of CUGBP1, which results in splicing abnormalities affecting some MBNL1-dependent transcripts. The CAGmut transcript is effectively translated into toxic polyQ protein (polyQ tract is indicated by a red line). In FXTAS cells (fourth panel), the CGGmut transcript colocalizes in the nucleus with SAM68 and MBNL1, but this colocalization does not occur via direct RNA–protein interaction. The consequence of this interaction is the aberrant splicing of only SAM68-dependent transcripts (orange). Moreover, FMR1 mRNA containing long CGG repeats in its 5′-UTR is poorly translated.
Cellular toxicity mediated by expanded CUG repeats in DM1 and SCA8
DM1 was the first neurological disorder in which the nuclear retention of transcripts containing expanded CUG repeats was detected (57,58). Although the reduced expression of both DMPK and the neighboring gene SIX5 accompany the mutant transcript retention (115), the main pathogenic mechanism is a deleterious gain-of-function of mutant RNA harboring CUG repeats. Over the years, as the number of DM1 model organisms has increased, the evidence for an RNA-dominant mechanism has grown stronger. The characteristic changes associated with DM1 pathogenesis in skeletal and cardiac muscles have been reproduced in transgenic mice and flies by expressing expanded CUG repeats (97,116,117). These similarities have been correlated with an adverse influence of mutant transcripts on RNA-binding proteins, i.e. MBNL1 and CUGBP1, that causes misregulated alternative splicing in DM1 (Figure 4B) (83,101).
To date, several aberrantly spliced transcripts have been identified that explain some of the phenotypic features of DM1 pathogenesis. For example, skeletal muscle hyperexitability (myotonia) and weakness have been associated with the mis-splicing of the chloride channel (CLCN1) (113,118–120) and the bridging integrator 1 (BIN1) (121) transcripts. Additionally, splicing alteration of the insulin receptor (INSR) contributes to the insulin resistance in DM1 muscle fibers (112,122). Defects in the cardiac functions are thought to be associated with the aberrant splicing of the troponin T type 2 (cTNT) transcript (123–125). Spliceopathy also features a number of central nervous system (CNS) transcripts including those for the glutamate receptor, ionotropic N-methyl d-aspartate 1 (NAMDAR1/GRIN1), amyloid beta precursor protein (APP) and microtubule-associated protein tau (MAPT) (68,126). Studies have shown that the DM-specific aberrant splicing pattern is reproduced not only in mice (102,127) and flies (128,129) expressing the DM1 mutation, but also in Mbnl1ΔE3/ΔE3 knockout mice (102) and in CUGBP1 overexpressing mice (127,130,131). These findings provide further evidence for the prominent roles played by these two splicing factors in DM1 pathogenesis.
An RNA gain-of-function mechanism by mutant CUG repeats is also implicated in SCA8 pathogenesis. This neurodegenerative disease primarily affects the cerebellum and is caused by a CTG•CAG repeat expansion that is transcribed in both directions and gives rise to the antisense non-coding CUG-harboring transcripts and translated CAG-bearing transcripts (Figure 2). The later transcripts undergo conventional translation that results in polyQ protein and non-ATG translation (13) that occurs across expanded CAG repeats in all reading frames and gives rise to the homopolymeric proteins of long polyglutamine, polyserine and polyalanine tracts [reviewed in (73,132)]. Whereas protein products of the sense transcripts build up as intranuclear inclusions that are detected in human brain autopsy tissue and in the brains of transgenic SCA8 mice (69), the nuclear retention of mutant CUG transcripts is manifested by the formation of RNA inclusions that colocalize with MBNL1 in selected neurons in the cerebellum. This event is thought to affect the alternative splicing of a number of CNS transcripts including APP, MAPT, NMDAR1 and MBNL1, which mimics aberrations observed in DM1 (68). In addition, the SCA8 CUG repeat expansion transcripts trigger splicing changes and increased expression of the GABA-A transporter 4 (GAT4/Gabt4) due to the dysregulation of MBNL/CELF regulated pathways in the brain (69).
Toxicity mediated by expanded CGG and CAG repeat RNAs
Strong evidence has been provided for the contribution of an RNA gain-of-function mechanism also in the pathogenesis of FXTAS and some of the polyglutamine disorders (11,109). In these diseases, specific proteins are sequestered into nuclear foci containing expanded CGG and CAG repeats that cause the deviated splicing of specific transcripts, which resembles DM1 pathogenesis (Figure 4B). In FXTAS, the mis-splicing might affect only transcripts regulated by proteins recruited early to CGG repeat foci as shown by Charlet-Berguerand and colleagues (108). In fact, in mutant CGG-expressing cells, the only foci protein whose functional activity is compromised is splicing factor SAM68, which is recruited to RNA foci before hnRNP G and MBNL1. Consequently, the misregulation of pre-mRNA alternative splicing controlled by SAM68 is observed in CGG-transfected cells and in FXTAS patients. An analysis of alternative splicing of the phospho-type-4 ATPase-11B (ATP11B) pre-mRNA showed a splicing misregulation in SAM68-depleted cells as measured by a significant decrease of exon-28B inclusion in comparison with control samples (108).
Experimental evidence has shown that expansions of CAG repeats in coding exons may convey pathogenic potential not only to polyQ proteins, but also to transcripts that harbor the repeat mutation. Initially, it was demonstrated by Cooper and colleagues that in COSM6 cells the transient expression of 960 CAG repeats causes nuclear retention of the expanded repeats (104). Subsequent experimental evidence provided support for the toxic capacity of mutant CAG repeat RNA in transgenic mice (11), worms (133) and the SCA3 Drosophila model (62) as well as in human SCA3 and HD fibroblasts (12). However, because mutant CAG repeat-triggered alternative splicing abnormalities were discovered very recently, the scale of these effects and their roles in pathogenesis has yet to be determined (Figure 4B). The most recent results from our laboratory show that in human HD and SCA3 fibroblasts, the expression of endogenous HTT and ATXN3 mutant transcripts causes the formation of CAG repeat foci and MBNL1 sequestration (12,88), which in turn trigger the aberrant splicing of endogenous sarco/endoplasmic reticulum Ca2+ ATPase 1 (SERCA1) and INSR transcripts. Similarly, expanded but exogenous CAG repeats mimic CUG repeats in the misregulation of alternative splicing, and in HeLa and neuroblastoma cells both types of repeat RNA cause defects in the processing of SERCA1, INSR, LIM domain binding 3 (LDB3) and CLCN1 pre-mRNAs (12). These results show that transcripts containing expanded CAG repeats may contribute to the pathogenesis of HD, SCA3 and perhaps other polyglutamine diseases.
EXPERIMENTAL THERAPIES DIRECTED AGAINST EXPANDED REPEAT RNA
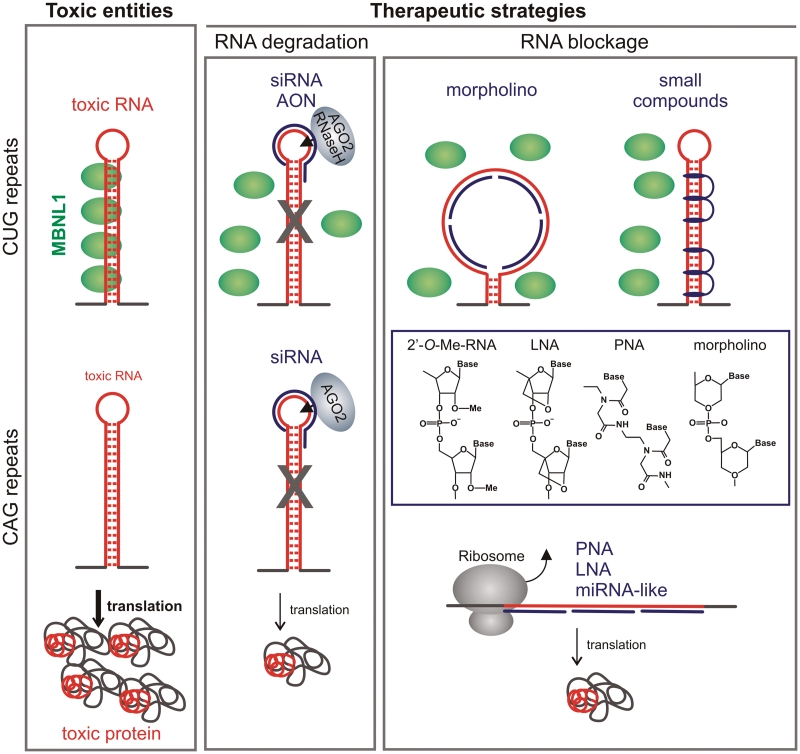
To date, three main strategies which use expanded repeat RNA as a target have been tested as experimental therapies for TREDs. The first is based on degrading mutant transcripts with RNA interference (RNAi) tools or antisense oligonucleotides (AON); the second utilizes repeat hairpin-specific small compounds or antisense oligomers to inhibit pathogenic interactions between repeat RNAs and nuclear proteins; and the third is aimed at blocking toxic protein synthesis by binding chemically modified antisense oligomers to repeats or by targeting repeats with mutant siRNAs acting as miRNAs (Figure 5). These strategies are aimed at destroying the toxic agents of pathogenic pathways associated with TREDs which involve either RNA, protein or both.
Figure 5.
Therapeutic strategies aimed at targeting expanded triplet repeats in TRED transcripts. The left panel shows the main pathogenic agent in diseases caused by expanded CUG or CAG repeats, which is either toxic RNA structure or toxic polyQ protein (both marked red). The middle and right panels show the application of therapeutic AON, RNAi reagents and small compounds (indicated by a blue line) that specifically bind to CUG or CAG repeat hairpins. Antisense reagents (AON) or siRNA called ‘cutters’ induce RNA degradation via RNaseH or AGO2 (middle panel), whereas ‘blockers’ (PNA, LNA, morpholino and miRNA-like duplexes) bind to repeat RNA alone or within protein complexes and inhibit either pathogenic RNA/protein interaction or translation of toxic polyQ proteins (right panel). See text for more details.
An important issue in therapies for TREDs that involve repeat targeting reagents is the requirement for gene and allele selectivity. Despite the fact, that there are numerous mRNAs containing triplet repeats in human transcriptome, the specific inhibition of mutant gene expression by targeting repeat regions is a promising therapeutic strategy for diseases caused by CAG and CUG repeat expansion. Among the features used in designing selective reagents are differences between normal and mutant transcripts related to their repeat sequence lengths and structural properties as well as, in the case of DM, cellular localization. The intended targets i.e. expanded repeats are likely to form hairpin structures in cells which make them both more prone (increased repeat length) and more resistant (more stable structure) to interaction with targeting reagents in comparison with normal repeats. The necessity of selective inhibition of mutant allele expression may not be equally important for all polyQ diseases; nevertheless, preserving the expression of the protein from normal allele seems to be an advantageous feature of any therapeutic strategy.
Below, we describe the use of AON and RNAi reagents as well as that of small compounds that specifically bind to some repeat hairpins. According to their mechanism of action, antisense reagents can be divided into two categories: ‘cutters’, which bind to complementary targets and induce their cleavage taking advantage of RNaseH or Argonaute 2 (AGO2) activities (AON or siRNA, respectively), and ‘blockers’, which are oligomers that bind to complementary targets, either alone or within protein complexes, but do not induce the cleavage of their complementary target. Examples of ‘blockers’ include peptide nucleic acids (PNAs), locked nucleic acids (LNAs), morpholino and miRNA-like acting duplexes (Figure 5). The exact mechanisms by which different reagents described below exert their activity are not yet known in detail.
The degradation of mutant transcripts induced by antisense oligonucleotides and RNAi triggers
Therapies for polyQ expansion diseases are aimed primarily at reducing the level of the mutant protein. In addition, mutant transcript downregulation may be desirable because of its involvement in pathogenesis. RNAi reagents require ~20 nt of complementary sequence for efficient silencing, which constitutes only 7 CAG repeats. While the normal alleles of CAG-bearing transcripts usually have 10–20 repeats, their mutant versions contain 40–100 CAG repeats meaning that transcripts from both alleles can be targeted by triplet repeat siRNA duplexes. Moreover, only among annotated human mRNAs, there are at least 50 transcripts containing CAG repeats and 30 with CUG repeats (29) being potential targets for complementary AONs and RNAi reagents.
RNA duplexes composed of CAG and CUG repeat strands have been tested in cell culture for their ability to silence HTT, AR, ATXN1 and ATXN3 transcripts. The reagents used included 81-bp long synthetic CAG/CUG RNA duplexes (134), 21-bp siRNA duplexes (135–137) and shRNA (138), and all showed only a slight silencing preference for mutant allele versus normal allele. Both strands of CAG/CUG siRNA were found to be active and silenced other transcripts also containing complementary repeats causing a considerable loss of the viability of human fibroblasts (137). The CAG/CUG siRNA induced some toxicity also in two Drosophila models co-expressing elevated levels of expanded CTG and CAG repeats (77,78). Interestingly, the cell culture experiments showed that hairpin structures which are likely formed in cells by expanded CAG repeats did not inhibit the activity of RNAi machinery directed at the repeat region (88,137). As a result, mutant HTT transcripts were efficiently targeted by CAG/CUG repeat siRNAs and the repeated CAG sequence was cleaved at numerous positions by AGO2 loaded with CUG repeat guide strand (88).
In DM1, the degradation of expanded CUG repeat transcripts was taken into consideration as possible therapy (139). To destroy toxic RNA in cells, several types of antisense reagents directed against CUG repeats have been successfully tested. Retroviral expression of 149-nt RNA complementary to part of the DMPK 3′-UTR in DM1 myoblasts resulted in the 80% silencing of the mutant DMPK transcript and a 50% reduction of the normal transcript leading to the partial restoration of some myoblast normal functions (140). Recently, Furling and colleagues (141) engineered AONs by the covalent linking of RNA sequences composed of 7, 11 or 15 CAG repeats to human U7 small nuclear RNA for their delivery exclusively to the cell nucleus. The lentiviral transduction of DM1 muscle cells with such constructs caused the specific silencing of ~60–70% of mutant DMPK mRNA without affecting normal DMPK transcripts. In the treated cells, the number of nuclei containing toxic mutant CUG repeat foci was reduced and the alternative splicing of several DM1-affected genes was significantly corrected (141).
In another study, Wansink and colleagues (142) designed a short single-stranded (CAG)7 2′-O-methyl phosphorothioate oligonucleotide (PS58) to degrade transcripts containing expanded CUG repeats. Such chemical modification of antisense oligomers is known to stabilize duplex formed with a target sequence and increase the resistance of AONs to cellular nucleases. PS58 decreased the level of mutant DMPK transcripts by 50–90% in patient myoblasts and in two DM1 mouse models, DM500 and HSALR. Interestingly, the levels of other transcripts containing short CUG repeats remained almost unchanged. Local administration of this AON into the mouse skeletal muscles reduced CUG repeat foci formation, partially restored the alternative splicing of DM-specific exons and significantly reduced myotonia (142).
Antisense ‘blockers’ targeting triplet repeat RNA
Another straightforward therapeutic strategy is targeting expanded CAG repeats in mutant transcripts to decrease the production of toxic polyQ protein. Corey and colleagues have shown the allele-selective inhibition of toxic protein translation in HD and SCA3 cells, using antisense PNA and LNA oligomers composed of 7 CTG repeats (136). After strong binding to complementary sequences, these oligomers formed an impassable translational barrier only in mutant transcripts (Figure 5). REP19N, containing 19 PNA residues and modified by addition of lysine residues at both termini, was the best allele-discriminating reagent, with a specificity of inhibition at least 10 times greater for mutant HTT protein translation than for normal protein. Moreover, the translation of other transcripts containing CAG repeats was not inhibited significantly in PNA- and LNA-treated cells. PNA oligomers were also tested for mutant ATXN3 translation inhibition; however, the maximum selectivity achieved (≤5) was lower than that observed for HTT (136). Oligomers with other chemical modifications such as 2′, 4′-constrained ethyl (cEt), carba-LNA, 2′-O-methoxyethyl (2′-MOE) or 2′-fluoro have also been shown to be promising blockers of mutant huntingtin translation (143).
Another approach to RNA repeat-targeted inhibition of mutant protein synthesis is the application of CAG/CUG siRNA duplexes containing base substitutions at specific positions causing mismatches with their mRNA targets. This approach was tested in cultured HD fibroblasts (137,144) where several duplexes caused the efficient translational inhibition of mutant huntingtin without downregulation of its transcript. CAG/CUG duplexes that form one or more mismatch with target sequences most likely act as miRNAs despite the fact that their target is located in the ORF. In most efficient and selective duplexes, the mismatched bases were present in the central positions of the duplex (144) or in its 3′ half (137). Interestingly, transcriptional activation of the normal HTT allele triggered by some of these duplexes occurred concomitantly with the silencing of mutant allele (137). This approach was also successfully used to target the CAG repeat region of the mutant ATXN3 transcript, although the selectivity of the reagents was lower than that observed for HTT inhibition (145). Efforts are being continued to develop this strategy further and to obtain even more selective reagents.
The primary pathogenic effect of expanded CUG repeats in DM1 is the sequestration of nuclear proteins, including MBNL1. Therefore, preventing pathogenic RNA–protein interactions is another approach to reducing the toxic effects of mutant RNA. Proof of the principle for this concept came from two independent studies (146,147). Swanson and colleagues showed that the overexpression of MBNL1 in an expanded CUG repeat expressing HSALR mice can reverse the DM-like phenotype (146). Thornton and colleagues using an antisense morpholino oligomer composed of 25 CAG repeats (CAG-25) inhibited the MBNL1/RNA interaction and disrupted the complexes preformed in vitro (147) (Figure 5). The morpholino modification provides resistance to cellular nucleases, high-stability duplexes with complementary targets and does not induce the cleavage of targeted RNA. These features made it a promising agent for in vivo therapy. The local administration of CAG-25 into the skeletal muscles of DM1 HSALR mice expressing (CUG)250 RNA resulted in some molecular and phenotypic changes manifested through the significant reduction or elimination of nuclear CUG repeat foci and nearly a complete reversion of abnormal splicing. These changes were driven by the release of MBNL1 protein from sequestration and the restoration of ion channel function leading to a significant reduction of myotonia. Moreover, in treated muscles, the mutant transcript was efficiently exported from the nucleus and translated in the cytoplasm. Importantly, the researchers found that the in vivo beneficial molecular effects were observed for as long as 14 weeks after a single injection and the expression of other transcripts containing short CUG repeats was not affected in skeletal muscle treated with CAG-25 (147).
Among the advantages of using repeat-targeting reagents, both ‘cutters’ and ‘blockers’, the most important is their potential application in several expansion-driven diseases. In contrast, widely exploited gene-specific and SNP-based strategies are applicable only for a fraction of patients suffering from specific disorders. On the other hand, the main challenge for repeat-targeting strategies is directing the reagent specifically to the mutant allele and leaving other repeat-containing transcripts intact. At this time, all potential mRNA off-targets are known (29), as discussed in ‘Triplet Repeats are Frequent Motifs in Human Transcripts’ section of this review, and their representatives may be tested along with testing allele selectivity of silencing disease-causing genes.
Small compounds that bind specifically to repeat RNA hairpins
Another strategy considered for the treatment of some TREDs is based on the identification of small compounds that interfere with pathogenic interactions between expanded RNA repeats and proteins. Different groups have used various approaches to identify ligands that specifically bind CUG and CAG repeat hairpins (148–153). Screening a library of ~11 000 compounds yielded a few molecules that showed selectivity for binding to either short or expanded CUG repeat hairpins (148). These ligands were able to prevent the CUG repeat/MBNL1 interaction in vitro with a low micromolar inhibition constant. In another work, pentamidine and neomycin B were shown to inhibit the interaction of short CUG repeat RNA with MBNL1 in vitro (152). However, only the former drug reversed the mis-splicing of two DM1-affected transcripts and released MBNL1 from nuclear inclusions in cells expressing 960 CUG repeats. Additionally, high doses of pentamidine administered by intraperitoneal injection into HSALR mice partially reversed the mis-splicing of Serca1 and Clcn1 pre-mRNAs (152). In the most recent work, d-amino acid hexapeptide (ABP1) was selected from a combinatorial peptide library screen in DM1 Drosophila model (153). In vitro, ABP1 induced a switch of CUG hairpins to a single-stranded conformation, whereas in Drosophila it reduced CUG foci formation and suppressed CUG-induced lethality. An intramuscular injection of ABP1 into DM1 HSALR mice reversed muscle histopathology and mis-splicing of Serca1 and Tnnt3.
The important issue regarding such compounds is their binding specificity for RNA structures formed by repeated sequences. To address this issue, Disney and colleagues designed several multivalent ligands containing between two and five Hoechst 33258 or kanamycin A derivatives attached to a peptoid backbone (Figure 5) (150,151,154). The modularly assembled ligands differed in their distance between RNA-binding modules. All ligands were tested both for their binding to CAG and CUG repeat transcripts and for the inhibition of repeat RNA/MBNL1 interactions. The best multivalent ligands inhibited the formation of RNA–protein complexes with inhibition constant falling within the low nanomolar range. These promising compounds have not yet been tested in cellular or animal systems expressing expanded repeats; however, their cell permeability has been demonstrated (151).
FINAL REMARKS AND FUTURE PERSPECTIVES
In this article, we reviewed several aspects of research on triplet repeat RNA. The occurrence of TNRs in human transcripts was presented and the structural features of both normal and pathogenic repeats were described. The role of mutant TNRs as triggers of disease pathogenesis and targets in experimental therapies for TREDs was discussed wherever relevant, from the perspective of RNA structure.
At present, all annotated human mRNAs containing triplet repeat tracts have been identified. However, only a fraction of these transcripts is relatively well characterized in terms of their TNR length polymorphism and tissue expression. There is a much larger gap in our knowledge of TNR RNAs if we take into consideration various antisense transcripts from human genes as well as sense and antisense transcripts from intergenic regions. Nevertheless, even this gap may be filled soon as relevant data may be mined by bioinformatics from several recently completed and on-going large-scale sequencing and expression profiling projects. We foresee that clearer and a nearly complete view of human TNR RNA repeatome is just on the horizon and this information may become available soon for more focused research.
The efforts of several laboratories over the past decade resulted in highly advanced structural characteristics of TNR RNA, which was achieved using biochemical and biophysical methods. TNRs may now be considered as a part of human RN-ome that belongs to the best-characterized structurally. We know which TNR types form G-quadruplex structures, which tend to form more stable and less stable hairpins and which are reluctant to form any higher order structures. We also know that repeats having hairpin structure forming potential are overrepresented in exons and therefore are likely implicated in some specific biological functions. At present, however, the normal functions of TNRs in transcripts are very poorly understood.
The specific functions of TNRs in RNA need to be expressed via interactions with proteins. But only a few proteins that interact directly with normal length TNR RNA have been identified thus far and their interactions characterized. As described in this review, the length of TNR RNA influences its protein binding properties and only mutant TNRs are efficient in specific protein sequestration. This raises a more general question about the protein binding potential of normal TNR sequences. One possible answer is that this potential is low and only few proteins are capable of getting involved in such interaction. The alternative answer is that this potential is higher, but no systematic study was conducted thus far to disclose it. In our opinion, further research focused on finding additional proteins that act on relatively short normal repeats and on expanded pathogenic repeats, may help in a better understanding of the normal roles of these sequences, and possibly in identifying new pathways of pathogenesis in TREDs. High-throughput analyses of triplet repeat RNAs associating with proteins are also encouraged to identify their transcriptome-wide interaction maps in cells.
Looking forward to any future structural studies of TNR RNA, we predict that recent successes at resolving crystal structures of short TNR duplexes may stimulate efforts to determine high-resolution structures of longer repeat TNRs and their complexes with various biologically and therapeutically relevant ligands. Further progress in this direction may help to better understand the RNA-triggered pathogenic mechanisms in TREDs, and promote a rational design of repeat-targeting therapeutic agents.
Recent years have witnessed rapid progress in developing experimental therapies for TREDs in various cellular and animal model systems. Several different concepts and approaches have been successfully tested, making clinical trials on humans a realistic prospect. Repeat-targeting antisense reagents and RNA interference reagents acting in miRNA fashion seem to be the most promising treatment options. Efforts are now under way to better characterize such reagents and optimize their gene selectivity and allele selectivity, and minimize sequence-specific and non-specific off-target effects.
FUNDING
European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-00-098/08 to W.J.K.); Ministry of Science and Higher Education (N N301 569340 to W.J.K.; N N302 278937 to P.K.; N N302 260938 to K.S. and N N401 572140 to M.W.). Funding for open access charge: European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-00-098/08).
Conflict of interest statement. None declared.
REFERENCES
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Li LB, Bonini NM. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. Trends Neurosci. 2010;33:292–298. doi: 10.1016/j.tins.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 4.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 6.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 7.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 8.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 9.Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 10.Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 11.Hsu RJ, Hsiao KM, Lin MJ, Li CY, Wang LC, Chen LK, Pan H. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS One. 2011;6:e16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr608. (doi:10.1093/nar/gkr1608; epub ahead of print; 27 July 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudreau RL, Rodriguez-Lebron E, Davidson BL. RNAi medicine for the brain: progresses and challenges. Hum. Mol. Genet. 2011;20:R21–27. doi: 10.1093/hmg/ddr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denovan-Wright EM, Davidson BL. RNAi: a potential therapy for the dominantly inherited nucleotide repeat diseases. Gene Ther. 2006;13:525–531. doi: 10.1038/sj.gt.3302664. [DOI] [PubMed] [Google Scholar]
- 16.Scholefield J, Wood MJ. Therapeutic gene silencing strategies for polyglutamine disorders. Trends Genet. 2010;26:29–38. doi: 10.1016/j.tig.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 19.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309:1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 20.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasinska A, Krzyzosiak WJ. Repetitive sequences that shape the human transcriptome. FEBS Lett. 2004;567:136–141. doi: 10.1016/j.febslet.2004.03.109. [DOI] [PubMed] [Google Scholar]
- 22.Krzyzosiak WJ, Sobczak K, Napierala M. In: Genetic Instabilities and Neurological Diseases. Wells RD, Ashizawa T, editors. 2006. 2nd edn. Elsevier Academic Press, San Diego, CA, pp. 705–713. [Google Scholar]
- 23.Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, Krzyzosiak WJ. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Astolfi P, Bellizzi D, Sgaramella V. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene. 2003;317:117–125. doi: 10.1016/s0378-1119(03)00659-0. [DOI] [PubMed] [Google Scholar]
- 27.Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borstnik B, Pumpernik D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002;12:909–915. doi: 10.1101/gr.138802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen BE, Villesen P, Wiuf C. Short tandem repeats in human exons: a target for disease mutations. BMC Genomics. 2008;9:410. doi: 10.1186/1471-2164-9-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc. Natl Acad. Sci. USA. 2009;106:17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pumpernik D, Oblak B, Borstnik B. Replication slippage versus point mutation rates in short tandem repeats of the human genome. Mol. Genet. Genomics. 2008;279:53–61. doi: 10.1007/s00438-007-0294-1. [DOI] [PubMed] [Google Scholar]
- 33.Stallings RL. Distribution of trinucleotide microsatellites in different categories of mammalian genomic sequence: implications for human genetic diseases. Genomics. 1994;21:116–121. doi: 10.1006/geno.1994.1232. [DOI] [PubMed] [Google Scholar]
- 34.Clark RM, Bhaskar SS, Miyahara M, Dalgliesh GL, Bidichandani SI. Expansion of GAA trinucleotide repeats in mammals. Genomics. 2006;87:57–67. doi: 10.1016/j.ygeno.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Wren JD, Forgacs E, Fondon JW, III, Pertsemlidis A, Cheng SY, Gallardo T, Williams RS, Shohet RV, Minna JD, Garner HR. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 2000;67:345–356. doi: 10.1086/303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozanska M, Sobczak K, Jasinska A, Napierala M, Kaczynska D, Czerny A, Koziel M, Kozlowski P, Olejniczak M, Krzyzosiak WJ. CAG and CTG repeat polymorphism in exons of human genes shows distinct features at the expandable loci. Hum. Mutat. 2007;28:451–458. doi: 10.1002/humu.20466. [DOI] [PubMed] [Google Scholar]
- 37.Butland SL, Devon RS, Huang Y, Mead CL, Meynert AM, Neal SJ, Lee SS, Wilkinson A, Yang GS, Yuen MM, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobczak K, Krzyzosiak WJ. Patterns of CAG repeat interruptions in SCA1 and SCA2 genes in relation to repeat instability. Hum. Mutat. 2004;24:236–247. doi: 10.1002/humu.20075. [DOI] [PubMed] [Google Scholar]
- 39.Fondon JW, III, Hammock EA, Hannan AJ, King DG. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci. 2008;31:328–334. doi: 10.1016/j.tins.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Fondon JW, III, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc. Natl Acad. Sci. USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richards RI, Holman K, Yu S, Sutherland GR. Fragile X syndrome unstable element, p(CCG)n, and other simple tandem repeat sequences are binding sites for specific nuclear proteins. Hum. Mol. Genet. 1993;2:1429–1435. doi: 10.1093/hmg/2.9.1429. [DOI] [PubMed] [Google Scholar]
- 42.Raca G, Siyanova EY, McMurray CT, Mirkin SM. Expansion of the (CTG)(n) repeat in the 5′-UTR of a reporter gene impedes translation. Nucleic Acids Res. 2000;28:3943–3949. doi: 10.1093/nar/28.20.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 44.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsugami A, Mashima T, Nishikawa F, Murakami K, Nishikawa S, Noda K, Yokoyama T, Katahira M. Structural analysis of r(GGA)4 found in RNA aptamer for bovine prion protein. Nucleic Acids Symp. Ser. 2008:179–180. doi: 10.1093/nass/nrn091. [DOI] [PubMed] [Google Scholar]
- 47.Nishikawa F, Murakami K, Matsugami A, Katahira M, Nishikawa S. Structural studies of an RNA aptamer containing GGA repeats under ionic conditions using microchip electrophoresis, circular dichroism, and 1D-NMR. Oligonucleotides. 2009;19:179–190. doi: 10.1089/oli.2008.0167. [DOI] [PubMed] [Google Scholar]
- 48.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 49.Pinheiro P, Scarlett G, Rodger A, Rodger PM, Murray A, Brown T, Newbury SF, McClellan JA. Structures of CUG repeats in RNA. Potential implications for human genetic diseases. J. Biol. Chem. 2002;277:35183–35190. doi: 10.1074/jbc.M202235200. [DOI] [PubMed] [Google Scholar]
- 50.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ofer N, Weisman-Shomer P, Shklover J, Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009;37:2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zumwalt M, Ludwig A, Hagerman PJ, Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 53.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U-U wobble': implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl Acad. Sci. USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr368. (doi:10.1093/nar/gkr1368; epub ahead of print; 19 May 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman PJ. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 61.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 62.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 64.Kim E, Napierala M, Dent SY. Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr542. (doi:10.1093/nar/gkr1542; epub ahead of print; 10 July 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J. Biol. Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Punga T, Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 68.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 69.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 71.Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 72.Marsh JL, Walker H, Theisen H, Zhu YZ, Fielder T, Purcell J, Thompson LM. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 73.Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 2011;7:e1002018. doi: 10.1371/journal.pgen.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilburn B, Rudnicki DD, Zhao J, Weitz TM, Cheng Y, Gu X, Greiner E, Park CS, Wang N, Sopher BL, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 2011;20:3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu Z, Teng X, Bonini NM. Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy. PLoS Genet. 2011;7:e1001340. doi: 10.1371/journal.pgen.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawlor KT, O'Keefe LV, Samaraweera SE, van Eyk CL, McLeod CJ, Maloney CA, Dang TH, Suter CM, Richards RI. Double stranded RNA is pathogenic in Drosophila models of expanded repeat neurodegenerative diseases. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr292. (doi:10.1093/hmg/ddr1292; epub ahead of print; 15 July 2011) [DOI] [PubMed] [Google Scholar]
- 79.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 80.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krzyzosiak WJ, Marciniec T, Wiewiorowski M, Romby P, Ebel JP, Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- 82.Ciesiolka J, Michalowski D, Wrzesinski J, Krajewski J, Krzyzosiak WJ. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J. Mol. Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 83.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sobczak K, Krzyzosiak WJ. Imperfect CAG repeats form diverse structures in SCA1 transcripts. J. Biol. Chem. 2004;279:41563–41572. doi: 10.1074/jbc.M405130200. [DOI] [PubMed] [Google Scholar]
- 85.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 86.Napierala M, Michalowski D, de Mezer M, Krzyzosiak WJ. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sobczak K, Krzyzosiak WJ. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 88.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, Nelson DL, Sinden RR. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 93.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 94.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 97.de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 98.Michalowski S, Miller JW, Urbinati CR, Paliouras M, Swanson MS, Griffith J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 101.Cardani R, Mancinelli E, Rotondo G, Sansone V, Meola G. Muscleblind-like protein 1 nuclear sequestration is a molecular pathology marker of DM1 and DM2. Eur. J. Histochem. 2006;50:177–182. [PubMed] [Google Scholar]
- 102.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 103.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 105.Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peel AL, Rao RV, Cottrell BA, Hayden MR, Ellerby LM, Bredesen DE. Double-stranded RNA-dependent protein kinase, PKR, binds preferentially to Huntington's disease (HD) transcripts and is activated in HD tissue. Hum. Mol. Genet. 2001;10:1531–1538. doi: 10.1093/hmg/10.15.1531. [DOI] [PubMed] [Google Scholar]
- 107.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 110.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr299. (doi: 10.1093/hmg/ddr1299; epub ahead of print; 16 July 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 113.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 114.Ohsawa N, Koebis M, Suo S, Nishino I, Ishiura S. Alternative splicing of PDLIM3/ALP, for alpha-actinin-associated LIM protein 3, is aberrant in persons with myotonic dystrophy. Biochem. Biophys. Res. Commun. 2011;409:64–69. doi: 10.1016/j.bbrc.2011.04.106. [DOI] [PubMed] [Google Scholar]
- 115.Frisch R, Singleton KR, Moses PA, Gonzalez IL, Carango P, Marks HG, Funanage VL. Effect of triplet repeat expansion on chromatin structure and expression of DMPK and neighboring genes, SIX5 and DMWD, in myotonic dystrophy. Mol. Genet. Metab. 2001;74:281–291. doi: 10.1006/mgme.2001.3229. [DOI] [PubMed] [Google Scholar]
- 116.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 117.Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, Willer JC, Ourth L, Duros C, Brisson E, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 118.Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 120.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 122.Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am. J. Hum. Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 124.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dhaenens CM, Schraen-Maschke S, Tran H, Vingtdeux V, Ghanem D, Leroy O, Delplanque J, Vanbrussel E, Delacourte A, Vermersch P, et al. Overexpression of MBNL1 fetal isoforms and modified splicing of Tau in the DM1 brain: two individual consequences of CUG trinucleotide repeats. Exp. Neurol. 2008;210:467–478. doi: 10.1016/j.expneurol.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 127.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Machuca-Tzili L, Thorpe H, Robinson TE, Sewry C, Brook JD. Flies deficient in Muscleblind protein model features of myotonic dystrophy with altered splice forms of Z-band associated transcripts. Hum. Genet. 2006;120:487–499. doi: 10.1007/s00439-006-0228-8. [DOI] [PubMed] [Google Scholar]
- 129.Garcia-Lopez A, Monferrer L, Garcia-Alcover I, Vicente-Crespo M, Alvarez-Abril MC, Artero RD. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One. 2008;3:e1595. doi: 10.1371/journal.pone.0001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 131.Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wojciechowska M, Krzyzosiak WJ. CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol. 2011;8:565–571. doi: 10.4161/rna.8.4.15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang LC, Chen KY, Pan H, Wu CC, Chen PH, Liao YT, Li C, Huang ML, Hsiao KM. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell Mol. Life Sci. 2011;68:1255–1267. doi: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Caplen NJ, Taylor JP, Statham VS, Tanaka F, Fire A, Morgan RA. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum. Mol. Genet. 2002;11:175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
- 135.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fiszer A, Mykowska A, Krzyzosiak WJ. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 139.Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 140.Furling D, Doucet G, Langlois MA, Timchenko L, Belanger E, Cossette L, Puymirat J. Viral vector producing antisense RNA restores myotonic dystrophy myoblast functions. Gene Ther. 2003;10:795–802. doi: 10.1038/sj.gt.3301955. [DOI] [PubMed] [Google Scholar]
- 141.Francois V, Klein AF, Beley C, Jollet A, Lemercier C, Garcia L, Furling D. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011;18:85–87. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- 142.Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hu J, Liu J, Corey DR. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hu J, Gagnon KT, Liu J, Watts JK, Syeda-Nawaz J, Bennett CF, Swayze EE, Randolph J, Chattopadhyaya J, Corey DR. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J. Am. Chem. Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl Acad. Sci. USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J. Am. Chem. Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. The role of flexibility in the rational design of modularly assembled ligands targeting the RNAs that cause the myotonic dystrophies. Chembiochem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Garcia-Lopez A, Llamusi B, Orzaez M, Perez-Paya E, Artero RD. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl Acad. Sci. USA. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]