Abstract
RNA editing enhances the diversity of gene products at the post-transcriptional level. Approaches for genome-wide identification of RNA editing face two main challenges: separating true editing sites from false discoveries and accurate estimation of editing levels. We developed an approach to analyze transcriptome sequencing data (RNA-seq) for global identification of RNA editing in cells for which whole-genome sequencing data are available. We applied the method to analyze RNA-seq data of a human glioblastoma cell line, U87MG. Around 10,000 DNA–RNA differences were identified, the majority being putative A-to-I editing sites. These predicted A-to-I events were associated with a low false-discovery rate (∼5%). Moreover, the estimated editing levels from RNA-seq correlated well with those based on traditional clonal sequencing. Our results further facilitated unbiased characterization of the sequence and evolutionary features flanking predicted A-to-I editing sites and discovery of a conserved RNA structural motif that may be functionally relevant to editing. Genes with predicted A-to-I editing were significantly enriched with those known to be involved in cancer, supporting the potential importance of cancer-specific RNA editing. A similar profile of DNA–RNA differences as in U87MG was predicted for another RNA-seq data set obtained from primary breast cancer samples. Remarkably, significant overlap exists between the putative editing sites of the two transcriptomes despite their difference in cell type, cancer type, and genomic backgrounds. Our approach enabled de novo identification of the RNA editome, which sets the stage for further mechanistic studies of this important step of post-transcriptional regulation.
RNA editing is a post-transcriptional process that alters the RNA sequences by base modifications, insertions, and deletions, thereby enhancing the diversity of gene products (for reviews, see Gott and Emeson 2000; Bass 2002; Maydanovych and Beal 2006; Farajollahi and Maas 2010; Nishikura 2010). The most prevalent type of known RNA editing in higher eukaryotes is A-to-I editing, where adenosine (A) residues are converted into inosine (I). The ADAR (adenosine deaminase acting on RNA) enzymes are the main players known to mediate A-to-I editing by binding to double-stranded RNAs (dsRNAs) ,which serve as the substrate for editing (Bass 2002; Nishikura 2010). However, target recognition by ADARs and the mechanisms of substrate interaction are not well understood. Since I is interpreted as guanosine during translation, A-to-I changes in protein-coding sequences may lead to codon changes and altered functional properties of the proteins (Maas 2010). In addition, A-to-I editing can play important roles in regulating gene expression (Maas 2010), such as by altering alternative splicing (Rueter et al. 1999; Laurencikiene et al. 2006; Schoft et al. 2007), miRNA sequences (Kawahara et al. 2007, 2008; Reid et al. 2008; Dupuis and Maas 2010), or miRNA target sites in the mRNA (Liang and Landweber 2007; Borchert et al. 2009). Other types of putative RNA editing events are also known, for example, C-to-U editing and U-to-C and G-to-A conversions (Nutt et al. 1994; Sharma et al. 1994; Villegas et al. 2002; Klimek-Tomczak et al. 2006), but with much less prevalence.
To identify RNA editing sites on a genome-wide scale, new approaches were developed in recent years built upon bioinformatic analyses and high-throughput sequencing methods (Wulff et al. 2010). Bioinformatic methods were often used to identify disparities between DNA and RNA sequences (likely due to RNA editing) by analyzing cDNA, expressed sequence tag (EST), and genomic sequences (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004; Gommans et al. 2008; Zaranek et al. 2010). To reduce false positives due to sequencing errors or somatic mutations, it was often necessary to use a priori knowledge of editing patterns to restrain the search, such as the known feature of clustering of putative editing sites or the presence of dsRNA structure. However, incorporation of such constraints often limits the results to editing sites with the corresponding characteristics. Taking advantage of the recently available high-throughput sequencing technology, Li et al. (2009a) developed an approach to verify 36,000 editing-site candidates by designing padlock probes to amplify the corresponding cDNA and genomic DNA (gDNA) regions, followed by sequencing of the amplification products. Others also designed similar approaches where editing-site candidates were specifically amplified and sequenced (Wahlstedt et al. 2009; Abbas et al. 2010).
The above approaches depend on a priori knowledge of editing-related features or candidate editing sites. Another desirable feature that is not afforded by some of the methods is the estimation of RNA editing levels. RNA editing levels (or editing ratios) represent the proportion of edited RNA molecules among all RNA molecules of a particular gene. Knowledge of editing levels can have profound biological significance. Recently, de novo identification of editing sites was made possible by whole-transcriptome sequencing (RNA-seq) (Picardi et al. 2010; Rosenberg et al. 2010; Ju et al. 2011; Li et al. 2011). Quantitative estimation of editing levels may be achieved by sequencing a large number of reads via high-throughput sequencing.
In analyzing RNA-seq data, a significant challenge lies in the mapping of the sequencing reads. At an RNA editing site, some or all RNA-seq reads contain the nucleotide that is different from the one in the reference genome. Mapping of such reads via commonly used approaches can suffer from a bias favoring reads harboring the reference base, a similar problem as previously reported for read-mapping in the presence of expressed single nucleotide polymorphisms (SNPs) (Degner et al. 2009; Heap et al. 2009). Here, we developed new mapping and analysis strategies to study RNA editing based on RNA-seq. We show that this approach is associated with a false-discovery rate of ∼5%, much lower than those reported by previous methods (Wulff et al. 2010). In addition, our method allows relatively accurate estimation of editing levels that correlate well with those derived by the traditional clonal sequencing method. Enabled by the large number of events identified in our study, we conducted a detailed characterization of sequence, evolutionary, and structural features related to A-to-I editing, and revealed novel insights about potential regulatory mechanisms and functional roles of editing.
Results
Mapping of RNA-seq reads for analysis at single-nucleotide level
To identify candidate RNA editing sites, we developed an approach that can accurately distinguish single-nucleotide differences in one set of RNA-seq data (Fig. 1A). A key step in this approach is the mapping of short sequencing reads containing the edited bases. Incorrect mapping of such reads may lead to not only inaccurate estimates of editing levels but also false-positive predictions of editing sites (see Discussion). By using the RNA-seq data described below (ADAR [also known as ADAR1] knockdown and control experiments), we estimated that the false-discovery rate could be as high as 28% if read-mapping was carried out in the nominal way used by many previous studies (e.g., allowing three of four mismatches in a 60-nt read).
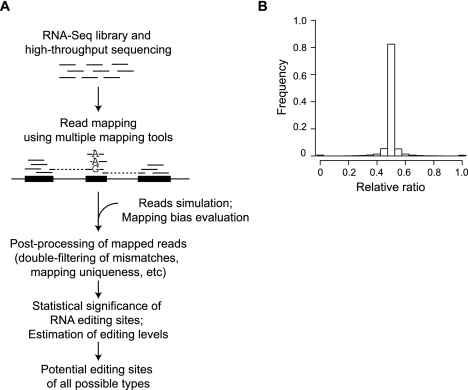
Figure 1.
Identification of RNA editing sites. (A) Generative process of the pipeline. (B) Evaluation of mapping bias using simulated data. Histogram shows the distribution of relative ratios of all simulated genomic sites with alternative alleles. Relative ratio is defined as follows: (N_mapped_ref/N_simulated_ref)/(N_mapped_ref/N_simulated_ref + N_mapped_edit/N_simulated_edit), where N_mapped_ref is the number of reads mapped to the reference base (e.g., A for A-to-I editing) and N_mapped_edit is the number of reads mapped to the edited base. N_simulated_ref and N_simulated_edit are defined similarly, but for the originally simulated reads. The average of all relative ratios is 0.499 and median is 0.500, neither of which is significantly different from the expected ratio 0.5 (P = 0.1, P = 0.3, respectively).
It is expected that the mapping accuracy of reads originating from regions with alternative bases in the RNA is sensitive to the treatment of mismatches in the mapped reads. Problems related to mapping can be exuberated if the sequence alignment tool does not provide 100% accuracy, as is the case for all available tools. To this end, we developed a strategy that combines the power of multiple read-mapping tools and stringently filters the mapping results according to the number of mismatches, uniqueness, and relative mapped locations of read pairs (Methods). In this strategy, we applied “double-filtering” of mismatches in the mapped reads such that only reads that mapped uniquely with ≤n1 mismatches and did not map to other genomic loci with ≤n2 mismatches are retained (n2 > n1). This method effectively removes reads with ambiguous mappings and those overlapping homologous regions in the genome.
Evaluation of mapping bias for single-nucleotide differences
To evaluate the mapping strategy, we simulated 870,280 reads (60 nt in length) covering 21,757 heterozygous genomic sites assumed to have alternative alleles (1:1 ratio). Paired-end reads were generated to be consistent with the actual RNA-seq data in our study (see below). Nevertheless, the methods presented in this work apply to single-end reads as well. Forty pairs of reads were generated to overlap each genomic site with a random insert size in the range of [60, 240] bp and random start position relative to the site, both sampled from a uniform distribution. The base at the heterozygous site was chosen as one of the alternative alleles with equal probability. Sequencing errors and base quality scores were simulated based on read position–specific Gaussian distributions parameterized using our actual RNA-seq data. With correct mapping, it is expected that the ratios between the numbers of aligned reads of the two alleles would be similar to those in the original reads. As shown in Figure 1B, the relative ratios (defined in the figure legend) of most of the sites are close or equal to the expected ratio 0.5, and the average and median values of the distribution are not different from 0.5. In this simulation, we chose the values of n1 and n2 to be 5 and 12, respectively, to achieve the best mapping accuracy. This result confirms the effectiveness of our mapping strategy in eliminating mapping biases associated with RNA editing or other types of single-nucleotide differences.
Identification of putative RNA editing sites in U87MG cells
Following read-mapping, we designed a statistical framework to analyze uniquely mapped reads, identify significant candidate RNA editing sites, and estimate their editing levels (Methods). Note that mechanisms generating DNA–RNA differences other than the A-to-G or C-to-U types are not well known and may not be related to RNA editing. However, we generally refer to all differences as candidate RNA editing events and their levels as editing levels or editing ratios for convenience. We first applied the method to study RNA editing in the U87MG cells. U87MG is a commonly used cell line derived from a human grade IV glioma, one of the most deadly types of brain cancer. The genome of this cell line was sequenced recently using high-throughput sequencing (Clark et al. 2010), which easily enables a distinction between editing sites and expressed genetic variants such as SNPs. In addition, using a cell line facilitates convenient molecular perturbations and experimental validations of RNA editing. As one means of validation, we conducted ADAR knockdown followed by RNA-seq, in parallel to RNA-seq of control cells. Note that the other family members of ADAR are expressed at very low levels in U87MG, as reported by Cenci et al. (2008) and calculated from our RNA-seq data of control cells (RPKM [Mortazavi et al. 2008] values: ADAR, 53.2; ADARB1 [also known as ADAR2], 5.2; ADARB2 [also known as ADAR3], 0.1). Given the known function of ADAR in A-to-I editing, these data enabled us to evaluate the results of our methods.
We obtained RNA samples from U87MG cells transfected with either a siRNA that targets the ADAR gene or a control siRNA (Supplemental Methods). The ADAR siRNA led to significant reduction of the protein expression to a barely detectable level (Supplemental Fig. 1). Two biological replicates were collected for each type of transfection. Each replicate was sequenced in one lane of the Illumina GA IIx sequencer. A total of ∼115 million paired-end reads (2 × 60 nt long) were obtained. By using the mapping strategy described above, about 59 million pairs (51.5%) of reads were uniquely mapped, most of which overlap known genes and known exons (Supplemental Table 1).
Our initial analyses showed that putative editing sites and their editing levels identified using individual samples are highly concordant between biological replicates (Supplemental Fig. 2; Supplemental Table 2). Therefore, in the subsequent analyses, we combined data from the two replicates to maximize the statistical power. In the control sample, 9636 DNA–RNA differences were identified using our method (Fig. 2A, red bars). Strikingly, 5965 (62%) of these sites correspond to A-to-G differences, consistent with A-to-I editing. This observation supports the existing knowledge that A-to-I editing is the primary type of RNA editing in human. Other types of differences are much less abundant. We found that 854 A-to-G differences resulted from our study are already included in the DARNED database (Kiran and Baranov 2010), a comprehensive repository of predicted or validated RNA editing events (mostly A-to-G types). The overlaps of our results with other published works are shown in Supplemental Table 3.
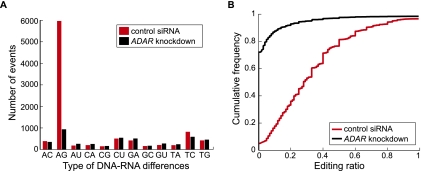
Figure 2.
DNA–RNA differences identified via RNA-seq. (A) Number of events for the 12 types of differences between RNA reads and genomic DNA sequences in samples transfected with control siRNA and ADAR siRNA, respectively. Labels of x-axis denote DNA and RNA nucleotides (e.g.: “AC” denotes “A” in DNA and “C” in RNA). (B) Empirical cumulative distribution function of editing ratios of putative A-to-I editing events identified from RNA-seq. A union of editing events identified in the two samples is included (6422 in total) in each curve. For nonediting events in one sample (those that failed the statistical identification procedure), the editing ratio was calculated as the number of reads with the “G” nucleotide at the predicted editing position divided by the total number of reads at that position.
As expected for results without significant false positives, the positions of putative editing sites in the reads are distributed relatively uniformly (Supplemental Fig. 3). If we assume that all the G-to-A differences reflect sequencing errors, then the false-discovery rate in the A-to-G identification is ∼7%, because sequencing errors are expected to produce at least as many G-to-A and A-to-G differences. This false-discovery rate may be an overestimate because there might exist authentic G-to-A differences due to RNA editing or other mechanisms. If we further filter the events by requiring a minimum editing level of 20%, then a total of 4141 sites (75% of all 5505 potential editing events) support A-to-I editing with an estimated false-discovery rate of 3.6% (see below for experimental validations of individual editing sites) (Supplemental Fig. 4).
Upon ADAR knockdown, the number of A-to-G differences significantly decreased (Fig. 2A), and it is the only type of difference with a considerable change in the number of events. This finding indicates that ADAR is indeed the main enzyme involved in A-to-I editing in the studied cell line. It also suggests that the other types of differences, if being bona fide editing events, are not likely affected by ADAR. Among the A-to-G differences identified in the control and knockdown samples (5965 and 938, respectively), 294 sites were in common, with 5671 unique to the control samples and 644 unique to the knockdown samples. In the subsequent sections, we only used the sites resulted from the control samples given the already large number of events in this group. Next, we examined the RNA editing level (i.e., editing ratio) calculated for each predicted site. Putative A-to-I editing events showed the largest degree of reduction in the editing level among all types of events upon ADAR knockdown (Fig. 2B; Supplemental Fig. 5). Note that the T-to-C events also demonstrated a significant change in editing level, which is discussed later. Importantly, the response of A-to-G differences to ADAR knockdown supports the validity and effectiveness of our method to identify RNA editing sites.
Validation of predicted A-to-I editing events
For a predicted editing site, it is desirable to validate two aspects of the prediction: whether it is a true event or not and the accuracy of the estimated editing level. For this purpose, we first used conventional Sanger sequencing to analyze the gDNA and cDNA sequences of the editing sites amplified with polymerase chain reaction (PCR) (Supplemental Fig. 6; Supplemental Table 4). The gDNA sequencing aims to confirm that the putative editing site is not a heterozygous SNP. The cDNA sequences can enable detection of edited nucleotides in the corresponding RNA. However, cDNA sequencing is not sensitive and quantitative enough to detect low-level editing or to provide accurate estimates of editing ratios (Supplemental Fig. 6). Instead, we used the traditional clonal sequencing approach to analyze the cDNA sequences and used PCR sequencing to confirm the gDNA sequences only.
We randomly picked four genes where a number of A-to-I editing sites are located within 400 bases (Supplemental Table 4). Their cDNA sequences were amplified and cloned into a TOPO vector. Twenty clones for each gene were randomly picked and analyzed by Sanger sequencing. A total of 93 A-to-I editing sites were detected by either RNA-seq or clonal sequencing or both. Among all the sites, four sites each were detected by only one method (Fig. 3A). The false-discovery rate of our predicted editing events is thus up to 4.5% (four of 89 sites). In addition, the estimated editing ratios by the two methods correlate relatively well (r = 0.76). The results in Figure 3A included all predicted editing sites regardless of the level of read coverage in RNA-seq (for more discussion about read coverage, see Supplemental Material; Supplemental Fig. 7). If we require at least 20 RNA-seq reads (same as the number of clones for Sanger sequencing) covering each editing site, the four false positives are not present in the predicted set (Supplemental Fig. 8) and the false-discovery rate is close to 0.
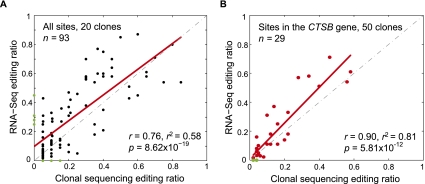
Figure 3.
Validation of predicted A-to-I editing events identified via RNA-seq. (A) Scatterplot of editing ratios for the full set of 93 A-to-I editing events identified by RNA-seq and the traditional clonal sequencing method (20 clones were picked for each editing site). Pearson correlation coefficient is shown. Data points corresponding to false-positive or false-negative predictions are shown as green dots. (B) Same as A, but for the 29 editing events in the CTSB gene (read coverage, 35–69 reads per site). A total of 50 clones were picked for each site.
The above result suggests that a modest increase in read coverage may facilitate better accuracy in editing identification. Many of our tested A-to-I sites had relatively low read coverage (31 reads or less per site) except those in the CTSB gene (35–69 reads per site). To further confirm the impact of read coverage on the estimation of editing ratios, we randomly picked 30 more clones for this gene so that the coverage of RNA-seq and clonal sequencing on the putative editing sites are comparable. Improved correlation and linear regression were observed between the editing ratios estimated by the two methods using 50 clones (Fig. 3B) compared with the original 20 clones. (Note that collecting 20 clones for Sanger sequencing limits the validation accuracy similarly as having only 20 reads per site.) Thus, our method can enable relatively high level of accuracy in the quantification of editing ratios, which can be demonstrated for sites with high RNA-seq read coverage.
The above validation focused on genes where a number of predicted A-to-I editing sites are clustered together, which is a feature found for the majority of the predicted sites (Supplemental Fig. 9) and is consistent with the known properties of A-to-I editing. In addition to such sites, we tested 10 randomly picked A-to-G events to represent those that are distant from other predicted sites (distance to closest neighbor, >1500 nt) and to encompass a wide range of predicted editing levels (Supplemental Table 5). This validation was carried out by a combination of Sanger sequencing and clonal sequencing methods. All 10 sites were confirmed as authentic A-to-G differences between the DNA and RNA sequences.
Characterization of predicted A-to-I editing events
Since sites with relatively low editing levels may not be associated with significant functional consequences, all analyses in this section focused on the 4141 A-to-I editing sites with a minimum editing level of 20% identified from the control siRNA samples (Supplemental Fig. 4; Supplemental Table 6). Among these sites, a large fraction is located in noncoding regions (introns or untranslated regions [UTRs]) (Supplemental Table 7). Only 45 (1.1%) sites reside in coding sequences, 31 of which change amino acids. The relative enrichment of synonymous and nonsynonymous sites is not significantly different (p ≈ 1). Consistent with the functional properties of ADAR, A-to-I editing sites were more often located in double-stranded regions compared with random controls (Supplemental Methods). This observation holds whether the sites are located in Alu sequences (88% of all 4141 sites) or not (Fig. 4A). As reported previously (Lehmann and Bass 2000; Athanasiadis et al. 2004; Li et al. 2009a), the nucleotides 5′ and 3′ to the editing site (−1 and +1 positions) have a strong preference for G depletion and enrichment, respectively (Fig. 4B). Moreover, we observed strong sequence biases at other positions (+12, +18, P < 3 × 10−9) (Fig. 4B) as well.
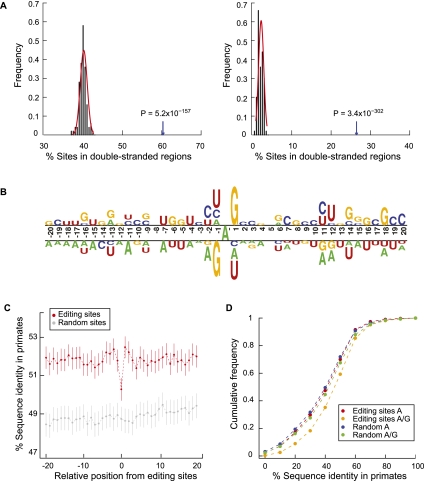
Figure 4.
Sequence features of predicted A-to-I editing sites and the flanking regions. (A) Double-stranded regions in the neighborhoods of predicted A-to-I editing sites. (Left) Editing sites and controls are located in Alu elements. Controls were picked as random As in such regions with matched G+C content relative to the test regions (Supplemental Methods). Percentage of editing sites in double-stranded regions shown by arrow; percentage of control sites in double-stranded regions shown by black histogram. P-value was calculated by fitting a normal distribution to the control histogram. (Right) Same as the left panel, but editing sites and controls are outside of Alu elements. (B) Sequence preferences for base positions flanking predicted A-to-I editing sites. Editing sites (the A nucleotide at position 0) are aligned together. Sequence preference is represented using a two-sample logo program (Vacic et al. 2006). (C) Conservation of the immediate neighborhood of predicted A-to-I editing sites. Sequence conservation (percentage of identity) of each position flanking editing sites was calculated using the UCSC multiz46way alignments of primate genomes (Supplemental Methods). Random controls were picked for each editing site in the same type of regions (e.g., Alus in coding exons, Alus in introns). Vertical lines represent 95% confidence intervals. (D) Sequence conservation among primates at the edited sites before and after editing. Cumulative distribution functions are shown for percentage of identity at the editing sites assuming the nucleotide being A and (A or G) in human, respectively. Random controls were picked similarly as described in C.
The sequence neighborhood of the editing sites shows an enhanced conservation level in primates compared with random sequences in similar regions (Supplemental Methods) (Fig. 4C; Hoopengardner et al. 2003), indicating that editing function of ADAR may be affected by the immediate sequence neighborhood. In addition, the editing sites themselves are less conserved on average than the neighboring bases, which is consistent with previous findings (Yang et al. 2008). Interestingly, if conservation was evaluated assuming editing has occurred (i.e., both A and G are present) in human RNA, the sequence conservation is much higher than that of the original base A (P < 2.2 × 10−16) (Fig. 4D). This difference in conservation is more pronounced than that between randomly picked As and when these As were converted to As and Gs in the human genome (Fig. 4D). Therefore, our results suggest that RNA editing may increase the conservation of a gene relative to its homologs in primates, which may have important evolutionary implications (see Discussion).
A structural motif in ADAR editing
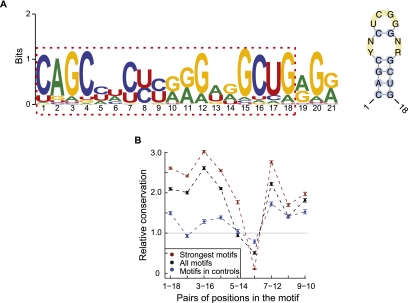
The large number of A-to-I editing sites allowed us to investigate common sequence or structural features important for editosome function. We analyzed the region in the vicinity of the editing sites (−100 to 100 nt) using the Multiple Em for Motif Elicitation (MEME) algorithm (Bailey and Elkan 1994). This method aims to detect motifs that are significantly enriched within this region, regardless of their relative location to the editing sites. Since most of the editing sites are located in Alu regions, we randomly picked other Alu sequences to generate a second-order Markov model to control for inherent Alu sequence background. (Results were similar if random Alu sequences were used as a background control sequence set.) This analysis revealed a significant 21-nt motif with a very small E-value (<10−100) (Fig. 5A). Interestingly, the first to 18th bases of the motif appear to be palindromic, indicating the existence of a possible RNA secondary structure (Fig. 5A).
Figure 5.
A novel motif with potential function in A-to-I editing. (A) Consensus motif (left) identified by MEME in the 201-nt neighborhood centered around each predicted A-to-I editing sites. (Right) Structure of the one to 18 bases of the consensus motif (RNAalifold). Y = U or C, R = G or A, N = A, C, G or U. (B) Conservation of the base-pairing patterns of the motif in primates based on multiz46way alignments. Strong motifs, motif score >24.4; all motifs, motif score >6.6; controls (motif score >6.6) were randomly picked from Alu elements in coding exons devoid of A-to-I editing sites. Error bars represent 95% confidence intervals. The conservation levels were normalized against expected levels calculated using random controls (Supplemental Methods).
The sequences of the motifs occurring within 100 nt of the editing sites are not very significantly conserved among primates (Supplemental Fig. 10A). However, considerable co-conservation was observed (Fig. 5B; Supplemental Fig. 10B) for the five pairs of positions with base-pairing patterns in the structure shown in Figure 5A. In addition, two motifs in the same gene potentially forming intermotif dsRNAs are much less co-conserved than the intramotif co-conservation (Supplemental Fig. 11). Thus, the motif by itself likely represents a functional structural unit under evolutionary selection. Since the consensus motif resembles part of a typical Alu sequence (Kriegs et al. 2007), we evaluated whether the motif is specifically relevant to RNA editing or it is an artifact due to the prevalence of Alus near editing sites. For the motif occurring in Alu sequences in coding exons without A-to-I editing in the vicinity, the structural conservation is much less than those near editing sites (Fig. 5B, “motifs in controls”). In addition, strong motifs are also significantly enriched near editing sites far away from Alus (Supplemental Table 8). These results suggest that this structural motif is likely functionally relevant to A-to-I editing, although its exact role is not yet clear.
Functional relevance of A-to-I editing to cancer
The set of 1167 genes with A-to-I editing sites in this study significantly overlaps the list of genes related to cancer as annotated by the NCI Cancer Gene Index project (341 genes in common, P < 2.2 × 10−16; P = 0.009 if only highly expressed genes were analyzed) (see Supplemental Material). Among the 341 genes, many are associated with processes critical to malignancy (Supplemental Table 9), including tumor suppressor and cancer marker genes and genes involved in apoptosis, metastasis, DNA repair, and signaling pathways. Editing sites in these genes are often located in the 3′ UTR regions, such as for the MDM4, MALT1, ERCC4, and TEP1 genes, which may affect regulation of gene expression via miRNA targeting or other mechanisms. A-to-I editing sites also induce nonsynonymous changes to the coding sequences of the cancer-related genes PRKCSH and CHD3. The PRKCSH gene encodes a substrate for protein kinase C, an important player in signal transduction cascades and many cancer-related processes (Reyland 2009). The CHD3 gene, possibly associated with leukemia (Camos et al. 2006), is one of the components of a histone deacetylase complex, which participates in chromatin remodeling. Thus, although hypo-editing was reported in cancer for some genes subject to A-to-I editing in normal tissues (Paz et al. 2007; Gallo and Galardi 2008), our results showed that editing of cancer-related genes might be prevalent in tumors. In addition, a number of RNA-binding proteins were found to harbor A-to-I editing sites (e.g., APOBEC3D, DDX58, EIF2AK2, FXR1, INTS1, MED28, and RBM5), suggesting that RNA editing may affect various steps of post-transcriptional gene regulation.
Other types of DNA–RNA differences
The other types of DNA–RNA differences shown in Figure 2A may be due to RNA editing (e.g., for C-to-U events) or other unknown mechanisms, which is not a focus of our study. The number of each type of event is much smaller than that of the A-to-G events, and most are not affected by ADAR knockdown (Fig. 2A; Supplemental Fig. 4). The categories of genomic regions harboring each type of events are shown in Supplemental Table 7. Interestingly, the other types of DNA–RNA differences tend to co-occur in genes with putative A-to-I editing (Supplemental Table 10). In addition, their “editing ratios” are sometimes correlated with those of the A-to-G events in the same genes (Supplemental Fig. 12), especially for the C-to-U, G-to-A, and T-to-C events. Different from A-to-I editing, the other types of events (except T-to-C, see below) are not enriched in Alu regions (<20% located in Alus).
We carried out clonal sequencing to validate a set of randomly selected events among all non–A-to-G events. To be cost-effective, we sequenced only five to 12 clones for each event because here we aim to validate the presence of the DNA–RNA differences, but not the quantitative values of the “editing ratio.” DNA sequences were confirmed by Sanger sequencing of PCR products. A total of 37 events were tested encompassing all types of non–A-to-G events. The success rate of this validation was 49% (18/37) (Supplemental Fig. 13; Supplemental Table 11). This result suggests that about half of the non–A-to-G differences are authentic, many of which may have been generated by novel mechanisms to be discovered in the future. Nevertheless, the validation rate is lower than that for the A-to-G differences. Events that failed validation may be due to the relatively small number of clones we analyzed. Alternatively, they may be resulted from mistakes introduced during RNA library preparation or other steps of data acquisition and processing. For example, the reverse transcriptase is known to have a relatively high error rate that can introduce base substitutions (Roberts et al. 1988). Although such errors may introduce different types of events at roughly the same rate, their impact on the accuracy of the A-to-G events can be relatively small because a much larger number of authentic A-to-G differences exist as a result of RNA editing compared to the other types.
The T-to-C events demonstrated some interesting features differing from the other types of events, including reduced “editing ratios” and a smaller number of events identified upon ADAR knockdown (Supplemental Figs. 4, 5). These observations bring up the question whether the T-to-C difference is also regulated by ADAR. However, since the RNA-seq libraries were prepared using the standard Illumina protocol that is nonstrand specific, the exact type of event (e.g., A-to-G vs. T-to-C) was determined using the strand information of known genes (Methods). DNA–RNA differences in regions with both sense and antisense transcription were excluded from the final list. However, regions with unknown sense-antisense transcription may lead to confusion of an actual A-to-G event as a T-to-C event, and vice versa. For this reason, it cannot be concluded that the T-to-C events are ADAR-dependent. Nevertheless, we expect that many of the identified T-to-C events are valid, as evidenced by the fact that five out of eight tested T-to-C events were confirmed in the validation experiments (Supplemental Table 11). Indeed, if most T-to-C events were resulted from A-to-I editing on the opposite strand, then they are expected to be as highly enriched in Alus as the A-to-G events. Yet, 63% of T-to-C events occur in Alus, significantly lower than the 88% among A-to-G events (P < 1 × 10−10).
Comparison to DNA–RNA differences identified in another cancer sample
We analyzed the genome-wide profile of DNA–RNA differences in another cancer sample, primary human breast cancer, for which RNA-seq and whole-genome sequencing data have been published (Shah et al. 2009). The genomic data were used to identify homozygous DNA sites and heterozygous SNPs. The RNA-seq data were analyzed in the same manner as for the U87MG data. The numbers of various types of DNA–RNA differences in the breast cancer samples are shown in Supplemental Figure 14. Similar to the U87MG results, the A-to-G type (Supplemental Table 12) represents the largest category of potential RNA editing in breast cancer, accounting for 82% (9722 out of 11,791) of all predicted events. Next, we examined whether there exists a significant overlap between the results from the two data sets. For this purpose, the background set consisted of all genomic homozygous sites in known genes that are common to the two data sets. In addition, we required at least five RNA-seq reads overlapping each homozygous site in each data set because this was the minimum read coverage requirement in identifying editing sites. As shown in Supplemental Table 13, a significant overlap was found between the results of the two cell types. Interestingly, a larger extent of overlap exists for genes undergoing RNA editing if the exact match of the sites themselves was not required. For example, 218 genes are common (P < 2.2 × 10−16) between those with predicted A-to-G sites in U87MG (647 genes) and breast cancer (379 genes) data. Our results suggest that cancer cells may have significant overlap in their editing profiles, despite the difference in cell types, cancer types, and genomic backgrounds.
Discussion
Here we presented an approach to identify and study genome-wide RNA editing events in the human transcriptome. This RNA-seq–based approach enables de novo identification of RNA editing of all possible types. Different from many previous methods, it does not assume any prior knowledge about RNA editing or require predefined candidate editing sites. To distinguish RNA editing sites from expressed genetic polymorphisms, this method requires knowledge of the genomic homozygous sites, often available based on genomic sequencing data. Experimental validations via traditional approaches confirmed a low false-discovery rate and relatively accurate estimation of editing ratios. This is the first report, to our knowledge, where the quantitative A-to-I editing levels derived from a genome-wide approach were shown to be estimated relatively accurately in a mammalian transcriptome.
Mapping of RNA-seq reads is critical to the accuracy of identified editing sites and estimated editing levels. In a related problem where expression of alternative alleles of SNPs was estimated, previous studies observed a bias favoring the reference allele in reads mapped to heterozygous SNPs (Degner et al. 2009; Heap et al. 2009). This bias is possibly due to the fact that mapping is usually conducted using genome or transcriptome sequences where only the reference allele of a SNP is included. However, the bias was not completely eliminated when mapping was carried out against genome sequences in which degenerate bases were used at the locations of known SNPs (Degner et al. 2009), a method applicable only to loci with known variants. This mapping bias will lead to inaccurate estimate of editing levels, assuming the corresponding candidate editing sites are true positives (Supplemental Fig. 15). However, it is also possible that an editing site is a false-positive prediction due to mapping errors (Supplemental Fig. 15). Such false positives arise due to the existence of highly homologous regions in mammalian genomes. This problem may not significantly affect the estimated results of overall gene expression levels based on RNA-seq. Yet, it is particularly alarming when the data are examined to identify single-nucleotide differences. Our mapping strategy stringently removes ambiguously mapped reads, and filters the reads according to the number of mismatches relative to the genome. This strategy was necessary to ensure unbiased mapping of reads containing the reference nucleotide versus those containing the edited nucleotide. In addition, we showed that increased read coverage at putative editing sites enabled better accuracy in the estimation of editing ratios (Fig. 3; Supplemental Fig. 8).
Genes harboring predicted A-to-I editing sites are enriched with cancer-related genes in the U87MG cells. In addition, we observed a significant overlap of the profiles of DNA–RNA differences (a majority being putative A-to-I editing events) between the U87MG cells and primary breast cancer samples. This high level of preservation of putative editing profiles supports the notion that RNA editing may contribute to important functional pathways that are common to many different (cancer) cell types. Our results also have different extents of overlap with the putative editing sites reported in the DARNED database (Kiran and Baranov 2010) and other previous studies (Supplemental Table 3), although many of such sites resulted from noncancer cells. In particular, two recent studies reported DNA–RNA differences identified using RNA-seq data (Supplemental Table 3; Ju et al. 2011; Li et al. 2011). Among the 1809 events reported by Ju et al. (2011), 172 sites were also identified in our U87MG data. However, the overlap between our study and that of Li et al. (2011) was small (73 out of 10,210 reported in their study). Since different cell types, reads mapping, and analysis approaches were used in the three studies, the accuracy of results from each study needs to be further evaluated and compared in the future.
We found that the predicted A-to-I editing sites are often associated with lower genomic conservation compared with their flanking regions. However, changing the A to I (G) via editing increases sequence conservation in primates. This observation indicates that G-to-A genomic mutations may be corrected by post-transcriptional RNA editing. Alternatively, an ancestral editing event may have been fixed in some genomes through genetic mutations (Tian et al. 2008). Regardless of the evolutionary origin, RNA editing contributes to transcriptome diversity similarly as genetic variants. Moreover, RNA editing enables such diversity at a low evolutionary cost because it is a reversible and regulated process. Since the level of editing can range from nearly zero to one, RNA editing creates a wide spectrum of expression variation. The combination of multiple editing events in the same gene can potentially generate tremendous diversity in the expressed transcripts.
Although spanning the range of [0, 1], editing levels of the A-to-I editing sites tend to be relatively low (mean, 0.35; median, 0.33). Among all 5965 A-to-G sites in U87MG cells, 31% have editing levels no more than 0.2, whereas only 5% have values greater than or equal to 0.8. The enrichment of low-level editing is consistent with the continuous probing (COP) hypothesis proposed by Maas and colleagues (Gommans et al. 2009). According to this hypothesis, low-level editing is prevalent due to COP of the transient and dynamic RNA secondary structures by the editing machinery. Widespread low-level editing generates transcript diversity that may render the organism survival advantage upon environmental changes and enhances evolvability.
In summary, we demonstrated that RNA editing can be accurately identified from high-throughput RNA sequencing data. As genome and transcriptome sequencing of a large number of organisms and human individuals is carried out, our method can enrich the analysis of such data sets and add an additional dimension to the underlying mechanisms of gene expression diversity.
Methods
Reads mapping
We first mapped each end of the paired-end reads to the genome (hg19) using a combination of tools, including Bowtie (Langmead et al. 2009), BLAT (Kent 2002), and TopHat (Trapnell et al. 2009). The latter two methods allow mappings across exon–exon junctions. We combined the results of different tools because of the observation that they could differ significantly for some reads due to the different algorithms involved. To minimize the mapping bias for the edited versus unedited bases, we allowed a relatively large number of mismatches per read in the initial mapping. This procedure can diminish the apparent mismatch contribution by one editing event in a read since it only creates one mismatch to the genome. Based on simulations described in the Results, we chose to allow a maximum of 12 mismatches in each 60-nt read in the initial mapping. The mapping parameters are as follows: BLAT (version 3.4): -minIdentity=75 -tileSize=11; Bowtie (version 0.12.3): -k 80 -e 140 -n 3 -l 20; and TopHat (version 1.0.13): -F 0–segment-mismatches 3. Subsequently, all mappings of each pair of reads were examined to determine if they pair correctly, specifically with the expected orientations and the distance between the pair being <500,000 bp in the genome. For reads that passed the above filters, we further required that the pair of reads map uniquely (as a pair, not necessarily individually) with five or less mismatches on each reads, and importantly, they do not map to anywhere else in the genome as a pair with 12 or less mismatches each. This stringent filter eliminates potentially ambiguous mappings to similar genomic regions and mapping errors due to sequencing errors, editing, or SNPs.
Identification of RNA editing sites
For homozygous sites derived from the U87MG genome sequencing data (Clark et al. 2010), we piled up reads overlapping these sites and examined whether mismatches to the genome sequence existed in the RNA reads. In this step, we removed all duplicate reads within each RNA-seq library except the one with the highest-quality score at the mismatch position. Duplicate reads were defined here as pairs of reads mapped to exactly the same genomic locations. Since RNA molecules were randomly fragmented during library construction, some duplicate reads are likely the results of amplification bias in the RT-PCR process (Pepke et al. 2009). This kind of bias can significantly affect the accuracy of the estimated editing ratio since the statistical power will be artificially augmented and the bias related to the edited and the original bases may be different.
We next determined the type of DNA–RNA differences. To distinguish between complementary types (e.g., A-to-G vs. T-to-C), the strand of the reads (i.e., the genomic strand from which the RNA was produced) must be known. Given the standard library preparation protocol used in our study, the resulted RNA-seq reads do not preserve strand information of the original mRNA. However, since the human transcriptome is well annotated, we inferred the strand of the reads based on the strand of the genes they were mapped to. Reads mapped to regions with bidirectional transcription (sense and antisense gene pairs) were discarded. To get a most comprehensive gene annotation, we combined gene structures defined by the following databases: Ensembl, RefSeq, UCSC KnownGenes, Gencode genes, and VegaGenes. Since the 5′ and 3′ ends of the genes may not be accurately annotated yet, we further extended the gene boundaries by 1 kb each beyond the two ends.
Next, we used a statistical approach to determine whether the DNA–RNA differences are likely authentic events or sequencing errors. For a position with sequence differences (e.g., DNA being A, RNA being a mix of A and G), we calculated the probability of observing the specific nucleotide (n) assuming that the site is “edited” with the true editing ratio r, the quality score of the observed n is q, and the position of n in the read is i; that is,
for A-to-I editing.
In this model, we assumed that the base quality and the position of the base in a read affect the likelihood of a base-call being a sequencing error or not, which is similar as used by SNP calling algorithms (Li and Durbin 2009; Li et al. 2009b). The optimal editing ratio r is calculated as the one that maximizes the above likelihood function. We then calculated a log-likelihood ratio (LLR) to evaluate the significance of a predicted event, similarly as by Li et al. (2009a):
The LLR represents a comparison of the likelihood of the site being “edited” at r to the likelihood that the DNA-RNA difference is not real (r = 0) but a possible sequencing error. We used a LLR cutoff of 2 to select significant candidates (Li et al. 2009a), which indicates that the site is 100 times more likely being a true locus with DNA–RNA difference than a result of sequencing error. To impose further stringency, we required at least two edited reads and at least five reads in total for each considered site. In addition, mismatches within the first and last five bases of a read were discarded.
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE28040.
Acknowledgments
We thank D.L. Black and L. Simpson for helpful discussions, Z. Wang and members of the Xiao laboratory for helpful comments on this manuscript, and B. O'Connor, Z. Chen, and S.F. Nelson for providing cells and helping with the genome sequence data of U87MG. This work was supported in part by NIH grant R21DA027039, and an Alfred P. Sloan Foundation Research Fellowship and Research Grant no. 5-FY10-486 from the March of Dimes Foundation to X.X, and an American Heart Association Postdoctoral Fellowship to J.H.L. This manuscript was prepared using a limited access data set (of the human primary breast cancer samples) obtained from BCCA and does not necessarily reflect the opinions or views of BCCA.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124107.111.
References
- Abbas AI, Urban DJ, Jensen NH, Farrell MS, Kroeze WK, Mieczkowski P, Wang Z, Roth BL 2010. Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method. Nucleic Acids Res 38: e118 doi: 10.1093/nar/gkq107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391 doi: 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bass BL 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Gilmore BL, Spengler RM, Xing Y, Lanier W, Bhattacharya D, Davidson BL 2009. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet 18: 4801–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedeu J, Montserrat E, et al. 2006. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res 66: 6947–6954 [DOI] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A 2008. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 283: 7251–7260 [DOI] [PubMed] [Google Scholar]
- Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF 2010. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet 6: e1000832 doi: 10.1371/journal.pgen.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis DE, Maas S 2010. MiRNA editing. Methods Mol Biol 667: 267–279 [DOI] [PubMed] [Google Scholar]
- Farajollahi S, Maas S 2010. Molecular diversity through RNA editing: a balancing act. Trends Genet 26: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Galardi S 2008. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol 5: 135–139 [DOI] [PubMed] [Google Scholar]
- Gommans WM, Tatalias NE, Sie CP, Dupuis D, Vendetti N, Smith L, Kaushal R, Maas S 2008. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA 14: 2074–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Mullen SP, Maas S 2009. RNA editing: a driving force for adaptive evolution? Bioessays 31: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB 2000. Functions and mechanisms of RNA editing. Annu Rev Genet 34: 499–531 [DOI] [PubMed] [Google Scholar]
- Heap GA, Yang JH, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, Dobson RJ, et al. 2009. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet 19: 122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836 [DOI] [PubMed] [Google Scholar]
- Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, Lee S, Lee WC, Yu SB, Park SS, et al. 2011. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of 18 Korean individuals. Nat Genet 43: 745–752 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A, Baranov PV 2010. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics 26: 1772–1776 [DOI] [PubMed] [Google Scholar]
- Klimek-Tomczak K, Mikula M, Dzwonek A, Paziewska A, Karczmarski J, Hennig E, Bujnicki JM, Bragoszewski P, Denisenko O, Bomsztyk K, et al. 2006. Editing of hnRNP K protein mRNA in colorectal adenocarcinoma and surrounding mucosa. Br J Cancer 94: 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J 2007. Evolutionary history of 7SL RNA-derived SINEs in supraprimates. Trends Genet 23: 158–161 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M 2006. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep 7: 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann KA, Bass BL 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39: 12875–12884 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM 2009a. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K 2009b. SNP detection for massively parallel whole-genome resequencing. Genome Res 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Landweber LF 2007. Hypothesis: RNA editing of microRNA target sites in humans? RNA 13: 463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S 2010. Gene regulation through RNA editing. Discov Med 10: 379–386 [PubMed] [Google Scholar]
- Maydanovych O, Beal PA 2006. Breaking the central dogma by RNA editing. Chem Rev 106: 3397–3411 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nishikura K 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79: 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Hoo KH, Rampersad V, Deverill RM, Elliott CE, Fletcher EJ, Adams SL, Korczak B, Foldes RL, Kamboj RK 1994. Molecular characterization of the human EAA5 (GluR7) receptor: a high-affinity kainate receptor with novel potential RNA editing sites. Receptors Channels 2: 315–326 [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. 2007. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17: 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A 2009. Computation for ChIP-seq and RNA-seq studies. Nat Methods 6 (11 Suppl): S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G 2010. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res 38: 4755–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JG, Nagaraja AK, Lynn FC, Drabek RB, Muzny DM, Shaw CA, Weiss MK, Naghavi AO, Khan M, Zhu H, et al. 2008. Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/ cleavage/anchor regions and stabilize predicted mmu-let-7a:mRNA duplexes. Genome Res 18: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyland ME 2009. Protein kinase C isoforms: multi-functional regulators of cell life and death. Front Biosci 14: 2386–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JD, Bebenek K, Kunkel TA 1988. The accuracy of reverse transcriptase from HIV-1. Science 242: 1171–1173 [DOI] [PubMed] [Google Scholar]
- Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN 2010. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol 18: 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80 [DOI] [PubMed] [Google Scholar]
- Schoft VK, Schopoff S, Jantsch MF 2007. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res 35: 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Sharma PM, Bowman M, Madden SL, Rauscher FJ 3rd, Sukumar S 1994. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes Dev 8: 720–731 [DOI] [PubMed] [Google Scholar]
- Tian N, Wu X, Zhang Y, Jin Y 2008. A-to-I editing sites are a genomically encoded G: implications for the evolutionary significance and identification of novel editing sites. RNA 14: 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Iakoucheva LM, Radivojac P 2006. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 22: 1536–1537 [DOI] [PubMed] [Google Scholar]
- Villegas J, Muller I, Arredondo J, Pinto R, Burzio LO 2002. A putative RNA editing from U to C in a mouse mitochondrial transcript. Nucleic Acids Res 30: 1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Enstero M, Ohman M 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 19: 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff BE, Sakurai M, Nishikura K 2010. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat Rev Genet 12: 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lv J, Gui B, Yin H, Wu X, Zhang Y, Jin Y 2008. A-to-I RNA editing alters less-conserved residues of highly conserved coding regions: implications for dual functions in evolution. RNA 14: 1516–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaranek AW, Levanon EY, Zecharia T, Clegg T, Church GM 2010. A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing. PLoS Genet 6: e1000954 doi: 10.1371/journal.pgen.1000954 [DOI] [PMC free article] [PubMed] [Google Scholar]