Abstract
Precise DNA replication is crucial for genome maintenance, yet this process has been inherently difficult to study on a genome-wide level in untransformed differentiated metazoan cells. To determine how metazoan DNA replication can be repressed, we examined regions selectively under-replicated in Drosophila polytene salivary glands, and found they are transcriptionally silent and enriched for the repressive H3K27me3 mark. In the first genome-wide analysis of binding of the origin recognition complex (ORC) in a differentiated metazoan tissue, we find that ORC binding is dramatically reduced within these large domains, suggesting reduced initiation as one mechanism leading to under-replication. Inhibition of replication fork progression by the chromatin protein SUUR is an additional repression mechanism to reduce copy number. Although repressive histone marks are removed when SUUR is mutated and copy number restored, neither transcription nor ORC binding is reinstated. Tethering of the SUUR protein to a specific site is insufficient to block replication, however. These results establish that developmental control of DNA replication, at both the initiation and elongation stages, is a mechanism to change gene copy number during differentiation.
Precise DNA replication is crucial for genome maintenance, and most cancer cells contain regions of increased gene copy number (Kallioniemi 2008). The mechanisms controlling metazoan replication origin activation and fork progression have been difficult to investigate not only because few origins have been delineated but also because initiation and elongation are hard to capture and mechanistically distinguish in vivo. Recent genomic approaches defined sites and timing of replication initiation in cell culture (Schubeler et al. 2002; Lucas et al. 2007; Cadoret et al. 2008; Hiratani et al. 2008; Schwaiger et al. 2009; Sequeira-Mendes et al. 2009; Karnani et al. 2010; MacAlpine et al. 2010; Cayrou et al. 2011; Eaton et al. 2011). These studies have established an association between replication initiation, active transcription, and, in the case of Drosophila cells, Origin Recognition Complex (ORC) binding. Comparison of different cell culture lines suggests that there is plasticity in origin timing activation (Schwaiger et al. 2009; Hiratani et al. 2010; Mesner et al. 2011; for reviews, see Gilbert 2010; Mechali 2010). To understand the relationship between DNA replication and differentiation, however, it is crucial to investigate replication in vivo in tissues undergoing differentiation. For example, ORC binding has yet to be mapped in a differentiated metazoan tissue.
Developmental modification of the parameters of DNA replication can provide powerful models both to identify metazoan replication origins and to elucidate their regulation. Increased DNA content at a genomic level is common throughout the plant and animal kingdoms, resulting in polyploid cells such as mammalian megakaryocytes or polytene cells such as rodent placental trophoblasts (Edgar and Orr-Weaver 2001; Lee et al. 2009; Ullah et al. 2009). Polyploid and polytene cells differ in that in the latter the replicated sister chromatids remain physically aligned. In many polyploid or polytene cells, the genome doubling is not integral, and there is differential replication, i.e., genomic regions whose copy number is amplified above the overall ploidy of the genome or under-replicated with reduced copy number at certain genomic regions. Because this differential replication occurs in response to developmental cues, the activation or repression of DNA replication can be analyzed at its time of occurrence and regulatory mechanisms delineated. For example, analysis of developmentally regulated gene amplification in Drosophila ovarian follicle cells revealed that there are multiple mechanisms by which origins can be activated (Park et al. 2007; Xie and Orr-Weaver 2008; Kim et al. 2011).
Heterochromatin is known to be under-replicated in Drosophila polytene tissues (Spradling and Orr-Weaver 1987). Constitutive heterochromatic regions, localized in large blocks surrounding the centromeres, make up about a third of the Drosophila genome, but because these regions are composed of highly repetitive DNA, they are not molecularly tractable. Belyaeva and coworkers defined intercalary heterochromatin in the larval salivary gland as regions dispersed on the chromosome arms that appear condensed and constricted, and therefore likely to be under-replicated (Belyaeva et al. 2008). They identified a fascinating chromatin protein, SUUR (Suppressor of Under-Replication), whose function is required for under-replication of the centric and intercalary heterochromatin (Belyaeva et al. 1998). By using microarrays of Drosophila EST sequences, genomic regions whose copy number is affected by SUUR were identified by array–based comparative genomic hybridization (aCGH), comparing copy number in strains with overexpressed SUUR relative to a SuUR mutant (Belyakin et al. 2005). These studies uncovered 52 SUUR-dependent under-replicated regions, and it was shown that the protein localizes broadly to the intervals (Pindyurin et al. 2007). Although these regions contain transcription units, they have heterochromatic characteristics of late replication in diploid cell culture (Belyakin et al. 2005; Pindyurin et al. 2007).
A recent analysis of transcription factor and chromatin protein binding, measured by DamID in transfected Drosophila Kc167 cell culture, defined five chromatin states (Filion et al. 2010). This study showed that chromatin can exist in a state associated with repression of transcription and these regions have reduced gene density, but this is distinct from heterochromatin. Almost 50% of the genome is in such a “BLACK” chromatin state, bound by the proteins SUUR, histone H1, D1, and IAL, with Lamin, the Su(Hw) insulator, and the Effete protein frequently also associated. BLACK chromatin is likely to be repressive for replication, as these domains undergo late replication and have reduced levels of ORC binding in cell culture (MacAlpine et al. 2010). Whether the genomic localization of BLACK chromatin is conserved in differentiated cells in vivo remains to be determined.
Here we use tiled genomic microarrays to identify all euchromatic regions of differential replication in Drosophila salivary glands and high-throughput RNA sequencing to quantify transcript levels across the genome. We have identified 34 under-replicated regions that correspond to a subset of BLACK chromatin and are repressed for transcription. We investigated the mechanism by which replication is repressed in these domains by analyzing the localization of ORC in wild-type and SuUR mutants. We find that ORC is absent from these domains and that loss of SUUR function results in replication across the domains, apparently by increased replication fork progression from flanking regions.
Results
Euchromatic genomic regions are under-replicated throughout salivary gland development
We used tiled microarrays spanning the entire Drosophila euchromatin to compare the copy number from isolated third-instar larval salivary glands with that of early-stage diploid embryos, in wild-type strains in which replication was not perturbed. The criteria for calling a sequence probe on these arrays euchromatic is that it is not present in pericentric or telomeric heterochromatin blocks, corresponds to chromosomal arm regions visible on polytene chromosomes, and is a nonrepetitive sequence. We identified 34 domains ranging from 100–450 kb that were up to 10-fold under-represented in copy number (Fig. 1A; Supplemental Fig. S1; Supplemental Table 1). In addition to these euchromatic regions, there were five regions with pericentric heterochromatin probes present on the arrays that were under-replicated. We defined a region as being under-replicated if the copy number was at least twofold reduced in regions of ≥10 kb in two independent aCGH experiments. The MA2C peak calling analysis software (Song et al. 2007) also identified these domains as significantly under-replicated, as well as 19 others that are less than twofold under-replicated. We focused on the 34 domains in euchromatin that were at least twofold under-replicated for our studies. Quantitative PCR was used as an independent means to confirm that the copy number was indeed reduced (Supplemental Fig. S2). Twenty-five of the under-replicated regions identified in this study were also found to be under-replicated in the study by Belyakin et al. (2005), in which SUUR was overexpressed. Of the nine unique regions, three were not probed by the microarrays used in the previous study, as these contained solely EST sequences.
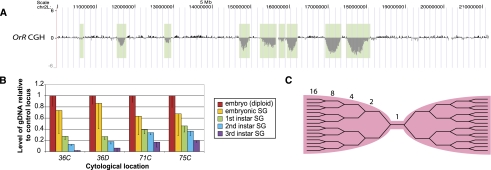
Figure 1.
Euchromatic regions are repressed for replication early in development. (A) aCGH of the proximal euchromatic half of chromosome 2L of Drosophila Oregon-R (OrR) third-instar salivary glands indicates the presence of euchromatic under-replicated regions. The genome profile shows the log2 ratio of copy number in the third-instar salivary gland genome relative to that of diploid embryonic DNA. Under-replicated loci are indicated by vertical light green bars. OrR flies are the wild-type strain used throughout this study. For this and other figures, UCSC Genome Browser (build dm3) was used (http://genome.ucsc.edu) (Rhead et al. 2010; Fujita et al. 2011). (B) Under-replication begins with the first endocycle in the embryonic salivary gland. Levels of DNA at four under-replicated loci relative to a fully replicated control locus were measured by qPCR in embryonic and first-, second-, and third-instar salivary glands. The diploid whole-embryo sample shows a 1:1 ratio between any two DNA loci. Error bars, SD of three biological replicates. By a one-sample t-test, the embryonic salivary gland sample is significantly different from the diploid embryo sample, P = 0.012. (C) Model of replication fork structure at under-replicated region.
To ask when during salivary gland development this under-replication begins, we dissected salivary glands from embryos and from each larval instar, and measured copy number at four under-replicated loci relative to a fully replicated control region by qPCR. For dissection from embryos and first-instar larvae, salivary glands were GFP-labeled with a Forkhead GAL4 driver to activate a UAS-GFP reporter. From the earliest endocycles, the under-replicated domains are already established and begin to be under-represented (Fig. 1B; for model of replication forks at under-replicated regions, see Fig. 1C).
Transcription is repressed in under-replicated domains
Given the positive relationship previously observed between replication and active promoters in metazoan cells, we speculated that genes within the under-replicated domains would show reduced transcript levels and that under-replication might be a mechanism to reduce gene expression in polytene cells (Schubeler et al. 2002; MacAlpine et al. 2004, 2010; Dutta et al. 2008; Karnani et al. 2010). Using Illumina-based RNA sequencing (RNA-seq) of RNA isolated from third-instar larval salivary glands, we performed a genome-wide analysis of transcription levels. We also measured the presence of RNA polymerase II (Pol II) by ChIP-chip (chromatin immunoprecipitation from dissected salivary glands followed by hybridization of the labeled precipitated DNA to microarrays).
We found that genes not only at the bottom of under-represented (UR) troughs but also at their sides are silent, with the exception of six in the center of the UR troughs and nine more at the edges of the troughs (Fig. 2B; Supplemental Fig. S1). These regions lack RNA Pol II (Fig. 2C), and thus the absence of transcripts is due to absence of transcription, as opposed to transcript instability. Genome-wide, the RNA Pol II signal is highly predictive of the presence of transcripts (Spearman's rank correlation coefficient = 0.7, P < 2 × 10−16). Therefore, the under-replicated regions in the salivary gland are transcriptionally repressed (Supplemental Fig. S3). For a list of the GO terms of repressed transcripts, see Supplemental Table 2.
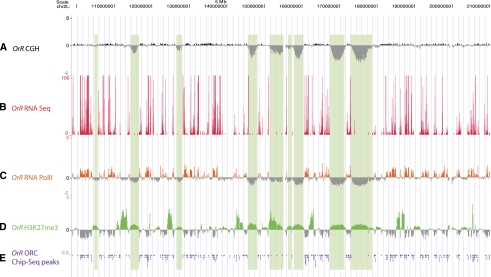
Figure 2.
Under-replicated regions are restrictive for ORC2 and RNA Pol II binding and are marked by weak binding of H3K27me3. (A) aCGH of proximal half of chromosome 2L, as described in Figure 1A. (B–E) RNA-seq, RNA Pol II ChIP-chip, H3K27me3 ChIP-chip, and ORC2 ChIP-seq peaks in Oregon-R (OrR) third-instar salivary glands, respectively. Total Illumina RNA sequencing reads are plotted; RNA Pol II ChIP was normalized to IgG control ChIP, and H3K27me3 ChIP data to input DNA. ORC2 ChIP-seq peaks were called by SPP compared with input sequencing lane; peaks for these data are organized downward by the UCSC Genome Browser.
Histone H3 trimethylated at lysine 27 (H3K27me3) is a mark associated with transcriptional repression of euchromatic regions (Schwartz and Pirrotta 2007) and has recently been shown to be the only repressive histone modification tested whose binding pattern parallels chromatin accessibility (Bell et al. 2010). Therefore, we checked, by ChIP-chip of third-instar salivary glands, whether the chromatin configuration in the under-replicated regions was likely in a repressed, inaccessible state and found that all under-replicated regions show the repressive H3K27me3 mark to varying extents (Fig. 2D, see Fig 6B; Supplemental Fig. S4). The under-replicated regions, however, were not the genomic intervals most intensely marked by the presence of H3K27me3.
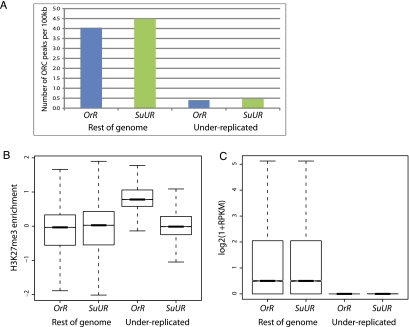
Figure 6.
Quantitative comparisons of ORC binding, H3K27me3 enrichment, and RNA expression levels between Oregon-R (OrR) and SuUR mutant in under-replicated regions and the remainder of the genome. Note that in the SuUR mutant these regions are no longer under-replicated, but we refer to them as UR. (A) Comparison of number of ORC binding sites per 100 kb of genome, as assayed by ChIP-seq, shows that in both OrR and SuUR mutants the under-replicated regions have many fewer ORC binding sites than the remainder of the genome. (B) Boxplots showing quantile-normalized H3K27me3 ChIP-chip data, comparing level of H3K27me3 in under-replicated regions with the rest of the genome, in OrR and SuUR mutant salivary glands. OrR UR was significantly enriched for H3K27me3 over the SuUR mutant UR with a P-value < 1 × 10−15 (unpaired Wilcoxon test). (C) Boxplots showing quantile-normalized RNA sequencing data (figure from one replicate; second is indistinguishable) comparing transcriptional levels in under-replicated regions with the rest of the genome in OrR and SuUR mutant salivary glands. Mutation of SuUR does not restore transcription in UR regions. In both strains, the transcription levels in the under-replicated regions were significantly lower than the remainder of the genome (P < 1 × 10−15, Wilcoxon test). The difference between the under-replicated regions in the two strains was not statistically significant (P = 0.3234, Wilcoxon test).
The ORC is absent in under-replicated regions
The reduction in copy number in the under-replicated domains could be due to a lack of origin firing within under-replicated regions, a block to replication fork progression through these regions, or a combination of the two. Therefore, we wanted to ascertain whether initiation complexes, marking potential origins of replication, were present in the under-replicated regions. We performed immunoprecipitations with anti-ORC2 antibody from dissected third-instar larval salivary glands and analyzed the samples either by ChIP-chip or by Illumina sequencing (ChIP-seq). Strikingly, both ChIP-seq (Fig. 2E) and ChIP-chip (Supplemental Fig. S5) of ORC2 indicate that the number of ORC binding sites per 100 kb in under-replicated regions was greatly diminished compared with that in the remainder of the fully replicated genome (see Fig. 6A). Drosophila origins tend to be spaced ∼40 kb apart (Spradling and Orr-Weaver 1987; MacAlpine et al. 2010); the under-replicated regions, some of which extend several hundred kilobases in length, harbor extremely sparse ORC binding sites or are devoid of ORC binding sites and therefore likely do not initiate replication by origin activation from a pre-RC complex.
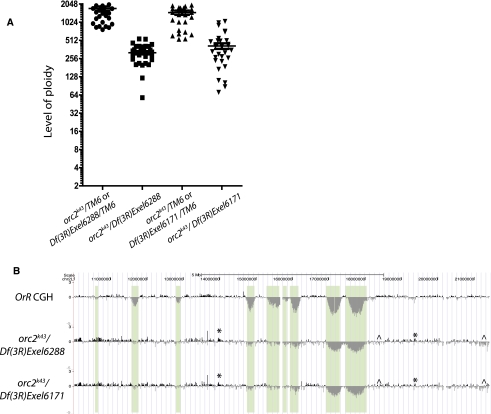
Park and Asano (2008) argued that in the salivary gland cells, ORC is dispensable for DNA replication. This conclusion implies that there would be no consequence of an absence of ORC in the under-replicated regions. Thus we examined the properties of genomic replication in the salivary glands of orc1- and orc2-null mutants (Landis et al. 1997; Park and Asano 2008). We find by microdensitometry that overall ploidy is reduced fourfold in orc2 and twofold in orc1 mutants (Fig. 3A for orc2; Supplemental Fig. S6 for orc1). Park and Asano (2008) also observed a twofold reduction in ploidy in orc1 mutants. Our aCGH analyses showed a markedly altered pattern of differential DNA replication in these mutants. We found dramatic changes in the copy number associated with a loss of ORC function. All but the most pronounced under-replicated regions became fully replicated, and there were interspersed regions of experimentally reproducible, but disorganized, amplification and under-replication (Fig. 3B; Supplemental Fig. S6). Thus, ORC is indeed functioning in polytene replication and affects the distribution of replication along the genome. The remaining replication in the mutants may have been allowed by maternal loading or residual activity in ORC complexes missing the mutated subunit. In addition, ORC-independent mechanisms of DNA replication may have resulted in the altered patterns of differential DNA replication in the orc mutants. Additional studies will be required to ascertain this point.
Figure 3.
orc2 mutants have reduced ploidy levels and show increased copy number of most under-replicated regions, as well as disorganized replication. (A) Ploidy levels of orc2 mutants and heterozygous sibling controls were quantified using DAPI microdensitometry. The intensity of DAPI staining was measured relative to diploid cells to calculate their ploidy. Each symbol represents the DAPI signal intensity from a single nucleus in two experimental replicates, and the bar is the mean of the measured nuclear values. (B) aCGH was performed comparing DNA of orc2k43/Df(3R)Exel6288 and orc2k43/Df(3R)Exel6171 third-instar salivary glands with that of diploid embryo. Oregon-R (OrR) aCGH is shown for comparison. The baseline of each aCGH is based on the average of all points on the array, so that the baseline of the orc mutant aCGHs is fourfold below that of the OrR aCGH shown. Plotted is the mean probe intensity. The asterisk marks examples of disordered but experimentally consistent overreplication, whereas the ^ symbol indicates disorganized but reproducible under-replication.
Comparison of ORC binding in the salivary gland and cell culture
As our study is the first in which ORC binding has been mapped in a primary tissue, we compared the sites of ORC binding across the entire genome in the nontransformed, differentiated salivary glands, with data sets from studies previously performed on several Drosophila cell lines (Fig. 4A; Eaton et al. 2011). Whereas 1618 sites are utilized by all cell types, there is a cell specificity to ORC binding, with the salivary gland tissue differing most in its ORC binding sites relative to the cell lines, perhaps due to its more differentiated state. Of the 5253 ORC binding sites in the salivary gland, 1501 (28%) are unique to salivary gland and are not present in any of the cell lines (Fig. 4A).
Figure 4.
Comparison of ORC binding sites in salivary gland cells (SG) to those in Kc, S2, and Bg3 Drosophila cell lines. (A) Origin binding sites were compared between the different cell types (cultured cell lines or salivary gland) indicated on the left. (Red boxes) Origins unique to a single cell type; (orange) origins shared by two cell types; (green) origins shared by three cell types; (purple) origins shared by all four cell types studied. Bars in blue above the boxes show the number of origins shared by the cell types indicated by the color-coded boxes below each bar. Less than 1 kb proximity of ChIP peaks was required for binding site conservation. The number of ORC binding sites differs slightly from that reported by Eaton et al. (2011) because our data merge some ORC binding sites into broader peaks. (B) Rectangular Venn diagram comparing the relationship between ORC binding and TSSs in salivary glands and Kc cell culture, with rectangles drawn to scale. After identifying the TSS nearest to each ORC site, the percentile rank of the corresponding transcript (in RPKM) was determined from the salivary gland and Kc cell RNA-seq data. From the difference in percentile ranks (DPR) between the salivary gland and Kc cell, each transcript was classified as salivary gland specific (SG >> Kc, for DPR > 40, black), higher in salivary gland or no difference (for DPR between 1 and 40, blue), Kc specific or no difference (for DPR ≤ 0, yellow). The percentile of transcripts not expressed in the salivary gland are in white. (C) ORC binding across the under-replicated regions in the salivary gland and the corresponding regions in Kc cells. Each of the 34 domains is displayed vertically, in the same order for ease of comparison. Each domain that is under-replicated in the salivary gland was divided into 100 windows, and each flanking region (the length of half the corresponding under-replicated region) was divided into 50 windows. Blue indicates the presence of an ORC binding site in the window.
In cell lines, ORC binding is associated with transcription start sites (TSSs) of actively transcribed genes (MacAlpine et al. 2010). Sixty-four percent of ORC sites are within a kilobase of an active TSS (MacAlpine et al. 2010). We examined the relationship between ORC binding and promoters in the salivary gland to test for a similar association and to determine whether differences in transcription could account for the tissue-specific ORC binding sites (Fig. 4B). In the salivary gland, 3819 ORC binding sites are within a kilobase of a TSS (73%), showing the same principle of ORC localization in this differentiated polytene tissue as in cell culture. Although 57% of the salivary gland–specific ORC sites are associated with a TSS, the genes controlled by these promoters are not uniquely expressed in the salivary gland (Fig. 4B). Thus the tissue specificity of ORC binding is not explained by active TSS sites.
We compared the pattern of ORC binding in the salivary gland and Kc cell culture in the regions that become under-replicated in the salivary gland (Fig. 4C). In Kc cells, these 34 regions have a lower density of ORC binding than in flanking intervals, but the levels are notably higher than in the salivary gland.
SuUR mutants restore replication, without reestablishing transcription or ORC binding
To study further mechanisms of active regulation of under-replicated regions, we turned to flies with mutated SUUR, a protein already implicated in controlling replication levels at UR sites (Belyaeva et al. 1998). We measured the copy number in SuUR mutants and found all the under-replicated domains to be fully replicated by aCGH, with slight partial under-replication only at cytological location 36D (2L: 16923976–17352361), the under-replicated domain with the lowest copy number and spanning the largest region in the entire Drosophila genome (Fig. 5B; for qPCR verification, see Supplemental Fig. S7; for other chromosome arms, see Supplemental Fig. S8).
Figure 5.
Loss of SuUR function restores genome replication without restoring ORC binding or transcription, but does lead to depletion of the repressive H3K27me3 mark. (A,B) aCGH of proximal half of chromosome 2L in OrR (A) and SuUR mutant (B) third-instar salivary glands, as described in Figure 1A. (C) ChIP-seq peaks of ORC in SuUR mutant third-instar salivary glands, called as described in Figure 2. (D) The ChIP-seq peaks of ORC from wild type for comparison. (E) Number of reads from Illumina RNA sequencing of SuUR mutant third-instar salivary glands. (F) RNA Pol II ChIP of SuUR mutant third-instar salivary glands, normalized to IgG control ChIP. (G) H3K27me3 ChIP-chip in SuUR mutant third-instar salivary glands, normalized to input DNA.
We next utilized the SuUR mutant as a tool to dissect mechanistically how SUUR affects differential replication using the parameters defined, namely, absence of ORC, transcriptional silencing, and the presence of the H3K27me3 repressive chromatin mark. We first looked at ORC localization by ChIP-seq and ChIP-chip from the salivary glands dissected from SuUR mutant larvae. Strikingly, we found that loss of functional SUUR protein allowed a restoration of copy number without a concomitant restoration of ORC binding within the previously under-replicated domains (Fig. 5C,D; Supplemental Fig. S8,9). Figure 6A quantitatively illustrates that at a genome-wide level, there is no restoration of ORC binding in the SuUR mutant.
Given that replication in SuUR mutants proceeds through previously under-replicated regions, we asked whether the chromatin accessibility of these regions was altered, looking both at transcription levels by RNA-seq and at the repressive H3K27me3 mark, which in wild-type flies marked all under-replicated domains, by ChIP-chip. In SuUR mutants, transcription was still repressed within the regions normally under-replicated, although copy number was restored (Figs. 5E, 6C). As it was possible that replication restored RNA Pol II binding but downstream factors were lacking to fully restore transcription, we performed a ChIP-chip with anti-RNA Pol II antibody and found that it did not bind the under-replicated regions in SuUR mutants (Fig. 5F). Whereas neither RNA Pol II binding nor transcription was restored in the salivary glands of SuUR mutants, the H3K27me3 mark was lost from most domains under-replicated in the wild-type salivary glands (Fig. 5G) and was restored to levels equal to those in the remainder of the genome (Fig. 6B). The exceptions are several genes within under-replicated regions that retain the H3K27me3 modification in SuUR mutants and an under-replicated region at 89E on chromosome 3R (12482529–12793969) that retains high levels of this mark in the mutant. The loss of the H3K27me3 mark in contrast to the continued transcriptional repression and absence of ORC binding in the SuUR mutant indicates that this histone modification is not causal for these effects.
SuUR mutants show enhanced rates of replication fork progression
Two mechanisms could explain how under-replicated regions become fully replicated in an SuUR mutant salivary gland. Either there is ORC-independent initiation in under-replicated regions normally repressed by SUUR, or ORC-initiated replication forks from outside these regions travel farther in the absence of SUUR. Although it is difficult to test for ORC-independent initiation, we tested the second hypothesis by devising an assay to determine whether SUUR affects replication fork progression.
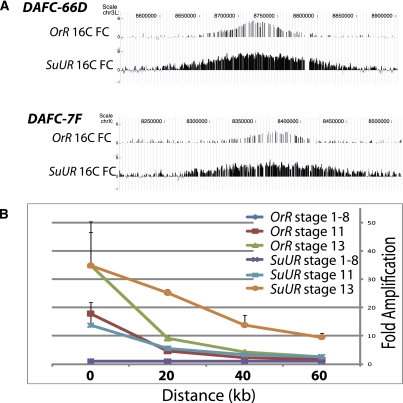
To test the rate of replication fork progression in the SuUR mutants, we exploited another model system of metazoan replication: gene amplification in the follicle cells of the Drosophila ovary. Developmentally regulated gene amplification occurs in these cells, producing 100-kb regions in which the DNA copy number is increased in a symmetrical gradient. From the peak of each amplicon, the copy number decreases out to ∼50 kb on either side. Mutants that enhance replication fork movement cause gradients extending to 200 kb (Park et al. 2007), and the gradients have been shown to reflect the rate of replication fork movement (Claycomb et al. 2002). We measured the copy number in the amplicons in wild-type and SuUR mutant follicle cells. In comparison with the wild type, SuUR mutant amplicons are broader but not taller (Fig. 7A), with forks traveling farther in the same developmental time window (Fig. 7B; Supplemental Fig. S10). These results indicate that absence of SUUR leads to increased fork movement but does not affect the number of rounds of initiation. Either SUUR may act at the replication fork or it could control chromatin across the domain to make a barrier to fork movement.
Figure 7.
SuUR mutant follicle cell amplicons exhibit enhanced replication fork progression. (A) aCGH of Oregon-R (OrR) and SuUR mutant FACS sorted 16C follicle cell nuclei at cytological locations encompassing Drosophila amplicon in follicle cells DAFC-66D and DAFC-7F. (B) SuUR mutant forks travel farther in the same developmental time window at DAFC-66D. SuUR mutant replication fork progression occurs between stage 11 and 13 in egg chamber development and is not due to alterations in egg chamber development timing. Quantitative PCR with primers 20 kb apart along the DAFC-66D amplicon was performed on hand-sorted egg chambers, relative to an unamplified control locus. The distances are from the peak of amplification (0 point). The experiment was performed in triplicate.
SUUR protein tethered to a single site is not sufficient to repress replication
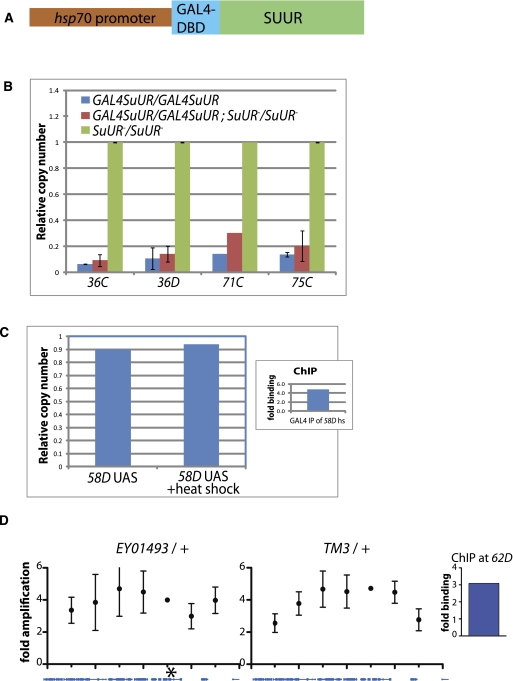
We wished to ascertain whether a single SUUR protein tethered to one site could block replication. To this end, we made a GAL4 DNA binding domain fusion to SUUR (GAL4DBD-SUUR) (Fig. 8A) and created heat-shock–inducible transgenic lines with this construct. Complementation tests of our construct in SuUR mutant lines demonstrated that the fusion protein was indeed functional (Fig. 8B), as it was able to rescue the SuUR mutant phenotype and restore under-replication.
Figure 8.
Tethering experiments indicate that tethering of a single SUUR is not sufficient to induce under-replication in salivary glands or to impede fork movement in a follicle cell amplicon. (A) Insertion constructed for tethering experiments, SUUR fused in frame to the DNA binding domain (DBD) of GAL4, with an hsp70 promoter. (B) Complementation assays showing that GAL4DBD-SUUR is capable of functioning as wild-type SUUR protein, restoring under-replication in SuUR mutant larvae. Copy number is relative to a control fully replicated locus. (C) Tethering of GAL4DBD-SUUR at a fully replicated locus is not sufficient to induce under-replication. (Inset) GAL4DBD-SUUR was bound to the UAS as analyzed by ChIP analysis relative to a negative control locus (pol α, where GAL4 is not bound). (D) Tethering of GAL4DBD-SUUR to the DAFC-62D amplicon in follicle cells does not affect amplification levels. The left panel shows the DNA copy number across the amplicon with GAL4DBD-SUUR tethered at the position designated by the asterisk. The right panel shows the sibling controls without the UAS binding site subjected to the same heat shock induction. The inset panel shows that GAL4DBD-SUUR was bound to the UAS, as determined by ChIP analysis.
We used the functional GAL4DBD-SUUR lines to tether SUUR via the GAL4 DNA binding domain to UAS sites at various locations along the Drosophila chromosomes and asked whether tethering of SUUR to a single site was sufficient to induce under-replication in the salivary gland at an ectopic locus. Figure 8C illustrates that tethering of SUUR to cytological location 58D8 is not sufficient to induce under-replication, despite the fact that ChIP-PCR demonstrates the presence of the SUUR protein at that locus. Similarly, tethering of SUUR to a UAS site adjacent to the origin of the amplicon at DAFC-62D shows SUUR bound there by ChIP-PCR but no decrease in amplification levels of that genomic locus in the follicle cells (Fig. 8D). Together, these results show that whereas SUUR marks under-replicated regions in the salivary gland, it either is in itself not sufficient to inhibit replication or needs to be present across a broad domain and not only at a discrete site to exert its repressive function on replication.
Discussion
Our study supports a model in which lack of ORC binding across broad regions combined with inhibition of progression of replication forks initiated from flanking origins can lead to chromosomal domains that are not replicated and thus under-represented in polytene cells. In the SuUR mutant, these regions are fully replicated, but ORC binding is not altered. Thus, the SUUR protein appears necessary to repress DNA replication by impeding replication fork progression.
The finding that under-replication begins from the onset of polytenization during embryogenesis has implications for the mechanism of replication repression and supports the model of inhibition of fork movement as well as initiation. The salivary glands undergo nine to 10 endocycles, rounds of alternating S and gap phases, to reach their ploidy of 1024C–2048C. The under-replicated regions are maximally 10-fold under-represented and thus are at least 100C. Given the onset of under-replication in embryogenesis, either these domains are fully replicated in some endocycles, there is variation in replication between cells in each salivary gland, or there is variation between DNA duplexes in the increasingly polytene cells. A parsimonious explanation is that SUUR is not able to block replication fork progression with 100% efficiency, and in some cells or on some DNA helices, the flanking forks replicate across the under-replicated domain. By measuring just a single pulse of CldU incorporation in the SuUR mutant, it was suggested that at the histone locus, nucleotide incorporation rates at the fork are not increased (Kolesnikova et al. 2009). Because only a subset of DNA duplexes are under-replicated each S phase, DNA combing studies are problematic for measuring fork rate, as they cannot distinguish replication forks being affected by SUUR and giving rise to under-replication.
A key conclusion is that although it is necessary for under-replication, the SUUR protein is not sufficient. The under-replicated domains identified in this comprehensive aCGH study are only a subset of the chromosomal regions to which SUUR was shown to bind on polytene salivary gland chromosomes (Zhimulev et al. 2003). Moreover, a comparison of the under-replicated regions in larval fat body, the midgut, and the salivary gland demonstrated that there is tissue-specific control of under-replication (Nordman et al. 2011). Some under-replicated regions are shared between the three tissues, but others are under-replicated in only one or two of the tissues. Despite the tissue specificity of the location of under-replication, replication is restored in all the domains when SUUR is mutated (Nordman et al. 2011), demonstrating that SUUR is necessary but not sufficient to block replication. Tethering of SUUR to a specific site in either the salivary gland or the follicle cells did not block replication or impede fork movement. Additional evidence that SUUR is not sufficient to repress replication comes from the analysis of ORC distribution. There are 22 euchromatic regions of ≥200 kb that lack ORC binding but are not significantly under-replicated. From the salivary gland immunofluorescence localization, it appears that at least nine of these are bound by SUUR (Makunin et al. 2002). Thus despite the absence of ORC and the presence of SUUR, replication occurred.
SUUR could act directly on replication proteins or indirectly by affecting chromatin configuration. Although it is conceivable that the SUUR protein acts directly at the replication fork to control processivity or rate of movement, we think it most likely that it affects replication by establishing a chromatin domain that impedes replication fork progression. The necessity but insufficiency of SUUR raises the possibility that it acts with other chromatin proteins to repress replication. In DamID studies in cell culture, the SUUR protein localizes across the domains of BLACK chromatin (Pindyurin et al. 2007; Filion et al. 2010). Immunostaining of SUUR on polytene larval salivary gland chromosomes also indicates that SUUR is bound across large regions (Makunin et al. 2002). Interaction between SUUR and specific chromatin proteins may establish a configuration inhibitory to fork movement.
This study provides the first map of ORC binding in differentiated, primary metazoan cells. As suggested by cell culture studies, ORC indeed displays different binding sites in different cell types. The positioning of ORC in polytene salivary glands is considerably more distinct than the differences observed between diploid Drosophila cell lines, as 28% of the ORC binding sites observed in the salivary gland are unique to that tissue, nearly triple the number of cell-type–specific origins seen in cell culture. The position of ORC binding sites in salivary glands supports the emerging paradigm that ORC localization frequently is linked to active TSSs (MacAlpine et al. 2004, 2010). Active transcription, however, does not appear responsible for directing ORC binding, and other factors must contribute. Thus ORC binding may require open chromatin configurations that show tissue specificity.
What could be the developmental function of under-replication? The domains are repressed for transcription, yet the effects on transcription and replication are separable by SuUR mutation. Thus under-replication is not required for silencing of transcription, and restoration of transcription is not needed for replication. The properties in cell culture of these regions that become under-replicated in the salivary gland and in some cases in the larval fat body and midgut suggest genomic hardwiring. There is less exon density across these domains, and in cell culture, transcription and ORC binding are reduced in these regions compared with flanking sequences. They also are late replicating. During salivary gland differentiation, this blueprint becomes more exaggerated because transcription is inhibited and ORC binding lost. The tissue specificity seen between the three polytene tissues, however, reveals that these under-replicated domains are not solely the passive consequence of late replicating regions established to control replication timing in diploid cells. In addition to tissue specificity with regard to the extent of replication, in the fat body many genes with the under-replicated regions are transcribed (Nordman et al. 2011). The genome structure appears to have hardwired these regions to be refractory to replication and transcription, but this can be increased or overcome in response to differentiation. It is interesting from an evolutionary perspective to ask what selective pressures drive the establishment of domains repressed for replication and the maintenance of the SUUR protein.
These results demonstrate mechanisms controlling gene copy number during the differentiation of polyploid cells. Strikingly, they operate by developmental control of both initiation and active control of fork movement. The recognition that SUUR controls fork movement and that it is present in diploid cells suggests that in diploid cells, proper regulation of DNA replication occurs not only at initiation but also via active regulation of replication elongation. Such regulation may be critical in diploid cells to maintain copy number, preventing changes associated with cancer.
Methods
Comparative genome hybridization
Seventy third-instar salivary glands from either Oregon-R (OrR) or homozygous SuUR mutant larvae were dissected in Ephrussi Beadle Ringer's (EBR) solution (Ephrussi and Beadle 1936). DNA from salivary glands or 0- to 2-h OrR embryos was phenol-chloroform–extracted and processed with Invitrogen's BioPrime Total for Agilent aCGH labeling kit. Slides were hybridized to a custom Agilent tiling array spanning the entire Drosophila euchromatin and were washed as per Agilent's recommendations.
Real-time qPCR
Real-time qPCR was performed with primers against under-replicated or amplified regions and normalized to DNA levels at a fully replicated control locus at cytological location 93F4 (primer sequences available upon request), using Perfect SYBR Green FastMix (Quanta Sciences) with an Applied Biosystems 7300 Real-Time PCR Instrument according to the manufacturer's instructions.
ChIP-chip
Salivary glands were dissected from wandering third-instar larvae in EBR for no longer than 30 min in the presence of Roche Complete Protease Inhibitor Cocktail, fixed, and processed according to the method described by Lee et al. (2006). Each ChIP was performed with 250 salivary glands, sonicated in lysis buffer 3 (MacAlpine et al. 2010) according to the method described by Austin et al. (1999). Immunoprecipitation was performed as described by Lee et al. (2006) with 2 μL (1:200 dilution) of anti-DmORC2 serum (kindly provided by Stephen Bell, Massachusetts Institute of Technology, Cambridge), anti-RNA Pol II (Upstate CTD4H8), anti-H3K27me3 (Abcam 6002), or normal IgG (Millipore 12-370 and 12-371). Unamplified DNA was processed for hybridization as described for aCGH, omitting the BioPrime restriction enzyme digestion step.
Array data analysis
Array intensities were median normalized across channels and smoothed by genomic windows of 1 kb using the Ringo package in R (Toedling et al. 2007). For ChIP experiments, enriched peaks were identified where at least three adjacent probes showed enrichment above the threshold determined by the upper bound of the null distribution, using the Ringo package in R.
ChIP-sequencing
Following ChIP, sequencing libraries were prepared with the Illumina ChIP-seq Sample Prep Kit according to the manufacturer's instructions and were sequenced as described. High-quality samples were confirmed by the appearance of a smooth smear of fragments with a peak distribution between 150 and 300 bp on an Agilent Bioanalyzer. After subsequent qPCR library quantification, 2–7 pM of linker-ligated DNA was applied to a flow cell using the Illumina Cluster Station fluidics device. Thirty-six-base sequencing was performed on an Illumina Genome Analyzer II (GAII) sequencer, according to Illumina's standard protocols. Images acquired were processed by the bundled Illumina image extraction pipeline version 1.6 and aligned to Drosophila NCBI build dm3 using ELAND (seed length 25).
ChIP peak calling
The ChIP-seq reads were mapped back to the Drosophila melanogaster genome (FlyBase version 5.2) using MAQ (version 0.7.1) (Li et al. 2008) with a maximum of three mismatches per read. For the salivary gland cells, we then used the SPP package (Kharchenko et al. 2008) to remove anomalous reads and detect peaks using the WTD method. We used maximum estimated enrichment cutoffs of 2.105 and 2.092 in the OrR and SuUR mutant samples, respectively, as determined by the SPP package's MSER function. For the Kc, BG3, and S2 cells, we used Peakseq3 (Rozowsky et al. 2009) to detect peaks in paired replicates with a maximum P-value of 0.05 and a minimum FDR of 0.05. The replicates were then combined by taking their intersection and throwing out any peaks that were not present in both samples. Additional information is available with associated metadata in the GEO repository (GSE20889, GSE20888, GSE20887).
RNA-sequencing
Third-instar salivary glands dissected in EBR were immediately transferred into TRIzol (Invitrogen) and were RNA-extracted according to the manufacturer's protocol. Ten micrograms of RNA was processed with Illumina mRNA Sample Preparation kit according to the manufacturer's protocol.
Developmental time course
Salivary glands from embryonic and larval stages were dissected from homozygous UAS-GFP; fkhIII-GAL4 lines (fkhIII-GAL4 flies kindly provided by Deborah Andrew, Johns Hopkins Medical School, Baltimore, MD; UAS-GFP from Bloomington) expressing GFP in salivary glands. DNA was amplified using Qiagen's RepliG kit. qPCR was performed in triplicate as described in the Supplemental Methods.
Staging of egg chambers
Drosophila ovaries were fixed and DAPI-stained and were analyzed by microscopy on a Nikon Eclipse Ti. At least 250 egg chambers from ovarioles from at least four different ovaries per genotype were analyzed. Shown is the ratio of egg chamber stages counted per each stage to the total number of chambers counted.
Follicle cell DNA preparation
Ovaries were isolated from OrR or SuUR mutant Drosophila ovaries fattened on wet yeast for 2 d. Genomic DNA was prepared from 16C FACS sorted follicle cell nuclei and prepared as for aCGH (Claycomb et al. 2004).
DAPI microdensitometry for ploidy quantification
Third-instar larval salivary glands were fixed with 4% formaldehyde and stained with 50 ng/mL DAPI for 1 h at room temperature. DAPI intensities of the experimental nuclei were measured and compared to the intensities of the 2C larval antennal disk nuclei to determine ploidy level, using a Nikon Eclipse Ti deconvolution microscope. At least 30 nuclei from two biological replicates were used for quantification.
SUUR tethering experiment
An SUUR fusion was created by cloning SUUR into the C terminus of the hsp70:GAL4 construct described by Aggarwal and Calvi (2004), and transgenic lines were generated by P element–mediated transformation. These were crossed to strains with UAS P elements inserted at 29C, 42A, 52D, 58D8, and 82F7. Larvae bearing both constructs were induced daily for 30 min at 37°C in a water bath, starting 6 h after egg laying, and were compared with non–heat-shocked controls. Salivary glands were dissected when larvae became committed wandering third instars and were processed for qPCR or ChIP as described above.
For the follicle cell experiment, flies with the GAL4DBD-SUUR fusion construct were crossed to the EY01493 UAS insertion in the yellow-g gene in the DAFC-62D amplicon. The fusion protein was induced by a 1-h heat shock at 37°C. After a 3-h recovery, amplification levels were tested across the amplicon in stage-13 egg chambers in the UAS-containing and balancer sibling controls by qPCR as described (Claycomb et al. 2002).
Data access
The data sets in this article have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under the reference series GSE31900. The specific experiments are as follows: (1) GSE31895-ChIP with anti-ORC2 antibody to identify regions of ORC binding in third-instar salivary glands of wild-type and SuUR mutant Drosophila; (2) GSE31896-RNAPol II ChIP to find differences between third-instar salivary glands of the wild type and SuUR; (3) GSE31897-ChIP with anti-H3K27me3 to compare binding in salivary glands of wild-type and SuUR Drosophila; (4) GSE31898-aCGH to ascertain levels of genomic DNA in third-instar salivary glands of various mutant Drosophila; (5) GSE31899-ChIP-seq of ORC2 bound to third-instar salivary gland DNA in wild-type and SuUR mutant Drosophila, analyzed by Illumina sequencing; (6) GSE33017-RNA-seq expression profiles of wild-type and SuUR mutant third-instar larval salivary gland tissue.
Acknowledgments
We thank Helena Kashevsky for doing the pilot aCGH studies, Yingdee Unhavaithaya for help with ploidy quantification, Tony Lee for advice, Tom Volkert and Sumeet Gupta of the Whitehead Genome Technology Core for help with ChIP-seq and RNA-seq, the Whitehead Genome Technology Core for help with arrays, and Tom DiCesare for help with graphics. Steve Bell generously provided the anti-ORC2 antibodies, the Bloomington Stock Center and Deborah Andrew (John Hopkins Medical School) provided stocks, Maki Asano (Ohio State University) provided the orc1 mutant, and Flybase was critical for genomic information (Tweedie et al. 2009). Steve Bell, Allan Spradling, Richard Young, and Daniel Sher provided helpful comments on the manuscript. N.S. was supported by a postdoctoral fellowship from the American Cancer Society; J.N. by the Damon Runyon Cancer Research Foundation. This work was initiated by support from the Mathers Charitable Foundation, and the aCGH studies were funded by NIH grant 1U01HG004279 as part of the modENCODE project. The project was supported by NIH grant GM57960 and by a Research Professor grant from the American Cancer Society to T.L.O.-W.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.126003.111.
References
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Austin RJ, Orr-Weaver TL, Bell SP 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev 13: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900 [DOI] [PubMed] [Google Scholar]
- Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci 95: 7532–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Andreyeva EN, Belyakin SN, Volkova EI, Zhimulev IF 2008. Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma 117: 411–418 [DOI] [PubMed] [Google Scholar]
- Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci 102: 8269–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. 2011. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL 2002. Visualization of replication initiation and elongation in Drosophila. J Cell Biol 159: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL 2004. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev Cell 6: 145–155 [DOI] [PubMed] [Google Scholar]
- Dutta C, Patel PK, Rosebrock A, Oliva A, Leatherwood J, Rhind N 2008. The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol Cell Biol 28: 5977–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, Macalpine HK, Tretyakov G, Kharchenko PV, Macalpine DM 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL 2001. Endoreplication cell cycles: more for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Ephrussi B, Beadle GW 1936. A technique of transplantation for Drosophila. Am Nat 70: 218–225 [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 doi: 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A 2008. CGH microarrays and cancer. Curr Opin Biotechnol 19: 36–40 [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Nordman J, Xie F, Kashevsky H, Eng T, Li S, MacAlpine DM, Orr-Weaver TL 2011. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev 25: 1384–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova TD, Demakov SA, Ivankin AV, Andreenkova NG, Zhimulev IF 2009. The mutation of the Suppressor of Underreplication gene does not affect the replication fork rate in the Drosophila melanogaster salivary gland polytene chromosomes. Dokl Biochem Biophys 427: 175–178 [DOI] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling AC, Tower J 1997. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci 94: 3888–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ 2009. Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM 2007. High-throughput mapping of origins of replication in human cells. EMBO Rep 8: 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF 2002. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160: 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, Bekiranov S 2011. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res 21: 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL 2011. Developmental control of the DNA replication and transcription programs. Genome Res 21: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Asano M 2008. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci 105: 12343–12348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EA, Macalpine DM, Orr-Weaver TL 2007. Drosophila follicle cell amplicons as models for metazoan DNA replication: a cyclinE mutant exhibits increased replication fork elongation. Proc Natl Acad Sci 104: 16739–16746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF 2007. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci 120: 2344–2351 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB 2009. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442 [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D 2009. Chromatin state marks cell-type– and gender-specific replication of the Drosophila genome. Genes & Dev 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 doi: 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS 2007. Model-based analysis of two-color arrays (MA2C). Genome Biol 8: R178 doi: 10.1186/gb-2007-8-8-r178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Orr-Weaver T 1987. Regulation of DNA replication during Drosophila development. Annu Rev Genet 21: 373–403 [DOI] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W 2007. Ringo: an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML 2009. Developmentally programmed endoreduplication in animals. Cell Cycle 8: 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Orr-Weaver TL 2008. Isolation of a Drosophila amplification origin developmentally activated by transcription. Proc Natl Acad Sci 105: 9651–9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, et al. 2003. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111: 377–398 [DOI] [PubMed] [Google Scholar]