Abstract
The structural basis for binding of the acidic transcription activator Gcn4 and one activator-binding domain of the Mediator subunit Gal11/Med15 was examined by NMR. Gal11 activator-binding domain 1 has a four-helix fold with a small shallow hydrophobic cleft at its center. In the bound complex, eight residues of Gcn4 adopt a helical conformation allowing three Gcn4 aromatic/aliphatic residues to insert into the Gal11 cleft. The protein-protein interface is dynamic and surprisingly simple, involving only hydrophobic interactions. This allows Gcn4 to bind Gal11 in multiple conformations and orientations, an example of a “fuzzy complex” where the Gcn4-Gal11 interface cannot be described by a single conformation. Gcn4 uses a similar mechanism to bind two other unrelated activator-binding domains. Functional studies in yeast show the importance of residues at the protein interface, define the minimal requirements for a functional activator, and suggest a mechanism by which activators bind to multiple unrelated targets.
Keywords: transcription, mediator complex, Gcn4, Gal11/Med15, acidic activator, activation domain, intrinsically disordered, NMR
Activation of transcription is the endpoint of many signal transduction pathways controlling cell growth, development and stress response. Most transcription activators enhance transcription by binding and recruiting coactivator complexes that directly interact with the transcription machinery (e.g., SAGA, TFIID, and Mediator) and/or function to remodel chromatin (e.g., SWI/SNF, p300, NuA4/Tip60, and SAGA) (Dyson and Wright, 2005; Ge et al., 2002; Green, 2005; Mittler et al., 2003; Prochasson et al., 2003; Stevens et al., 2002; Yang et al., 2004). In many instances, activation domains (ADs) interact with multiple unrelated coactivators. Likewise, coactivators can bind multiple seemingly unrelated ADs. (Dames et al., 2002; Freedman et al., 2003; Freedman et al., 2002; Herbig et al., 2010; Reeves and Hahn, 2005). What constitutes a functional AD and how these domains work with this apparent lack of binding specificity is unclear.
Although not conserved in primary sequence (Martchenko et al., 2007), ADs often have a simple and biased sequence composition and are enriched for specific residue types such as acidic residues, proline, or glutamine. Most known ADs are disordered in the absence of a binding target (Dyson and Wright, 2005). This property is not limited to ADs, as many functional eukaryotic protein segments, and in some cases entire proteins, lack a stable tertiary structure (Dunker et al., 2001). There are many examples of disordered proteins whose structure is stabilized to different extents upon interaction with their binding partners (Dyson and Wright, 2005; Tompa and Fuxreiter, 2008).
Acidic ADs are an important class of activators that universally stimulate transcription in all eukaryotes tested (Ptashne and Gann, 2002). Originally recognized in yeast Gal4 and Gcn4 (Hope et al., 1988; Ma and Ptashne, 1987), the acidic activators encompass most of the well-characterized yeast ADs and include strong mammalian and viral activators such as p53, E2F, and Vp16. P53 contains tandem ADs, and several structures containing these domains bound to coactivator targets reveal binding via one or two short α-helices. These interactions are mediated primarily by hydrophobic contacts and can be facilitated by charged and/or polar interactions (Di Lello et al., 2006; Feng et al., 2009; Kussie et al., 1996; Langlois et al., 2008; Uesugi et al., 1997). However, these known protein interfaces have fairly specific geometry and the general basis for how activators and their targets can bind multiple unrelated partners is not understood.
Yeast Gcn4 contains tandem acidic ADs (Fig 1A) that act in conjunction with the coactivators Mediator, SAGA, and SWI/SNF (Brown et al., 2001; Fishburn et al., 2005; Herbig et al., 2010; Jedidi et al., 2010; Swanson et al., 2003; Yoon et al., 2003) and directly regulates >70 genes involved in diverse processes such as the response to metabolic stress and autophagy. The two Gcn4 ADs (residues 1–100 and 101–134) are unrelated in sequence apart from their acidic character. Site-specific cross-linkers positioned within either Gcn4 AD and incorporated into pre-initiation complexes (PICs) target three common coactivator subunits, Gal11/Med15 (Mediator), Tra1 (SAGA and NuA4), and Taf12 (TFIID and SAGA) (Fishburn et al., 2005; Herbig et al., 2010) while full length Gcn4 also binds two subunits of the chromatin remodeler SWI/SNF (Prochasson et al., 2003). Gal11 has 3 conserved Gcn4-binding domains (Fig 1A) that bind Gcn4 with micromolar affinity (Herbig et al., 2010; Jedidi et al., 2010; Majmudar et al., 2009; Park et al., 2000). These multiple weak Gcn4-Gal11 interactions additively contribute to overall transcription activation and illustrate an important principal of Gal11 recruitment by Gcn4; Gcn4 binds Gal11 not by a single high affinity, high specificity interaction but rather by multiple low affinity interactions.
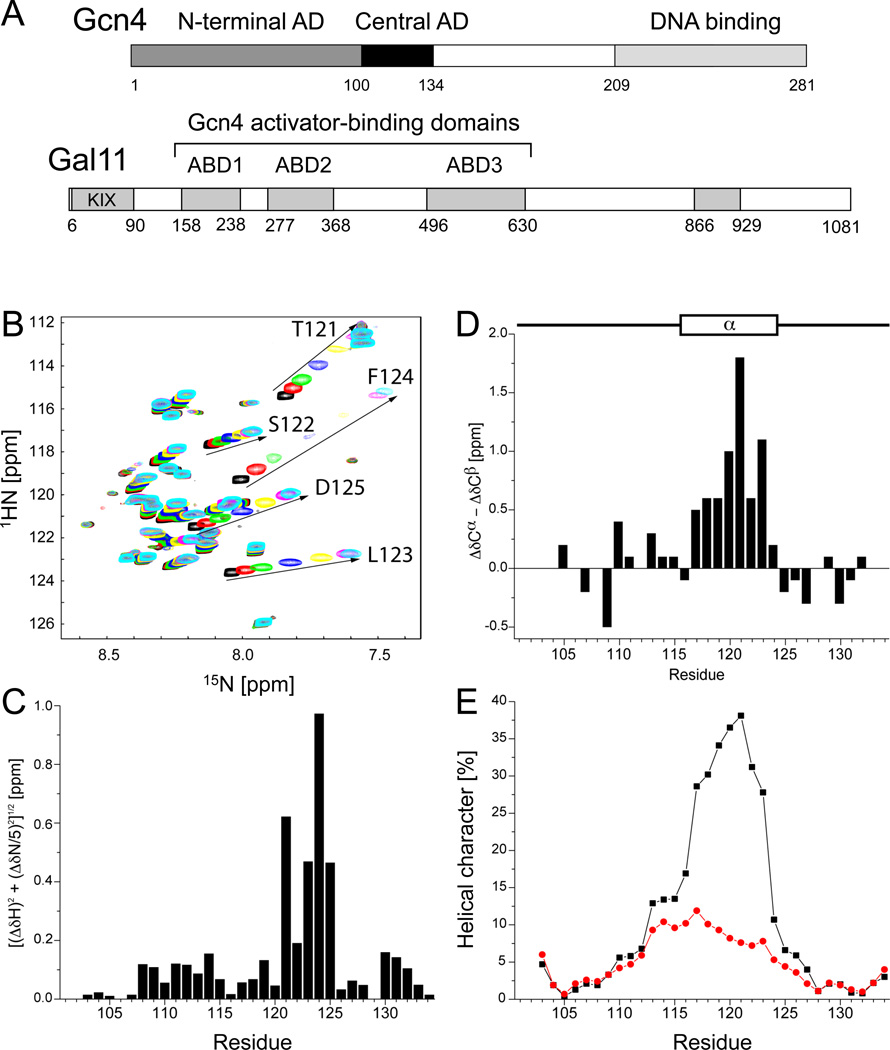
Fig 1. Gcn4 cAD Forms a Short α-helix Upon Binding to Gal11 (also see Fig S1).
(A) Position of the two Gcn4 ADs and three Gal11 domains (ABD-1, 2, 3) that bind Gcn4. Conserved regions of Gal11 are shown in grey. (B) 1H,15N-HSQC spectra of 0.3 mM 15N-labeled Gcn4 (101–134) in the absence (black) and presence of 0.125 (red), 0.25 (green), 0.5 (blue), 1 (yellow), 2 (magenta), or 3 equivalents (cyan) of Gal11 ABD1 (158–238). Amides with the largest chemical shift perturbations (residues 121–125) are labeled and highlighted by arrows. (C) Backbone amide chemical shift perturbations of Gcn4 upon addition of 3 equivalents of ABD1. The formula [(ΔδH)2-(ΔδN/5)2]1/2 was used to calculate the combined chemical shifts of 15N and 1HN. No 1HN-peaks were observed for residues 101 and 102. (D) Combined chemical shift perturbations of 13Cα and 13Cβ of Gcn4 (101–134) bound to ABD1 in reference to free Gcn4. The location of the cAD α-helix is indicated. (E) Probability for the formation of α-helical secondary structure elements predicted by CS-Rosetta (Shen et al., 2008) for Gcn4 (101–134) in the absence (red) and presence (black) of ABD1. NMR chemical shift assignments of 13Cα, 13Cβ, 13C’, 15N, and 1HN for free and bound Gcn4 were input and used for the generation of 100 9-residue fragments starting at each residue. The percentage of fragments showing helical secondary structure at each position is shown.
To understand the molecular basis for Gcn4-Gal11 complex formation and to investigate principles that govern how activators and their targets bind multiple, unrelated partners, we examined the structure and function of a representative Gcn4-Gal11 complex. We find that the Gcn4 AD adopts a helical conformation upon binding Gal11. Complex formation is driven primarily by relatively weak hydrophobic protein contacts that allow Gcn4 to bind Gal11 in multiple orientations. These findings, and the accompanying functional studies, suggest a mechanism for how activator-binding domains recognize seemingly unrelated activators and further define the minimal requirements for a functional AD.
RESULTS
The Gcn4 Tandem ADs are Intrinsically Disordered and Structurally Independent
The structural properties of the Gcn4 tandem ADs were investigated using Nuclear Magnetic Resonance (NMR) spectroscopy. The 1H,15N-HSQC spectra of each individual AD (Gcn4 1–100 and 101–134) overlay almost perfectly with the spectrum of a construct containing both ADs (residues 1–134), indicating that the two regions are structurally independent (Fig S1A). In all three spectra, the dispersion of resonances is over a narrow range in the 1HN-dimension, indicating that both Gcn4 ADs are intrinsically disordered in the absence of binding partners. Our observation of a lack of ordered secondary structure is consistent with a previous NMR analysis of a Gcn4 fusion protein containing residues 39–139 (Huth et al., 1997). Based on its smaller size, the 34-residue central AD of Gcn4 (cAD) was chosen for further structural characterization of its interactions with Gal11. Backbone 13C, 15N, and 1HN NMR resonances of the cAD (residues 101–134; Fig 1A) were assigned using conventional heteronuclear techniques (Sattler et al., 1999). Backbone resonance chemical shifts, particularly 1Hα, 13Cα, and 13Cβ shifts, depend on local backbone geometry and provide a means to identify regions of regular secondary structure (Wishart et al., 1991). No patterns could be discerned from the cAD chemical shifts, consistent with a lack of ordered secondary structure.
The Gcn4 Central AD Adopts a Helical Conformation on Binding to Gal11-158-238
Binding of the cAD to the first Gal11 activator-binding domain (ABD1, residues 158–238) (Herbig et al., 2010) was monitored by collection of a series of 1H, 15N-HSQC spectra of 15N-labeled cAD mixed with increasing concentrations of ABD1 (Fig 1B). Several cAD backbone amide resonances are strongly perturbed upon addition of ABD1. At the endpoint of the NMR titration (three-fold molar excess of ABD1), perturbations of up to 1.0 ppm are observed for residues 121–125 (Fig 1C). Importantly, similar shifts were observed in the spectrum of the tandem Gcn4-ADs when Gcn4 1–134 was titrated with ABD1, suggesting that the mode and affinity of interaction between the cAD and ABD1 is conserved in the context of the tandem Gcn4 ADs (Fig S1B).
To further characterize the structural properties of Gal11-bound Gcn4, backbone and side chain resonance assignments were determined for Gcn4-cAD in the presence of excess ABD1. Comparison of Gcn4 backbone resonances (1HN, 15N, 13Cα, 13Cβ, and 13C’) in the free and bound states indicates that the cAD adopts an α-helical conformation for binding to ABD1. The difference in chemical shift between cAD free and bound to ABD1 was calculated for each 13Cα (Δδ13Cα) and 13Cβ (Δδ13Cβ) resonance and combined (Δδ13Cα - Δδ13Cβ) to give a composite chemical shift difference for each residue (Fig 1D). The histogram shows a contiguous stretch of positive combined differences for residues S117-F124, strongly indicative of α-helical structure (Marsh et al., 2006). In contrast, the combined carbon chemical shift differences of residues 103–116 and 125–134 are much smaller and do not show any trends, indicating that these residues do not take on regular secondary structure in the bound state. 13C’ and 1Hα chemical shift values also indicate stabilization of helical structure upon binding: 13C’ resonances of residues 115–122 shift downfield upon addition of Gal11 (Fig S1C) and 1Hα resonances of residues 117–127 resonate upfield when compared to random coil values (Fig S1D; the 1Hα-chemical shifts for free Gcn4-cAD were not assigned).
The backbone resonance assignments of the free- and bound-states of the cAD were used as input for CS-ROSETTA, a protocol that uses NMR chemical shifts to predict protein structure (Shen et al., 2008; Shen et al., 2009). Up to 40% of the fragments generated by CS-ROSETTA exhibited helical character when the bound chemical shift values were input (Fig 1E, black), while no significant helical character was predicted for free Gcn4-cAD (Fig 1E, red). The prediction of helical character is particularly high for residues S117-L123. Thus, the chemical shift differences and the CS-ROSETTA output indicate that Gcn4-cAD residues S117-F124 adopt a helical conformation upon binding to ABD1.
Structure of Gal11-ABD1
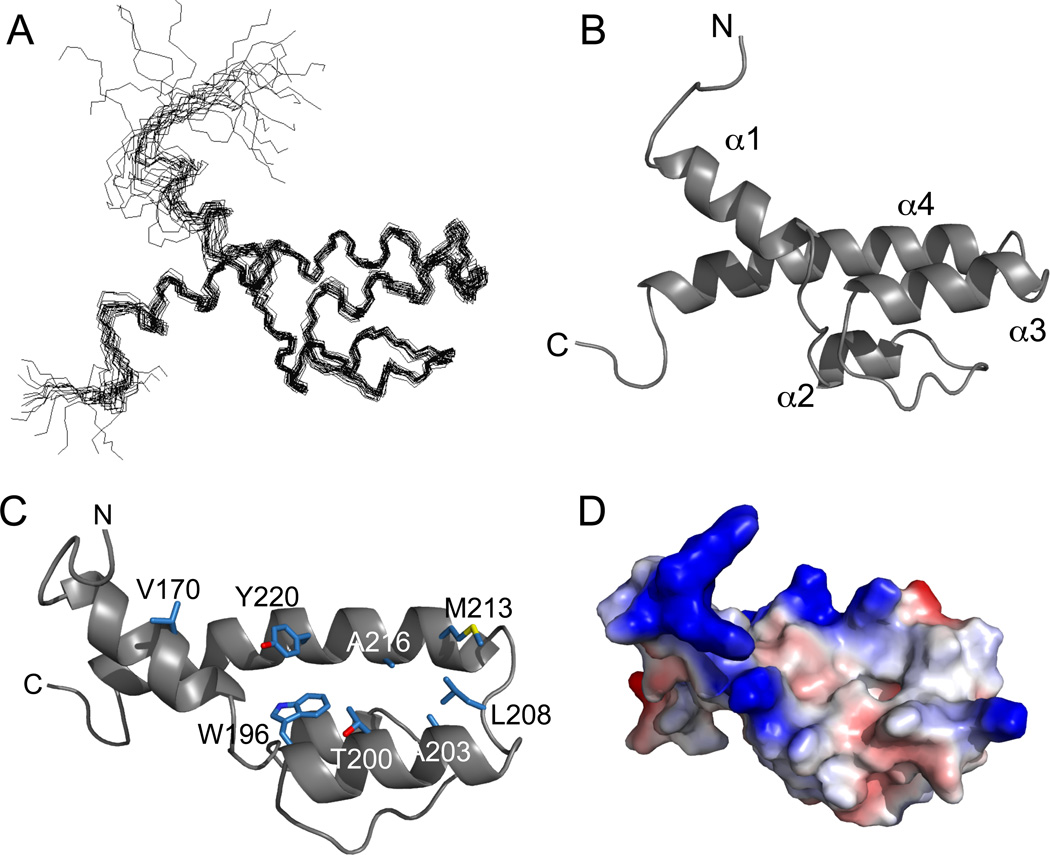
Backbone and side chain resonances (> 98% complete) for 13C,15N-labeled ABD1 were determined in the presence of excess Gcn4-cAD. Nearly complete assignments were obtained for the cAD in the presence of excess ABD1. Dihedral angle constraints were derived from backbone assignments using the program TALOS (Cornilescu et al., 1999). Distance restraints used for structure calculations were derived from analysis of aliphatic and aromatic 13C-NOESY-HSQC and 13C-edited, 13C-filtered NOESY spectra (Table 1). These data were used to first calculate the structure of ABD1 (Fig 2A), which is comprised primarily of four α-helices. A long 26-residue C-terminal helix, α4, spans the entire length of the domain and the remaining helices (α1, α2, and α3) are all oriented anti-parallel with respect to α4 (Fig 2B). Helix α3 runs directly anti-parallel to α4 and is linked to α4 via an extended 5-residue segment. Helix α1 is angled relative to the plane formed by helices α3 and α4. There is some minor variation in the angle of helix α1 as the NOE restraints were not sufficient to define a single orientation. The region between α1 and α3 (residues 175– 195) covers one face of the α3-α4 plane. This segment is mostly extended structure, but includes the short α2 helix (residues 180–185). The α1, α3, and α4 helices create a cleft with a largely hydrophobic floor formed by residues W196 and Y220 and flanked by V170, T200, A203, L208, M213, and A216 (Fig 2C). The electrostatic surface of ABD1 has a net overall positive potential and the hydrophobic cleft is bounded a number of positively charged residues (Fig 2D). A 3D structure search using the DALI (Holm et al., 2008) and COPS servers (Suhrer et al., 2009) failed to identify any structural homologues, indicating that the Gal11-ABD1 structure represents a unique domain fold.
Table 1.
Experimental NMR Restraints and Structural Statistics for the Gal11 Structure.
| Structural Restraints | |||
| Gal11 (residues 158–238) | |||
| NOE distance restraints | 472 | ||
| Short-range (|i–j| = 1) | 189 | ||
| Medium-range (|i<|i–j|<5) | 98 | ||
| Long-range (|i–j|>5) | 59 | ||
| NOE constraints per restrained residue | 6 | ||
| Dihedral-angle constraints | 132 | ||
| Restraint Violations | |||
| Distance violations per model (0.2–0.5Å) | 0.05 | ||
| Dihedral angle violations per model (1–10°) | 2.1 | ||
| Ensemble RMSD (50 models) | all | ordered | |
| All backbone atoms | 2.3Å | 0.9Å | |
| All heavy atoms | 2.9Å | 1.3Å | |
| (ordered residues: 163–187, 191–193, 195–232) | |||
| Ramachandran statistics | |||
| Most favored | 92.2% | ||
| Additionally favored | 7.8% | ||
| Generously allowed | 0.0% | ||
| Disallowed | 0.0% |
Fig 2. Solution Structure of Gal11-ABD1.
(A) NMR ensemble of 20 low-energy Gal11-ABD1 structures. Average pairwise RMSDs for the ordered backbone atoms of residues 163–187, 191–193, 195–232 is 0.9Å. (B) Ribbon representation of the Gal11-ABD1. (C) Orientation from (A) was rotated ~90 degrees about the x-axis to highlight the residues from α1, α3, and α4 that form the ABD1 hydrophobic cleft (shown in stick representation with carbons in blue, oxygens in red, and sulfur in yellow. (D) The surface electrostatic potential of ABD1 oriented as in (B). Red, negatively polarized; blue, positive; white, non-polar.
The Gcn4-Gal11 Interface
The 1H,15N-HSQC titration data is consistent with the Gcn4-cAD binding to the ABD1 hydrophobic cleft, but is not definitive as a large number of Gal11 resonances shift upon interaction with Gcn4 (Figs. S1E and F). To examine the intermolecular interaction we adopted a three-tiered approach involving 1) analysis of intermolecular NOEs, 2) modification of the Gcn4-cAD with paramagnetic spin-labels, and 3) computational docking of Gcn4-cAD onto the Gal11-ABD1. As described below, the sum of these approaches show that Gcn4 binds to the Gal11 hydrophobic cleft, although there is no single mode of interaction. Instead, the short Gcn4 helix binds in multiple orientations with respect to Gal11, a characteristic of intrinsically-disordered proteins that form “fuzzy” complexes (Tompa and Fuxreiter, 2008).
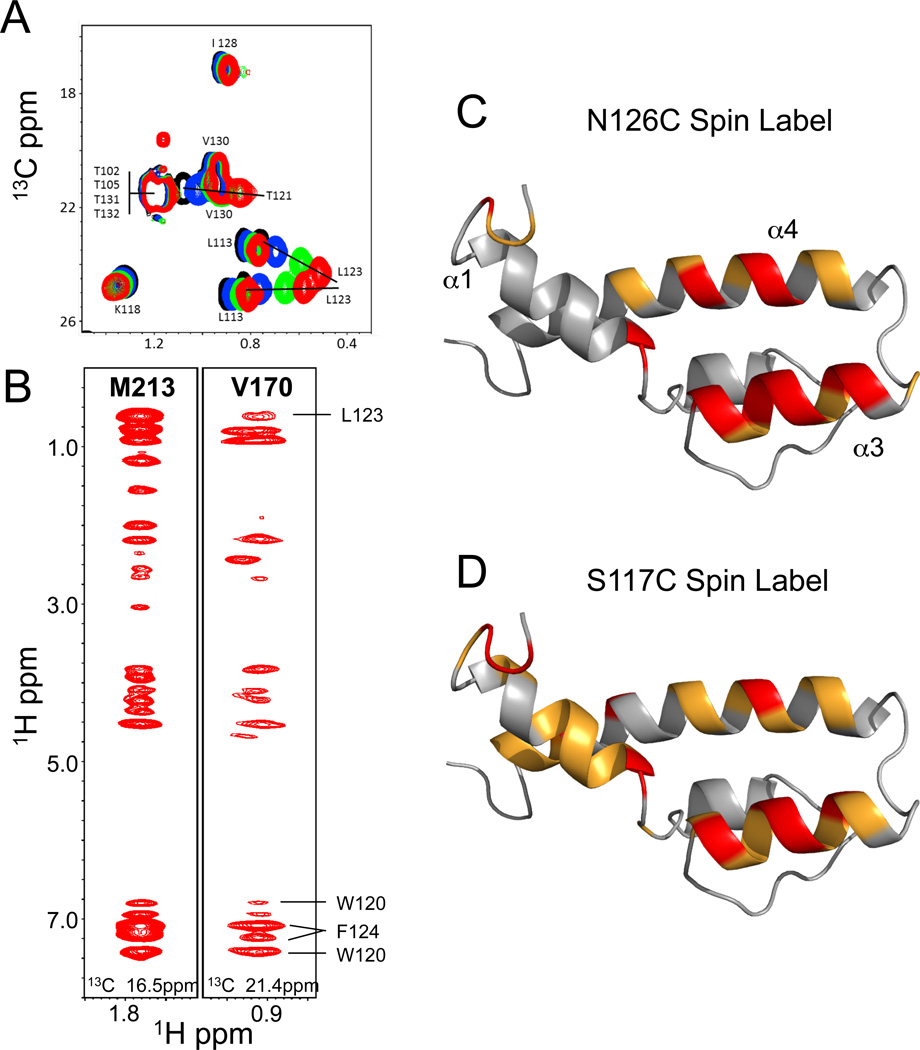
13C-edited and 13C-edited, 13C-filtered-NOESY spectra encompassing the aliphatic resonances of ABD1 were collected to identify intermolecular interactions. The 13C-edited, 13C-filtered-NOESY spectrum allows selective observation of magnetization transferred from the aliphatic side chains of 13C-labeled ABD1 to protons on 12C-labeled Gcn4-cAD. However, there are several features of the complex that impact interpretation of the intermolecular NOE data: 1) the ABD1-cAD complex is in intermediate-to-fast exchange on the NMR time scale (Fig 1B), indicative of a lifetime in the low millisecond range for the complex, 2) the cAD is present in excess and is unstructured in the unbound state, and 3) the molecular weights of the individual components and the protein complex are relatively small. Thus, multiple binding and dissociation events occur during the NOESY mixing time (140ms) and the observed NOESY cross peaks do not arise from a single static complex. These factors are often encountered in systems that give rise to transferred NOEs (Campbell and Sykes, 1993; Sykes, 1993). Therefore, the chemical shifts of cross peaks corresponding to cAD resonances represent a weighted average of chemical shifts in the free and bound states. To properly interpret the spectra, we collected a series of 13C-HSQC spectra of Gcn4-cAD at increasing concentrations of ABD1. The titrations define the trajectory of Gcn4 aliphatic and aromatic protons that shift upon complex formation. A representative section of the 1H,13C-HSQC aliphatic region is shown in Fig 3A. Substantial shifts are particularly evident for the Gcn4 methyl groups of L123 and T121, residues that adopt helical character upon binding to ABD1. Likewise, the aromatic resonances of Gcn4 W120 and F124 undergo large changes in chemical shift (Fig S2A). In contrast, the methyl resonances of L113 (Fig 3A) and aromatic resonances of F108 and Y110 (Fig S2A) are observed to shift, but to a much smaller extent.
Fig 3. NMR spectra of the Gal11-ABD1/Gcn4-cAD complex and effects of Gcn4 paramagnetic spin labels (also see Fig S2).
(A) Titration of 13C-labeled Gcn4-cAD with unlabeled ABD1. The portion of the 13C-HSQC shows the chemical shift perturbations of the T121 and L113 methyl groups upon binding to ABD1. The trajectories of these groups were used to assign resonances that arise from intermolecular interactions observed in NOESY spectra. (B) Portions of the 13C-edited, 13C-filtered NOESY spectrum showing crosspeaks that arise from M213 and V170. Labeled crosspeaks could be unambiguously assigned to specific Gcn4 residues. M213 and V170 are located at opposite ends of the ABD1 hydrophobic cleft (Fig 2B), yet show crosspeaks to the same Gcn4 residues, suggesting that Gcn4 binds to ABD1 in multiple orientations. (C,D) Paramagnetic spin labels were incorporated at four different positions of the cAD (104, 117, 126, 133), where positions 117 and 126 flank the nascent Gcn4-cAD helix. Observed intensity perturbations in ABD1 upon complex formation with Gcn4 spin-labeled at positions 126 and 117 are shown. Gal11 (gray ribbon), with strongly affected residues (intensity decrease > 80% relative to reference spectrum) in red and significantly affected residues (intensity decrease between 50–80%) in orange.
Examination of the NOESY spectra show that Gal11 residues L162, Q167, L169, V170, V199, T200, A203, M213, A216, K217, and Y220 are in NOE contact with Gcn4 residues W120, T121, L123, and F124. What is particularly striking is that side chains throughout the Gal11-ABD1 binding cleft interact with the same set of Gcn4 hydrophobic residues. For instance, both V170 and M213, located at opposite ends of the ABD1 hydrophobic cleft (Fig 2C), exhibit NOEs to same protons in the W120, T121, L123, and F124 side chains of Gcn4 (Fig 3B). These observations are consistent with a model in which the cAD binds to Gal11 in more than one orientation.
For the second tier of structural analysis, spin-labels were attached to specific sites on the cAD. Single cysteine residues were introduced at four locations: residues 117 and 126, flanking the Gcn4 helical segment, and residues 104 and 133, near the N- and C-terminus of the Gcn4-cAD. Each cAD Cys-derivative was modified with the spin label 4C-(2-Iodoacetamido)-TEMPO. Spin-labeled cAD induces peak broadening of Gal11 resonances that are in proximity to the unpaired electron in TEMPO (Fig S2B; Histograms of the residue-by-residue spin label effects are shown in Fig S2C). The spin label at position 126 produces large decreases in peak intensity (≥80%) in the Gal11 amide resonances of residues L162, Q172, K174, W196, Q197, V199, T200, A201, A203, Q204, A216, K217 and Y220 (red, Fig 3C). These residues are located at the C-terminal end of α1, helix α3, and the N-terminal portion of α4. Smaller but significant effects on resonance intensity (50–80% decrease) are observed in a second set of resonances, most of which are located on helix α4 in the region where it crosses helix α1 (orange, Fig 3C). Incorporation of a spin-label at position 117 yielded similar results (Fig 3D). Resonances in α1 and α3 are affected by the spin-label in this position, though to a lesser extent, while residues located in the region bounded by α1 and α4 and including the Gcn4-cAD N-terminal segment are perturbed to a greater extent (Fig S2C). Thus, placement of a spin-label at either end of the Gcn4 helix affects residues throughout the hydrophobic binding cleft. In contrast, placement of spin-labels near the cAD termini (residues 104 and 133) indicate that regions outside the cAD helix do not make close contact with ABD1. These spin-labeling experiments also support the model where the cAD binds to ABD1 in multiple orientations.
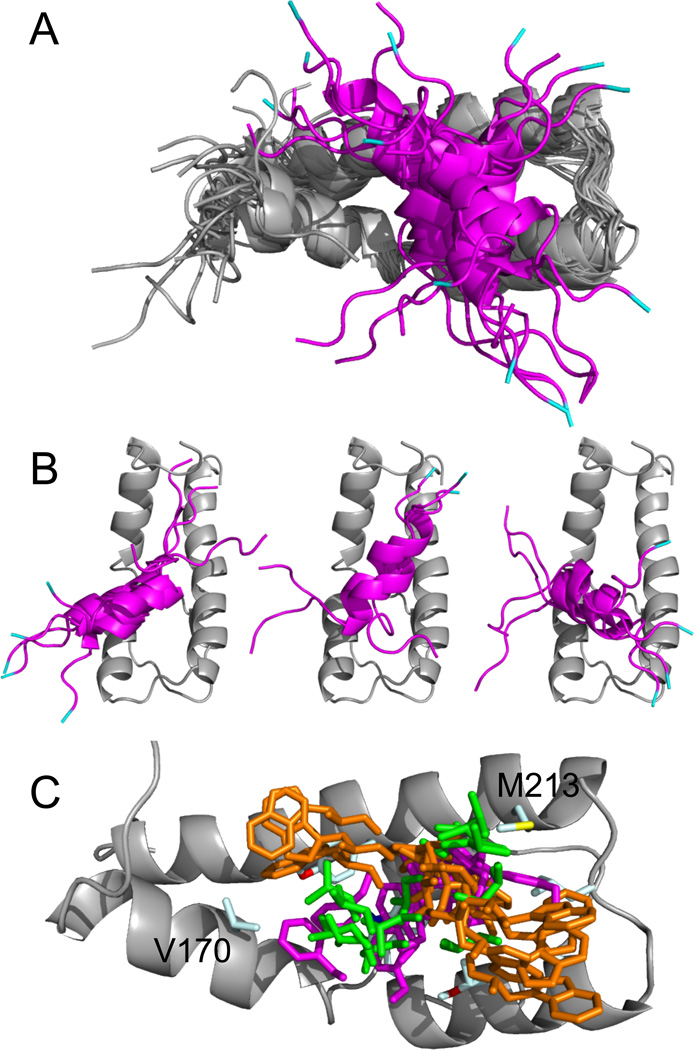
The finding that there is no single mode of interaction between the cAD and ABD1 precludes the determination of pairwise distance restraints to define the complex. The transferred NOEs observed in (13C-edited-, filtered)-NOESY spectra do define Gal11 residues involved in complex formation. Therefore, for the third tier of structural analysis we used ambiguous interaction restraints (AIRs) to calculate models of the complex. AIRs allow for any interacting Gcn4 residue to potentially contact any Gal11 residue located at the intermolecular interface (Dominguez et al., 2003). Because Gcn4 adopts helical character upon binding Gal11, loose dihedral angle constraints were generated for the bound form of the Gcn4-cAD using the program TALOS (Cornilescu et al., 1999). These were used to create a starting ensemble of cAD structures with helical character and, together with the ensemble of ADB1 structures previously calculated, were used as starting structures to perform docking calculations with the program HADDOCK (Dominguez et al., 2003). Dihedral angle constraints and unambiguous distance constraints used for the calculations of the ABD1 structure were also included. The ABD1 domain (residues 169–232) was aligned in the resulting ensemble of 200 structures. Structures were then filtered to select those most consistent with the spin-labeling data by requiring that both S117 and N126 are within 15–17Å of amides maximally affected (>80%) by spin-labels at these positions. The twelve resulting structures are shown in Fig 4A. Examination of the complexes reveals three different orientations of the Gcn4 helix on ABD1 (Fig 4B). These three clusters are representative of Gcn4 binding, but are not meant to be exclusive.
Fig 4. Models of the ABD1-cAD Complex Derived from NOE and Spin-Labeling Data.
(A) Ribbon representations for the ensemble of HADDOCK generated structures for Gcn4-cAD (magenta) binding to the ABD1 (gray). Gcn4 residues 101–112 and 131–134 have been removed for clarity and L113 (cyan) marks the N-terminus. (B) Three different orientations of the Gcn4 peptide are evident in the ensemble of structures depicted in (A). (C) Positions of key Gcn4 side chains W120 (orange), L123 (green), and F124 (magenta) relative to ABD1 (gray ribbon) are shown from the ensemble in (A). ABD1 residues V170 and M213 are labeled. The different modes of binding bring W120, L123, and F124 in proximity to both residues, consistent with observations derived from the (13C-edited, 13C-filtered)-NOESY (see Fig 3B).
In each of the clusters, the hydrophobic face formed by Gcn4 residues W120, T121, L123, and F124 binds to the ABD1 hydrophobic cleft (Fig 4C). There is a shallow depression in the ABD1 surface located between helices α3 and α4, bounded by A203, L208, and M213 on one side, Y220 on the other, and with A216 and W196, which is partly accessible, forming the base (Fig 2C,D). Typically, the aromatic side chain of either Gcn4 W120 or F124 inserts into this depression. In most models, these interactions position the nascent Gcn4 helix across the ABD1 α3 and α4 helices. Though the angle of intersection can range from nearly perpendicular to almost parallel, most models orient the helix in a way that can bring Gcn4 L123 in proximity to either Gal11 M213 or V170 (Fig 4C).
Importance of Residues at the Gcn4-Gal11 Interface
Alanine substitutions were made at Gcn4-cAD W120 and F124, residues making direct contact with Gal11-ABD1, Gal11 W196 positioned in the center of the activator-binding cleft, and Gal11 residues M213 and T200, located at the edge of the binding cleft. Binding affinities were measured using isothermal titration calorimetry (Table 2, Fig S3A–E). Wild-type cAD binds ABD1 with a Kd of 10.1 µM, similar to values measured using fluorescence polarization (Herbig et al., 2010). As expected from NMR analysis, Ala substitutions at Gcn4 120 or 124 decreased binding affinity for ABD1 by 5.6- and 6.5-fold, respectively.
Table 2.
Mutations within the Gcn4-Gal11 Interface Decrease the Affinity of interaction.
| Gal11 | Gcn4 AD | Kd (µM) |
ΔH (cal/mol) |
ΔS (cal/mol/deg) |
|---|---|---|---|---|
| 158–238 | 101–134 | 10.1 ± 1.4 | −5394 ± 150 | 4.58 |
| 158–238 | 101–134 W120A | 56 ± 6 | −1823 ± 80 | 13.3 |
| 158–238 | 101–134 F124A | 65 ±3 | −3880 ± 46 | 6.02 |
| 158–238 W196A | 101–134 | NM | NM | NM |
| 158–238 M213A | 101–134 | 76 ±5 | −2812 ± 78 | 9.32 |
| 158–238 T200A | 101–134 | 10.5± 0.5 | −5021 ± 62 | 5.81 |
Affinities were measured by ITC. NM, no measureable binding.
The Gal11-ABD1 W196 mutant did not bind the cAD. Circular dichroism spectra showed that this mutation disrupts ABD1 secondary structure with a significant decrease in α-helical content (Fig S3F). As W196 lies at the intersection of three Gal11 α-helices, it makes important contacts within the core of the domain and likely promotes cooperative folding in addition to forming part of the Gcn4-binding cleft. Mutation of Gal11 M213 to Ala decreased binding 7.5-fold without affecting the helical content of ABD1 (Fig S3F). However, the M213 side chain does contribute to the overall thermal stability of ABD1 as the melting temperature of this derivative decreased from 53° to 41° (not shown). In contrast, Gal11 T200A showed no change in Gcn4 binding affinity. Given the hydrophobic nature of the interaction, it is likely that loss of polar character in going from Thr to Ala has no significant effect.
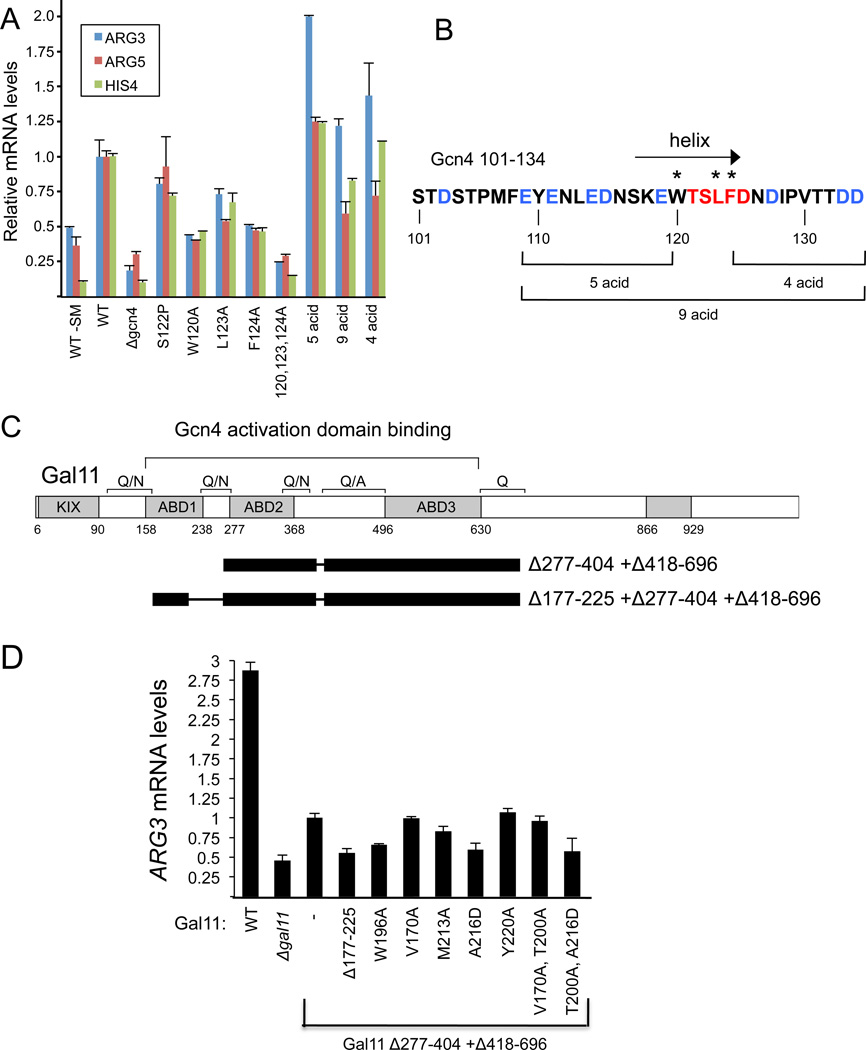
To determine the effect of Gcn4 mutations on transcription activation in vivo, a series of mutations were generated in a Gcn4 derivative lacking a functional N-terminal AD and assayed for function in a yeast strain where Gal11 430–680 was deleted, leaving ABD1 as the highest affinity Gcn4 binding site (Herbig et al., 2010). Cells were grown under starvation conditions to induce Gcn4 expression and mRNA levels from three Gcn4 and Gal11-dependent genes were measured by RT-qPCR (Fig 5). Two complications are encountered when measuring the in vivo effects of the Gcn4 mutants. First, activation by Gcn4 requires that the ADs directly interact with Gal11 (Mediator) as well as Tra1 (SAGA/NuA4) and possibly SWI/SNF so this assay does not strictly measure Gcn4 interaction with Gal11. Second, the stability of Gcn4 is directly related to its role in activation, as it is targeted for ubiquitylation and degradation while stimulating transcription (Hinnebusch, 2005; Irniger and Braus, 2003). Gcn4 is normally unstable and present at very low levels even under starvation-induced conditions. However, nearly all of the Gcn4 mutations used here show significantly elevated protein levels compared with the wild-type central AD (Fig S4A). Thus, the observed effects of these mutations on transcription activation may be partially obscured due to the elevated levels of these Gcn4 derivatives.
Fig 5. Effect of Mutations in the cAD-ABD1 Interface on Transcription Activation in vivo (also see Fig S4).
(A) Cells with the indicated Gcn4 mutations and Gal11 Δ418–696 were induced for 90 min with SM (sulfometuron methyl; except where noted, -SM) to induce starvation. mRNA was extracted and quantitated by RT qPCR. Error bars represent the SEM. (B) Sequence of the cAD. Residues with the largest chemical shift perturbations (Fig 1B) are red; acidic residues outside of this region are blue. The arrow indicates the position of the α-helix formed upon binding Gal11. * indicates the position of alanine substitutions at hydrophobic residues and brackets indicate the positions of acidic residues substituted with Ala. (C) Schematic of the Gal11 derivative used for mutagenesis of ABD1 where black bars represent regions deleted from Gal11. Conserved regions of Gal11 are shown by shaded boxes. (D) Cell grown as in (A) and mRNA quantitated by RT qPCR. Error bars represent the SEM.
Under these conditions, deletion of Gcn4 reduced transcription ≥ 4-fold from all three genes. A triple Ala substitution of three Gcn4 hydrophobic residues at the Gal11 interface (120, 123, and 124) was nearly equivalent to a Gcn4 deletion (Fig 5A) (Drysdale et al., 1995). Mutation of the individual aromatic residues W120 and F124 had the strongest effects, with transcription of ARG3, ARG5, and HIS4 reduced 2-3-fold. Mutation of Gcn4 L123 had a smaller effect. To test the sensitivity of the Gcn4 α-helix to mutation, proline was substituted for Gcn4 residue S122 and found to have only a minor effect on transcription, likely due to the flexibility of Gcn4 in binding to ABD1.
The cAD is highly enriched for acidic residues with 10 of its 34 residues acidic (Fig 5B, blue). To test if specific acidic residues or the overall negative charge is required for activity, groups of 4, 5, or 9 acidic residues within the cAD were substituted with Ala. Surprisingly, these mutations had little effect on transcription from ARG3 or HIS4 (Fig 5A). Transcription from ARG5 showed only modest reductions in Gcn4 constructs with 4 or 9 acidic residues changed to Ala while transcription of ARG3 was slightly elevated. Mutation of the one other acidic residue at position 103 also had no effect on transcription of any gene tested (not shown). Similar results were observed when cells containing wild type Gal11 were used (Fig S4B). From these results, we conclude that the acidic residues in the cAD are not required for function.
We also examined the in vivo role of ABD1 residues in the hydrophobic cleft using a Gal11 derivative deleted for both ADB2 and ADB3 (Fig 6C; Δ277-404 + Δ418-696). Because all three activator-binding domains contribute additively to activation by Gcn4 (Herbig et al., 2010; Jedidi et al., 2010), this derivative shows only a two-fold decrease in transcription of ARG3 when ABD1 is deleted (Herbig et al., 2010; Jedidi et al., 2010). ARG3 transcription was decreased nearly equivalently by deletion of ABD1 or by the mutations on the floor of the binding cleft W196A and A216D, both of which disrupt the folding of Gal11 (Fig 5D and data not shown). M213A, shown above to decrease the affinity for Gcn4, also decreased transcription, while Y220A, V170A, or the double mutant V170A, T200A showed no significant change in ARG3 expression. These in vivo results, combined with the above in vitro binding assays, show that Gal11 side chains within the binding cleft are not solely involved in activator recognition. Residues W196, 213, and 216 likely play a role in both stabilization of ABD1 structure and activator binding.
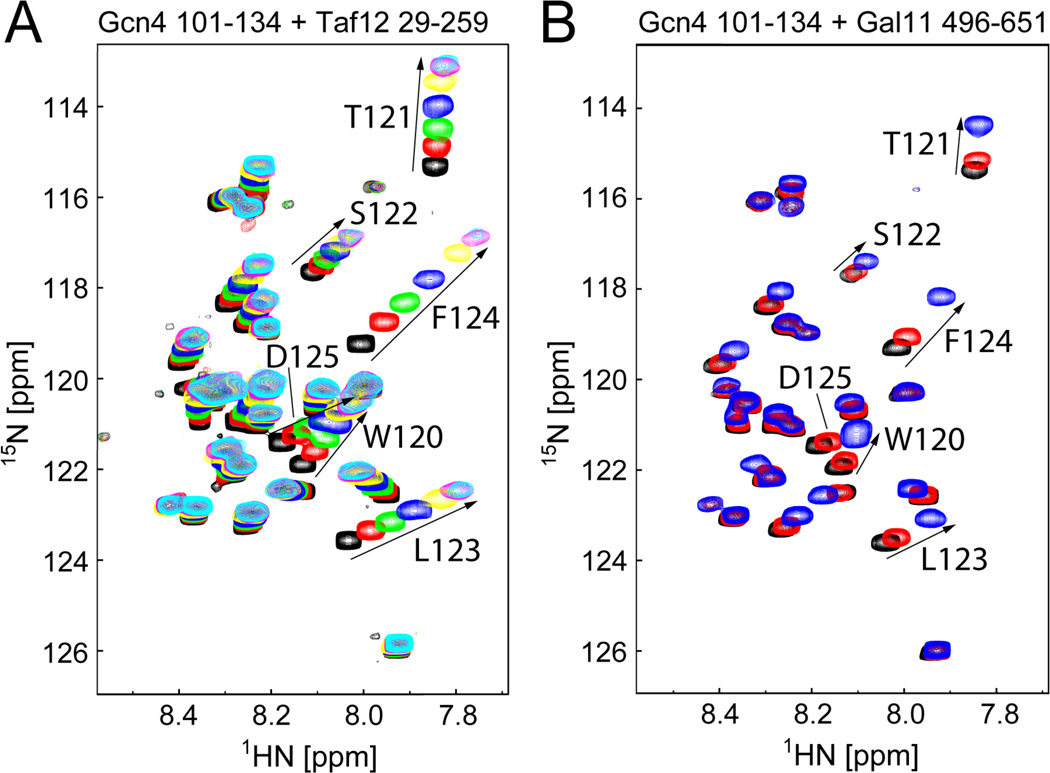
Fig 6. Gcn4 Uses Similar Mechanisms for Recognition of Taf12 and Gal11 ABD3 (also see Fig S5).
(A) 1H,15N-HSQC spectra of 0.3 mM 15N-labeled Gcn4 (101–134) in the absence of (black) and presence of 0.125 (red), 0.25 (green), 0.5 (blue), 1 (yellow), 2 (magenta), or 3 equivalents (cyan) of Taf12 (29–259). (B) 1H,15N-HSQC spectra of 0.3 mM 15N-labeled Gcn4 (101–134) in the absence of (black) and presence of 0.1 (red) or 0.5 equivalents (blue) of Gal11 (496–651). In each spectrum, amides with the largest chemical shift perturbations (residues 120–125) are labeled and highlighted by arrows. A complete titration could not be performed due to the limited solubility of ABD3.
Gcn4 Recognizes Other Activator-Binding Domains Using a Conserved Mechanism
The Gcn4-cAD specifically binds at least two other Gal11 activator-binding domains and a domain in Taf12 (Herbig et al., 2010; Jedidi et al., 2010; Majmudar et al., 2009). To test if Gcn4 uses a similar mechanism to bind to these other domains, we examined the HSQC spectrum of 15N-Gcn4-cAD upon binding to either Gal11 residues 496–651 (ABD3) or Taf12 residues 29–259 (Fig 6). cAD chemical shift perturbations observed upon addition of unlabeled Taf12 or ABD3 are remarkably similar to those seen with ABD1 (Fig 1B). The same Gcn4 residues undergo the largest backbone amide perturbations in each case, though the absolute magnitudes of resonance perturbations are smaller compared to Gcn4 binding to Gal11-ABD1 (compare Figs. 1C and S5). These observations indicate that the Gcn4-cAD uses a similar interface to interact with each of these domains and the ability of Gcn4 to adopt α-helical conformation may be a common element to the formation of each complex.
DISCUSSION
Assembly of the transcription pre-initiation complex (PIC) relies on a number of low affinity and relatively low specificity protein-protein and protein-DNA interactions (e.g., TBP-TATA, TAF-INR, TFIIB-DNA, Pol II-TFIIB). Even among these low affinity complexes, transcription activator/target interactions stand out as particularly enigmatic (Sigler, 1988). Critical structural elements are difficult to define as there is little sequence conservation among transcription ADs and these domains are often structurally disordered in the absence of a binding partner (Tompa and Fuxreiter, 2008). Mutational analysis can be ambiguous, as multiple mutations are often required to significantly alter activator activity. In addition, a single AD may bind to multiple unrelated activator-binding domains indicating that adaptability in the binding interface is an important structural characteristic. In cases where structures have been determined, the ADs use one or two amphipathic α-helices to form complexes with well-defined geometry. In these structures, the binding interface is composed primarily of interactions between hydrophobic residues with additional polar and/or charged contacts (Bochkareva et al., 2005; Dames et al., 2002; Di Lello et al., 2006; Langlois et al., 2008; Radhakrishnan et al., 1997; Uesugi et al., 1997; Zor et al., 2004). Importantly, the polar and ionic interactions contribute to binding specificity and orientation of the two molecules in the complex, and increase protein-protein affinity. In several cases, activator/coactivator binding is regulated by phosphorylation, where the phosphorylated residue makes specific contact with the activator-binding domain and greatly alters the affinity of interaction (Feng et al., 2009; Ferreon et al., 2009; Radhakrishnan et al., 1997). The interaction between the p53 AD and MDM2 represents one example where hydrophobic contacts predominate. Three hydrophobic residues of p53 bind in a deep hydrophobic cleft in MDM2 with high steric complementarity, forming a high affinity, stable complex (Kussie et al., 1996). From these static structures, it is difficult to extrapolate to other systems where unrelated activators interact with a common target protein or cases where a single activator binds multiple unrelated targets.
The Gcn4-cAD/Gal11-ABD1 complex shows two striking differences with previous activator-target complexes. First, the protein-protein interface is much simpler. Three Gcn4 residues, W120, L123, and F124, interact with hydrophobic residues in a shallow ABD1 cleft. Although the cAD and ABD1 have opposite electrostatic surface potentials, structural and mutational analysis demonstrate that there are no specific polar or ionic interactions at the interface. This likely contributes to the second major difference from previous activator-coactivator structures: the cAD helix binds to ABD1 in multiple orientations. These properties indicate that the Gcn4-Gal11 complex is an example of a “fuzzy complex” in which the protein interface cannot be described by a single conformational state (see Tompa and Fuxreiter, 2008 and references therein). “Fuzziness” has been proposed to be functionally important by providing adaptability and reversibility to protein-protein interactions, consistent with the biological function of many ADs.
Wild-type Gcn4 functionally interacts with wild-type Gal11 by binding three low affinity activator-binding sites rather than through a single high affinity, high specificity site (Herbig et al., 2010). Our NMR results show that this binding is in fast exchange on the NMR time scale, suggesting that Gcn4 can rapidly sample multiple Gal11 activator-binding domains as a mechanism to recruit Mediator to the enhancer/promoter region. This model explains why the Gal11 activator-binding domains act additively to increase activated transcription and why multimerization of transcription factor DNA binding sites often greatly stimulates transcription. These results would not be expected if activators have a single high affinity, high specificity target with a slow dissociation rate.
An unexpected result was the finding that acidic residues within the cAD are not essential for function. Since Gal11 has a net overall positive charge, it was expected that non-specific electrostatic interactions would contribute to long range attractions between Gcn4-Gal11 (Shoemaker et al., 2000). An additional possibility is that the electrostatic properties of Gcn4-ADs and Gal11-ABDs act as a screen to prevent unwanted interactions with proteins that have a suitable hydrophobic interface but have the wrong surface potential. In either case, the mutagenesis results emphasize the lack of highly specific electrostatic interactions in the Gcn4-cAD/Gal11-ABD1 complex, consistent with our model for multiple modes of Gcn4-Gal11 binding.
To activate transcription, Gcn4 must also interact with other coactivator subunits unrelated to Gal11. Our results show that the cAD uses the same key residues to interact with Gal11-ABD3, and the activator-binding domain of Taf12. Given the simple nature of the Gcn4/Gal11 interface, these other activator-binding domains likely bind Gcn4 using a similar mechanism. The low affinity and specificity of Gcn4/target interactions seems to require only a simple protein-protein interface where the activator can readily adapt to fit a fairly generic hydrophobic binding cleft.
What then defines the requirements for a minimal AD? Gcn4 consists of an inherently disordered polypeptide, with a segment able to adopt a helical fold when bound to its targets. This helical segment has hydrophobic residues at positions i, i +3, i +4 (residues 120, 123, 124) on the same face of the helix. This pattern has been noted previously in several activators (Chi et al., 2005; Uesugi et al., 1997) and includes the LXXLL interaction motif (Darimont et al., 1998; Nolte et al., 1998). Within Gcn4, the aromatic residues at positions 120 and 124 are the most important for activity as these two residues define the minimal pattern for Gcn4 activator function. Sequence motifs vary among well-characterized activators. The acidic activators VP16, p53, and EKLF (Chi et al., 2005; Mas et al., 2011; Uesugi et al., 1997) all contain sequences that match the motif described for Gcn4-cAD where at least one of these hydrophobic residues is functionally important (Cress and Triezenberg, 1991; Lin et al., 1994; Mas et al., 2011). In contrast, Gal4 has two overlapping matches to this pattern, but mutagenesis of nearly every residue within the Gal4 AD has not revealed any one residue critical for activator function (Ansari et al., 1998). The Gcn4-N-terminal AD contains two matches to this pattern but functional studies define this AD as nearly 100 residues in length, suggesting that this activator does not use the minimal motif (Jackson et al., 1996). Future studies to precisely define the minimal activator motif as well as examining the mechanism of more complex activators will be highly informative for defining different classes of activators and a better understanding of the diverse mechanisms used in eukaryotic transcriptional regulation.
EXPERIMENTAL PROCEDURES
See also Supplemental Experimental Procedures
NMR Experiments and Resonance Assignments
NMR experiments were performed at 25°C on a Bruker 500 MHz AVANCE or Varian INOVA spectrometers (600 MHz, 800 MHz, or 900 MHz instruments located at Pacific Northwest National Laboratories). Data were processed and analyzed using NMRPipe (Delaglio et al., 1995) and NMRView (Johnson and Blevins, 1994). NMR samples for Gal11-ABD1 structure determination consisted of 1.2 mM[13C,15N]-Gal11 – ABD1 plus 2.4 mM unlabeled Gcn4 –cAD in NMR buffer with 10% or 99% D2O. Likewise, 1.2 mM [13C,15N]-Gcn4-cAD with and without 2.4 mM unlabeled Gal11 –ABD1 were used for the assignment of the backbone and side chain resonances of free and bound Gcn4. Assignment of backbone and side chain resonances were accomplished by analysis of standard triple-resonance experiments (Sattler et al., 1999, see also Supplemental Experimental Procedures). Mixing times for NOESY-based experiments were 140ms. Chemical shift assignments for Gcn4-bound Gal11 and Gal11-bound Gcn4 are deposited at BioMagResBank (accession no. 16488)
Structure Calculations and Modeling
Distance restraints for calculation of the Gal11-ABD1 domain were obtained by manual inspection of 3D-NOESY experiments. Only unambiguous cross peaks with symmetry related resonances were used, interacting atoms were binned based on observed NOE intensities, and used to generate distance constraints. Constraints involving side chains of residues found to be involved in intermolecular interactions were excluded. Dihedral backbone angles restraints for Gal11 were predicted from backbone assignments and generated using TALOS (Cornilescu et al., 1999). Only dihedral angle restraints with good fits were included. These constraints were used as input for structure calculations in CNS 1.3. (Brunger et al., 1998).
The 20 lowest energy structures were used in calculations to model the complex formed with Gcn4-cAD. Edited-filtered NOESY experiments were used to define interacting Gal11 and Gcn4 residues. These data were then used to generate Ambiguous Interaction Restraints to model the complex using the docking program HADDOCK (Dominguez et al., 2003). The resulting models were compared with the results of spin-labeling experiments (see below) to identify structures that most closely reflect experimental observations. Models of the complex have been deposited at the Protein Data Bank (PDB code: 2KO4).
Paramagnetic Relaxation Enhancement Experiments with Spin-labeled Gcn4
Single cysteine mutants of Gcn4 (101–134) were prepared through site-directed mutagenesis (S104C, S117C, N126C, and D133C) and modified with 4-(2-Iodoacetamido)-TEMPO. For the measurement of paramagnetic relaxation enhancement, 0.6 mM spin-labeled Gcn4 mutant-protein was added to 0.3 mM 15N-labeled Gal11 (158–238). 1H,15N-HSQC spectra in the absence and presence of 3 mM ascorbic acid (to reduce the paramagnetic nitroxide) were collected. The intensity of each backbone amide resonance was measured to calculate the ratio of the intensity in the absence of ascorbic acid versus the intensity in the presence of ascorbic acid.
Supplementary Material
Acknowledgements
We thank Barry Stoddard and members of the Hahn and Klevit labs for comments and suggestions during the course of this work and Nadia Kulshina for help with ITC experiments. NMR experiments were performed, in part, in the Environmental Molecular Sciences Laboratories at the Pacific Northwest National Laboratories. DB is an HHMI investigator. This work was funded by NIH grant GM075114 to SH and a Boehringer Ingelheim Fonds Ph.D. Scholarship to CH; REK and PSB are supported by NIH R01 GM088055.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ansari AZ, Reece RJ, Ptashne M. A transcriptional activating region with two contrasting modes of protein interaction. Proc Natl Acad Sci U S A. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Campbell AP, Sykes BD. The two-dimensional transferred nuclear Overhauser effect: theory and practice. Annu Rev Biophys Biomol Struct. 1993;22:99–122. doi: 10.1146/annurev.bb.22.060193.000531. [DOI] [PubMed] [Google Scholar]
- Chi SW, Lee SH, Kim DH, Ahn MJ, Kim JS, Woo JY, Torizawa T, Kainosho M, Han KH. Structural details on mdm2-p53 interaction. J Biol Chem. 2005;280:38795–38802. doi: 10.1074/jbc.M508578200. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes & development. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Drysdale CM, Duenas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. Journal of molecular graphics & modelling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Feng H, Jenkins LM, Durell SR, Hayashi R, Mazur SJ, Cherry S, Tropea JE, Miller M, Wlodawer A, Appella E, et al. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure. 2009;17:202–210. doi: 10.1016/j.str.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, Eck MJ. Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nature structural biology. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol. 2010;30:2376–2390. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annual review of microbiology. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Bioinformatics (Oxford, England) Vol. 24. 2008. Searching protein structure databases with DaliLite v.3; pp. 2780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Mahadevan S, Struhl K. Structural and Functional Characterization of the Short Acidic Transcriptional Activation Region of Yeast GCN4 Protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Braus GH. Controlling transcription by destruction: the regulation of yeast Gcn4p stability. Curr Genet. 2003;44:8–18. doi: 10.1007/s00294-003-0422-3. [DOI] [PubMed] [Google Scholar]
- Jackson BM, Drysdale CM, Natarajan K, Hinnebusch AG. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol. Cell. Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, et al. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J Biol Chem. 2010;285:2438–2455. doi: 10.1074/jbc.M109.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRVIEW: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kussie P, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotien bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Langlois C, Mas C, Di Lello P, Jenkins LM, Legault P, Omichinski JG. NMR structure of the complex between the Tfb1 subunit of TFIIH and the activation domain of VP16: structural similarities between VP16 and p53. Journal of the American Chemical Society. 2008;130:10596–10604. doi: 10.1021/ja800975h. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes & development. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Majmudar CY, Wang B, Lum JK, Hakansson K, Mapp AK. A high-resolution interaction map of three transcriptional activation domains with a key coactivator from photo-cross-linking and multiplexed mass spectrometry. Angew Chem Int Ed Engl. 2009;48:7021–7024. doi: 10.1002/anie.200902669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Whiteway M. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryotic cell. 2007;6:291–301. doi: 10.1128/EC.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas C, Lussier-Price M, Soni S, Morse T, Arseneault G, Di Lello P, Lafrance-Vanasse J, Bieker JJ, Omichinski JG. Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF) Proc Natl Acad Sci U S A. 2011;108:10484–10489. doi: 10.1073/pnas.1017029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. The EMBO journal. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting Activity Is Required for SWI/SNF Function In Vivo and Is Accomplished through Two Partially Redundant Activator-Interaction Domains. Mol Cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34:93–158. [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler PB. Acid Blobs and Negative Noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- Suhrer SJ, Wiederstein M, Gruber M, Sippl MJ. COPS--a novel workbench for explorations in fold space. Nucleic Acids Res. 2009;37:W539–W544. doi: 10.1093/nar/gkp411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes BD. Determination of the conformations of bound peptides using NMR-transferred nOe techniques. Curr Opin Biotechnol. 1993;4:392–396. doi: 10.1016/0958-1669(93)90003-f. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends in biochemical sciences. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. Journal of molecular biology. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Qiu H, Swanson MJ, Hinnebusch AG. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol Cell Biol. 2003;23:8829–8845. doi: 10.1128/MCB.23.23.8829-9945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. Journal of molecular biology. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.