The authors conducted genetic, molecular, and functional analyses of the promoter region that controls the expression of LEC2, a key seed regulatory gene. They characterized three cis-elements, including RLE, which is necessary for histone H3 trimethylation. The results provide data about the transcriptional regulatory network that controls seed development and chromatin regulation of plant gene expression.
Abstract
LEAFY COTYLEDON2 (LEC2) is a master regulator of seed development in Arabidopsis thaliana. In vegetative organs, LEC2 expression is negatively regulated by Polycomb Repressive Complex2 (PRC2) that catalyzes histone H3 Lys 27 trimethylation (H3K27me3) and plays a crucial role in developmental phase transitions. To characterize the cis-regulatory elements involved in the transcriptional regulation of LEC2, molecular dissections and functional analyses of the promoter region were performed in vitro, both in yeast and in planta. Two cis-activating elements and a cis-repressing element (RLE) that is required for H3K27me3 marking were characterized. Remarkably, insertion of the RLE cis-element into pF3H, an unrelated promoter, is sufficient for repressing its transcriptional activity in different tissues. Besides improving our understanding of LEC2 regulation, this study provides important new insights into the mechanisms underlying H3K27me3 deposition and PRC2 recruitment at a specific locus in plants.
INTRODUCTION
In eukaryotes, complex developmental programs require tightly regulated expression of a specific set of genes at both spatial and temporal levels. Chromatin state dynamics and transcription factors are involved in the underlying molecular mechanisms (Beisel and Paro, 2011). Recent research has emphasized the importance of histone posttranslational modifications, such as acetylation, methylation, ubiquitination, or phosphorylation, in these regulatory processes (Berger, 2007; Kouzarides, 2007; Zhang et al., 2007a; Lee et al., 2010). In plants, genome-wide chromatin and transcriptomic analyses have confirmed the correlation existing between histone posttranslational modifications and transcriptional activity (Pfluger and Wagner, 2007; Roudier et al., 2009, 2011; Lauria and Rossi, 2011). For instance, Histone H3 Lys (residues 4, 9, 27, and 36) methylations are strongly associated with the transcriptional regulation of plant genes (Li et al., 2008; Liu et al., 2010; Lafos et al., 2011; Zheng and Chen, 2011). H3K4me3 and H3K36me2/me3 are predominantly linked with active chromatin, whereas H3K9me1/me2 and H3K27me3 modifications are considered to be repressive. More specifically, dynamic regulation of H3K27me3 marking has been shown to play a crucial role in the developmental regulation of gene expression in eukaryotes, including plants (Kouzarides, 2007; Zhang et al., 2007a; Schatlowski et al., 2008; Lafos et al., 2011; Zheng and Chen, 2011). H3K27me3 is extensively distributed over the transcribed and promoter regions of genes. It extends over large intergenic regions in animals, whereas the distribution seems to be more gene centered in plants (Turck et al., 2007; Zhang et al., 2007b; Schatlowski et al., 2008).
H3K27me3 deposition is catalyzed by Polycomb-group (PcG) proteins with a SET [for Su(var)3-9, Enhancer-of-zeste, Trithorax] domain that belong to Polycomb Repressive Complex2 (PRC2) (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009; Morey and Helin, 2010; Margueron and Reinberg, 2011). On this line, it has been recently shown that PRC2 activity is necessary for H3K27me3 deposition in plants (Schubert et al., 2006; Bouyer et al., 2011). PcG proteins were first identified in Drosophila melanogaster and are evolutionary conserved among higher eukaryotes, including plants (Ringrose et al., 2004; Hennig and Derkacheva, 2009; Chen et al., 2010; Sawarkar and Paro, 2010; Margueron and Reinberg, 2011; Zheng and Chen, 2011). The PRC2 complex is composed of four core proteins, including ENHANCER OF ZESTE [E(Z); a PcG protein with a SET domain and methyltransferase activity], EXTRA SEX COMBS (ESC; a WD40 domain protein), SUPRESSOR OF ZESTE-12 (a zinc finger protein), and NUCLEOSOME REMODELING FACTOR55. PcG proteins act in establishing and maintaining inactive chromatin states at numerous target genes throughout development (Francis and Kingston, 2001; Schwartz et al., 2006; Schwartz and Pirrotta, 2008; Müller and Verrijzer, 2009; Köhler and Hennig, 2010; Margueron and Reinberg, 2011).
Recruitment of PRC2 at a specific locus likely relies on multiple mechanisms (Margueron and Reinberg, 2011). Genomic domains named Polycomb Repressive Element (PRE) have been described mainly in Drosophila (Ringrose and Paro, 2007; Schuettengruber et al., 2007). PRE domains are characterized by short DNA motifs that include binding sites for regulatory proteins like PLEIOHOMEOTIC (PHO), ZESTE (Z), and GAGA factors (GAFs) (Schuettengruber and Cavalli, 2009). PHO and GAFs would be directly involved in recruiting PRC2. Recently, PRE-like elements have also been identified in mice and humans (Sing et al., 2009; Woo et al., 2010). Nevertheless, our understanding of PRC2 recruitment is still scarce, and no PRE elements have been identified in plants to date (Schatlowski et al., 2008; Morey and Helin, 2010; Sawarkar and Paro, 2010; Smith and Shilatifard, 2010; Enderle et al., 2011; Margueron and Reinberg, 2011).
In Arabidopsis thaliana, the three E(Z) homologs, namely MEDEA (MEA) and the two partially redundant proteins CURLY LEAF (CLF) and SWINGER (SWN), act throughout plant development (Kiyosue et al., 1999; Luo et al., 1999; Pien and Grossniklaus, 2007; Hennig and Derkacheva, 2009; Köhler and Hennig, 2010; Rodrigues et al., 2010; Zheng and Chen, 2011). MEA predominantly represses the MADS box gene PHERES1 and MEA itself in the endosperm (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006). CLF and SWN are required for the repression of floral homeotic genes, such as AGAMOUS (AG) or SHOOTMERISTEMLESS (STM) (Goodrich et al., 1997; Katz et al., 2004; Schubert et al., 2006; Farrona et al., 2008). PRC2 activity also represses the transcription of the master regulators of seed development during vegetative and/or endosperm development (Makarevich et al., 2006; Bouyer et al., 2011). These master regulators include LEAFY COTYLEDON1 (LEC1), a protein homologous to the HAP3 subunits of the CAAT Box binding Factors, and three B3-domain transcription factors, namely, ABA INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 (Braybrook and Harada, 2008; Santos-Mendoza et al., 2008). These AFL (for ABI3, FUS3, and LEC) regulatory proteins control a network of local and redundant gene regulations (Santos Mendoza et al., 2005; To et al., 2006). Among these regulators, LEC2 is specifically expressed in embryos within a short temporal window, from 4 d after pollination (DAP) to 14 DAP, peaking at 10 DAP, and is not expressed in any other tissues (Stone et al., 2001; Kroj et al., 2003). The ectopic expression of LEC2 in transgenic plants triggers the formation of somatic embryos as well as other organ-like structures and confers embryonic characteristics to seedlings (Stone et al., 2001).
Consistent with the involvement of proteins with a SET domain, other reports have confirmed that the expression of LEC2 is controlled, in a developmental manner, by PcG complexes. It has been shown recently that FERTILIZATION INDEPENDENT ENDOSPERM, the Arabidopsis ESC homolog of the PRC2 complex, negatively regulates the expression LEC2 and other AFL genes (Bouyer et al., 2011). PRC1-like ring finger proteins also repressed the expression of the AFL genes (Chen et al., 2010). In addition, PICKLE (PKL), a chromatin remodeler of the CHD3 family, and a closely related homolog (PKR2) are also involved in this regulation (Ogas et al., 1999; Zhang et al., 2008; Aichinger et al., 2009). PKL promotes indirectly H3K27me3 deposition at different loci, including LEC2, in vegetative tissues, through the induction of PcG gene expression (e.g., SWN or EMBRYONIC FLOWER2) (Aichinger et al., 2009, 2011). Other mechanisms involved in the regulation of LEC2 expression have been identified (Zhang and Ogas, 2009). For example, the inhibition of histone deacetylase activity triggers LEC2 expression upon germination (Tanaka et al., 2008). ASIL1 (for Arabidopsis 6b-interacting protein 1-like 1), a plant-specific trihelix factor, represses LEC2 during both germination and early maturation phase (Gao et al., 2009).
Besides the large amount of data accumulated regarding the regulation of LEC2 expression, the DNA cis-elements involved in the specific expression of LEC2 during seed development are unknown. More specifically, the means by which the LEC2 locus is marked by H3K27me3 during vegetative development remain to be determined. In this work, we identified and functionally characterized three cis-regulatory elements involved in the developmental regulation of LEC2 expression. Two of them were found to be essential for the transcriptional activation of LEC2. The first one presents some similarities with CArG regulatory elements recognized by MADS box transcription factors. The second one is similar to a GAFs box and bound by plant BASIC PENTACYSTEINE (BPC) proteins, both in yeast and in vitro. The third element, which we named Repressive LEC2 Element (RLE), is 50 bp long and is necessary for the repression and H3K27 trimethylation of LEC2 in vegetative tissues. Remarkably, the RLE cis-element was also sufficient to regulate the activity of an unrelated promoter (PF3H) and to promote H3K27me3 mark deposition in transgenic plants. Therefore, RLE acts as a functional homolog of the PRE element found in Drosophila. Taken together, the data presented here shed new light on the regulation of LEC2, a master regulator of seed development and bring important new insights into the regulation of H3K27me3 deposition and, thus, PRC2 recruitment in plants.
RESULTS
Identification of the cis-Regulatory Elements That Control the Specific Expression of LEC2
It has been previously shown that the 2.5-kb sequence located upstream of the LEC2 open reading frame recapitulates the embryo-specific pattern of LEC2 expression when fused to the β-glucuronidase (GUS) reporter gene (Stone et al., 2001; Kroj et al., 2003). Here, it was shown that a 500-bp-long fragment, hereafter denoted as PLEC2, was sufficient to faithfully confer a similar expression pattern (Figure 1A). In addition, the expression of the LEC2 cDNA under the control of PLEC2 fully complemented the lec2-4 mutation (see Supplemental Figure 1 online).
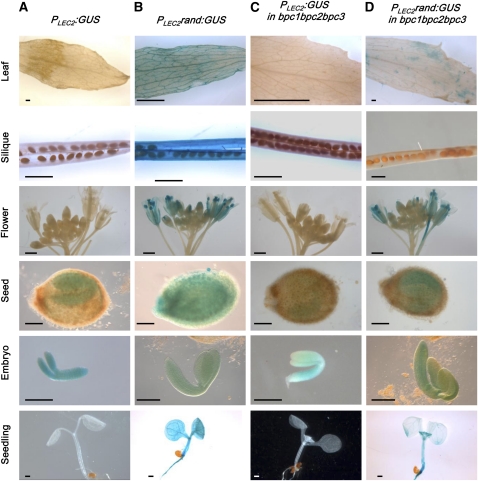
Figure 1.
Expression Patterns Using the Wild Type and Mutated LEC2 Promoter.
Representative GUS patterns are presented for PLEC2:GUS (A) and PLEC2rand:GUS (B) in the wild-type Columbia-0 background and for PLEC2:GUS (C) and PLEC2rand:GUS (D) in the bpc1 bpc2 bpc3 triple mutant background, respectively. Bars = 1 mm in leaf, silique, and flower, and 0.1 mm in seed, embryo, and seedling. Independent transgenic lines (n > 12 for each construct) were assayed for GUS activity in leaves, siliques, flowers, seeds, embryos, and seedlings (10 d after sowing).
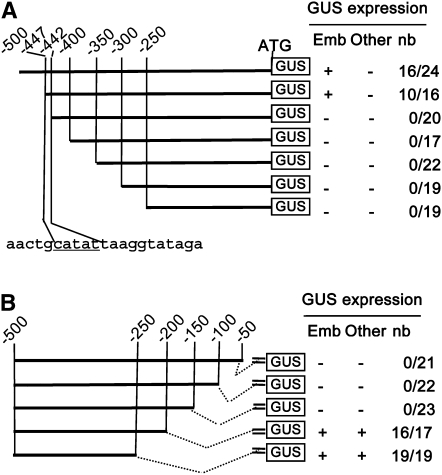
To gain further insight into the cis-regulatory elements necessary for LEC2 expression during embryogenesis, a series of 5′-end deletions was performed on PLEC2, and the resulting DNA fragments were fused to the GUS reporter gene to test their respective transcriptional activity in transgenic plants (Figure 2A). The analysis of GUS activity identified a region between −447 and −400 bp upstream of the first codon that is necessary for the expression of LEC2 in the embryo. Deletions every 5 bp narrowed this region down to a 5-bp motif (CATAT; between −447 and −442 bp). This 5-bp regulatory sequence showed no similarity to any known consensus cis-regulatory element described in public databases.
Figure 2.
Molecular Dissection Analysis of the LEC2 Promoter.
Schematic representation of the deletion series of the LEC2 promoter (PLEC2) fused to the GUS reporter gene. The presence or lack of GUS expression is indicated by (+) or (–), respectively. The ratio (nb) indicates the number of transgenic lines expressing GUS (numerator) and the total number of transgenic lines analyzed (denominator). GUS expression was tested in embryos (emb), valves of siliques, flowers, rosettes, cauline leaves, and stems (other).
(A) The 5′-end deletions. The nucleotide sequence underlined is essential for PLEC2 activity.
(B) The 3′-end deletions were fused to a minimal promoter derived from the 35S of the cauliflower mosaic virus (=).
In parallel, a series of 3′-end deletions of PLEC2 was performed and fused to a minimal 35S promoter carrying a transcription start site derived from the cauliflower mosaic virus that was placed upstream of the GUS reporter gene (Debeaujon et al., 2003). Analysis of the GUS activity in transgenic plants indicated that the 50-bp region upstream of the first codon, which corresponds to the 5′-untranslated region of LEC2, was required for the activity of PLEC2 in this context (Figure 2B). Surprisingly, further deletion of 150 bp resulted in the recovery of GUS expression. In fact, the promoter fragment extending from −500 to −250 bp was sufficient to confer ubiquitous GUS expression throughout plant and seed development. This analysis suggested that the 50-bp domain comprised between −200 and −150 bp contained sufficient information to repress the transcriptional activity of PLEC2 (Figure 2B).
LEC2 Expression Relies on a CT-Rich Element Bound by BPC Proteins
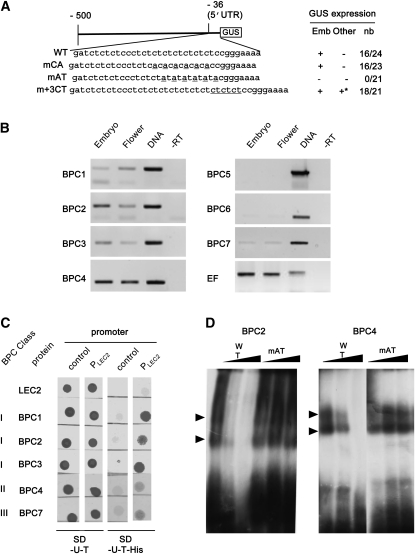
In silico analysis of the 50-bp sequence located upstream of the first codon revealed a GAGA box motif (inverted CT repeats; Figure 3A) previously found in different plant promoters (Meister et al., 2004; Monfared et al., 2011; Wanke et al., 2011). The conversion by site-directed mutagenesis of CT to CA repeats (mCA) did not affect the transcriptional activity of PLEC2 in planta, whereas changing CT to AT repeats (mAT) fully abolished the activity of PLEC2 (Figure 3A). Previous analyses have shown the positive effect of adding CT repeats on promoter activity (Pauli et al., 2004). Here, the addition of three CT repeats to the GAGA box led to the ectopic activation of PLEC2 in pollen grains (see Supplemental Figure 2 online). These data indicated that the GAGA box made of eight C-T (or A) dinucleotide repeats is essential for the correct expression of LEC2.
Figure 3.
The Promoter of LEC2 Contains a CT-Rich Repeat Bound by BPC Proteins.
(A) The wild-type (WT) PLEC2 sequence is shown in the top row (WT). Mutated nucleotides are underlined in the next three rows (mCA, mAT, and m+3CT). GUS activity in pollen is indicated with an asterisk. Presence or lack of GUS expression is indicated by (+) or (–), respectively. The ratio (nb) indicates the number of transgenic lines expressing GUS (numerator) and the total number of transgenic lines analyzed (denominator). GUS expression was tested in embryos (emb), valves of siliques, flowers, rosettes, cauline leaves, and stems (other). UTR, untranslated region.
(B) Analysis of BPC expression in embryos and flowers. Genomic DNA, embryo RNA untreated by the superscript II enzyme (-RT), and EF1αA4 (EF) were used as control.
(C) One-hybrid analysis. The interactions of the different full-length BPC proteins, fused to GAL4, with PLEC2 were tested. The ability to grow in the absence of His indicates an interaction.
(D) EMSA competition assays. BPC2 and BPC4 proteins were tested with labeled wild-type CT repeat probe, and competitions were performed with wild-type or mutated CT repeat (mAT) unlabeled probes. Shifts are indicated with arrowheads.
In plants, GAGA motifs are bound by BPC proteins (Meister et al., 2004; Monfared et al., 2011; Wanke et al., 2011). In Arabidopsis, the BPC family contains seven members grouped into three classes. The accumulation of BPC mRNA in embryos shown in the databases (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) was confirmed by RT-PCR experiments (Figure 3B). All BPC genes but BPC5 (which likely is a pseudogene) (Meister et al., 2004; Monfared et al., 2011) were expressed in 10-d-old embryos.
Then, the ability of BPC proteins to interact with PLEC2 was tested in yeast one-hybrid experiments (Figure 3C). A strong interaction was detected for the three BPCs of class I (BPC1, 2, and 3), whereas a weak interaction was detected for BPC proteins that belong to class II (BPC4) and III (BPC7). To define the ability of BPC to bind directly to the GAGA motif found in PLEC2, in vitro electrophoretic mobility shift assays (EMSAs) were performed with recombinant BPC2 (class I) and BPC4 (class II) proteins. Both proteins BPC2 and BPC4 proteins were able to interact with the poly-CT double-stranded probe (Figure 3D). Competition experiments with wild-type and mutated (mAT) probes demonstrated the specificity of these interactions. Finally, the accumulation of LEC2 mRNA was measured in seeds of available bpc mutants (i.e., bpc1, bpc2, and bpc3 single or the corresponding multiple mutants). No significant differences were observed in comparison to wild-type seed mRNA level (see Supplemental Figure 3 online).
Taken together, these results showed that LEC2 expression relies on a GAGA box and that BPC proteins are probably involved in the transcriptional regulation of LEC2. Further analyses of additional bpc mutants are required to determine which of the BPC proteins are involved. Interestingly, the functional analysis of a modified LEC2 promoter, active in vegetative tissues (see the results presented below), strongly supported the role of the BPC proteins in the control of LEC2 expression.
Characterization of a cis-Regulatory Element (RLE) That Restricts LEC2 Expression to the Embryo
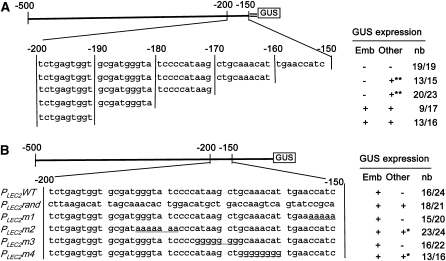
To further characterize the domain located between 200 and 150 bp upstream of the first codon (Figure 2B), a new series of deletions fused to the 35S minimal promoter upstream of the GUS reporter gene was constructed and tested in transgenic plants (Figure 4). This experiment suggested that at least the sequences between −185/−178 and −168/−160bp were necessary to repress PLEC2 transcriptional activity in seed and vegetative tissues, respectively (Figure 4A). To supplement this deletion analysis, mutagenesis experiments were performed using the entire PLEC2 promoter. The 50-bp element associated with the repression activity was replaced by a random 50-bp nucleotide sequence, with no similarity within the Arabidopsis genome, providing the PLEC2rand promoter. In comparison to the embryo-specific GUS staining observed with PLEC2:GUS lines (Figure 1A), the PLEC2rand:GUS lines showed ectopic expression in leaves, stems, flowers, pollen, siliques, and seed coat (Figures 1B and 4B). Similar results were obtained in the Columbia-0 background (see Supplemental Figure 4 online), and these results confirmed the role of the repressive element in the developmental regulation of LEC2 expression. Thus, we named this element RLE. In addition, site-directed mutagenesis was performed to support and refine the structure of the repressive cis-regulatory element. Some of the mutations (m1 and m3) did not modify the pattern of GUS activity, whereas other mutations (m2 and m4) led to the ectopic activity of GUS in pollen (Figure 4B).
Figure 4.
Molecular Dissection Analysis of the RLE Domain.
Presence or absence of GUS activity is indicated by (+) or (–), respectively. The ratio to the right (nb) indicates the number of transgenic lines that display GUS activity (numerator) and the total number of transgenic lines analyzed (denominator). GUS activity was tested in embryos (emb), valves of siliques, flowers, rosettes, cauline leaves, and stems (other). The detection of GUS activity in pollen (+*) or in all organs with the exception of the seed coat and embryo (+**) is indicated.
(A) Analysis using 3′-end deletion constructs fused to the minimal 35S promoter (=) and to the GUS reporter gene.
(B) Analysis using modifications of the RLE domain by replacement with a random 50-bp sequence (PLEC2rand) or by site-directed mutagenesis (PLEC2m1, m2, m3, and m4).
Taken together, these results indicated that the 50-bp element located between −150 and −200 bp upstream of the first codon has a negative effect on the transcriptional activity of PLEC2 in planta. In addition, they suggested that RLE likely contains different functional motifs that are necessary to confer this repressive activity.
Finally, to test if the BPC proteins are involved in the ectopic activity of PLEC2 when RLE is mutated, PLEC2rand:GUS was introduced into the triple bpc1 bpc2 bpc3 mutant by transformation. In this genetic background, GUS activity was completely lost in siliques and seed integument and was strongly downregulated in leaves and cotyledons, in comparison to the wild type (Figure 1C). These results strongly suggested that class I BPCs were involved in the regulation of LEC2 expression.
The RLE Element Identified in PLEC2 Is Necessary for H3K27me3 Deposition
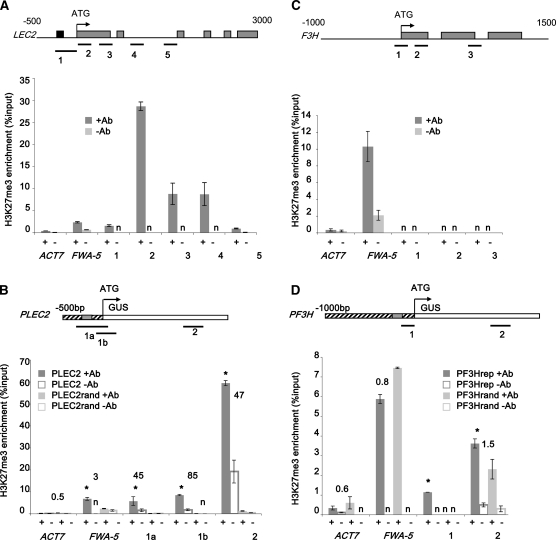
Previous reports have shown that PRC2, which catalyzes H3K27 trimethylation, is involved in the transcriptional repression of LEC2 in seedlings (Makarevich et al., 2006; Chen et al., 2010; Bouyer et al., 2011). To determine if the RLE element identified in PLEC2 was involved in this chromatin-based regulatory process, H3K27me3 enrichment was investigated by chromatin immunoprecipitation (ChIP) and subsequently analyzed by quantitative PCR. The H3K27me3 mark was detected over the LEC2 locus, in the promoter and more strongly right downstream of the first codon. These results are consistent with the data obtained by genome-wide analyses (Zhang et al., 2007b; Roudier et al., 2011).
Then, the level of H3K27me3 at the PLEC2:GUS and PLEC2rand:GUS loci was investigated in 10-d-old seedlings of transgenic plants. The distribution of H3K27me3 at the PLEC2:GUS loci was similar to the pattern found at the endogenous LEC2 locus (Figure 5A). Importantly, the level of H3K27me3 over the PLEC2rand:GUS locus was significantly lower than over PLEC2:GUS locus (at P < 0.002, according to a Student’s unpaired t test). The level was between 45- and 85-fold lower, depending on the probes considered (Figure 5B). The analyses were performed using several (n > 10) independent transgenic lines for each construct. Therefore, the result indicated that the difference between PLEC2rand:GUS and PLEC2:GUS loci was strictly dependent on the presence of RLE cis-element, independent of the transgene insertion sites within the genome. These observations therefore demonstrated that H3K27me3 enrichment over LEC2 depends on the RLE sequence. Taken together, these data and the ectopic GUS activity detected in the PLEC2rand:GUS transgenic plants (Figure 1B) strongly suggest that the RLE cis-element is a major determinant of PRC2-mediated repression of LEC2 outside of embryogenesis (Figure 1B).
Figure 5.
ChIP-PCR Analysis of H3K27me3 Enrichment over the Endogenous and Transgenic LEC2 and F3H Loci.
The H3K27me3 enrichments in 10-d-old seedlings at LEC2 (A), PLEC2:GUS (B), F3H (C), and PF3H:GUS (D) loci are presented. The enrichment is expressed relative to chromatin input (%input). All the experiments were repeated at least twice on a pool of five independent transgenic lines. ACT7 and FWA genes were used as negative and positive controls, respectively. Bars denote se, and n = not detectable. For (A) and (C), the positions of the primers used (thick lines), RLE element (black box), introns (thin line), and exons (dark boxes) are indicated on the top. For (B) and (D), the positions of the primers (thick lines), LEC2 promoter (striped box), RLE element (dark box), and GUS coding sequence (white box) are indicated on the top. The numbers above the histogram bars represent the ratio of the H3K27me3 enrichment when comparing constructs with and without the RLE element (i.e., PLEC2 with PLEC2Rand [B] and PF3Hrle with PF3HRand [D]). The asterisks indicate that the difference (with or without RLE) is significant at P < 0.002 according to Student’s unpaired t test.
Insertion of RLE in pF3H, an Unrelated Promoter, Is Sufficient to Repress Its Transcriptional Activity in Seed and Can Trigger H3K27me3 Deposition
The repressive function and the role of RLE in the initiation of H3K27me3 marking were tested by a gain-of-function experiment with an unrelated promoter. The RLE sequence or a mutated version (random) were introduced at similar position (i.e., 150 bp upstream of the first codon) into the promoter of the FLAVONONE 3-HYDROXYLASE (F3H) gene to generate PF3Hrle:GUS and PF3Hrand:GUS constructs, respectively. The F3H promoter was chosen for two reasons. First the cis-regulating elements that control its activity have been described, and no motifs were located at the insertion site (Hartmann et al., 2005). Furthermore, no H3K27me3 was detectable in ChIP-quantitative PCR analyses (Figure 5C), in agreement with previous analyses (Zhang et al., 2007b). Similar analyses were performed to establish the H3K27me3 landscape at the PF3Hrle:GUS and PF3Hrand:GUS loci in transgenic seedlings. It revealed a weak enrichment of H3K27me3 at PF3Hrle:GUS loci when compared with PF3Hrand:GUS (Figure 5D).
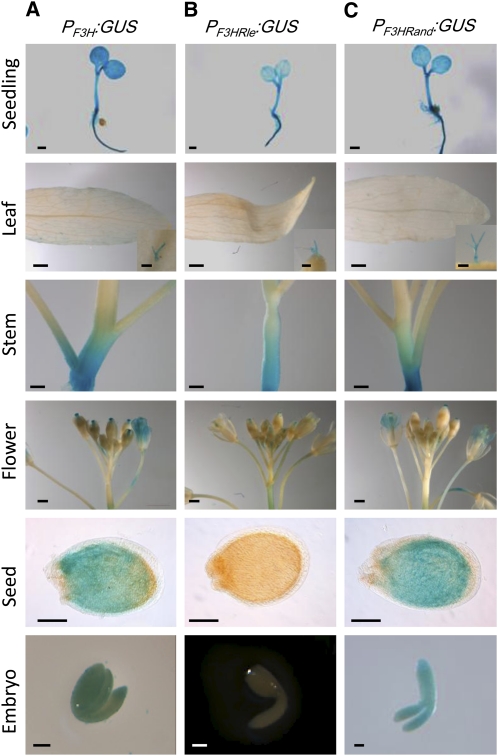
The effect of RLE insertion on PF3H transcriptional activity was investigated. The endogenous F3H promoter was fused to the GUS reporter gene (PF3H:GUS) and introduced into Arabidopsis as a control. GUS activity was analyzed in transgenic plants carrying one of the three different constructs, namely, PF3H:GUS, PF3Hrle:GUS, and PF3Hrand:GUS. For each construct, 24 independent transformants were analyzed. Whereas a strong GUS activity was detected in seeds of 19 transgenic lines carrying PF3H:GUS and 17 PF3Hrand:GUS lines, it was undetectable in seeds of the 14 PF3Hrle:GUS transgenic lines showing GUS activities in other tissues (Figure 6). These results demonstrated that the insertion of RLE is sufficient to repress PF3H transcriptional activity in the seed (embryo and seed coat). On the contrary, similar GUS activity was found in vegetative tissues for the three constructs, suggesting that H3K27me3 marking was not sufficient to fully inhibit gene expression in these tissues. Taken together, these observations demonstrate that the RLE cis-element is sufficient to trigger H3K27me3 deposition and control transcriptional activity in a developmentally regulated manner.
Figure 6.
Insertion of the RLE Element Inhibits PF3H Activity in Seed.
Representative patterns of GUS staining are presented for PF3H:GUS (A), PF3Hrle:GUS (B), and PF3Hrand:GUS (C). Independent transgenic lines (n = 24 for each construct) were analyzed for GUS activity in seedling, leaf, trichome, stem, flower, seed, and embryo. Bars = 1 mm for seedling, leaf, and flower and 0.1 mm for trichome, seed, and embryo.
DISCUSSION
Both a CArG and a CT-Rich Activating cis-Element Are Essential for PLEC2 Activity
The molecular and functional dissection of the LEC2 promoter in planta revealed two essential activating sequences that are necessary for embryo-specific expression (Figure 7). The first cis-regulatory element is made of five nucleotides (5′-CATAT-3′) and located 447 bp upstream of the translational start codon. This very short motif did not present any obvious similarities with known cis-regulatory sequences. Nevertheless, when a wider frame is considered (i.e., GCATATTAAGG), a similarity was found with the consensus CArG motif [i.e., CC(A/T)6GG] (Riechmann and Meyerowitz, 1997). The CArG motifs are known to be recognized by MADS box proteins, a family of transcription factors that control multiple developmental processes, ranging from root to floral or embryo development (Becker and Theissen, 2003). Interestingly, the AGAMOUS-like15 (AGL15) MADS box protein preferentially binds CArG motifs with longer A/T-rich region, such as C(A/T-rich)8G and C(A/T-rich)7GG sequences (Tang and Perry, 2003; Hill et al., 2008; Zheng et al., 2009). This later motif is similar to the CArG sequence found in PLEC2 [i.e., C(A/T)7GG]. In addition, genetic and genome-wide ChIP analyses have demonstrated that LEC2 is a direct target of AGL15 (Zheng et al., 2009). Taken together, these results strongly suggested that the C(A/T-rich)7gg motif found in PLEC2 is one of the AGL15 binding sites involved in the regulation of LEC2 expression. Furthermore, AGL15 can interact with histone deacetylase HDA19 (Hill et al., 2008) and thus could be involved in histone deacetylase recruitment at the LEC2 locus.
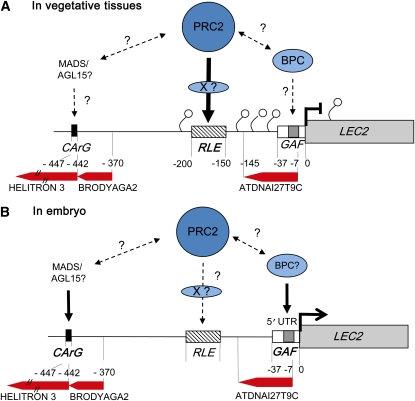
Figure 7.
Schematic Structure and Regulation of the LEC2 Promoter.
Putative model of regulation in vegetative tissues (A) and in the embryo (B). The CArG box, GAF box, and RLE element are represented as well as the transposon marks (MuDR and HEL:Helitron) and putative functional interactions between the proteins. Small circles represent H3K27me3 marks.
The second cis-regulatory element essential for LEC2 expression is a CT-rich domain (containing eight CT repeats) located in the 5′-untranslated region of LEC2 (Figure 3A). This domain displays strong similarities with GAGA elements (inverted CT repeats). Consistent with functional data previously obtained (Meister et al., 2004; Kooiker et al., 2005), the results presented here demonstrated that the CT-rich element of PLEC2 is bound by BPC proteins (preferentially of class I), both in yeast and in vitro. The quantitative differences in DNA binding observed between the BPC in vitro and yeast (Figures 3C and 3D) may be related to the target sequences used [i.e., the entire 500-bp-long promoter for the one-hybrid experiment and a short repetition of three CT motifs, 3 × (CT)8, in vitro] (see Supplemental Table 1 online). It has been shown that, in vitro, BPC1 binding was affected by modification of the stretch of purine residues (i.e., by introducing any pyrimidine residue) (Kooiker et al., 2005). Our results suggested that, in planta, the replacement of cytosine by adenine residues strongly affected PLEC2 activity (Figure 3A). This suggested that the strong base pairing between complementary C-G nucleotides is important for the function of the CT element. Interestingly, the addition of three extra CT repeats was sufficient to trigger ectopic PLEC2:GUS activity in pollen grains (see Supplemental Figure 2 online). This result was consistent with evidence indicating that CT repeats have an enhancer effect on promoter activity (Pauli et al., 2004). The three additional CT repeats inserted in PLEC2 may increase transcriptional activation, up to the threshold of detection of GUS activity, in pollen. A second nonexclusive possibility is that the addition of these repeats increased the affinity for another BPC protein expressed in pollen. This later hypothesis is supported by the fact that the promoter of BPC2, BPC3, BPC4, and BPC7 are all active in pollen (Monfared et al., 2011).
The BPC mRNAs of class I (BPC1, 2, and 3) together with BPC4 (class II) were the most abundant in embryo (Figure 3). Other BPC mRNAs were also detected (with the exception of BPC5) but at very low levels. These expression profiles together with the results of DNA binding assays suggested that BPC factors are likely to regulate LEC2 through binding to the GAGA cis-element found in PLEC2. Consistent with this hypothesis, the ectopic GUS activity driven by PLEC2rand:GUS was fully abolished in siliques and seeds integuments and was strongly affected in leaves and cotyledons of the triple class I mutant (Figure 1C).
Altogether, these results suggested that BPC proteins that belong to class II or III act redundantly with class I BPCs to activate LEC2 expression through its specific GAGA cis-regulatory motif. This hypothesis is in agreement with the high genetic redundancy recently reported for BPCs (Monfared et al., 2011). Moreover, it has been shown that BPCs of group II can form homodimers or heterodimers with group I BPCs (Wanke et al., 2011). Therefore, obtaining new bpc single and multiple mutants of the different classes will be necessary to unravel which of the BPC proteins are involved in the regulation of LEC2 expression.
RLE Is Sufficient to Repress Transcriptional Activity and Is Necessary to Trigger H3K27me3 Deposition
The molecular and functional dissection of PLEC2 presented here has identified a new and negative regulatory cis-element (RLE) involved in restricting the expression of LEC2 to the seed (Figure 7). This 50-bp-long cis-acting sequence located 150 bp upstream of the LEC2 first codon is required for repressing LEC2 expression in leaves, stems, flowers, pollen, seed coat, and siliques (Figures 1 and 2B). Additional experiments (i.e., deletions of 10 bp and site-directed mutagenesis approaches) confirmed these observations and suggested that RLE is made of different functional stretches of nucleotides that are necessary to confer this repressive activity (Figure 4B).
Remarkably, the presence of RLE in PLEC2:GUS is sufficient to trigger H3K27me3 deposition in transgenic seedlings (Figures 5C and 5D). In addition, the insertion of RLE into the F3H promoter (PF3HRLE) was sufficient to inhibit its transcriptional activity in both the seed coat and embryo but not in vegetative tissues after germination (Figure 6B). The detection of GUS activity in vegetative tissues is not totally unexpected. Indeed, it may be related to the environmental regulation of F3H (Pelletier and Shirley, 1996; Blödner et al., 2007; Dubos et al., 2008; Zhang et al., 2011) and to counteracting chromatin-based regulation (Liu et al., 2010; Schmitges et al., 2011). Consistent with this hypothesis, genome-wide analyses showed that in seedlings the F3H endogenous locus is largely enriched in H3K4me3, a mark usually associated with transcriptionally active chromatin (Roudier et al., 2011). In conclusion, the RLE cis-element is necessary to trigger H3K27me3 enrichment and concomitant inhibition of transcriptional activity, although additional factors are likely involved in maintaining or alleviating this repression.
RLE Functions as PREs Identified in Drosophila
It has been shown that PcG complexes that control the H3K27me3 level are involved in the transcriptional repression of LEC2 in seedlings (Makarevich et al., 2006; Chen et al., 2010; Bouyer et al., 2011). Our results demonstrated that the RLE element found in PLEC2 is sufficient to initiate H3K27me3 deposition at the LEC2 locus. In addition, some PREs have been found in the proximal promoter of target genes in Drosophila (Morey and Helin, 2010; Enderle et al., 2011), as RLE in LEC2. Last, it has been demonstrated recently that H3K27me3 deposition is abolished in fie mutant lacking PRC2 activity (Bouyer et al., 2011). These results suggest that the RLE element functions as Drosophila PRE. Nevertheless, the weak effect of RLE on H3K27me3 deposition at pF3H suggests that it may not be sufficient in any genomic contexts.
PREs contain a few short transcription factor binding sites, including a GAGA box (Hagstrom et al., 1997; Horard et al., 2000). Similarly, RLE is associated with a CT-rich element in PLEC2. In addition, some other plant promoters that are regulated by PcG also display CT-rich elements and are regulated by BPC proteins. For instance, SEEDSTICK and INNER NO OUTER (INO) are two targets of BPC1 and BPC2, respectively (Villanueva et al., 1999; Meister et al., 2004; Kooiker et al., 2005). Both loci are associated with strong H3K27me3 enrichment (Zhang et al., 2007b; Roudier et al., 2011). GA repeats can be found in the regulatory sequences of many other PcG targets, such as AG, FLOWERING LOCUS C (FLC), and STM (Schatlowski et al., 2008). In Drosophila, the GAGA factors were shown to be involved in the regulation of gene expression in relation with chromatin packaging (Adkins et al., 2006; Nakayama et al., 2007). Therefore, it is very tempting to speculate that there is an interaction between BPC and PcG silencing in plants. The availability of new and multiple bpc mutants will give the opportunity to further investigate this hypothesis, by looking at the chromatin landscape, at different loci containing both type of elements, such as LEC2, but also INO, STM, FLC, or STM.
No sequences showing similarity with RLE were detected genome-wide. This is not surprising considering that PREs are made of different short transcription factor binding sites. These motifs are usually small, degenerate, and difficult to detect by simple DNA sequence comparison (Ward and Bussemaker, 2008; Badis et al., 2009; Moyroud et al., 2011). Additional functional analyses, similar to those presented in this report, will be necessary to establish a model allowing a more systematic identification of putative RLE elements in plant genomes. Sequence analysis of PLEC2 revealed the presence of three transposable elements (Figure 7). The transcription start site and the GAGA box are included in a class II transposable element sequence of the MuDR superfamily (Lisch, 2002). The CArG motif is located at the beginning of a HELITRON element (Kapitonov and Jurka, 2007; Yang and Bennetzen, 2009), which is itself juxtaposed with another MuDR sequence. Finally, RLE is flanked by the two MuDR sequences. It can be reasonably speculated that insertion of these transposable elements likely shaped the specific expression pattern of LEC2. It has been shown that transposons can be involved in transcriptional regulation of nearby genes, including PRE, although the precise mechanisms remain unknown and seem to be rather gene specific (Valenzuela and Kamakaka, 2006; Weil and Martienssen, 2008; Comet et al., 2011). In addition, selected transposons are specifically targeted by the PCR2 complex in Arabidopsis endosperm (Weinhofer et al., 2010). However, the hypothesis that transposons have a general function in recruiting or regulating PcG activity in plants requires further experimental data.
To conclude, the data presented here demonstrate that the developmental regulation of LEC2 expression results from a combination of activating and repressive mechanisms involving histone H3 methylation. This regulation involves at least three distinct cis-regulatory elements located within the proximal PLEC2 promoter, including repressive RLE element. Remarkably, RLE is sufficient to initiate H3K27me3 deposition and to repress transcription in a developmentally regulated manner. This study provides important new insights into the mechanism by which PRC2 might be recruited at specific target loci and paves the way for a more comprehensive understanding of PcG-mediated gene silencing in plants.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana, accession Wassilewskija, was used for plant transformation. The mutant lines bpc1 (SALK_072966.43.30.x), bpc2 (SALK_090810C), and bpc3 (N89719) were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000) and have all been described by Monfared et al. (2011). Plants were grown, transformed, and selected as previously described (Harscoët et al., 2010).
Constructs and Plant Transformation
All the primers used in this work are described in Supplemental Table 1 online. For 5′-end deletions of the LEC2 promoter (PLEC2), genomic DNA was PCR amplified with the Phusion DNA polymerase (Finnzymes) using primers carrying B1 and B2 GATEWAY recombination sites (Invitrogen). PCR products were cloned by BP recombination (according to the manufacturer’s recommendations) into the entry vector pDONR207 and then LR recombined into the destination vector pBI101R1R2 for plant transformation (Baudry et al., 2004). Modified CT repeat constructs were performed similarly using 3′ primers as follows: AttB2mCA, AttB2mAT, and AttB2m+3CT.
The 3′-end deletion series constructs were PCR amplified using 5′ and 3′ oligonucleotides carrying KpnI and XhoI restriction sites, respectively. The 5′ primer used was P500KpnI, and the 3′ primers were P250XhoI, P200XhoI, P190XhoI, P180XhoI, P170XhoI, P160XhoI, P150XhoI, P100XhoI, and P50XhoI. PCR products were digested with KpnI-XhoI and cloned into KpnI-XhoI–digested pBS35sminGUS (Debeaujon et al., 2003). PLEC2 fragments fused to the minimum 35S promoter and the UidA reporter gene were transferred into pCAMBIA1300 linearized by HindIII-KpnI digestion and ligated with T4 DNA ligase (New England Biolabs).
For the PLEC2rand construct, the fragment P500-200 was amplified with the Phusion DNA polymerase using the 5′ primer P500KpnI and the 3′ primer P200randXhoI. The sequence (5′-CTTAAGACATTAGCAAACACTGGACATGCTGACCAAGTCAGTATCCGCAT-3′) was randomly chosen, and in silico analysis was performed to ensure that no known cis-regulatory elements were found in the sequence. The PCR product was digested with KpnI-XhoI and cloned into KpnI-XhoI–digested pBS35sminGUS (Debeaujon et al., 2003). The minimum 35S promoter was discarded after XhoI-NcoI digestion of the p200rand-pBS35sminGUS and replaced by the PCR amplified fragment between –150 and the translational start site, with P150XhoI and PATGNcoI 5′ and 3′ primers, respectively. The F3H promoter sequence was PCR amplified with Phusion DNA polymerase and primers AttB1PF3H and AttB2PF3H carrying B1 and B2 GATEWAY recombination sites. PF3Hlre and PF3Hrand were amplified with three successive PCRs using AttB1PF3H/PF3Hrev1, AttB1PF3H/PF3Hrev2, and AttB1PF3H/AttB2PF3H and were cloned using the approach described for wild-type PF3H. All these promoter fragments were recombined in the entry vector pDONR207 and then LR recombined into pBI101R1R2 for plant transformation, as described above.
One-Hybrid Assays
Reporter constructs containing the different promoters were made in the pHIS-i plasmid (Clontech), which carries the URA3 selection marker and the minimal HIS3 promoter upstream of the HIS3 reporter gene (Li and Herskowitz, 1993). The 500-bp-long LEC2 promoter (PLEC2) was PCR amplified with the Phusion DNA polymerase with P500KnpI and PATGNcoI 5′ and 3′ primers, respectively, and blunt cloned into pHIS-i digested with SmaI. The control (−500 to −250 fragment of the LEC2 promoter) was cloned using the same method with 5′ primer P500KpnI and the 3′ primer P250LEC2 (see Supplemental Table 1 online). LEC2 and BPCs cDNAs were amplified with the Phusion DNA polymerase using primers carrying B1 and B2 GATEWAY recombination sites. PCR products were BP recombined according to the manufacturer’s (Invitrogen) recommendations into pDONR207 and then LR recombined into pDEST22 vector that allows fusion with the GAL4-activating domain. pHIS-i constructs were stably transformed into the EGY 48 yeast (Saccharomyces cerevisiae) strain and selected on SD-Uracil medium prior to transformation with pDEST22 vectors for protein expression. The interactions were tested on SD-Uracil-Trp-His medium. To increase the stringency of the test, various concentrations of 3-aminotriazol were used, from 5 to 50 mM.
EMSA Experiments
The cDNAs obtained for one-hybrid experiments were introduced into pDEST15 (Invitrogen), which produces an N-terminal glutathione S-transferase tag fusion. BPCs proteins were produced in Escherichia coli and purified with the Glutathione Sepharose 4B system (Amersham Biosciences) according to the manufacturer’s instructions. One hundred nanograms of both forward and reverse primers (EMSACT/mAT forw and rev) were boiled for 5 min and annealed at room temperature. Double-stranded probes were 32P labeled after Klenow treatment (Invitrogen). Binding conditions have been previously described (Kooiker et al., 2005).
Histochemical GUS Assays
Arabidopsis tissues were immersed in GUS staining solution (100 mM sodium phosphate buffer, pH 7.2, 10 mM EDTA, 0.1% Triton, and 1 mM X-GlcA) and then placed under vacuum for 1 h. After incubation at 37°C overnight, the staining solution was removed and samples were cleared by sequential changes of 75 and 95% (v/v) ethanol.
ChIP Experiments
The experiments were performed as previously described (Gendrel et al., 2002). Briefly, all the experiments have been duplicated and performed using pools of five independent transformants. Relative enrichment was measured by comparison between input and ChIP value. ACT7 was used as a negative control (H3K27me3 modification is barely detectable over the ACT7 locus), and FWA was used as a positive control for H3K27me3 enrichment (Roudier et al., 2011). Primers used in this analysis are described in Supplemental Table 1 online.
Analysis of Gene Expression
RNA extractions were performed using the total mammalian RNA kit (Sigma-Aldrich) according to the manufacturer’s instructions. cDNAs were synthetized using the SuperScriptII first-strand synthesis system for RT-PCR (Invitrogen) and dT oligonucleotides. The PCR experiments were conducted as previously described (Baud et al., 2005), with specific primers described in Supplemental Table 1 online. The data presented are the results of three biological and three technical repeats.
Accession Numbers
Sequence and mutant data from this article can be found in the GenBank/EMBL data libraries or the Arabidopsis Genome Initiative database under the following accession numbers: bpc1 (SALK_072966.43.30), bpc2 (SALK_090810C), bpc3 (N89719), BPC1 (At2g01930), BPC2 (At1g14685), BPC3 (At1g68120), BPC4 (At2g21240), BPC5 (At4g38910), BPC6 (At5g42520), BPC7 (At2g35550), LEC2 (At1g28300), ACT7 (At5g09810), FWA-5 (At4g25530), and EF1α4 (AT5G60390).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of the lec2-4 Mutation by PLEC2:LEC2.
Supplemental Figure 2. Activation of PLEC2 in Pollen by Adding 3CT Repeats in the GAF Box of PLEC2.
Supplemental Figure 3. LEC2 Expression Is Not Modified in bpc Mutants.
Supplemental Figure 4. Expression Patterns of PLEC2:GUS and PLEC2rand:GUS in the Col-0 Ecotype Background.
Supplemental Table 1. List of Primers Used in This Study.
Acknowledgments
Part of this work was supported by grants from the Agence Nationale de la Recherche to L.L., namely, TF-code (BLAN-07-185566) and CERES (BLAN-10-123801). We thank Charles S. Gasser for providing bpc mutant seeds. Our laboratory belongs to the Laboratoire d’Excellence Saclay Plant Sciences (https://www6.inra.fr/saclay-plant-sciences).
AUTHOR CONTRIBUTIONS
N.B., B.D., and C.D. performed the experiments. All the authors designed the research, analyzed data, and wrote the article.
References
- Adkins N.L., Hagerman T.A., Georgel P. (2006). GAGA protein: A multi-faceted transcription factor. Biochem. Cell Biol. 84: 559–567 [DOI] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B., Di Mambro R., Sabatini S., Köhler C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B., Farrona S., Reyes J.C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., et al. (2009). Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C., Gagliardini V., Page D.R., Grossniklaus U. (2006). Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Wuillème S., Lemoine R., Kronenberger J., Caboche M., Lepiniec L., Rochat C. (2005). The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Beisel C., Paro R. (2011). Silencing chromatin: Comparing modes and mechanisms. Nat. Rev. Genet. 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Blödner C., Goebel C., Feussner I., Gatz C., Polle A. (2007). Warm and cold parental reproductive environments affect seed properties, fitness, and cold responsiveness in Arabidopsis thaliana progenies. Plant Cell Environ. 30: 165–175 [DOI] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook S.A., Harada J.J. (2008). LECs go crazy in embryo development. Trends Plant Sci. 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Chen D., Molitor A., Liu C., Shen W.H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20: 1332–1344 [DOI] [PubMed] [Google Scholar]
- Comet I., Schuettengruber B., Sexton T., Cavalli G. (2011). A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl. Acad. Sci. USA 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Enderle D., Beisel C., Stadler M.B., Gerstung M., Athri P., Paro R. (2011). Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 21: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Coupland G., Turck F. (2008). The impact of chromatin regulation on the floral transition. Semin. Cell Dev. Biol. 19: 560–573 [DOI] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E. (2001). Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2: 409–421 [DOI] [PubMed] [Google Scholar]
- Gao M.J., Lydiate D.J., Li X., Lui H., Gjetvaj B., Hegedus D.D., Rozwadowski K. (2009). Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21: 54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Hagstrom K., Muller M., Schedl P. (1997). A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146: 1365–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harscoët E., Dubreucq B., Palauqui J.C., Lepiniec L. (2010). NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS ONE 5: e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Horard B., Tatout C., Poux S., Pirrotta V. (2000). Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20: 3187–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P.E., Katz A., Oliva M., Ohad N., Berger F. (2006). Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2007). Helitrons on a roll: Eukaryotic rolling-circle transposons. Trends Genet. 23: 521–529 [DOI] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L. (2010). Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 20: 541–547 [DOI] [PubMed] [Google Scholar]
- Kooiker M., Airoldi C.A., Losa A., Manzotti P.S., Finzi L., Kater M.M., Colombo L. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kroj T., Savino G., Valon C., Giraudat J., Parcy F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M., Rossi V. (2011). Epigenetic control of gene regulation in plants. Biochim. Biophys. Acta 1809: 369–378 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Smith E., Shilatifard A. (2010). The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.J., Herskowitz I. (1993). Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262: 1870–1874 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2008). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. (2002). Mutator transposons. Trends Plant Sci. 7: 498–504 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., Grossniklaus U., Köhler C. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister R.J., Williams L.A., Monfared M.M., Gallagher T.L., Kraft E.A., Nelson C.G., Gasser C.S. (2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37: 426–438 [DOI] [PubMed] [Google Scholar]
- Monfared M.M., Simon M.K., Meister R.J., Roig-Villanova I., Kooiker M., Colombo L., Fletcher J.C., Gasser C.S. (2011). Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant J. 66: 1020–1031 [DOI] [PubMed] [Google Scholar]
- Morey L., Helin K. (2010). Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35: 323–332 [DOI] [PubMed] [Google Scholar]
- Moyroud E., Minguet E.G., Ott F., Yant L., Pose D., Monniaux M., Blanchet S., Bastien O., Thevenon E., Weigel D., Schmid M., Parcy F. (2011). Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23: 1293–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Verrijzer P. (2009). Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Nishioka K., Dong Y.X., Shimojima T., Hirose S. (2007). Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 21: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli S., Rothnie H.M., Chen G., He X., Hohn T. (2004). The cauliflower mosaic virus 35S promoter extends into the transcribed region. J. Virol. 78: 12120–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M.K., Shirley B.W. (1996). Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol. 111: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J., Wagner D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Grossniklaus U. (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta 1769: 375–382 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378: 1079–1101 [PubMed] [Google Scholar]
- Ringrose L., Ehret H., Paro R. (2004). Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16: 641–653 [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R. (2007). Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232 [DOI] [PubMed] [Google Scholar]
- Rodrigues J.C., Luo M., Berger F., Koltunow A.M. (2010). Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex. Plant Reprod. 23: 123–133 [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., Teixeira F.K., Colot V. (2009). Chromatin indexing in Arabidopsis: An epigenomic tale of tails and more. Trends Genet. 25: 511–517 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M., Dubreucq B., Baud S., Parcy F., Caboche M., Lepiniec L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M., Dubreucq B., Miquel M., Caboche M., Lepiniec L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Sawarkar R., Paro R. (2010). Interpretation of developmental signaling at chromatin: The Polycomb perspective. Dev. Cell 19: 651–661 [DOI] [PubMed] [Google Scholar]
- Schatlowski N., Creasey K., Goodrich J., Schubert D. (2008). Keeping plants in shape: polycomb-group genes and histone methylation. Semin. Cell Dev. Biol. 19: 547–553 [DOI] [PubMed] [Google Scholar]
- Schmitges F.W., et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Scholl R.L., May S.T., Ware D.H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz Y.B., Kahn T.G., Nix D.A., Li X.Y., Bourgon R., Biggin M., Pirrotta V. (2006). Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. (2008). Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. (2009). Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Sing A., Pannell D., Karaiskakis A., Sturgeon K., Djabali M., Ellis J., Lipshitz H.D., Cordes S.P. (2009). A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897 [DOI] [PubMed] [Google Scholar]
- Smith E., Shilatifard A. (2010). The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Perry S.E. (2003). Binding site selection for the plant MADS domain protein AGL15: An in vitro and in vivo study. J. Biol. Chem. 278: 28154–28159 [DOI] [PubMed] [Google Scholar]
- To A., Valon C., Savino G., Guilleminot J., Devic M., Giraudat J., Parcy F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R.T. (2006). Chromatin insulators. Annu. Rev. Genet. 40: 107–138 [DOI] [PubMed] [Google Scholar]
- Villanueva J.M., Broadhvest J., Hauser B.A., Meister R.J., Schneitz K., Gasser C.S. (1999). INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke D., Hohenstatt M.L., Dynowski M., Bloss U., Hecker A., Elgass K., Hummel S., Hahn A., Caesar K., Schleifenbaum F., Harter K., Berendzen K.W. (2011). Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS ONE 6: e16070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L.D., Bussemaker H.J. (2008). Predicting functional transcription factor binding through alignment-free and affinity-based analysis of orthologous promoter sequences. Bioinformatics 24: i165–i171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C., Martienssen R. (2008). Epigenetic interactions between transposons and genes: Lessons from plants. Curr. Opin. Genet. Dev. 18: 188–192 [DOI] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. (2010). A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Bennetzen J.L. (2009). Structure-based discovery and description of plant and animal Helitrons. Proc. Natl. Acad. Sci. USA 106: 12832–12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ogas J. (2009). An epigenetic perspective on developmental regulation of seed genes. Mol. Plant 2: 610–627 [DOI] [PubMed] [Google Scholar]
- Zhang H., Rider S.D., Jr, Henderson J.T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H.J., Romero-Severson J., Muir W.M., Ogas J. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283: 22637–22648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Sridhar V.V., Zhu J., Kapoor A., Zhu J.K. (2007a). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2: e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007b). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zheng S., Liu Z., Wang L., Bi Y. (2011). Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 168: 367–374 [DOI] [PubMed] [Google Scholar]
- Zheng B., Chen X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]