Abstract
Though membrane-associated proteins are ubiquitous within all living organisms and represent the majority of drug targets, a general method for direct, label-free measurement of ligand binding to native membranes has not been reported. Here we show backscattering interferometry (BSI) to be a viable technique for quantifying ligand-receptor binding affinities in a variety of membrane environments. By detecting minute changes in the refractive index of a solution, BSI allows binding interactions of proteins with their ligands to be measured at picomolar concentrations. Equilibrium binding constants in the micromolar to picomolar range were obtained for small- and large-molecule interactions in both synthetic- and cell-derived membranes without the use of labels or supporting substrates. The simple and low-cost hardware, high sensitivity, and label-free nature of BSI should make it readily applicable to the study of many membrane-associated proteins of biochemical and pharmacological interest.
Cellular membranes are complex two-dimensional fluid structures comprised of stringently regulated combinations of phospholipids and proteins. Many critical cellular processes are triggered through the transduction of binding-dependent molecular signals across the membrane.1 Membrane-associated proteins and their interactions are therefore of paramount interest in the design of clinical therapies, accounting for almost 70% of existing drug candidate targets.2 Though many assays exist to examine this class of molecular interactions, targets of interest are typically removed from the native membrane environment or undergo substantial modification prior to observation. Often the membrane protein is truncated or functionalized via genetic modification in an attempt to engineer experimental compatibility with investigative techniques that rely primarily on either covalent labeling or surface coupling strategies. Though necessary for quantitative analysis, such modifications can affect a target’s function in unpredictable ways, pose experimental hazards, and are not uniformly applicable in all systems.3
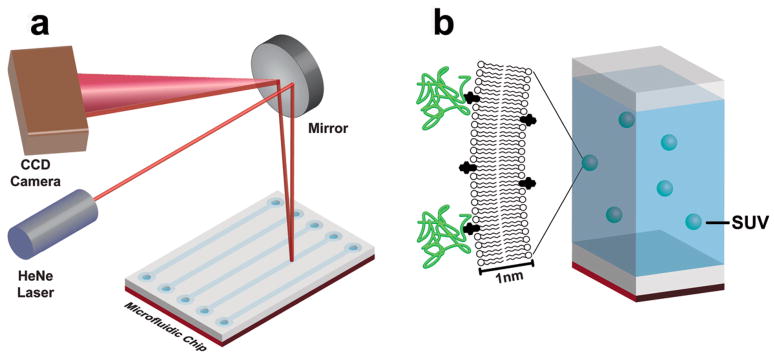
We have recently found backscattering interferometry (BSI) to be remarkably sensitive to the minute changes in refractive index of a solution or surface that occur when dissolved or adsorbed molecular agents bind analytes.4–7 By introducing a sample into a microfluidic device, properly configured to create a resonance cavity and a long effective path length, incident coherent light is converted into an interferometric fringe pattern that can be captured on a standard CCD camera (Figure 1a). Fourier analysis of this fringe pattern reports on very small changes in refractive index that can be correlated in real time with receptor-ligand interactions occurring in solution or with species tethered to the microfluidic channel surface without the use of contrast-enhancing label molecules of any kind. We describe here the extension of this methodology to several, increasingly heterogeneous, membrane-supported species, in an attempt to mirror more closely the relevant functionality of biological molecules in their native environment.
Figure 1.
(a) Major components of backscattering interferometry instrumentation. (b) Schematic representation of the small unilamellar vesicles (SUVs) on which BSI measurements were made in this work.
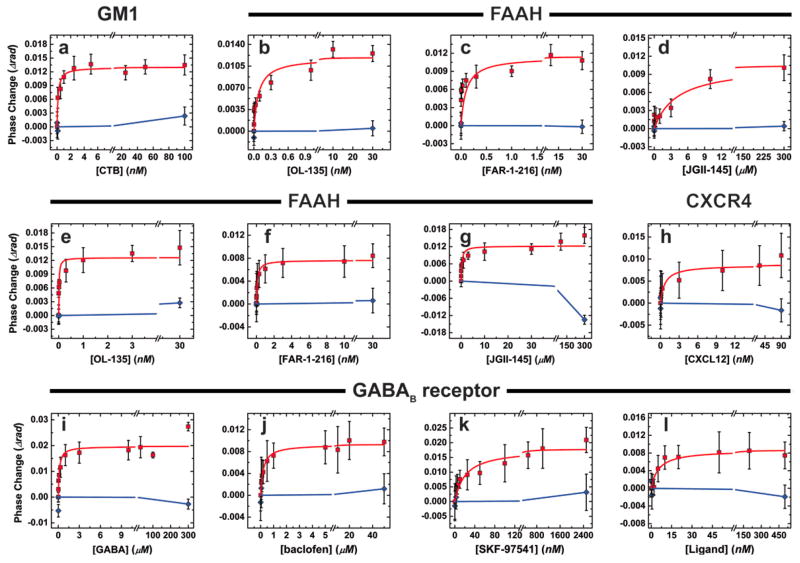
In the simplest case, binding interactions of integral membrane components were examined using fully synthetic membrane components, allowing for well-defined display of the desired ligands within the context of a fluid lipid bilayer. For this example the monosialoganglioside GM1 was combined with phosphatidylcholine and phosphatidylserine to form small, unilamellar vesicles (SUV’s, Figure 1B) averaging 40 ± 3 nm in diameter, as determined by dynamic light scattering (DLS). GM1 is implicated in a variety of neurological developmental processes and diseases,8 and is the primary lipid component to which cholera- and related toxins bind in order to gain entry into the cell.9 Separate samples of the GM1-containing vesicles were incubated with varying concentrations of the cholera toxin B (CTB) subunit to reach equilibrium. These solutions were analyzed by BSI in comparison to mixtures lacking the GM1 ligand, but of otherwise identical composition to each test sample. The observed change in refractive index, manifested as a shift in phase of the interference fringe pattern, was plotted against the concentration of added analyte, giving a sigmoidal curve that fit well to a simple single-site binding model (Figure 2a), yielding an equilibrium binding constant of 129 ± 27 pM. This value is between the published determinations of 4.6 pM (SPR measurements on immobilized GM110) and 20 nM (fluorescence microscopy on supported lipid bilayers11), and represents conditions that are significantly closer to the natural environment of this interaction than either of the previously described experiments. As a negative control, the same GM1-containing vesicles were treated with full-length tetanus toxin, which binds other membrane receptors than GM1. Little or no signal was observed in these experiments, showing that the BSI signal reflected the specific GM1-CTB interaction rather than a nonspecific association with the membrane, and that sub-nM binding affinity can be determined in free solution with this technique.
Figure 2.
Representative plots of BSI signal vs. ligand concentration for the determination of binding constants for the following pairs of molecules (membrane-bound species + ligand). The “control” ligand in each case was tested at the highest concentration in order to verify its nonbinding nature (plotted in blue). Unless otherwise indicated, the membrane-bound partner was displayed in SUVs as described in the text. (a) GM1 + cholera toxin B subunit, control = full-length tetanus toxin; (b–f) FAAH + small-molecule inhibitors atpH 7.4, control = cholesterol; b = OL-135, c = FAR-1-216, d = JGII-145; (e–g) as in b–d, at pH 9.0; (h) vesicles derived from the membrane of SUP-T1 cells, natively expressing CXCR4. Ligand: CXCL12. Control: fully denatured CXCL12. (i–l) vesicles derived from the membrane of CHO cells overexpressing the GABAB receptor + small-molecule substrates, agonists, or antagonists. Ligands: i = GABA, j = baclofen, k = SKF-97541, l = CGP-54626. Controls: (i–k) L-alanine, (l) cholesterol. Each data point represents the average of at least four independent measurements; error bars are plus and minus the full value of standard error in each direction. Repeat determinations of the binding curves gave very similar KD values.
In the above study, a uniformly distributed small molecule ligand (GM1) was incorporated into the membrane, which was then addressed by a soluble protein (CTB). More generally useful, however, is quantification of the binding of soluble ligands with membrane-bound proteins. Using detergent solubilization and dialysis, it is relatively straightforward to incorporate labile, functional membrane proteins, from both natural and recombinant sources, into synthetic membranes to form proteoliposomes. Binding measurements are most commonly made on proteoliposomes in free solution with fluorescence spectroscopy or by radioligand displacement. Techniques such as total internal reflection fluorescence microscopy and atomic force microscopy, though highly sensitive, require a supported lipid bilayer to allow compatibility with the optical geometries involved.12 In such cases, transmembrane proteins usually suffer from restricted movement, presumably due to interactions with the underlying surface.13,14 It is common to truncate such proteins, attaching a membrane-compatible insertion tag such as a glycosylphosphatidylinositol anchor to allow free lateral diffusion within the membrane leaflet. As BSI is capable of detecting binding events on free vesicles in solution, we postulated that such modifications would be unnecessary.
We examined three integral membrane protein systems to test this hypothesis, fatty acid amide hydrolase (FAAH), the CXCR4 receptor, and the GABAB receptor. FAAH is a membrane protein important in neurological function and a drug target for pain management and other indications.15,16 Purified samples were incorporated into synthetic SUV’s in the manner described above for GM1. Upon treatment of these vesicles with several small-molecule inhibitors, dose-dependent signals were observed by BSI (Figure 2b–d, Table 1). The compounds OL-135 and FAR-I-216 gave apparent Kd values of 0.26 ± 0.04 nM and 0.13 ± 0.02 nM, significantly different from the Ki values reported for these reversible covalent inhibitors17 (4.7 nM and 20 nM, respectively18,19). The closely related analogue JG-II-145, a less potent inhibitor of FAAH by more than three orders of magnitude,20 gave a correspondingly weakened Kd of 4.1 ± 1.7 μM (Table 1), illustrating the dynamic range of the BSI assay to be at least several logs. When performed at the same pH used for the reported enzyme inhibition assay (pH 9.0), BSI measurements showed somewhat tighter binding of the inhibitors, and again mirrored the relative differences between observed Ki values (Figure 2, Table 1). No BSI signals were obtained under either set of conditions when the same FAAH-containing vesicles were treated with high concentrations of cholesterol, which is likely to be incorporated into the lipid bilayer when introduced to the solution. Taken together, the data indicate that a specific binding event is detected by BSI rather than a change induced by the nonspecific interaction of hydrophobic molecules with lipid bilayers.
Table 1.
Binding constants determined by BSI from the plots shown in Figure 2
| Membrane-bound binding partner | Solution-phase ligand | Kd (BSI)a | Literature valueb |
|---|---|---|---|
| GM1 | Cholera toxin B subunit | 0.13 ± 0.03 nM | Kd = 4 pMc Kd=20 nMd |
| FAAH | OL-135 | 0.26 ± 0.04 nM (pH 7.4) 13±7.6 pM (pH 9.0) |
Ki = 4.7 nM |
| FAAH | FAR-I-216 | 0.13 ± 0.02 nM (pH 7.4) 0.10 ± 0.03 nM (pH 9.0) |
Ki=20 nM |
| FAAH | JG-II-145 | 4.1 ± 1.7 μM (pH 7.4) 310±170 nM (pH 9.0) |
Ki=10 μM |
| CXCR4 | CXCL12 | 0.69 ± 0.33 nM | IC50 = 1.8 nMe |
| GABAB receptor | GABA | 140 ± 65 nM | IC50=140 nMf |
| GABAB receptor | Baclofen | 210±34 nM | IC50 = 210, 250 nMf |
| GABAB receptor | SKF-97541 | 20±7.1 nM | IC50 = 66 nMf |
| GABAB receptor | CGP-54626 | 5.4±1.7 nM | IC50 = 2.2 nMf |
Error limits are derived from the statistical error of curve fitting the curves shown in Figure 2.
References given in Supplementary Information.
Determined by SPR.
Determined by fluorescence microscopy.
Determined by radioligand displacement on human T-lymphoblasts.
Determined by radioligand displacement on rat cerebral cortex extracts.
The mismatch in absolute values between the Kd values observed here for the FAAH inhibitors and Ki values reported in the literature reflects the fundamentally different nature of these parameters, especially for this type of system in which inhibition is the result of both a binding step and a covalent capture step in the enzyme active site.21 While direct comparisons are rare, other examples of differences in binding constant and inhibitory activity of similar magnitudes have been described (Supplementary Information). That there exist no values of binding affinities in the literature for antagonists of FAAH and many other important enzymes shows how much more difficult it is to make such measurements than it is to measure the effects of inhibitors on enzyme function.
It is especially troublesome to perform direct, label-free measurements of binding to proteins in native membranes, although such data would be the most relevant to in vivo considerations. Ligand binding events have therefore typically been inferred from cellular responses in whole cells or with the use of radioactive assays on cell lysates. Because the refractive index changes detected by BSI on the above membrane-associated samples should not be affected by the presence of non-interacting species in the mixture, we tested the ability of BSI to detect binding events in native cell extracts. To provide the required type of isotropic matrix that scatters little light yet is still suitable for BSI analysis, we separated the outer membranes of cultured cells from their intracellular components and converted the membranes into vesicle-like suspensions of uniform size.
We chose two representative targets for this study. The CXCR4-CXCL12 system represents a receptor-chemokine pair of great fundamental and applied interest in cell biology and medicine.22 Using a variation of existing methods for the preparation of red blood cell “ghosts”, human SUP-T1 lymphoma T cells expressing the CXCR4 receptor were subjected to hypotonic lysis, sonication and purification to produce outer membrane-derived vesicles (Supplementary Figure 2) with an approximate diameter of 180 ± 20 nm, measured by dynamic light scattering. It is assumed that these vesicles display the cell’s associated membrane proteins in a manner closely analogous to their native environment. The vesicle preparations were interrogated with increasing concentrations of the soluble chemokine CXCL12 to derive the binding curve shown in Figure 2H, and a Kd value (0.7 nM) very similar to the reported value for 50% saturation of the receptor (1.8 nM, Table 1). SUP-T1 cells with the CXCR4 receptor knocked out showed no interaction with added SDF1 (data not shown), illustrating the specificity of the BSI detection of the binding event amidst a variety of other membrane-bound components.
The heterodimeric23 γ-aminobutyric acid (GABA) receptor represents an even more complex target.24 This G-protein coupled receptor is implicated in many neurological disease phenotypes, and is characteristically difficult to isolate and purify. Outer membrane derived vesicles from Chinese hamster ovary (CHO) cells genetically modified to overexpress the B(1b) and B2 components of the GABAB receptor were prepared as above. The presence of the GABAB receptor in these lysis-derived vesicles was verified by Western blotting (Supplementary Figure 3). As shown in Figure 2I–L, the receptor ligand (GABA), two agonists (baclofen25 and SKF-9754126,27) and one antagonist (CGP-5462628) all gave BSI binding curves, in contrast to the negative control compounds (L-alanine and cholesterol). Again, the Kd values obtained in these experiments agreed remarkably well with previously reported values, which were derived from radioligand displacement and competitive binding assays using membrane fractions from similar cells (Table 1). While it did not compromise the overall accuracy of the measured values, the error bars for BSI measurements on the native membrane derived samples were larger than for vesicles incorporating purified proteins, presumably caused by sample heterogeneity and nonspecific adsorption to the channel walls.
Backscattering interferometry is essentially a detector of differences in molecular structures, with binding measurements relying on the comparison of signals between free and bound states. Modeling of the BSI phenomenon, to be described elsewhere, indicates that much of the signal derives from changes in conformation and solvation that occur when a molecule such as a protein interacts with a ligand. It may be difficult to detect the replacement of one ligand by another using BSI if the bound structures have very similar levels of hydration and/or radii of gyration. Therefore, direct measurements of competitive binding are potentially more challenging with BSI than are individual measurements of binding affinities for ligands being compared.
In any event, BSI is shown here for the first time to provide direct and quantitative insight into binding events involving integral membrane proteins and membrane-bound species: no other label-free solution-phase technique is as generally and straightforwardly applicable with similar sensitivity. We used a convenient “end-point” mode in these studies, wherein liposome samples and ligands are simply mixed at the desired concentrations and measurements are made after equilibrium is reached, but real-time measurements should be feasible, as have been performed previously on aqueous-soluble systems. The need to isolate the membrane-bound protein of interest often requires assays to be done in the presence of significant quantities of organic solvents, in detergent micelles, or in other non-native environments. The BSI-based methodology provides data that are more relevant to the true binding abilities of the membrane-bound species of interest, without the need for protein alteration or, in some cases, isolation, and will therefore be of significant value in drug discovery, biochemistry, and related fields.
Methods
Materials
Lipids were obtained from Avanti Polar Lipids. 1,2-Dimyristoleoyl-sn-glycero-3-phosphocholine(DMOPC) and 1,2-dimyristoyl-sn-glycero-3-[phospho-L-serine] (sodium salt) (DMPS), were received in chloroform and stored at −20°C for up to two weeks. Monosialoganglioside GM1, bovine-ammonium salt (GM1) was received from Matreya Inc. in powder form and dissolved in 2:1 chloroform/methanol to 1 mg/mL for storage at −20°C. Cholera toxin subunit B (CTB) was purchased from Sigma, dissolved in 0.5×PBS to a concentration of 0.5 mg/mL and stored at 4°C. Full-length tetanus toxin (TT), was purchased from Sigma, reconstituted in sterile H2O to a concentration of 0.1 mg/mL TT and 10 mM sodium phosphate buffer, and stored at 4°C. Full-length, purified, lyophilized, fatty acid amide hydrolase was provided by the laboratory of Prof. R.C. Stevens (The Scripps Research Institute) and was reconstituted in 1% w/v n-octyl-beta-D-glucopyranoside (n-OG) in 1X PBS. Compounds FAR-1-216, OL-135, and JG-II-145 were provided by Prof. D. Boger and Ms. J. Garfunkel (The Scripps Research Institute) as lyophilized solids and dissolved in pure dimethylformamine (DMF) immediately before use. SKF 97541, CGP 54626, and R-baclofen were purchased from Tocris Biosceince and reconstituted in either 1×PBS or DMF immediately before use. Anti-GABAB polyclonal IgG and corresponding secondary (horseradish peroxidase conjugated) antibodies were purchased from Santa Cruz Biotechnology Inc. Stromal derived cell factor 1 alpha (SDF-1α) was provided by Prof. Tracy M. Handel and Dr. Melinda S. Hanes (University of California, San Diego) and kept frozen until use.
Synthetic Membranes
Small unilamellar vesicles (SUV) were formed using standard techniques. A lipid solution in chloroform was evaporated in small round-bottom flasks and hydrated for an hour at 4°C in Milli-Q deionized (18.2 MW-cm) water, 0.5×PBS or 1×PBS at ~3.3 mg/mL. The lipids were probe-sonicated to clarity in an ice-water bath and transferred to a 100 nm Millipore Ultrafree-MC centrifuge tube filter. Samples were centrifuged for 2 h at 16,000 g and 4°C. All solution that passed through the centrifuge tube filter was collected and stored at 4°C for up to one week. Full-length FAAH was incorporated into synthetic lipid vesicles by mixing FAAH and SUVs to a final concentration of 100 μg of protein per mL of centrifuged SUV solution. The resulting mixture was then dialyzed extensively against either 1×PBS, pH 7.4 or 100mM Tris pH 9.0 to facilitate complete removal of detergent. The resulting proteoliposomes were approximately 150 nm in diameter, as measured by DLS, with an estimated lipid:protein ratio of approximately 3300:1. Proteoliposomes were stored at 4°C for up to one week.
Mammalian Cell Cultures
Two different lines of adherent Chinese hamster ovary (CHO-K1) cells were used; one wild-type, and one engineered to express the full length transmembrane B-forms of both the rat and human gamma-aminobutyric acid receptor (GABAB). Wild-type cells (ATCC cell line CCL-61) were kindly provided by Prof. Kevin Morris (The Scripps Research Institute) and the engineered cells were kindly provided by Dr. Klemens Kaupmann of Novartis, Inc. (internal designation b12.224). Two different lines of suspension human T-lymphocytes (SUP-T1) cells were also used; one wild-type (CXCR4-positive) and one engineered to express a zinc finger nuclease (CXCR4-negative). Both wild-type and engineered cells were kindly provided by Prof. Bruce Torbett (The Scripps Resarch Institute). Adherent cells were grown at 37°C and 5% ambient CO2 to near 100% confluence over three days from initial addition to 175 cm2-area flasks. Adherent cells were harvested by removing all growth medium from the flask and incubating with 4 mL of Detachin solution for 5 min at 37°C. Incubation buffer (48 mL) was then added to the flask and the contents removed and transferred to two 50 mL centrifuge tubes. Suspension cells were grown to an approximate concentration of 300,000 cells/mL. In both cases, the cells and media were centrifuged for 5 min at 300 g to pellet the cells. Following centrifugation, the media was removed from the centrifuge tubes, the cells were re-suspended in 1×PBS, and the cell/PBS suspension re-centrifuged. Cell pellets were rinsed three times in 1×PBS and used immediately. The expression of the GABAB receptor in membranes derived from the b12.2 cells was verified by Western immunoblotting (Supplementary Figure 3).
Native Membrane Vesicles
A cell pellet containing approximately 106 cells of either type was re-suspended in 20 mL of ice-cold lysis buffer (2.5 mM NaCl, 1 mM Tris, 1x EDTA-free, broad-spectrum protease inhibitors, pH 8.0) and placed on a rotator for 45 minutes at 4°C. The resulting solution was then centrifuged at 40,000 g for 60 min at 4°C. The supernatant was removed and re-suspended in 4 mL of ice-cold 1×PBS and transferred to a 5 mL glass dram vial. The pellet and buffer were then probe-sonicated to clarity in an ice bath and transferred to a 220 nm Millipore Ultrafree-MC centrifuge tube filter. The resulting solutions were centrifuged for 1 h at 16,000 g and 4°C. All solution that passed through the centrifuge tube filter was collected and stored at 4°C for up to two days. Sizes of both synthetic and native membrane vesicles was determined using a Wyatt Technologies DynaPro dynamic light scattering apparatus.
BSI instrumentation.6,29
BSI utilizes a red helium-neon (HeNe) laser (λ= 632.8 nm) to illuminate the microfluidic channel in a simple optical train. The laser is coupled to a collimating lens through a single-mode fiber, producing a 100 μm diameter beam and yielding probe volumes in the 300 picoliter range. When the laser beam intersects the fluid contained in the channel, a set of high contrast interference fringes is produced, the spatial position of which depends on the refractive index (RI) of the fluid within the channel. The fringes were monitored in the direct backscatter region at relatively shallow angles (typically less than 7 degrees) using a Garry 3000 linear array CCD camera (Ames Photonics). The camera was positioned so that the centroid of the interference pattern was located just above the lens on the end of the single-mode optical fiber to ensure that the alignment was along a central plane. Under these conditions the fringe pattern contains a dominant Fourier frequency; the phase of this dominant frequency is the BSI fringe shift signal, in radians.16
BSI chips were isotropically etched in borosilicate glass to give a cross section described by two quarter-circles of 40 μm radius connected by a 10 μm flat region, manufactured by Micronit, Inc. For BSI measurements, the chip was maintained in the instrument at 25°C using a feedback-controlled peltier system.
Very little sample is required: 1 μL of solution (approximately 5 μM in protein) is introduced into the microfluidic channel for each measurement, of which 290 picoliters are interrogated by the laser. Five 1-μL aliquots containing different amounts of ligand were used to generate a complete binding curve, requiring a total of approximately 20 pmoles of protein.
Detection of Ligand Binding
Ligand binding to the SUVs was accomplished by incubating a fixed amount of SUV suspension with varying concentrations of ligands (bacterial toxins, FAAH ligands, GABAB modulators) for 8 hours in the dark at 4°C. Such a long incubation period is not necessary, and was done here only for experimental convenience and to ensure that equilibrium had been reached at the cold temperature which slows membrane fluidity significantly. Channels used with synthetic membrane systems were pre-treated by incubating with a solution of non ligand-containing SUVs followed by extensive rinsing with Milli-Q deionized (18.2 MW-cm) water. Channels used with cell-derived SUVs received no pre-treament; the signal-to-noise ratio for all experiments showed no change for either pre-treatment condition. The chip surfaces were regenerated between uses by brief (5 min) incubation with sulfuric acid followed by extensive rinsing with Milli-Q deionized (18.2 MW-cm) water.
For each sample, a solution of SUVs and the “control” ligand was introduced into the channel and the BSI signal was measured for 15 seconds. The channel was rinsed and the mixture of SUV and ligand of interest was introduced into the channel and the signal measured, and this procedure was repeated iteratively for increasing ligand concentrations. The binding signal was calculated as the difference in phase between the control SUV–ligand solution and the analyte SUV–ligand complex. The background signal due to the presence of the SUVs was subtracted from all measurements. This corrected binding signal was then plotted versus concentration to form a saturation binding curve and fitted to a square hyperbolic function to calculate the affinity.
While we did not explore the detection limits for membrane-bound protein receptors, we note that the signal intensities observed here are very similar to those observed for soluble proteins in buffer. We therefore suspect that binding events to membrane-bound molecules will be detectable by BSI to very low concentrations, since binding to solution-phase proteins has been characterized down to picomolar concentrations and proteins bound in a monolayer to the microfluidic channel are similarly detectable.4–7,30
Data Analysis
Spatial changes in interference fringes are measured in near real-time using high-resolution interference fringes and a fast Fourier transform FFT. In the general case, a Fourier transform is defined as
| (1) |
Where A(x) is the complex Fourier transform of the function a(x), F[x] denotes the Fourier transform operation, y is a spatial variable, and f is a spatial frequency in the Fourier domain. For a particular set of characteristic frequencies, it is possible to calculate the observed phase change by evaluating the real and imaginary parts of the Fourier transform at a given frequency. As a result, differences in phase changes can be calculated for different ligand-receptor binding pairs. It was observed that only cognate ligand-binding interactions created substantial differences in phase when compared to binding-nonbinding pairs. For example, the maximal binding phase change for a particular sample is typically approximately 0.01, while non-binding phase changes are about 10 times lower.
By plotting phase changes as a function of varying ligand concentration and fitting the resulting points to a simple single site binding model given by
| (2) |
where y is the observed, calculated phase change, ymax is the observed, maximal, calculated phase change, x is the ligand concentration and Kd is the equilibrium dissociation constant.
Supplementary Material
Acknowledgments
This work was supported by the NIH (RO1 EB003537-01A2; U01 MH069062; and the Joint Center for Innovative Membrane Protein Technologies, Roadmap Grant GM073197), and The Skaggs Institute for Chemical Biology. We are grateful to Ms. Joie Garfunkel and Prof. Dale Boger for samples of the FAAH inhibitors, Prof. Raymond Stevens for samples of the FAAH protein, Dr. Melinda Hanes and Prof. Tracy Handel for the samples of the SDF-1α chemokine, and to Dr. Klemens Kaupmann of Novartis for the GABAB-transfected CHO cell line.
Footnotes
Author Contributions
M.M.B. developed methods for sample preparation, prepared samples for analysis, and processed the raw BSI data; A.K.K. performed BSI measurements and processed the raw data; M.M. prepared the FAAH protein; M.M.B., A.K.K., M.F., and D.J.B. designed the project and wrote the manuscript.
References
- 1.Krummel MF, Davis MM. Dynamics of the immunological synapse: finding, establishing and solidifying a connection. Curr Opin Immunol. 2002;14:66–74. doi: 10.1016/s0952-7915(01)00299-0. [DOI] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Disc. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Wise A, Gearing K, Rees S. Target Validation of G-Protein Coupled Receptors. Drug Disc Today. 2002;7:235–246. doi: 10.1016/s1359-6446(01)02131-6. [DOI] [PubMed] [Google Scholar]
- 4.Markov DA, Swinney K, Bornhop DJ. Label-free molecular interaction determinations with nanoscale interferometry. J Am Chem Soc. 2004;126:16659–16664. doi: 10.1021/ja047820m. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen HS, Larsen NB, Latham JC, Bornhop DJ, Andersen PE. Highly sensitive biosensing based on interference from light scattering in capillary tubes. Appl Phys Lett. 2006;89:151108. [Google Scholar]
- 6.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sorensen HS. Free-Solution, Label-Free Molecular Interactions Studied by Back-Scattering Interferometry. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 7.Kussrow A, Kaltgrad E, Wolfenden ML, Cloninger MJ, Finn MG, Bornhop DJ. Measurement of Monovalent and Polyvalent Carbohydrate-Lectin Binding by Back-Scattering Interferometry. Anal Chem. 2009;81:4889–4897. doi: 10.1021/ac900569c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnino S, Mauri L, Chigorno V, Prinetti A. Gangliosides as components of lipid membrane domains. Glycobiology. 2007;17:1R–13R. doi: 10.1093/glycob/cwl052. [DOI] [PubMed] [Google Scholar]
- 9.Fishman PH, Pacuszka T, Orlandi PA. Gangliosides as Receptors for Bacterial Enterotoxins. Adv Lipid Res. 1993;25:165–187. [PubMed] [Google Scholar]
- 10.Kuziemko GM, Stroh M, Stevens RC. Cholera Toxin Binding Affinity and Specificity for Gangliosides Determined by Surface Plasmon Resonance. Biochemistry. 1996;35:6375–6384. doi: 10.1021/bi952314i. [DOI] [PubMed] [Google Scholar]
- 11.Fang Y, Frutos A, Lahiri J. Ganglioside Microarrays for Toxin Detection. Langmuir. 2003;19:1500. [Google Scholar]
- 12.Cannon B, Weaver N, Pu Q, Thiagarajan V, Liu S, Huang J, Vaughn MW, Cheng KH. Cholesterol Modulated Antibody Binding in Supported Lipid Membranes as Determined by Total Internal Reflectance Microscopy on a Microfabricated High-throughput Glass Chip. Langmuir. 2005;21:9666–9674. doi: 10.1021/la0502645. [DOI] [PubMed] [Google Scholar]
- 13.Brian AA, McConnell HM. Allogenic Stimulation of Cyto-Toxic T-Cell by Supported Planar Membranes. Proc Natl Acad Sci USA. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 15.Cravatt BF, Lichtman AH. Fatty acid amide hydrolase: an emerging therapeutic target in the endocannabinoid system. Curr Opin Chem Biol. 2003;7:469–475. doi: 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]
- 16.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and Structure of a Brain Constituent that Binds to the Cannabinoid Receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 17.Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Binding and inactivation mechanism of a humanized fatty acid amide hydrolase by α-ketoheterocycle inhibitors revealed from co-crystal structures. J Am Chem Soc. 2009;131:10497–10506. doi: 10.1021/ja902694n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boger DL, Miyauchi H, Du W, Hardouin C, Fecik RA, Cheng H, Hwang I, Hedrick MP, Leung D, Acevedo O, Guimaraes CRW, Jorgensen WL, Cravatt BF. Discovery of a potent, selective, and efficacious class of reversible alpha-ketoheterocycle inhibitors of fatty acid amide hydrolase effective as analgesics. J Med Chem. 2005;48:1849–1856. doi: 10.1021/jm049614v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero FA, Du W, Hwang I, Rayl TJ, Kimball FS, Leung D, Hoover HS, Apodaca RL, Breitenbucher JG, Cravatt BF, Boger DL. Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. J Med Chem. 2007;50:1058–1068. doi: 10.1021/jm0611509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garfunkle J, Ezzili C, Rayl TJ, Hochstatter DG, Hwang I, Boger DL. Optimization of the Central Heterocycle of alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase. J Med Chem. 2008;51:4392–4403. doi: 10.1021/jm800136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Prusoff WH. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor which Causes 50 Per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 22.Hesselgesser J, Liang M, Hoxie J, Greenberg MM, Brass LF, Orsini MJ, Taub D, Horuk R. Identification and Characterization of the CXCR4 Chemokine Receptor in Human T Cell Lines: Ligand Binding, Biological Activity, and HIV-1 Infectivity. J Immunol. 1998:877–883. [PubMed] [Google Scholar]
- 23.Pin J-P, Kniazeff J, Binet V, Liu J, Maurel D, Galvez T, Duthey B, Havlickova M, Blahos J, Prézeau L, Rondard P. Activation mechanism of the heterodimeric GABAB receptor. Biochem Pharmacol. 2004;68:1565–1572. doi: 10.1016/j.bcp.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Urwyler S, Mosbacher J, Lingenhoehl K, Heid J, Hofstetter K, Froestl W, Bettler B, Kaupmann K. Positive Allosteric Modulation of Native and Recombinant gamma-Aminobutyric AcidB Receptors by 2,6-Di-tert-butyl-4-(3-hydroxy-2,2-dimethyl-propyl)-phenol (CGP7930) and its Aldehyde Analog CGP13501. Mol Pharm. 2001;60:963–971. [PubMed] [Google Scholar]
- 25.Bowery NG, Hill DR, Hudson AL. [3Hl(−)Baclofen: an improved ligand for GABAB sites. Neuropharmacology. 1986;24:207–210. doi: 10.1016/0028-3908(85)90075-9. [DOI] [PubMed] [Google Scholar]
- 26.Howson W, Mistry J, Broekman M, Hills JM. Biological Activity of 3-Aminopropyl (Methyl) Phosphinic Acid, a Potent and Selective GABAB Agonist with CNS Activity. Bioorg Med Chem Lett. 1993;3:515–518. [Google Scholar]
- 27.Froestl W, Mickel SJ, Hall RG, Von Sprecher G, Strub D, Baumann PA, Brugger F, Gentsch G, Jaekel J, Olpe H-R, Rihs G, Vassouta A, Waldmeier PC, Bittiger H. Phosphinic Acid Analogs of GABA. 1. New Potent and Selective GABAB Agonists. J Med Chem. 1995;38:3297–3312. doi: 10.1021/jm00017a015. [DOI] [PubMed] [Google Scholar]
- 28.Brugger F, Wicki U, Olpe HR, Froestl W, Mickel S. The action of new potent GABA-B receptor antagonists in the hemisected spinal cord preparation of the rat. Eur J Pharmacol. 1993;235:153–155. doi: 10.1016/0014-2999(93)90836-7. [DOI] [PubMed] [Google Scholar]
- 29.Kussrow AK, Enders CS, Castro AR, Cox DL, Ballard RC, Bornhop DJ. The potential of backscattering interferometry as an in vitro clinical diagnostic tool for the serological diagnosis of infectious disease. Analyst. 2010;135:1535–1537. doi: 10.1039/c0an00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kussrow A, Baksh MM, Bornhop DJ, Finn MG. Universal Sensing by Transduction of Antibody Binding using Backscattering Interferometry. ChemBioChem. 2010 doi: 10.1002/cbic.201000671. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.