Abstract
Myelodysplastic syndromes (MDS) are hematopoietic stem cell disorders that often progress to chemotherapy-resistant secondary acute myeloid leukemia (sAML). We used whole genome sequencing to perform an unbiased comprehensive screen to discover all the somatic mutations in a sAML sample and genotyped these loci in the matched MDS sample. Here we show that a missense mutation affecting the serine at codon 34 (S34) in U2AF1 was recurrently mutated in 13/150 (8.7%) de novo MDS patients, with suggestive evidence of an associated increased risk of progression to sAML. U2AF1 is a U2 auxiliary factor protein that recognizes the AG splice acceptor dinucleotide at the 3′ end of introns and mutations are located in highly conserved zinc fingers in U2AF11,2. Mutant U2AF1 promotes enhanced splicing and exon skipping in reporter assays in vitro. This novel, recurrent mutation in U2AF1 implicates altered pre-mRNA splicing as a potential mechanism for MDS pathogenesis.
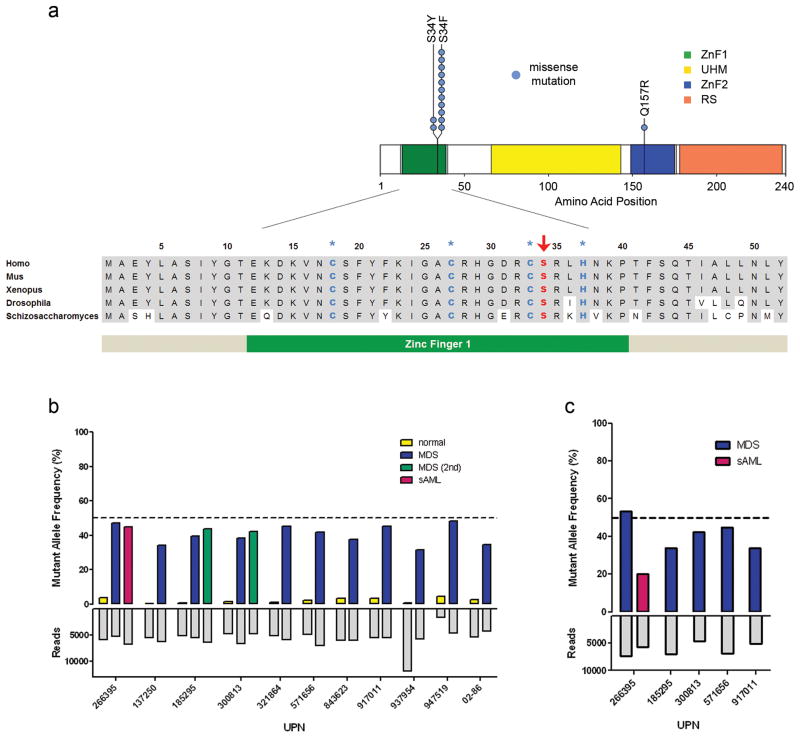
Myelodysplastic syndromes (MDS) are a heterogenous group of hematopoietic stem cell disorders characterized by dysplastic blood cell formation and peripheral blood cytopenias. Up to 30% of patients with MDS will progress to highly chemotherapy-resistant secondary acute myeloid leukemia (sAML). Whole genome sequencing (WGS) offers an unbiased approach to discover all the genetic mutations present in cancer genomes and has been used to identify novel mutations in de novo and therapy-related AML genomes3–7. Here we report the results of WGS of an MDS-derived sAML sample and the matched normal (skin) sample. We performed WGS using 100 base pair paired-end reads and obtained 39.1x and 38.2x haploid and 99.3% and 98.9% diploid coverage of the sAML and normal samples, respectively (see Supplementary Note, Supplementary Table 1). We divided the genome into non-overlapping tiers, as previously described4, and validated putative mutations using deep sequencing of captured DNA isolated from the sAML, normal, and MDS samples. We validated 507 somatic single nucleotide variants (SNVs) in the sAML sample, including 30 SNVs in protein coding regions (tier 1 mutations). 505/507 SNVs preexisted in the MDS sample, including 30 tier 1 mutations (Supplementary Fig. 2, Supplementary Tables 2, 3). The same codon in U2AF1 (U2AF35) was mutated in 2 additional MDS-derived sAML cases analyzed by whole genome sequencing (data not shown). This was the sole recurrent mutation in these cases. To determine the frequency of this mutation in MDS, we sequenced the entire coding region of U2AF1, including 9 exons, in diagnostic bone marrow and paired normal (skin) samples from 150 consecutively accrued de novo MDS patients (including the index case) and identified 13 patients (8.7%) with missense mutations affecting the highly conserved serine at amino acid position 34 (S34) in U2AF1 (Fig. 1a). The same nucleotide was mutated in all samples, resulting in either a S34F (n=11) or S34Y (n=2) substitution (Supplementary Table 4). One patient with an S34F mutation (UPN 947519) also had a U2AF1 Q157R mutation located in the second zinc finger (Fig. 1a). Sequencing of the U2AF1 cDNA from this patient revealed that both mutations occur on the same allele. No other somatic SNVs affecting U2AF1 were detected in these samples. Subsequent analysis focused on the highly recurrent S34 mutations.
Figure 1. U2AF1 mutations found in patients with myelodysplastic syndromes (MDS).
(a) Missense mutations were detected in codons 34 and 157 of U2AF1. The ZnF1 (zinc finger 1), UHM (U2AF homology motif), ZnF2 (zinc finger 2), and RS (arginine-serine rich) domains are shown. The amino acid sequence of the ZnF1 domain is highly conserved (shaded). The zinc coordinating and mutated residue are shown in blue (asterisks) and red (arrow), respectively. (b) Deep sequencing of U2AF1 using DNA collected from paired normal, MDS, or secondary AML (sAML) samples. Mutant allele frequencies represent the proportion of sequencing reads supporting the mutant allele reads/total reads. Total read counts are shown below (mean 5,651 reads/sample). The mutation is present in the majority of cells (mutant allele frequency 31.4–48.2%) in all cases. (c) Deep sequencing of cDNA from MDS or sAML samples. The mutant allele is expressed in all cases tested. UPN, unique patient number.
U2AF1 is the small (35 kDa) subunit of U2 snRNP auxiliary factor (U2AF) that is involved in pre-mRNA processing (splicing), and it forms a heterodimer with the larger subunit U2AF2 (U2AF65)2. U2AF1 binds the 3′ AG splice acceptor dinucleotide of the pre-mRNA target intron2 and U2AF2 binds the adjacent polypyrimidine tract. PCR amplicons spanning the S34 codon were generated using genomic DNA and cDNA templates from unpurified MDS bone marrow cells from 11 patients with confirmed U2AF1 mutations, and subjected to deep sequencing to obtain mutant allele frequencies. Importantly, there was no deletion or uniparental isodisomy (UPD) that spanned the U2AF1 locus (chromosome 21q22.3) based on SNP arrays and whole genome sequencing data for the index case. Read counts from the genomic DNA samples (including the sAML sample from the index case and serial MDS samples from two other patients) showed that the S34 mutant allele frequencies were ~40–50%, indicating that the majority of cells in the samples contained a heterozygous mutation, even though the myeloblast counts ranged from 0–21% in the MDS samples (Fig. 1b). Similar results were obtained from cDNA deep sequencing (~30–50% mutant allele frequency), indicating that both the S34 mutant and wild-type alleles were expressed in all samples tested, regardless of the myeloblast count (Fig. 1c). In addition, there was no difference in the total levels of U2AF1 mRNA or the dominant U2AF1 isoform that was expressed in unfractionated MDS bone marrow samples from patients with and without U2AF1 mutations (Supplementary Fig. 4a). Collectively, these results suggest that U2AF1 mutations are an early, initiating genetic event in MDS pathogenesis.
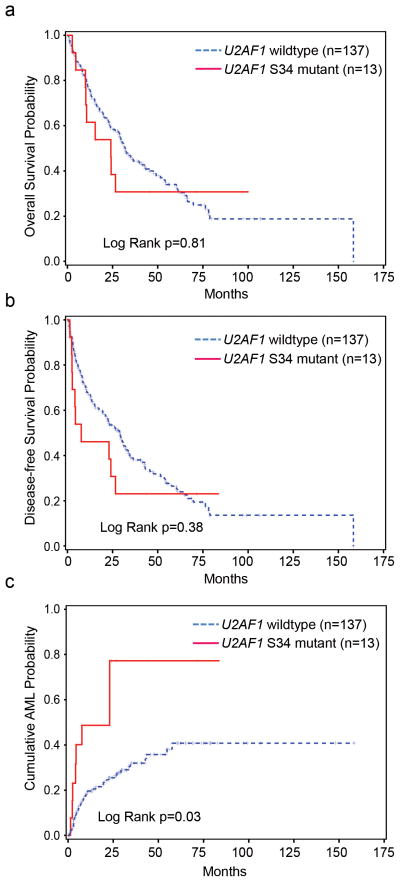
Although eight of the mutant samples had myeloblast counts > 5%, patients with U2AF1 mutations were not restricted to a particular International Prognostic Scoring System (IPSS) category and had a median IPSS score of 1 (range 0–3) (Supplementary Table 5). Patients with a del(20q) or −20 karyotype were more likely to harbor a U2AF1 mutation (P=0.03), although the number of patients with mutations and del(20q) or −20 is small (n=4) (Table 1). No difference in event-free or overall survival was observed in patients with or without U2AF1 mutations (Fig. 2a–b). However, the 2 mutant patients with the longest overall survival had received hematopoietic stem cell transplants (Supplementary Fig. 1). Patients with U2AF1 mutations had an increased probability of progression to sAML (P=0.03) (Fig. 2c), an observation that will require confirmation in a larger cohort. This corresponds to a U2AF1 mutation frequency of 15.2% (7/46 patients) in the subset of MDS patients that progress to sAML vs. 5.8% (6/104 patients) in the subset that did not. Since there was no statistical difference in the myeloblast count or IPSS distribution of patients with or without U2AF1 mutations (Table 1), the mutant genotype does not appear to be a surrogate for these well-established predictors of sAML risk.
Table 1.
Patient Characteristics
| U2AF1 wild-type | U2AF1 S34F/Y mutant | p-valuea | |
|---|---|---|---|
| n (%) | 137 (91.3) | 13 (8.7) | |
|
| |||
| Age at diagnosis – yr ±SD | 61 ±14 | 59 ±10 | 0.37b |
| range (median) | 20–87 (62) | 42–79 (59) | |
|
| |||
| Median survival | 975 days | 729 days | 0.81c |
|
| |||
| Gender | |||
| Male – no.(%) | 82 (59.9) | 10 (76.9) | 0.37 |
|
| |||
| Blood counts | |||
| WBC (K/mcL) | 6 ±11 | 6 ±11 | 0.22b |
| ANC (K/mcL) | 3 ±7 | 3 ±6 | 0.43b |
| Hb (g/dL) | 10 ±2 | 10 ±2 | 0.94b |
| PLT (K/mcL) | 91 ±88 | 78±66 | 0.94b |
|
| |||
| Bone marrow blasts (%) | 6 ±6 | 8 ±7 | 0.41 b |
|
| |||
| FAB subtype – no.(%) | 0.44 | ||
| RA | 63 (46.0) | 4 (30.8) | |
| RARS | 5 (3.7) | 0 (0) | |
| RAEB | 64 (46.7) | 8 (61.5) | |
| RAEB-T | 5 (3.7) | 1 (7.7) | |
|
| |||
| Cytogenetics – no.(%) | |||
| normal | 65 (47.4) | 3 (23.1) | 0.14 |
| -Y only | 1 (0.7) | 0 (0) | 1.00 |
| −5, del(5q) | 31 (22.6) | 1 (7.7) | 0.30 |
| −7, del(7q) | 17 (12.4) | 1 (7.7) | 1.00 |
| −17, del(17q) | 5 (3.7) | 0 (0) | 1.00 |
| −20, del(20q) | 11 (8.0) | 4 (30.8) | 0.03 |
| +8 | 18 (13.1) | 3 (23.1) | 0.39 |
| complex (≥ 3) | 35 (25.6) | 2 (15.4) | 0.52 |
| other | 15 (11.0) | 2 (15.4) | 0.64 |
| not available | 3 (2.2) | 0 (0) | 1.00 |
|
| |||
| IPSS – no.(%) | 0.56 | ||
| low | 22 (16.1) | 1 (7.7) | |
| INT−1 | 54 (39.4) | 6 (46.2) | |
| INT−2 | 36 (26.3) | 2 (15.4) | |
| high | 20 (14.6) | 4 (30.8) | |
| not available | 5 (3.7) | 0 (0) | |
WBC, white cell blood count; ANC, absolute neutrophil count; Hb, hemoglobin; PLT, platelet count; FAB, French American British; RA, refractory anemia; RARS, refractory anemia with ringed sideroblasts; RAEB, refractory anemia with excess blasts; RAEB-T, refractory anemia with excess blasts, in transformation; IPSS, International Prognostic Scoring System.
Fisher’s exact, except where indicated;
Wilcoxon test;
Log-rank
Figure 2. Impact of U2AF1 mutations on clinical outcome.
(a) Overall and (b) Event-free survival are not impacted by U2AF1 genotype. (c) The probability of secondary AML (sAML) progression is increased in patients with U2AF1 mutations (P=0.03).
Splicing involves cleavage of intronic sequences from pre-mRNA, followed by ligation of the remaining exons together to produce a mature mRNA product8. Inclusion and exclusion of different exons or utilization of alternative 3′ splice sites during pre-mRNA processing produces multiple protein isoforms that can have different functions within a cell, and alternative splicing can be affected by the levels of U2AF1 in a cell9–12. It is unknown which domain of U2AF1 binds the pre-mRNA. Interestingly, the S34 and Q157 residues are located within zinc finger domains (Fig. 1a) that may be important for RNA binding activity. Indeed, the U2AF1 zinc fingers are structurally similar to the murine and human ZFP36 family zinc fingers (both CX8CX5CX3H zinc fingers)13–15 that bind RNA and the noncanonical RNA recognition motif (RRM; also known as U2AF homology motif, UHM) in U2AF1 only weakly binds RNA16.
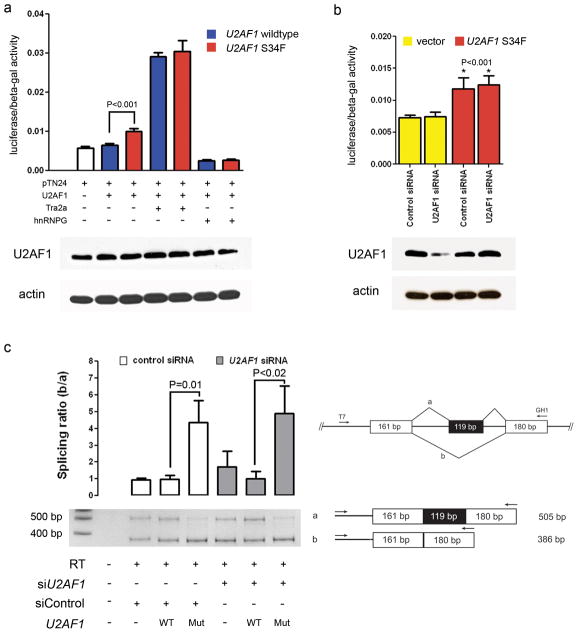
To examine the effects of the S34F mutation on U2AF1 splicing activity, we utilized previously described and validated in vitro double-reporter splicing and minigene reporter assays9,17. The double-reporter plasmid constitutively expresses β-galactosidase, while luciferase is expressed only if appropriate splicing removes an upstream intron that contains translational stop codons. Transient co-expression of the double-reporter plasmid pTN24 and the S34F mutant U2AF1 cDNA in 293T cells resulted in a significant increase in splicing (as detected by an increase in the luciferase/β-galactosidase ratio), compared to co-expression of wild-type U2AF1, despite similar total U2AF1 protein levels in all samples (Fig. 3a, P<0.001). The level of splicing was similar in cells depleted of endogenous U2AF1 compared to control cells (Fig. 3b, left column), and the increase in splicing observed with the S34F mutant U2AF1 is independent of endogenous U2AF1 levels (Fig. 3b, right column, P<0.001 when compared to vector alone). This suggests that splicing activity in this assay is insensitive to U2AF1 levels, and increased splicing mediated by the mutant protein is attributable to a novel gain-of-function activity.
Figure 3. U2AF1 S34F mutation induces splicing alterations.
(a) Transient coexpression of the pTN24 double-reporter splicing construct with or without a splicing enhancer (Tra2α), splicing inhibitor (hnRNPG), wild-type U2AF1, or mutant (S34F) U2AF1 cDNA in 293T cells. The double-reporter construct constitutively expresses β-galactosidase, while luciferase is expressed only if an upstream intron which contains multiple stop codons is spliced out. In the absence of the Tra2α splicing enhancer or hnRNPG splicing inhibitor, expression of the mutant U2AF1 increases splicing of the pTN24 construct resulting in an increase in the luciferase/β-galactosidase expression compared to expression of wild-type U2AF1 (P<0.001). Tra2α (positive control) and hnRNPG (negative control) cause increased or decreased splice efficiency, respectively. A representative Western blot of U2AF1 levels in the same cells used for luciferase assays is shown below each combination of plasmids. (b) Transient coexpression of the pTN24 double-reporter splicing construct with a control plasmid (vector) or mutant (S34F) U2AF1 cDNA in the presence of a control siRNA or siRNA targeting the endogenous U2AF1 in 293T cells. Expression of the S34F mutant U2AF1 results in an increase in splicing of the pTN24 construct with an increase in the luciferase/β-galactosidase expression compared to a control plasmid (P<0.001). The result is independent of endogenous U2AF1 expression levels. A representative Western blot of U2AF1 is shown below each condition. (c) The GH1 minigene was transiently transfected into 293T cells with a control plasmid (pcDNA3.1-YFP), wild-type U2AF1 cDNA, or mutant (S34F) U2AF1 cDNA in the presence of a control siRNA or siRNA targeting the endogenous U2AF1. RNA was harvested 48 hours later and a reverse transcriptase (RT) reaction was performed to create cDNA. PCR using the indicated primers resulted in a fully spliced 505 base pair amplicon or a 386 base pair amplicon that skips the middle exon shaded in black (exon skipping). A representative PCR gel image is shown and the ratio of the lower band (exon skipping = amplicon b) relative to the fully spliced upper band (amplicon a) is shown above each condition. Expression of the S34F mutant U2AF1 results in an increase in exon skipping compared to control or wild-type U2AF1 (P<0.02). bp, base pair; WT, wild-type; Mut, mutant; T7, T7 primer. Error bars, s.d.
Next, we examined exon skipping using a minigene reporter plasmid (a human gene fragment containing an upstream and downstream exon surrounding an intron-flanked exon). Appropriate splicing produces an mRNA with all 3 exons, while exon skipping fuses the upstream and downstream exons only. We measured the levels of exon skipping using a GH1 minigene reporter plasmid in cells transiently co-transfected with either wild-type or S34F mutant U2AF1 cDNA. The proportion of transcripts with a skipped exon (lower PCR band) relative to the appropriately spliced GH1 minigene (upper PCR band) is increased in cells expressing the S34F U2AF1 mutant compared to control or wild-type U2AF1 expression (Fig. 3c, P=0.01). This increase in exon skipping mediated by mutant U2AF1 remained significant after depletion of endogenous U2AF1 in 293T cells (Fig. 3c, P<0.02). We also observed an increased utilization of alternative 3′ cryptic splice sites in the FMR1 gene in clinical MDS samples with U2AF1 mutations compared to MDS samples without U2AF1 mutations (Supplementary Fig. 3a). The alternative splicing of FMR1 was confirmed to be mutant U2AF1-dependent using a FMR1 minigene splicing reporter assay in vitro (Supplementary Fig. 3b).
Collectively, these results suggest that U2AF1 S34 mutations may result in subtle increases in splicing efficiency (Fig. 3a) -- or possibly altered isoform expression (Fig. 3c) --which could induce gene expression changes. To test whether U2AF1 mutations alter global gene expression levels, we analyzed mRNA microarray data (Affymetrix U133plus2) obtained from bone marrow CD34+ cells purified from 6 MDS patients with a U2AF1 mutation, 9 MDS patients without a mutation, and 4 normal donors18. The U2AF1 mutant samples did not segregate together using an unsupervised hierarchical clustering algorithm with all 19 samples, however, the 6 U2AF1 mutant samples did segregate together when compared to the normal control samples (Supplementary Fig. 4b). Next, we identified the genes that were significantly different between control and mutant patients using Significance Analysis of Microarrays (SAM)19. SAM identified 401 dysregulated probesets (50 up-regulated and 351 down-regulated) in U2AF1 mutant versus control samples (FDR<0.005) (Supplementary Fig. 4b, Supplementary Table 6). Three of the most enriched functional annotation categories for genes that are down-regulated in U2AF1 mutant samples are splicing and RNA recognition motif (RRM) genes (enrichment scores 2.5–4.3) (Supplementary Fig. 4c, Supplementary Table 6). These results suggest that a compensatory down-regulation of splicing genes may exist in U2AF1 mutant samples. These gene categories were not down-regulated in U2AF1 wild-type MDS patients compared to controls (data not shown), suggesting that down-regulation of splicing and RRM genes is not common to all MDS samples, but instead is associated with U2AF1 mutations.
Mutations in U2AF1 represent a novel mechanism that could alter gene expression in MDS and expand the list of commonly mutated genes in MDS that may affect transcription or translation, including RPS14, TET2, EZH2, ASXL1, and DNMT3A20–26. Only two patients in our cohort of 150 de novo MDS samples had both a DNMT3A and U2AF1 mutation (Supplementary Table 7). Both of these mutations appear to be early genetic events in MDS, given their high mutant allele burdens in patients with early stage disease26.
U2AF1 is highly conserved (Fig. 1a)1, and homozygous loss is lethal in many organisms27–29. We did not observe any nonsense, frameshift or missense mutations affecting the coordinating CCCH residues in the zinc fingers, again suggesting the S34 mutations are not loss-of-function mutations. The corresponding amino acid in the human ZFP36L2 (a ZFP36 family member) protein interacts with RNA through a hydrogen bond, further suggesting that the S34 position may be important for RNA binding13. Additionally, interactions between conserved aromatic amino acids in ZFP36 family members and RNA bases stabilize the protein RNA complexes formed13. The two amino acid substitutions we identified at S34 add a bulky aromatic ring (phenylalanine or tyrosine) to the zinc finger, which may alter, or even enhance, binding of the zinc finger to RNA. We suggest that the S34F/Y mutations in U2AF1 alter the specificity of U2AF1-dependent splicing. Pre-mRNAs with strong polypyrimidine tracts can splice independently of U2AF1 in vitro, whereas weak polypyrimidine tracts are more dependent on U2AF1 for appropriate splicing2,11. Therefore, the pattern of U2AF specificity (determined by both U2AF1 and U2AF2) may be influenced by the nucleotide sequence in pre-mRNAs and may be an important factor in determining which genes are altered in cells expressing mutant U2AF1.
Alternative splicing has been described for a wide range of cancers30,31, although the underlying mechanisms that influence cancer pathogenesis remain largely unknown. Alterations in the transcriptome mediated by alternative splicing may contribute directly to cancer, or indirectly by engaging some other pathway. The identification of somatic mutations in spliceosome genes in MDS by our group and others32–34, raises the possibility that mutations in splicing factors, including U2AF1, may be responsible for the observed alterations of splicing in cancer. Ultimately, cancer cells may generate genetic diversity in a large number of genes by selecting cells with mutations in U2AF1 or other spliceosome proteins. Identification of key target genes affected by U2AF1 mutations will be critical for our understanding of how these mutations contribute to MDS pathogenesis.
ONLINE METHODS
Flow sorting of bone marrow samples
Bone marrow cells from the secondary AML (sAML) sample, cryopreserved in 10% DMSO, were rapidly thawed at 37°C, washed, and stained with PE-Cy7 conjugated hCD45, clone J.33 (Beckman Coulter), and FITC conjugated anti-hCD34, clone 581 (Beckman Coulter). The blast population (low SSC/CD45 dim) was sorted using a Reflection high speed cell sorter (Sony iCyt) directly into lysis buffer and genomic DNA was prepared by column purification (Qiagen DNeasy).
Whole genome sequence production
Four DNA libraries were generated for paired-end sequencing: two from the tumor sample (flow-sorted sAML myeloblasts), and two from the normal sample (punch biopsy of unaffected skin). Sequence data was generated using both Illumina GAIIx and Illumina Hiseq platforms in 2 × 100 paired-end reads. Reads were aligned individually to NCBI Build 36 of the human reference sequence using BWA 0.5.5 and SAMtools r544. Alignments were merged into a single BAM file and marked for duplicates using Picard 1.17 (http://picard.sourceforge.net). Only non-duplicate reads were used for all downstream analyses.
Somatic mutation detection
Candidate point mutations were predicted using SomaticSniper (D. Larson et al, in press), previously referred to as glfSomatic4,35. Putative single nucleotide variants (SNVs) with somatic score of 40 and average mapping quality of 40 were considered high-confidence (HC); all others were deemed low-confidence (LC). Small (<100 bp) insertion/deletion events (indels) were called using a combination of GATK36, IndelGenotyper, Pindel37, and a modified version of SAMtools38. Both SNVs and indels were annotated using gene structure and conservation information, and classified by tier as previously described4. Briefly, tier 1 contains all changes in the amino acid coding regions of annotated exons, consensus splice-site regions, and RNA genes (including microRNA genes). Tier 2 contains changes in highly conserved regions of the genome or regions that have regulatory potential. Tier 3 contains mutations in the nonrepetitive part of the genome that do not meet tier 2 criteria, and tier 4 contains mutations in the remainder of the genome. High confidence tier 2 and tier 3 mutations, and all tier 1 mutations (regardless of confidence) were selected for validation (see below).
To identify somatic DNA copy number changes from whole genome sequencing (WGS) data, reads aligned by BWA39 were binned into contiguous, non-overlapping 1 kb windows. Copy number for each bin was normalized to the median copy number for each chromosome in tumor and normal separately. A Hidden Markov Model algorithm40 was used to generate a list of segments with copy number expressed as log2 (tumor/normal). Copy number changes were also supported if loss of heterozygosity (LOH) was observed in the affected regions. In brief, heterozygous SNPs were identified in WGS data from the normal sample (>10 reads of >q10 quality with non-reference allele frequencies of 0.4–0.6). The variant allele frequencies at these positions were then averaged in bins of 20 consecutive SNPs and visualized for the normal and tumor samples separately. Deletions, amplifications, inter-, and intrachromsomal rearrangements were also predicted using the BreakDancer algorithm41.
Mutation validation
To comprehensively evaluate tier 1–3 predictions, we utilized a custom solid-phase capture platform. We selected all tier 1 SNV predictions (HC and LC) and all HC tier 2–3 SNVs. Tier 1–3 indel predictions were also included. In addition, we used this approach to validate SV predictions (deletions and rearrangements). We identified 8–16 SNPs that were heterozygous in the normal DNA sample (determined using the WGS and SNPa data) that were located within the affected segments and 8 SNPs from flanking normal regions. The genomic positions of SNVs and indels (with a 200 bp margin) and SVs (with a 400 bp margin) were submitted for probe design. Probes were synthesized on custom HD2.1 long oligonucleotide arrays (Roche NimbleGen). Whole genome amplified DNA (REPLI-g, Qiagen) from the normal (skin), unfractionated MDS, and unfractionated sAML samples was used as bait for capture on the arrays and the recovered DNA (enriched for target sequences) was resequenced on the Illumina GAIIx platform.
At least 10x coverage was obtained for ~87.16% of the target sequence for all samples (see Supplementary Note, Supplementary Table 1). Reads were mapped using BWA39, deduplicated, and merged into BAM files. The reference or somatic status at the nucleotide of interest was then determined for each sample using VarScan242 with the following parameters: min-coverage=10, min-var-freq=0.05, somatic-p-value<0.01, validation=1. To validate low-frequency (2–5%) SNVs, we re-ran VarScan with adjusted parameters: min-coverage=100, min-var-freq=0.02, somatic-p-value<0.01, validation=1. In validation mode, VarScan reads data from tumor and normal samples simultaneously, performing pair-wise comparisons at every position covered in both samples. Each position is classified as Reference (wild-type), Germline, LOH, or Somatic, based upon a comparison of the consensus genotypes and supporting read counts (Fisher’s Exact test). Positions called Somatic are further subjected to our internally-developed false-positive filter which removes sequencing- and alignment-related artifacts using several criteria (read count, mapping quality, average read position, strand representation, homopolymer-like sequence context, mismatch quality sum difference, trimmed read length, Q2 distance) and were manually reviewed. Chromosome X and Y somatic positions are determined using the false-positive filter and manual review. SIFT and PolyPhen2 computational algorithms were used to predict whether U2AF1 mutations were damaging, as previously described43,44.
Sanger sequencing
To screen for recurrence of U2AF1 mutations, we performed Sanger sequencing using whole genome amplified DNA extracted from unfractionated bone marrow aspirates and paired normal tissue (skin) from 150 individual patients with de novo MDS. PCR amplicons covering all 9 exons and splice sites in U2AF1 were sequenced using BigDye chemistry and analyzed on an ABI 3730 sequencer (primer sequences in Supplementary Table 8). Sequence variants were called by The Genome Institute’s mutational profiling pipeline and manually reviewed. Potential somatic mutations (present in the bone marrow sample and not detectable in skin) were confirmed by independent PCR and sequencing.
Deep sequencing of U2AF1 mutations in DNA and cDNA
Unfractionated bone marrow samples from 11 patients with validated U2AF1 mutations were selected for deep sequencing to estimate clone size. Whole genome amplified DNA from normal (skin), MDS, and sAML samples were amplified by PCR individually using barcoded primers (Supplementary Table 8). The products were then pooled and sequenced on the Roche/454 platform. In parallel, RNA was extracted from MDS and sAML samples (Trizol, Invitrogen), converted to cDNA using the Ovation RNA-seq Kit (NuGEN), and amplified with barcoded primers spanning intron/exon boundaries (Supplementary Table 8). Reads were aligned to Hs36 using BWA-SW39. Following alignment, BAM and pileup files were generated using SAMtools and analyzed by Picard to remove duplicates. Only uniquely mapped bases with >q20 scores were retained. Reads supporting the reference or variant allele were identified by VarScan2.
SNP array analysis
Genomic DNA samples (not subjected to whole genome amplification) from the normal, MDS, and sAML specimens (not flow-sorted) were hybridized to Affymetrix 6.0 SNP arrays (Affymetrix, Inc.). Analysis of copy number alterations and copy neutral loss of heterozygosity was performed using the Partek Genomics Suite (Partek, Inc).
mRNA expression profiling
Total RNA was harvested from unfractionated bone marrow cells (69% myeloblast) from the secondary AML sample from UPN 266395 and hybridized to the Affymetrix Exon 1.0 ST array. Raw data was extracted using the Affymetrix Expression Console (Affymetrix, Inc.) and analyzed in Prism 5.04 (GraphPad Software, Inc.).
Total RNA was harvested from CD34+ purified MDS bone marrow samples (n=15) and control bone marrow (n=4) and hybridized to the Affymetrix U133plus2 array, as previously reported18. Supervised hierarchical clustering was performed using Ward’s clustering algorithm with a Euclidean distance similarity measure in Spotfire (TIBCO Software Inc). Significance Analysis of Microarrays (SAM), Gene Set Enrichment Analysis (GSEA), and Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7b were performed as previously described19,45,46.
Reverse transcriptase PCR
cDNA was made from RNA using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase or Superscript III kit (Invitrogen). Quantitative real-time RT-PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems) (primer and probe sequences for GAPDH are provided in47). The U2AF1 primer and probe set spans exons 2–3 (Hs00739599_m1, Applied Biosystems). All samples were run in duplicate on a 7300 Real-Time PCR system (Applied Biosystems) and analyzed using the relative standard curve method. Non-quantitative RT-PCR for U2AF1 mRNA isoform expression was performed and loaded on a 10% polyacrylamide gel (Forward: 5′-GCCTCCATCTTCGGCACCGA-3′, Reverse: 5′-GGCATGGCTCAGAATCGCCC-3′).
Generation of U2AF1 expression plasmids
RNA from a patient bone marrow biopsy (UPN 571656) was used to generate both U2AF1 (wild-type) and U2AF1 (S34F mutant) expression vectors. cDNA was generated from patient bone marrow RNA using Superscript III kit (Invitrogen). Both wild-type and mutant U2AF1 cDNAs were obtained via PCR amplification, cloned with the Topo Cloning kit (Invitrogen), and sequenced for verification. The U2AF1 cDNAs were then shuttled into the EcoRI site of the pcDNA3.1+ vector (Invitrogen) for transient transfection experiments.
Luciferase-β-galactosidase double-reporter assay
293T cells were seeded in a 6-well plate (1 × 106 per well) and cultured in DMEM (Gibco/Invitrogen) supplemented with 10% FBS and L-glutamine. Following overnight culture, cells were co-transfected with the expression vectors (U2AF1 wild-type or S34F mutant, or empty vector) and the pTN24 splicing reporter plasmid (containing a constitutively expressed β-galactosidase reporter for transfection normalization and a luciferase reporter that is conditional on removal of a translational stop codon by splicing) with or without splicing modulators hnRNPG and Tra2α17. In some experiments, cells were also co-transfected with 30nM U2AF1-specific siRNA (5′-CGUAGAAAGUGUUGUAGUUGAUUGA-3′; IDT, Inc.) or 30nM siRNA scramble control (Dharmacon). Cells were harvested 48 hours following transfection, and reporter expression was detected, as previously described17 using the Dual Light Reporter System (Applied Biosystems) and analyzed by calculating the ratio of luciferase to β-galactosidase signal. Changes in U2AF1 levels were confirmed by Western blot using antibodies specific for U2AF1 (SAS1300700, Sigma-Aldrich) or β-actin (A5441, Sigma-Aldrich) as a loading control. Three independent experiments were performed and the data was analyzed using a Student’s t-test.
Minigene constructs and transfection
293T cells were cultured and transfected with U2AF1 expression vectors, as above. Cells were also co-transfected with a GH1 minigene splicing reporter construct9. In other experiments, cells were co-transfected with a FMR1 minigene splicing reporter construct containing partial sequence from exons 14 and 15 and the complete intronic sequence. Amplification of the FMR1 DNA fragment (including the intron) was achieved using the FMR1-201 E15 set of primer sequences previously published10. The amplified fragment was cloned into the TopoTA vector (Invitrogen), purified following BamH1 and XhoI digestion, and subsequently cloned into pcDNA3.1 (Invitrogen). Co-transfection of U2AF1 or control siRNAs was also performed, as above. Cells were harvested 48 hours following transfection, and RNA was extracted using the RNeasy reagent (Qiagen) following the manufacturer’s instructions. The RNA was used as a template for cDNA synthesis via RT-PCR with random hexamers and oligo(dT) primers. Changes in minigene splicing were then measured by PCR of the cDNA using a T7 forward primer and gene-specific reverse primers as previously described9,10 and quantified by densitometry. U2AF1 knockdown by siRNA and U2AF1 reconstitution were confirmed by Western blot analysis, as above. Three to four independent experiments were performed and the data was analyzed using a Student’s t-test.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01HL082973 (Graubert), RC2HL102927 (Graubert), U54HG003079 (Wilson), P01CA101937 (Ley), and a Howard Hughes Medical Institute Physician-Scientist Early Career Award (Walter). Technical assistance was provided by the Alvin J. Siteman Cancer Center High Speed Cell Sorting Core, the Molecular and Genomic Analysis Core, the Biomedical Informatics Core, and the Tissue Procurement Core which are supported by an NCI Cancer Center Support Grant P30CA91842. Additional technical assistance was provided by Masayo Izumi. We thank Dr. Kinji Ohno (Nagoya University Graduate School of Medicine, Japan) for minigene constructs. We thank Dr. Kathleen Hall (Washington University School of Medicine) for helpful scientific discussions.
Footnotes
AUTHORS CONTRIBUTIONS
Timothy A. Graubert: project leader, study design, execution and analysis, manuscript preparation.
Dong Shen: project leader, sequence analysis.
Li Ding: project leader, supervisor data analysis team.
Theresa Okeyo-Owuor: in vitro splicing assays.
Cara L. Lunn: quantitative reverse transcriptase PCR, in vitro splicing assays.
Jin Shao: microarray data analysis and PCR assays.
Kilannin Krysiak: gene expression analysis.
Christopher C. Harris: sequence analysis.
Dan C. Koboldt: capture validation data analysis.
David E. Larson: mutation analysis and annotation.
Michael D. McLellan: auto-analysis and manual review of validation data.
David J. Dooling: IT and data management, data analysis automation leader.
Rachel M. Abbott: variant validation production.
Robert S. Fulton: variant validation oversight.
Heather Schmidt: manual review of variants.
Joelle Kalicki-Veizer: manual review of variants.
Michelle O’Laughlin: variant validation production.
Marcus Grillot: clinical data management and specimen acquisition.
Jack Baty: statistical analysis of clinical variables and outcomes.
Sharon Heath: clinical data management and specimen acquisition.
John L. Frater: clinical hematopathology review.
Talat Nasim: design of in vitro dual reporter splicing assay.
Daniel C. Link: study design, execution and analysis and manuscript preparation.
Michael H. Tomasson: study design, execution and analysis.
Peter Westervelt: clinical data and specimen acquisition, study design, execution and analysis.
John F. DiPersio study design, execution and analysis and manuscript preparation.
Elaine R. Mardis: project conception, analysis coordination and manuscript preparation.
Timothy J. Ley: project conception, study design, manuscript preparation.
Richard K. Wilson: project conception and oversight, manuscript preparation.
Matthew J. Walter: project leader, study design, analysis coordination and manuscript preparation.
DATA ACCESS SECTION
Sequence and SNPa data have been deposited in dbGAP under accession number phs000159.v3.p2. Gene expression profiling data have been deposited in GEO under accession number GSE30195.
Please see Supplementary Note for Text, Results, Figures (4), Tables (9), and References.
COMPETING INTERESTS STATEMENT
The authors have no competing interest to declare.
LITERATURE CITED
- 1.Webb CJ, Wise JA. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol. 2004;24:4229–40. doi: 10.1128/MCB.24.10.4229-4240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–5. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 3.Welch JS, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–84. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link DC, et al. Identification of a Novel TP53 Cancer Susceptibility Mutation Through Whole-Genome Sequencing of a Patient With Therapy-Related AML. JAMA. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Masuda A, Ito M, Shinmi J, Ohno K. AG-dependent 3′-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res. 2011;39:4396–404. doi: 10.1093/nar/gkr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kralovicova J, Vorechovsky I. Allele-specific recognition of the 3′ splice site of INS intron 1. Hum Genet. 2010;128:383–400. doi: 10.1007/s00439-010-0860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol Cell Biol. 2006;26:8183–90. doi: 10.1128/MCB.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell. 2006;17:4187–99. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–64. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 14.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem. 2002;277:9606–13. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 15.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 17.Nasim MT, Eperon IC. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat Protoc. 2006;1:1022–8. doi: 10.1038/nprot.2006.148. [DOI] [PubMed] [Google Scholar]
- 18.Graubert TA, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS ONE. 2009;4:e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 21.Ebert BL, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–9. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–42. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 23.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 24.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 25.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 26.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–8. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golling G, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 28.Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci U S A. 1996;93:10333–7. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA. 1999;5:487–94. doi: 10.1017/s1355838299982225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grosso AR, Martins S, Carmo-Fonseca M. The emerging role of splicing factors in cancer. EMBO Rep. 2008;9:1087–93. doi: 10.1038/embor.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visconte V, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2011 Sep 2; doi: 10.1038/leu.2011.232. 10.1038. [DOI] [PubMed] [Google Scholar]
- 33.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011 Sep 26; doi: 10.1056/NEJMoa1103283. 10.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 35.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baum LE, Eagon JA. An Inequality with Applications to Statistical Estimation for Probabilistic Functions of a Markov Process and to a Model for Ecology. Bulletin of the American Mathematical Society. 1967;73:360–363. [Google Scholar]
- 41.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koboldt DC, et al. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortier JM, et al. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia. 2010;24:950–7. doi: 10.1038/leu.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.