Abstract
Before a tooth erupts into the oral cavity, the mineralized enamel and dentin layers begin to develop. During these early stages of enamel formation, an abundant group of proteins known as amelogenins are secreted by ameloblast cells within the developing tooth. These proteins are required for the enamel layer to reach its normal thickness and attain its intricate structure. Human patients with amelogenin gene mutations have a condition referred to as amelogenesis imperfecta, and we have analyzed human gene defects so that we can recreate them in mice. We have generated mice with a null amelogenin mutation where no amelogenin is produced, mice that over-express normal and mutated amelogenins, and over-expressors have been mated to null mice for rescue experiments. Because there are at least 15 messages that are alternatively spliced from a single amelogenin primary RNA transcript, these approaches have begun to reveal the functions of individual amelogenin proteins during enamel development. Finally, amelogenins are processed by carefully regulated proteolytic digestion leading to many additional amelogenin peptides and it is likely that protein function is altered during this developmental process. We have also had some surprises, as one of our mouse models develops odontogenic tumors, and we know now that some of the amelogenins are expressed in other regions of the body outside of the oral cavity, and may have a role in signal transduction.
Keywords: amelogenin, enamel development, amelogenesis imperfecta, transgenic mice
Introduction
During a series of interactions between epithelial and mesenchymal cell layers, teeth begin to develop within the vertebrate mandible or maxilla1. Some of the ectomesenchymal cells differentiate into dentin-producing odontoblasts, and adjacent epithelial cells elongate and differentiate into the ameloblasts that produce dental enamel2. These two mineralized tissues are found in the crown of the tooth, and are joined by an unusual structure, the enamel-dentin junction, which firmly binds these two mineralized layers with fundamentally different characteristics. Dentin is also produced by root odontoblasts, and forms a layer covered by another mineralized tissue, the cementum, on the surface of the root of the tooth.
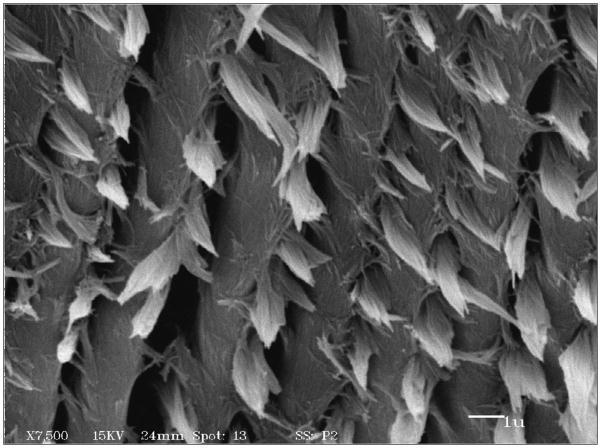
This review will describe the secreted proteins that are synthesized by ameloblasts and that orchestrate development of the intricate rod-like structure found in dental enamel (Fig. 1)3. This image illustrates the mineralized end-products of individual ameloblast cells, and shows enamel rods that are interwoven in order to produce enamel structural integrity. Enamel composition begins as a secreted organic matrix, into which mineral ions are added while proteins are degraded by the action of several enamel proteases. By the end of enamel maturation as the tooth begins to erupt, the composition of the enamel layer is approximately 95% mineral with less than 2% of the organic material remaining4. Enamel cannot be regenerated following eruption as the ameloblast cell layer is no longer present.
Fig. 1.
Scanning electron micrograph of murine enamel showing ends of enamel rods
Enamel Development and the Principal Proteins Involved
Amelogenesis can be divided into two stages. The first is the Secretory Stage, during which ameloblasts differentiate into tall secretory cells and develop a cellular extension known as Tomes’ process. The role of this structure is to organize the development of the enamel rods. During the Secretory Stage, most of the enamel proteins are produced and secreted into the developing enamel matrix, while mineralization and protein processing begin. During the subsequent Maturation Stage, additional proteases are produced that remove most of the remaining organic material as mineralization continues.
The proteins secreted by ameloblasts are predominantly amelogenins, but also include lesser amounts of other structural proteins, such as ameloblastin and enamelin (Table 1). The MMP-20 (matrix metalloproteinase-20) protease is secreted along with the structural proteins during the Secretory Stage and this enzyme begins to cleave the enamel proteins while another protease, kallikrein-4 (KLK4), is produced during the Maturation Stage to further process the remaining organic matrix13,14. Two recently described enamel proteins with unknown functions are also listed in Table 1, and references to reviews are provided for more recent information15,16.
Table 1.
Mutations leading to AI in humans
| Protein | Function | Gene | Mutations Reported | Authors (year) |
|---|---|---|---|---|
| Amelogenin | Structural | AMELX | 15 | Lagerstrom, et al. (1991)5 |
| Enamelin | Structural | ENAM | 10 | Rajpar, et al. (2001)6 |
| Ameloblastin | Structural | AMBN | * | |
| Matrix metalloproteinase-20 | Protease | MMP20 | 4 | Kim, et al. (2005)7 Ozdemir, et al. (2005)8 |
| Kallikrein 4 | Protease | KLK4 | 2 | Hart, et al. (2004)9 |
| FAM83H | Unknown | FAM83H | 15 | Kim, et al. (2008)10 Lee, et al. (2008)11 |
| WDR72 | Unknown | WDR72 | 4 | El-Sayed, et al. (2009)12 |
The website http://www.dentistry.unc.edu/research/defects, maintained by J.T. Wright, summarizes current AI information.
not yet identified in humans
Gene Mutations Lead to Amelogenesis Imperfecta (AI) in Humans
Amelogenesis imperfecta (AI) is the general term used to describe enamel defects that affect primarily dental enamel, and are not syndromic in nature. It is not surprising that mutations in the genes listed in Table 1 lead to an enamel defect, but there is heterogeneity in phenotypic appearance both among members of a kindred and even within a single dentition17. This is partly because mutations in different genes lead to different clinical phenotypes, and because mutation in either the N- or the C-terminus of enamel proteins may lead to differences in severity and appearance of the enamel17. The basic categories of AI have been divided into hypoplasia (enamel that is too thin because of a defect in secretion), hypocalcification (a defect in the mineral crystals) or hypomaturation (protein processing defect with reduced removal of the organic material) with autosomal or X-linked inheritance patterns18. In addition, there are many subcategories according to phenotypic subtleties and genetic inheritance patterns.
This review will focus on the effects of human amelogenin gene mutations, and on animal models generated to reproduce the human AI genotype and phenotype. Similarities between human and murine amelogenin genes are apparent at the levels of gene structure, DNA sequence and pattern of expression during development19.
Amelogenin Proteins
In humans, the amelogenin proteins are primarily encoded by the AMELX gene on the X chromosome. The AMELY gene on the Y chromosome in males is estimated to be only about 10% as active as AMELX in producing amelogenin proteins20. The AMELX gene is transcribed to produce a primary transcript, or RNA, which is then spliced to produce a number of mRNAs which are translated into the amelogenin proteins; these vary according to the encoding exons present. At least five mRNAs have been reported in humans and all have the potential to be translated to produce amelogenin proteins, which are present in different amounts according to Western blot analysis of extracts taken during early stages of enamel development. The mechanism responsible for generating the varying levels of different amelogenin messages is unknown.
AMELX gene deletions and deletions that affect only the N-terminal or C-terminal regions of the protein lead to AI, but several single amino acid substitutions have also been reported to cause AI210,15. Kindreds with N-terminal mutations tend toward hypomaturation AI while C-terminal mutations frequently lead to a hypoplastic phenotype17. In addition, an individual gene mutation within a spliced region can affect some amelogenin proteins and not others, leading to complexity in the enamel appearance in families carrying AMELX gene mutations. Males with an AMELX mutation frequently have a severe phenotype as insufficient AMELY protein is generated for development of normal enamel structure.
Because of the number of individual amelogenin proteins produced by alternative splicing of the primary transcript, the function of individual amelogenin proteins has been difficult to decipher using human samples.
Mouse Models for Amelogenesis Imperfecta
At least 15 amelogenin messages have been reported due to alternative splicing of the primary RNA transcript of the single murine X-chromosomal amelogenin gene 22, 23. A Y-chromosomal amelogenin gene has not been identified in mice. Mouse models for AI have been developed to test the function of the amelogenin protein group, and to begin to define the roles of the individual amelogenin proteins. Mice have been generated to express an overabundance of one amelogenin, to express a mutated amelogenin or to express only one amelogenin, but none of the other amelogenins 24,25,26.
The Amelx null (KO) mutation was engineered by cloning a segment of the mouse Amelx gene into an expression vector from which a segment that encoded the amelogenin N-terminus and part of intron 2 was removed, while replacing this gene segment with an antibiotic resistance gene expressed in the opposite orientation for clone selection. This vector was transferred into ES cells and selected clones were injected into blastocyststo produce viable offspring with germ-line transmission. This strategy allowed a shortened RNA to be expressed by the mice, but none of the amelogenin proteins could be detected27. The Amelx null mice have an enamel defect with hypoplasia and disorganization of the normal enamel structure similar to severe AI in humans, leading to the conclusion that amelogenin proteins are responsible for generating proper enamel thickness and prismatic structure in the enamel layer. A knock-in mouse has also been generated where a mutated Amelx gene replaced the endogenous gene, and this approach also altered the enamel phenotype28.
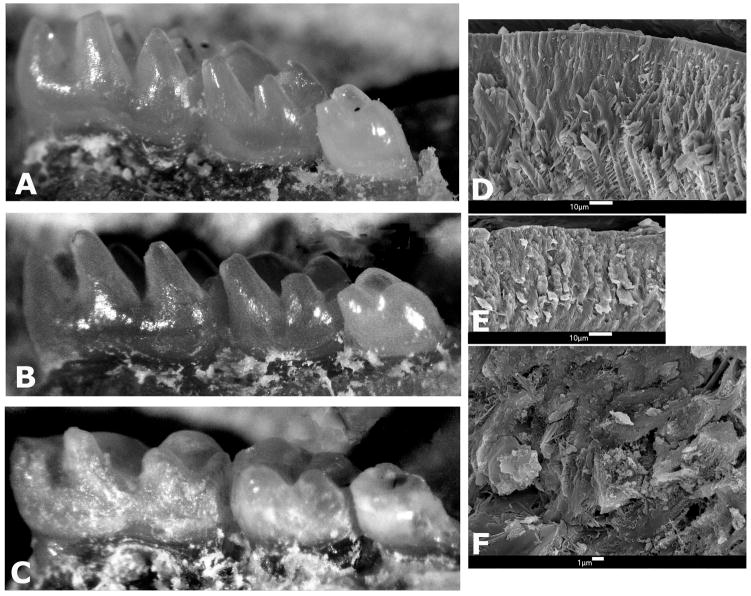
To begin to answer the question of the function of individual amelogenin proteins, another approach was used to generate a transgenic mouse that expressed the most abundant amelogenin protein, which includes 180 amino acids and is referred to as TgM180. This strategy began with generation of an expression vector with the coding region of M180 plus regulatory regions found upstream, downstream and within intron 1. After injection, strains with germ-line transmission were developed and their phenotype was indistinguishable from wild-type (WT) (Fig. 2A,B,D) as only normal amelogenin proteins were expressed29.
Fig. 2.
Molar phenotype of wild-type and transgenic molars. A–C, light microscopy of murine molars; D–F, scanning electron microscopy of fractured surfaces of murine enamel. A, WT; B, TgM180; C, TgP70T; D, TgM180; E,F, TgP70T.
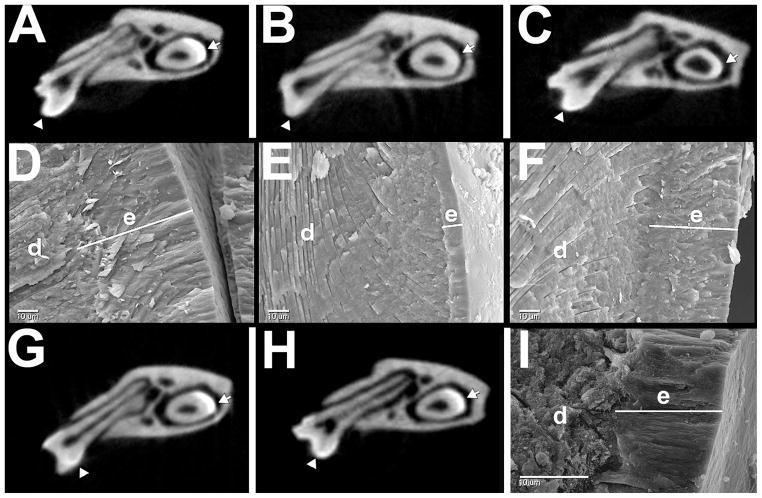
In order to ask whether the TgM180 had an important function by itself, without the assistance of the other amelogenin proteins, a male TgM180 mouse was mated with a female Amelx null mouse, and offspring with an Amelx null genetic background but that expressed only TgM180 were generated. TgM180KO offspring showed improvement in their enamel defect compared to the null (KO) mice, and were considered to have a partial rescue30 (Fig. 3). The microCT images show a dense white layer on the surface of WT and TgM180 molars and incisors (Fig. 3A, G), which is missing from amelogenin null mice (Fig. 3B), but is partially restored in the rescued TgM180KO mice (Fig. 3C). The rescued mice had a thicker molar enamel layer (Fig. 3D,E,F), and greater enamel volume and density compared to null (KO) mice30. However, the structure was not entirely normal as the hardness and elastic modulus were not improved and remained similar to that measured for the Amelx null enamel layer30. It was concluded that other amelogenins in addition to M180 were also required for normal enamel to form, as originally predicted.
Fig. 3.
MicroCT and scanning electron microscopy analysis of enamel layers. A–C,G,H microCT at the level of the lower 1st molar; D–F,I scanning electron microscopy. Shown are wild-type (A,D); Amelx null (B,E); TgM180KO (C,F). TgM180 (G); TgP70T (H); (I) scanning electron microscopy of fractured surface of TgM180KO incisor enamel, which lacks substantial rescue as the transgene is expressed primarily in molar ameloblasts.
In a second set of experiments, a transgene similar to TgM180 was generated, but with a single amino acid change, similar to that seen in human AI kindreds 21,29. This vector was used to generate transgenic mice, with the alteration of a single proline to a threonine at amino acid position 70, referred to as TgP70T29. TgP70T mice with all of the normal amelogenin proteins plus this mutated amelogenin had an enamel defect which included surface roughness and hypoplasia. (Fig. 2C,E,F; Fig. 3H).
We hypothesized that this mutated transgene would not rescue the enamel defect in the way that TgM180 had been able to when TgM180 males were mated to Amelx null females. However, we were surprised when the TgP70TKO mice had a very poor enamel appearance with a severe phenotype, including abnormal cellular proliferations and odontogenic tumors adjacent to the normal teeth31. The abnormal cells were identified within an unusual extracellular matrix that was positive for amelogenin protein using an anti-amelogenin antibody by immunohistochemistry. Some of the regions had similarity to the human odontogenic tumor, calcifying epithelial odontogenic tumor, or Pindborg’s tumor32. Some of the mice with tumors also had supernumerary teeth (Fig. 4A,B). Islands of epithelium containing ghost cells and calcified material are shown (Fig. 4C)31.
Fig. 4.
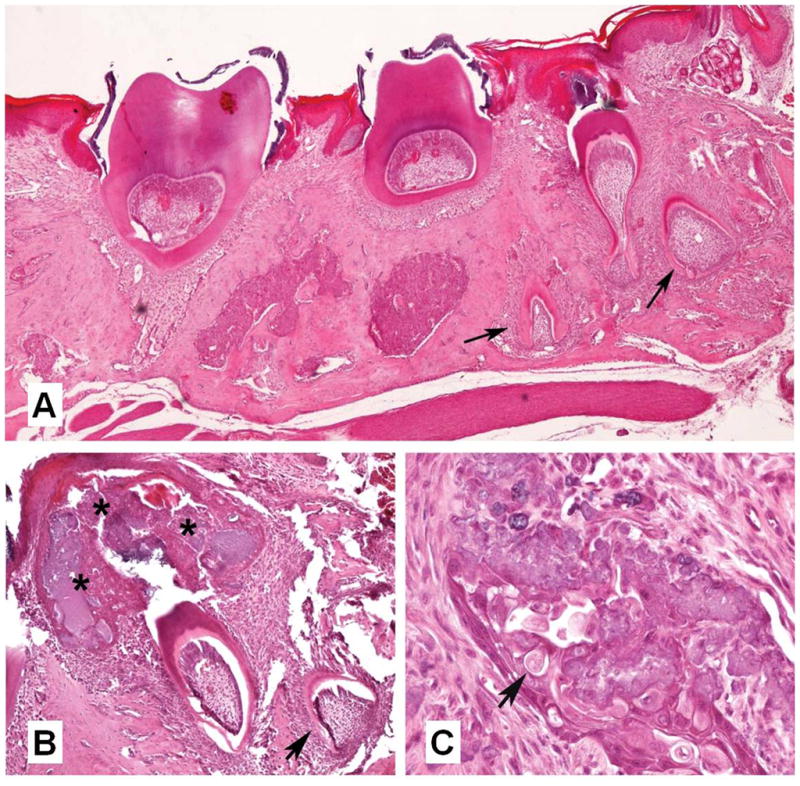
The P70T mutation disrupts normal odontogenetic development. A, supernumerary teeth (arrows) were observed in the right maxilla of a male TgP70TKO mouse; B, an aberrant benign epithelial proliferation (*) overlying areas of tooth development in a TgP70T het female; C, island of epithelium with ghost cells (arrow) and calcified substance from the mouse shown in B.
Further studies revealed a disregulation of the Notch signaling pathway31, which is involved in the development of the stratum intermedium and ameloblast cell layers during early tooth development33. The mechanism for this pathology is unknown but is likely related to inappropriate proliferation of at least one of these cell layers.
Amelogenin Expression in Other Tissues
Amelogenin message and protein have been detected by various strategies in other tissues, including dental pulp cells34–37, periodontium, bone, brain and other tissues38,39. The reader is directed to a recent review for discussion40, but major defects have not yet been reported outside of the enamel in either murine models or in humans with AI specific gene mutations.
Amelogenin’s Proposed Function as a Signaling Protein
In 2000, Arthur Veis reported that a small amelogenin protein referred to as leucine rich amelogenin protein, or LRAP, when injected into rat muscle, induced cartilage or bone specific gene expression41,42, and many investigators have reported similar results with other systems of cultured cells or animal studies. A product known as Emdogain is used clinically to assist with periodontal tissue regeneration and this tissue is primarily obtained from developing porcine teeth that are making enamel proteins, primarily amelogenin43. Small LRAP amelogenins have been reported to be expressed by stem cells during osteogenic differentiation, and stem cell treatment with LRAP leads to differentiation of these cells into the osteoblast lineage44. The varied functions of the amelogenin protein family is becoming a promising area of endeavor for regenerative medicine.
Summary and future directions
Since amelogenin proteins were first described in 19804, biochemical and molecular biology approaches have revealed that there are many amelogenin proteins, due to both alternative splicing of amelogenin RNA and proteolytic cleavages, which have been proposed to alter protein function during development. Several other enamel proteins have been proven to be important in enamel development through analysis of kindreds with gene mutations or because of relevant animal models. Amelogenin gene mutations and animal models reveal that this family of proteins is important for developing correct enamel thickness and organization, but we know little about how the various enamel proteins work together to generate an enamel layer with its exquisite decussated structure. More recent studies led to the acceptance that amelogenins are expressed by cells other than ameloblasts, and that several of the amelogenin proteins can participate in intercellular signaling. The amelogenin gene is embedded within another much larger ARHGAP6 gene with the opposite orientation45, and the relationship between these genes is currently unknown. There are many challenges ahead for understanding the multiple roles of amelogenin proteins, including their molecular mechanisms for generating a mineralized tissue, and potential roles in the other tissues where they are expressed, as well as potential use in therapy and regenerative approaches.
Acknowledgments
I acknowledge current and past members of the Gibson laboratory for their research accomplishments, Tim Wright and Cynthia Suggs from University of North Carolina for collaborative discussions and images, Ashok Kulkarni from NIDCR for collaboration on the amelogenin null mouse project, and Faizan Alawi (University of Pennsylvania) for discussions concerning mouse pathology. This work was supported by the National Institutes of Health through NIDCR grant DE011089.
References
- 1.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 2.Mitsiadis TA, Graf D. Cell fate determination during tooth development and regeneration. Birth Defects Res (Part C) 2009;87:199–211. doi: 10.1002/bdrc.20160. [DOI] [PubMed] [Google Scholar]
- 3.Pugach MK, Li Y, Suggs C, Wright JT, Aragon MA, Yuan ZA, Simmons D, Kulkarni AB, Gibson CW. The amelogenin C-terminus is required for enamel development. J Dent Res. 2010;89:165–169. doi: 10.1177/0022034509358392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- 5.Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 6.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant Amelogenesis imperfecta. Hum Molec Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JCC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Part JT, Seo BM, Hu JC, Simmer JP. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82:489–494. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, Hu JCC, Bartlett JD, Lee KE, Lin BP-J, Simmer JP, Kim JW. Mutational spectrum of FAM83H: The C-terminal portion is required for tooth enamel calcification. Hum Mut. 2008;29:E95–E99. doi: 10.1002/humu.20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 2009;85:699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith C. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 14.Simmer JP, Hu JCC. Expression, structure and function of enamel proteinases. Conn Tiss Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 15.Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet Part A. 2006;140A:2547–2555. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng FK, Messer LB. Dental management of amelogenesis imperfecta: a primer on genotype-phenotype correlations. Ped Dent. 2009;31:20–30. [PubMed] [Google Scholar]
- 17.Hart PS, Aldred MJ, Crawford PJM, Wright NJ, Hart TC, Wright JT. Amelogenesis imperfecta phenotype-genotype correlations with two amelogenin gene mutations. Archs Oral Biol. 2002;47:261–265. doi: 10.1016/s0003-9969(02)00003-1. [DOI] [PubMed] [Google Scholar]
- 18.Witkop CJ, Sauk JJ. Heritable defects of enamel. In: Stewart RE, Prescott GH, editors. Oral Facial Genetics. Mosby; St. Louis: 1976. pp. 151–226. [Google Scholar]
- 19.Delgado S, Ishiyama M, Sire JY. Validation of amelogenesis imperfecta inferred from amelogenin evolution. J Dent Res. 2007;86:326–330. doi: 10.1177/154405910708600405. [DOI] [PubMed] [Google Scholar]
- 20.Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and Y chromosomes. Am J Hum Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- 21.Collier PM, Sauk JJ, Rosenbloom J, Yuan ZA, Gibson CW. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Archs Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- 22.Simmer JP, Hu CC, Lau EC, Sarte P, Slavkin HC, Fincham AG. Alternative splicing of the mouse amelogenin primary RNA transcript. Calcif Tissue Int. 1994;55:302–310. doi: 10.1007/BF00310410. [DOI] [PubMed] [Google Scholar]
- 23.Hu CC, Ryu OH, Qian Q, Zhang CH, Simmer JP. Cloning, characterization and heterologous expression of exon 4-containing amelogenin mRNAs. J Dent Res. 1997;76:641–647. doi: 10.1177/00220345970760020401. [DOI] [PubMed] [Google Scholar]
- 24.Wright JT, Hart TC, Hart PS, Simmons D, Suggs C, Daley B, Simmer J, Hu J, Bartlett JD, Li Y, Yuan ZA, Seow WK, Gibson CW. Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs. 2009;189:224–229. doi: 10.1159/000151378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunglas C, Septier D, Paine ML, Zhu DH, Snead ML, Goldberg M. Ultrastructure of forming enamel in mouse bearing a transgene that disrupts the amelogenin self-assembly domains. Calcif Tissue Int. 2002;71:155–166. doi: 10.1007/s00223-001-2116-5. [DOI] [PubMed] [Google Scholar]
- 26.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 28.Zhu D-H, Paine ML, Luo W, Bringas P, Snead ML. Altering biomineralization by protein design. J Biol Chem. 2006;281:21173–21182. doi: 10.1074/jbc.M510757200. [DOI] [PubMed] [Google Scholar]
- 29.Gibson CW, Yuan ZA, Li Y, Daly B, Suggs C, Aragon MA, Alawi F, Kulkarni AB, Wright JT. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Suggs C, Wright JT, Yuan ZA, Aragon M, Fong H, Simmons D, Daly B, Golub EE, Harrison G, Kulkarni AB, Gibson CW. Partial rescue of the amelogenin null dental enamel phenotype. J Biol Chem. 2008;283:15056–15062. doi: 10.1074/jbc.M707992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Li Y, Alawi F, Bouchard JR, Kulkarni AB, Gibson CW. An amelogenin mutation leads to disruption of the odontogenic apparatus and aberrant expression of Notch1. J Oral Pathol Med. 2011;40:235–242. doi: 10.1111/j.1600-0714.2010.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pindborg JJ. A calcifying epithelial odontogenic tumor. Cancer. 1958;11:838–843. doi: 10.1002/1097-0142(195807/08)11:4<838::aid-cncr2820110423>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, Wakisata S. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Comm. 2006;340:611–616. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 34.Oida S, Nagano T, Yamakoshi Y, Ando H, Yamada M, Fukae M. Amelogenin gene expression in porcine odontoblasts. J Dent Res. 2002;81:103–108. [PubMed] [Google Scholar]
- 35.Nagano T, Oida S, Ando H, Gomi K, Arai T, Fukae M. Relative levels of mRNA encoding enamel proteins in enamel organ epithelia and odontoblasts. J Dent Res. 2003;82:982–986. doi: 10.1177/154405910308201209. [DOI] [PubMed] [Google Scholar]
- 36.Papagerakis P, MacDougall M, Hotton D, Mailleul-Forestier I, Oboeuf M, Berdal A. Expression of amelogenin in odontoblasts. Bone. 2003;32:228–240. doi: 10.1016/s8756-3282(02)00978-x. [DOI] [PubMed] [Google Scholar]
- 37.Ye L, Le TQ, Zhu L, Butcher K, Schneider RA, Li W, DenBesten PK. Amelogenins in human developing and mature dental pulp. J Dent Res. 2006;85:814–808. doi: 10.1177/154405910608500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haze A, Taylor AL, Blumenfeld A, Rosenfeld E, Leiser Y, Dafni L, Shay B, Gruenbaum-Cohen Y, Fermon E, Haegewald S, Bernimoulin JP, Deutsch D. Amelogenin expression in long bone and cartilage cells and in bone marrow progenitor cells. Anat Rec. 2007;290:455–460. doi: 10.1002/ar.20520. [DOI] [PubMed] [Google Scholar]
- 39.Deutsch D, Haze-Filderman A, Blumenfeld A, Dafni L, Leiser Y, Shay B, Gruenbaum-Cohen Y, Rosenfeld E, Fermon E, Zimmermann B, Haegewald S, Bernimoulin JP, Taylor AL. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci. 2006;114 (Suppl 1):183–189. doi: 10.1111/j.1600-0722.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- 40.Gibson CW. The amelogenin “enamel proteins” and cells in the periodontium. Crit Rev Euk Gene Expres. 2008;18:345–360. doi: 10.1615/critreveukargeneexpr.v18.i4.30. [DOI] [PubMed] [Google Scholar]
- 41.Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, Brownell AG, Jengh S-M, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 42.Veis A. Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammarstrom L. Enamel matrix, cementum development and regeneration. J Clin Periodontol. 1997;9(pt 2):658–668. doi: 10.1111/j.1600-051x.1997.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 44.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Comm. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson CW, Thomson NH, Abrams WR, Kirkham J. Nested genes: biological implications and use of AFM for analysis. Gene. 2005;350:15–23. doi: 10.1016/j.gene.2004.12.045. [DOI] [PubMed] [Google Scholar]