Background: When P fimbrial adhesins are expressed in uropathogenic E. coli, PapX represses motility.
Results: SELEX combined with high throughput sequencing, using PapX as bait, identified a novel binding site.
Conclusion: PapX represses motility by binding a unique nucleotide sequence within the flhDC promoter.
Significance: Combining SELEX and high throughput sequencing technology represents a powerful new method to identify bacterial transcription factor binding sites.
Keywords: Bacterial Adhesion, DNA Sequencing, DNA-Protein Interaction, Escherichia coli, Helix-Loop-Helix Transcription Factors, SELEX, High Throughput Sequencing, CAST, SAAB, Illumina, Adherence, UPEC, Reciprocal Control, UTI
Abstract
Most uncomplicated urinary tract infections (UTIs) are caused by uropathogenic Escherichia coli (UPEC). Both motility and adherence are integral to UTI pathogenesis, yet they represent opposing forces. Therefore, it is logical to reciprocally regulate these functions. In UPEC strain CFT073, PapX, a non-structural protein encoded by one of the two pap operons encoding P fimbria adherence factor, represses flagella-mediated motility and is a putative member of the winged helix transcription factor family. The mechanism of this repression, however, is not understood. papX is found preferentially in more virulent UPEC isolates, being significantly more prevalent in pyelonephritis strains (53% of isolates) than in asymptomatic bacteriuria (32%) or fecal/commensal (12.5%) strains. To examine PapX structure-function, we generated papX linker insertion and site-directed mutants, which identified two key residues for PapX function (Lys54 and Arg127) within domains predicted by modeling with I-TASSER software to be important for dimerization and DNA binding, respectively. To determine the PapX binding site in the CFT073 genome, systematic evolution of ligands by exponential enrichment (SELEX) in conjunction with high throughput sequencing was utilized for the first time to determine a novel binding site for a bacterial transcription factor. This method identified a 29-bp binding site within the flhDC promoter (TTACGGTGAGTTATTTTAACTGTGCGCAA), centered 410 bp upstream of the flhD translational start site. Gel shift experiments demonstrated that PapX binds directly to this site to repress transcription of flagellar genes.
Introduction
Urinary tract infection (UTI)2 is a costly and potentially dangerous malady in the United States, associated with an estimated healthcare cost of $3.5 billion in 2000 (1) and 479,000 hospitalizations in 2006 (2). The most common cause of uncomplicated UTI in humans is uropathogenic Escherichia coli (UPEC) (3). Typically, infection proceeds when bacteria colonize the periurethral area, gain access to the urinary tract, and ascend to the bladder, causing cystitis, or further ascend to the kidneys, causing the clinically more severe acute pyelonephritis.
To establish an infection of the bladder or kidneys during UTI, ascension of the urinary tract is an essential component of pathogenesis. To ascend, however, bacteria must first resist being expelled by the flushing mechanism of voiding while also engaging in upward motility. To avoid elimination, bacteria use fimbriae, cell surface appendages carrying adhesins, to bind to glycolipid or glycoprotein receptors expressed on the surface of uroepithelial cells, to immobilize themselves along the walls of the urinary tract. Although UPEC strains can encode more than 10 different types of fimbriae, certain fimbriae have been identified as critical to the development of a UTI. For example, mutations in genes encoding Type 1 fimbriae greatly reduce the ability of UPEC to cause UTI in a murine model of infection (4). P fimbria, encoded by the pap (pyelonephritis-associated pili) operon (5), binds to Galα(1–4)βGal moieties of P blood group antigen, a glycosphingolipid, present on the surface of kidney epithelial cells of most humans (6, 7). In epidemiological studies, the presence of P fimbria is strongly associated with UPEC strains in general and development of upper UTI in humans in particular (8, 9), although the importance of P fimbriae in a murine model of infection has not been firmly established (10). To ascend, UPEC relies on flagella, organelles that mediate motility. Flagellar expression is controlled by the master regulators of motility, flhD and flhC, whose gene products together regulate the first step of a tiered regulatory cascade that results in the production of flagella (reviewed in Ref. 11 and references therein). Flagella clearly also contribute to virulence of UPEC in a murine model of UTI (12) and are important in bacterial ascension via the ureters from the bladder to the kidneys (12, 13).
Motility, mediated by flagella, and adherence, mediated by fimbriae, are inherently antagonistic forces when simultaneously expressed. The immobilizing effect of fimbriae would limit the motility of flagella, and increased motility by flagella would reduce the ability of bacteria to adhere at one site, yet both motility and adherence contribute significantly to the development of UTI. Thus, it is logical that bacteria would have in place a mechanism to reciprocally control motility and adherence.
It has been shown previously in Proteus mirabilis, a UPEC-related Gram-negative enteric bacterium also capable of causing UTI, that MrpJ, a protein encoded by the MR/P fimbrial operon, inhibits both swarming and swimming motility (14). In UPEC, flagella are coordinately expressed during urinary tract infection (15). Recent studies in UPEC have demonstrated that PapX, a non-structural 183-amino acid protein of the highly conserved 17-kDa family and a functional homolog of MrpJ, which is encoded by the PAI-CFT073pheV but not PAI-CFT073pheU-associated pap operon, down-regulates motility by repressing the expression of flagella (16). The presumed mechanism of repression is the direct binding to the flhDC promoter, the master regulator of flagellar synthesis (17). However, the precise mechanism of repression and structure-function relationships within PapX are unknown.
To further our understanding of the mechanism of PapX-mediated repression of motility, we functionally characterized the PapX protein and used systematic evolution of ligands by exponential enrichment (SELEX) in conjunction with high throughput sequencing technology to identify a novel DNA binding site for PapX within the flhDC promoter. To the best of our knowledge, this is the first description of the use of SELEX in conjunction with high throughput sequencing technology in bacteria to study DNA-binding proteins and the first time these techniques have been used to identify a novel binding site instead of to confirm or refine known binding sites. Our application of SELEX with high throughput sequencing provides for novel advantages over previous techniques for identifying bacterial transcription factor binding sites and may serve as the paradigm for future high throughput screens. Because the 17-kDa family of genes is highly conserved, by elucidating the mechanisms underlying reciprocal control of motility by PapX in UPEC, we will be better equipped to manipulate and disrupt this important regulatory cascade and virulence property of a pathogenic microbe.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
For bacterial strains and plasmids used in this study, see supplemental Table 1. E. coli CFT073, a fully sequenced (18) strain of UPEC, was cultured from the blood and urine of a hospitalized patient with acute pyelonephritis and urosepsis (19). E. coli BL21(DE3) was used for overexpression of PapX-His6. All E. coli strains were cultured overnight at 37 °C in Luria broth with aeration or on Luria agar plates containing either ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), kanamycin (25 μg/ml), or no antibiotic. For Western blots, PapX was expressed from the isopropyl β-d-thiogalactopyranoside (IPTG)-inducible plasmid pPxWT in E. coli K12 strain MG1655 in the presence or absence of 300 μm IPTG. Growth curves were generated in triplicate using a Microbiology Reader Bioscreen C (Oy Growth Curves AB, Ltd.) in 0.2-ml volumes; A600 was recorded every 15 min for 24 h.
Motility Assay
Flagellum-mediated motility was measured as described previously (16). Briefly, soft Luria agar (0.25% (w/v)) plates containing ampicillin (100 μg/ml) were stabbed in the center using an inoculating needle with either Luria broth cultures normalized to A600 = 0.9–1.0 or colonies struck onto plates the day prior. Care was taken not to touch the bottom of the plate during inoculation to ensure that only swimming motility was assessed. Plates were incubated at 30 °C for 16 h, and swimming diameter was measured. Results were analyzed using a paired t test.
Construction of flhDC-lacZ Fusions and β-Galactosidase Assays
Nested deletions of the flhDC promoter were fused to lacZ using a promoterless lacZ-containing plasmid, pRS551, using a previously described methodology (20). Briefly, primers containing flanking BamHI or EcoRI overhangs were designed to amplify DNA fragments that were 130, 215, 360, 501, 597, and 718 bp upstream of the translational start site of flhD of E. coli CFT073 (supplemental Table 2). Fragments and the pRS551 vector were digested with EcoRI and BamHI and ligated to form constructs pRS551A to pRS551F (supplemental Table 1). Electrocompetent E. coli DH5α was electroporated with pRS551A to pRS551F, incubated for 1 h in 500–800 μl of Super Optimal broth with catabolite repression (SOC), and plated onto Luria agar plates containing kanamycin (25 μg/ml). Plasmids were extracted from KanR colonies using a Miniprep kit (Qiagen) and sequenced to confirm the proper fusion constructs. Plasmid constructs were transformed into E. coli CFT073 ΔpapXΔlacZ (CamR) or E. coli CFT073 ΔlacZ (CamR). β-Galactosidase activities of the constructs were measured using the modified Miller assay as described (21). Briefly, overnight cultures of each construct were diluted 1:100 in LB broth and incubated at 37 °C with aeration. A600 was measured after 2.5 h, and samples (5 μl) were added to 95 μl of 100 mm Na2HPO4, 20 mm KCl, 2 mm MgSO4, 0.8 mg/ml cetyltrimethylammonium bromide, 0.4 mg/ml sodium deoxycholate, 5.4 μl/ml β-mercaptoethanol to lyse bacterial cells. Lysates (100 μl) were mixed with 600 μl of 60 mm Na2HPO4, 40 mm NaH2PO4, 1 mg/ml ortho-nitrophenyl-β-galactoside, 2.7 μl/ml β-mercaptoethanol reaction buffer and incubated at 30 °C. Reactions were stopped after 5–25 min with 700 μl of 1 m sodium carbonate. Bacterial cells were pelleted by centrifugation (10,000 × g, 10 min, 23 °C), and A420 of the supernatant was measured. Miller units were computed using the formula, 1000 × (A420)/(A600 × 0.05 × time in min). Results were compared using a paired t test.
Construction of E. coli Mutants
Deletion mutants were constructed using the λ-red recombinase system (22). Briefly, flanking homologous primers were designed (supplemental Table 2) and used to generate amplicons from plasmid pKD3 or pKD4 to generate kanamycin or chloramphenicol resistance cassettes. Amplicons were treated with DpnI for 3 h at 37 °C to remove parental DNA and transformed into electrocompetent CFT073 bacteria expressing pKD46 induced in 10 mm (final concentration) arabinose. Resistance cassettes were removed by transforming resultant constructs with pCP20 and leaving cultures to incubate at 37 °C overnight.
Site-directed mutants were constructed in pPxWT using the QuikChange II kit (Stratagene) according to the manufacturer's instructions. Briefly, overlapping primers were designed to contain nucleotide substitutions and used for rolling PCR of pPxWT. PCR products were treated for 3 h with DpnI at 37 °C to remove parental DNA. Amplicons were transformed into electrocompetent CFT073 and recovered for 1 h in 500–800 μl of SOC, and then 200 μl was plated onto LB agar containing ampicillin (100 μg/ml). Resistant colonies were cultured overnight in LB broth at 37 °C, and plasmid DNA was extracted and sequenced as described above.
PapX Purification
Nucleotides encoding amino acids 13–183 of PapX were cloned into pMCSG7 in front of a His6-TEV cleavage site by ligation-independent cloning. pMCSG7 was transformed into E. coli BL21 (DE3). Bacteria were cultured in 500 ml of terrific broth in a 2-liter flask at 250 rpm at 37 °C to A600 ≅ 1.0 and then cooled to room temperature for 1 h. Bacteria were induced overnight with 300 μm IPTG. After induction, cells were centrifuged (13,000 × g, 20 min, 4 °C), and pellets were frozen at −80 °C. Frozen pellets were suspended in 20 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm DTT, 300 mm NaCl and ruptured by sonication using a blunt tip probe for seven cycles of 60% maximum energy for 30-s bursts with 1-min intervals to cool (GENEQ, 500-watt ultrasonic processor). The lysate was centrifuged (112,000 × g, 1 h, 4 °C), and the supernatant was passed through a 0.22-μm pore size filter (MillexGP, Millipore) and incubated for 1 h at 4 °C with 5–10 ml of Ni2+-NTA resin (Invitrogen). Resin was washed three times in 20 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm DTT, 300 mm NaCl, 20 mm imidazole, and protein was eluted with 20 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm DTT, 300 mm NaCl, 500 mm imidazole. Protein was concentrated to 2 mg/ml, and 50-μl aliquots were frozen at −80 °C. The presence of PapX was confirmed as the predominant species on a 12% acrylamide gel. Bands were analyzed using tandem mass spectroscopy (LC-MS/MS) by the Michigan State University Proteomics Core Facility.
Electrophoretic Motility Shift Assays
Gel shift assays were performed as described (16) with the following modifications. Recombinant PapX-His6, expressed from pMCGS7, was purified as described above and used for gel shift assays. The DNA fragment used was the −345 through −501 bp region upstream of the flhD translational start site in E. coli CFT073. Poly(dI-dC) and poly-l-lysine were substituted with tris-EDTA-sodium (TEN) buffer in the binding reaction. Competition was performed using four unlabeled fragments. Three fragments (TTACGGTGAGTTATTTTAACTGTGCGCAA, TTACGGTGAGTTATTTTAAC, and GTTATTTTAAC) were derived from the predicted PapX binding site from SELEX (see “Results”). The fourth fragment (ACTCCACTCACGGCCGTTTCGACGGTACCG) was derived from the gapA promoter, which has previously been demonstrated not to shift in the presence of PapX (16).
Genomic SELEX
A library of 80–100-bp sequences was generated from CFT073 genomic DNA and paired end-ligated as described (23) at the University of Michigan DNA Sequencing Core. PapX-His6 was purified as described above; however, before eluting bound protein, 200-μl samples of the resin were incubated with gentle agitation for 30 min at 37 °C in the presence of 150 ng of library DNA. The resin was washed with 20 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm DTT, 300 mm NaCl, 20 mm imidazole to remove unbound library fragments, and bound DNA-PapX complexes were eluted using the same buffer but containing 500 mm imidazole. DNA was isolated from the eluted complexes by two phenol/chloroform extractions. DNA was subjected to 12–15 rounds of PCR using primers specific to the ligated paired ends to ensure that no contaminating DNA was amplified. Libraries were quantified using quantitative PCR. The resultant library (150 ng) was run again over a fresh 200-μl sample of PapX-His6 bound to Ni2+-NTA resin, and the process was repeated four times. The unselected library, as well as enriched rounds two, three, and four, were sequenced using Illumina high throughput sequencing. A local BLAST comparison to the CFT073 genome was done for each short read per run, and the number of reads at each chromosomal position in CFT073 was recorded. Matlab (Student version 7.12.0) was used to identify peaks that had enriched competitively throughout the cycles of amplification to identify relevant peaks. 60-bp regions, centered on each relevant peak, were analyzed using the MEME suite (24) to generate the most likely motifs.
Protein Structural Predictions
PHYRE (version 0.2) (25) and I-TASSER (version 1.1) (26, 27) were employed to predict the most likely structure of the PapX protein. The model predicted by I-TASSER was rendered using the PyMOL Molecular Graphics System (version 1.3, Schrödinger, LLC). Recombinant purified PapX-His6 was also analyzed by size exclusion chromatography on a Sephacryl S-200 size exclusion column (GE Healthcare) and seminative gel electrophoresis in 12% SDS-polyacrylamide gel.
Antiserum Production
Antiserum to PapX was commercially generated in rabbits using a polyclonal antibody fast production protocol (Rockland Immunochemicals Custom Antibody Services) from a single-protein peak eluted from a Sephacryl S-200 size exclusion column (GE Healthcare) of nickel affinity-purified recombinant PapX-His6.
Transcriptional Profiling
Quantitative real-time PCR was performed as described (16). Briefly, total RNA, extracted from 200 μl of bacterial culture, was reverse-transcribed to generate cDNA and analyzed using SYBR Green and primers for 100-bp fragments of genes of interest (supplemental Table 2). gapA was used as a normalizer for all conditions.
DNase Footprinting
Primers, end-labeled with 6-carboxyfluorescein, were used to generate fluorescent probes from the first 130-, 215-, 360-, and 500-bp fragments of the flhDC promoter as well as a 300-bp fragment of the gapA promoter. Fragments were incubated with purified recombinant PapX-His6 and exposed to low levels of DNase I for 1–10 min. Sheared fragments were processed by electrophoresis (as for DNA sequencing) to produce chromatograms representing all sheared fragments and examined for stretches of troughs between peaks.
Multiplex PCR
Multiplex PCR was performed using methods and a 294-member subset of a 315-member strain collection as described (28, 29). Primers were designed to unique flanking sequences of papX and focX and common regions of both (supplemental Table 2).
Seminative Gel Electrophoresis
Non-reducing/seminative gel electrophoresis was performed as described previously (30). Briefly, protein was isolated as described above. SDS-PAGE loading buffer lacking dithiothreitol was added to samples, which were electrophoresed in the presence of 0.1% SDS on a 12% acrylamide gel (3.75% stacking gel) at 200 V for 1 h at room temperature. Gels were stained using Coomassie Brilliant Blue R-250 dissolved in a buffer composed of 10% acetic acid, 40% methanol, and 50% water. Gels were destained in a buffer composed of 10% acetic acid, 40% methanol, and 50% water.
RESULTS
PapX Has Homologs in Extraintestinal Pathogenic E. coli
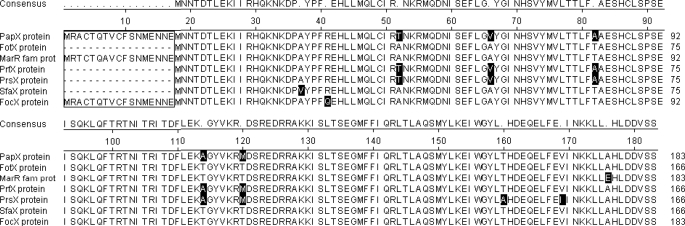
To assess the distribution and conservation of PapX, we compared the amino acid sequence of PapX homologs using ClustalW. PapX has six closely related homologs of the 17-kDa protein family among strains of UPEC and meningitis-associated E. coli, with over 93% amino acid identity between them (Fig. 1). Three of the 10 non-identical residues are substitutions between Ala and Val or Ile and Val. Four non-identical sites contain either Ala or Thr. These features result in pairs of PapX homologs sharing up to 99% amino acid sequence identity.
FIGURE 1.
Alignment of PapX homologs in the 17-kDa family. PapX homologs of the 17-kDa protein family were aligned using ClustalW (DNA star, Lasergene suite, Megalign, version 8.0.2(13), 412). Perfect consensus is shown above. Positions with imperfect amino acid identity between homologs are shown as dots. Amino acid residues that represented the minority for that position are shaded in black within each homolog's amino acid sequence. The seven family members are ≥93% identical to one another at the amino acid level past residue 17. The N-terminal start site annotation in homologs (boxed) is the result of in silico analysis and has not been experimentally verified.
Prevalence of PapX Homologs among UPEC
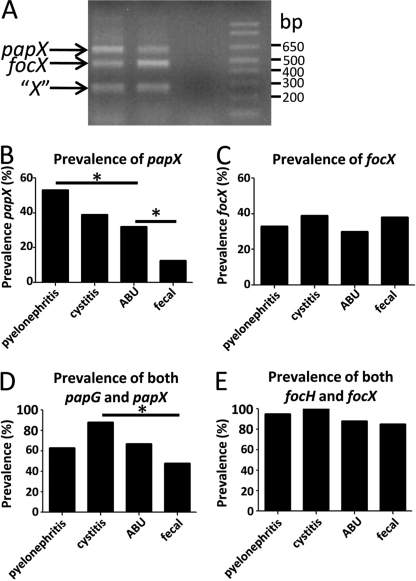
To assess the prevalence of papX in UPEC strains, a collection of 294 E. coli strains representing a range of isolates from fecal/commensal strains (n = 88) to strains isolated from clinical cases of asymptomatic bacteriuria (n = 54), complicated UTI (n = 39), uncomplicated cystitis (n = 37), and acute pyelonephritis (n = 76) was examined by multiplex PCR for the presence of papX homologs. Primers were designed to unique flanking sequences of papX and focX and to common regions of both to amplify fragments of a specific size (Fig. 2A, supplemental Table 2). Within this collection, 53% of pyelonephritogenic strains of E. coli contained at least one copy of papX, the highest prevalence within the strain collection and significantly more prevalent than the 32% of asymptomatic bacteriuria strains (p = 0.0192). Only 12.5% of fecal/commensal E. coli contained at least one copy of papX, the lowest prevalence within the strain collection and significantly less prevalent than in asymptomatic bacteriuria strains (p = 0.0081) (Fig. 2B). focX, a close homolog of papX, did not vary significantly in prevalence within the UPEC strain collection (p = 0.7927) (Fig. 2C). This was not surprising because focX is closely associated with genes encoding F1C fimbria, which appears more closely associated with meningitis-associated strains than with UTI-associated strains (31).
FIGURE 2.
Multiplex PCR to analyze a 294-member library of UPEC and fecal/commensal isolates for the presence of papX, focX, and nonspecific 17-kDa family genes. A, three primer sets were designed to amplify a 607-bp fragment of papX and flanking region, a 449-bp fragment of focX and flanking region, and a 249-bp region common to all papX homologs. All primer sets shared a common reverse primer (supplemental Table 2, papXMltplxRev) with a unique forward primer (supplemental Table 2, papXMltPlxfwd, focXMltPlxfwd, and XMltPlxfwd). papX and focX contain nearly identical nucleotide sequence, so unique primers for each originated in the sequence flanking each gene. Fragments were sequenced to confirm their composition. Each fragment was generated from extracted, pooled CFT073 WT genomic DNA and separated on 2% agarose gel. The prevalence of papX (B) and focX (C) within the library was scored by the appearance of their respective amplicon during multiplex PCR within each strain type. The percentage of papX-containing (D) or focX-containing (E) strains within papG- or focH-containing strains, respectively, is shown.
The association of the pap operon, which encodes P-fimbriae, with pyelonephritogenic strains of UPEC is well documented, with 77% of pyelonephritogenic isolates containing P fimbriae, as compared with only 23% of cystitis, 20% of asymptomatic bacteriuria, and 16% of fecal/commensal isolates (8, 9). 59% of all strains tested carrying at least one copy of papG also carried at least one copy of papX. Therefore, to differentiate the influence of the increased representation of the pap operon in UTI-causing UPEC from the role of papX alone, we quantified the prevalence of papX in strains containing pap operons for each type of strain within the collection, using the prevalence of papG as a proxy for the prevalence of the pap operon (29).
Within strains that contained both the pap operon and papX, the relative prevalence of papX compared with papG was significantly higher in cystitis-causing strains of E. coli as compared with fecal/commensal strains (p = 0.0165) (Fig. 2D). The prevalence of strains containing both focX and focH, a proxy for the presence of F1C fimbriae, was not significantly different between UTI-causing strains of E. coli and fecal/commensal strains of E. coli (p = 0.2222) (Fig. 2E). These data indicate that the presence of papX, but not focX, is strongly associated with UPEC. Taken together with the highly conserved nature of the protein sequence between PapX homologs, these data suggest that PapX is a contributor to UPEC virulence but that encoding papX alone is not sufficient to convey virulence to E. coli strains.
Functional Characterization of PapX
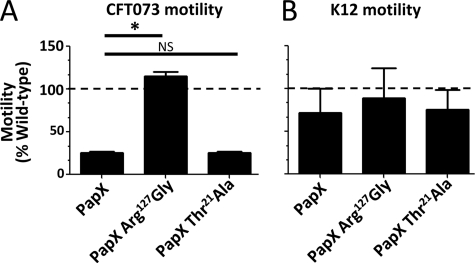
We previously demonstrated that PapX down-regulates motility (16, 32). In strain CFT073, deletion of the single copy of papX, which resides on the PAI-CFT073pheV- but not PAI-CFT073pheU-associated pap operons (16), results in increased motility as compared with wild type. On the contrary, overexpressing papX from the leaky, inducible plasmid pPxWT repressed motility. papX overexpression in CFT073 (pPxWT) has a specific transcriptional effect, with 38 of 42 differentially expressed genes being associated with motility and the downstream effects of flhD and flhC regulation, the master regulator of motility (16). Coupling this finding with the finding that papX appears to be strongly associated with uropathogens, we introduced papX, carried on pPxWT, into strain CFT073 and the prototypical non-pathogenic fecal/commensal E. coli K12 strain MG1655. Motility assays, performed on the resulting transformants, demonstrated that CFT073 had reduced motility in the presence of papX when compared with vector control (Fig. 3A) but that papX had no effect on the motility of the non-pathogenic K12 strain MG1655 (Fig. 3B). This informed us that the role of papX is pathogen-specific.
FIGURE 3.
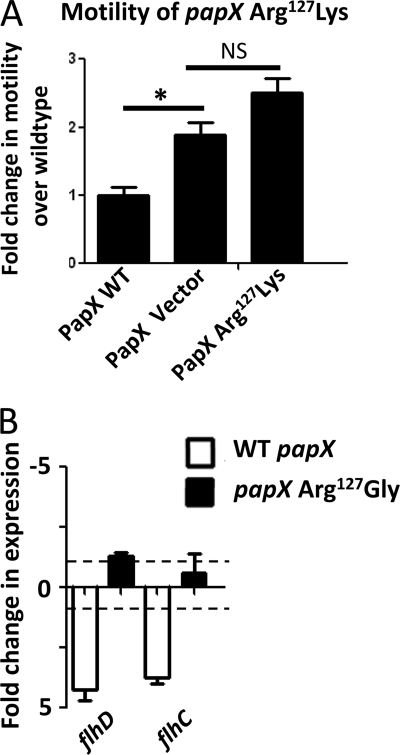
Effect of papX and papX R127G mutant on motility of UPEC and a non-pathogenic E. coli. E. coli CFT073 and K12 MG1655 were transformed with pPxWT (WT papX), pR127G (papX R127G mutant) or pT21A (papX T21A mutant). Constructs were then stabbed into soft agar plates and incubated at 30 °C. The diameter of swimming motility was recorded after 16 h. For each construct, assays were performed on four biological replicates in triplicate. A, compared with vector control (dashed line) in CFT073, pPxWT and pT21A reduced motility significantly, but pR127G did not. B, none of the three constructs influenced motility significantly differently from vector control in K12 MG1655. Error bars, S.E.
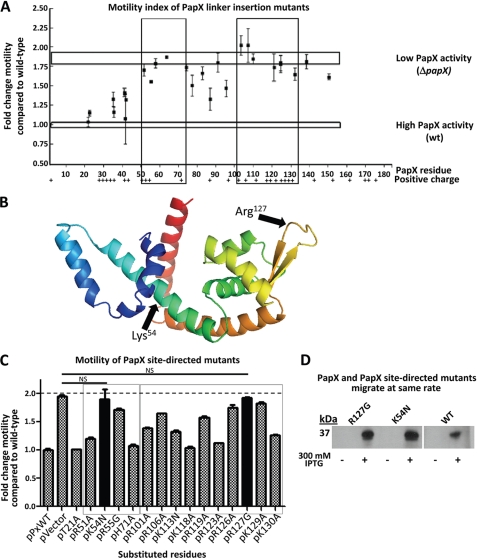
To investigate structure-function relationships within PapX, we constructed a library of linker insertion mutations within papX in pPxWT. 15-nucleotide insertions were randomly introduced along the coding sequence of papX. 30 unique in-frame insertions were generated, covering amino acid residues 15–162 of PapX. Each of the 30 insertional mutants was electroporated into CFT073 ΔpapX. The growth rates of the transformants were not significantly different from CFT073 ΔpapX complemented with either wild-type papX or vector control (supplemental Fig. 1). Motility was measured for each construct in soft agar, side-by-side with motility assays of CFT073 ΔpapX complemented with either pPxWT (which encodes papX) or pVector (vector control) (Fig. 4A). We found two regions of papX (residues 51–74 and 103–133) that, when mutated, rendered PapX unable to repress motility (Fig. 4A, boxes). Insertions into either of these regions abolished PapX activity, resulting in motility similar to that observed for the CFT073 ΔpapX construct containing vector alone. These results suggested that these two regions were critical for PapX function.
FIGURE 4.
Motility index of 15-bp insertional mutants of papX. A, 30 in-frame 15-bp insertions were made in papX in pPxWT. The 30 constructs, pPxWT (WT papX), and pVector (empty vector) were electroporated into CFT073 ΔpapX. Motility was assessed in soft agar and compared relative to CFT073 ΔpapX electroporated with pPxWT. Vertical boxes indicate regions of loss of function. Top and bottom horizontal boxes represent motility of CFT073 ΔpapX electroporated with pVector or with pPxWT, respectively. The presence of positively charged residues is indicated along the bottom. B, I-TASSER software was used to generate a predicted structural model based on the amino acid sequence of PapX. Arrows indicate putative DNA binding (Arg127) and dimerization (Lys54) regions of the protein. C, site-directed mutants of papX were made in pPxWT and transformed into a CFT073 ΔpapX background. Soft agar motility assays were performed on the resulting constructs, performed in triplicate using biological triplicates. Boxes indicate regions sensitive to mutation determined in A by linker-insertional mutagenesis. Solid bars are not significantly different from vector control (dashed line). D, Western blot showing pPxWT (WT PapX), pR127G (PapX R127G mutant), and pK54N (PapX K54N mutant) transformed into E. coli strain K12 either uninduced or induced with 300 mm IPTG. Bands migrate with the same electrophoretic mobility for mutants and wild-type PapX by seminative SDS-PAGE (12% acrylamide). Error bars, S.E.
We reasoned that structural modeling would provide insight into the structure and function of this DNA-binding protein. Indeed, structural models of PapX generated using I-TASSER (26, 27) and PHYRE (25) suggested that PapX is likely to be a member of the MarR family of helix-wing-helix homodimeric transcriptional regulators (Fig. 4B). Size exclusion chromatography and seminative SDS-PAGE confirmed that PapX is a dimer (supplemental Fig. 2). The MarR family of transcription factors tends to rely on specific positively charged residues, often Arg, to mediate DNA binding interactions (33–37) and dimerization. When the predicted structure is compared with existing MarR family proteins (33, 34), the two regions of PapX sensitive to insertional mutagenesis (amino acid residues 51–74 and 103–133) were predicted to occur in dimerization and DNA-binding domains, respectively (Fig. 4B, arrows).
To investigate the role of the two regions in PapX highlighted by insertional mutagenesis, site-directed mutants were constructed, in which positively charged residues were substituted with non-positively charged residues in pPxWT (supplemental Table 1). Site-directed mutant constructs were transformed into CFT073 ΔpapX, and motility assays were performed for the resulting transformants (Fig. 4C). We found that two mutants, K54N and R127G, abolished PapX activity, producing the same motility phenotype as vector control (solid bars). We confirmed that each of these mutants produced PapX of the predicted apparent molecular weight as assessed by Western blot (Fig. 4D) by overexpressing each construct from an inducible vector in the K12 background, which lacks a native papX or papX homolog. These results indicated that either of these single amino acid substitutions alone is capable of ablating PapX activity, suggesting that Lys54 and Arg127 are key residues necessary for PapX function.
By homology modeling, Lys54 and Arg127 appear to occur in the predicted dimerization domain and DNA binding domain of PapX, respectively. In MarR family proteins, often an Arg in the wing of the winged helix domain is implicated in DNA binding (33–36, 38). Our alignment suggested that this conserved Arg corresponds to Arg127 in PapX. To further investigate the Arg127 mutant, we used site-directed mutagenesis to construct an R127K mutant containing a different positively charged residue. Surprisingly, the R127K mutant also disrupted PapX function, as assessed by motility assay (Fig. 5A), suggesting that Arg127 is a key residue in PapX that is characteristic of the MarR family's DNA-binding protein fold (33, 34) and that positive charge at position 127 alone is not sufficient for activity.
FIGURE 5.
PapX R127G mediates motility and flagellar transcription. A, CFT073 ΔpapX was complemented with pVector, pPxWT (WT papX), or pR127K (PapX R127K mutant), and motility was assayed for each construct using soft agar motility assays. Motility of CFT073 ΔpapX electroporated with either pVector or pR127K was not significantly different from one another (NS, p > 0.05), and both were significantly more motile (*, p < 0.05) than CFT073 ΔpapX electroporated with pPxWT. B, pR127G (PapX R127G mutant), pPxWT (WT papX), and pVector (empty vector) were electroporated into CFT073 ΔpapX. RNA was isolated and analyzed using quantitative PCR, using gapA as the normalizer. Bars indicate -fold change in expression of flhD and flhC compared with CFT073 ΔpapX electroporated with vector control. Dashed lines indicate 1- and −1-fold (no differential change in expression). Compared with the presence of pVector, both flhD and flhC transcript levels were decreased in the presence of pPxWT, but levels were unchanged in the presence of pR127G. Error bars, S.E.
To better understand the motility phenotype of the Arg127 mutant, we compared the transcriptional profile of our R127G mutant expressed from pR127G, wild-type PapX expressed from pPxWT, and vector control expressed from pVector in CFT073 ΔpapX by quantitative real-time PCR (Fig. 5B). Expression of flhD and flhC in the presence of the pR127G mutant was not significantly different from vector control, whereas pPxWT significantly reduced transcription of flhD and flhC as well as downstream genes (p = 0.0091 and p = 0.0469, respectively) (Fig. 5B). Taken together, these results suggest that Arg127 is a key residue required for full PapX function and plays a residue-specific role in the repression of motility by regulating expression of flhD and flhC.
PapX Binds to a Specific DNA Motif within the flhDC Promoter
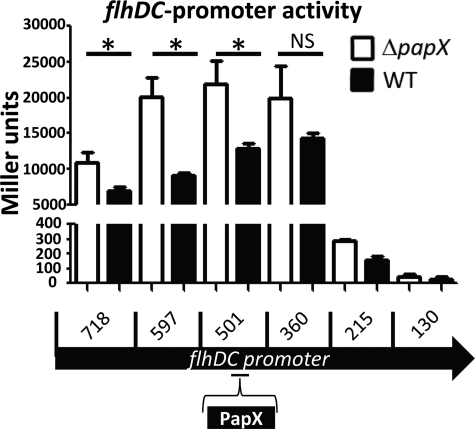
Transcriptional data assessed by microarray, quantitative real-time PCR, and homology modeling suggested that PapX is a DNA-binding protein that acts directly on the flhDC promoter (16) (Figs. 4B and 5B). As assessed previously by electrophoretic mobility shift assay (EMSA), PapX interacts with the 500 bases upstream of the flhD translational start site (16). However, we had no data regarding sequence specificity of the target. To validate these results and to hone in on the region of the flhDC promoter recognized by PapX, we constructed nested deletions of the flhDC promoter and fused them to a promoterless lacZ in pRS551. Fusion constructs were transformed into CFT073 ΔlacZΔpapX and CFT073 ΔlacZ. β-Galactosidase activity was measured using a modified Miller assay as a surrogate for promoter activity (21) (Fig. 6). A significant PapX-dependent difference in β-galactosidase activity was observed in all fragments containing at least 501 bases of the flhDC promoter (p < 0.025) but not in the fragment containing ≤360 bases of the flhDC promoter (p > 0.13). This suggests that PapX exerts its effect between −360 and −501 bases upstream of the flhDC start site. Additionally, DNase protection and EMSAs on the first 360 bases of the flhDC promoter failed to demonstrate sequence-specific effects (supplemental Fig. 3). Although MarR family members often act near the −10 and −35 sequences of promoters to sterically regulate gene expression, this is not always the case (37). For example, it has also been shown previously in EHEC, a relative of UPEC, that the flhDC promoter has a drastically reduced ability to drive gene expression when fewer than 300 bases of the 3′-end of the promoter are present (20), as is the case with our fusions. This suggests that positive factors exist upstream of 300 bases that could be antagonized by a suppressor of flhDC transcription, such as PapX.
FIGURE 6.
PapX repression of flhDC-lacZ promoter fusions. Nested deletions of the flhDC promoter were fused to promoterless lacZ in pRS551. Deletions contained progressively less of the 5′-end of the promoter. pRS551-flhDC promoter constructs were transformed into CFT073 WT (black bars) or CFT073 ΔpapX (white bars). No significant difference (NS, p > 0.05) was observed in β-galactosidase activity as assessed by a Miller assay between CFT073 WT and CFT073 ΔpapX using a one-tailed t test when only the first 360 bases of the flhDC promoter was included. Significant differences (*, p < 0.025) were observed when 501 bases or more of the flhDC promoter was included in the fusion construct. Error bars, S.E.
PapX Binds to a Short Motif in the CFT073 Chromosome
To determine the exact binding site of PapX, we used SELEX methodology, an unbiased whole-genome approach (39), in conjunction with high throughput sequencing technology. This combined two robust methods for identifying subtle but specific, binding motifs (40). To identify such a motif, wild-type CFT073 genomic DNA was sheared to generate 80–100-bp fragments. The resulting pool represented every possible 80–100-bp fragment from the CFT073 genome (confirmed after construction by high throughput sequencing), or roughly 5 × 106 unique members. Adaptors of ∼50 bp in length were paired end-ligated onto each fragment to complete library synthesis.
Recombinant PapX-His6 was purified and analyzed by mass spectroscopy to confirm that it was the predominant species after purification (Table 1). PapX-His6 was then bound to Ni2+-NTA resin, and a sample (200 μl) of resin-bound PapX-His6 was incubated with library DNA (150 ng, 30 min, 37 °C). Unbound DNA was removed with three washes in 20 mm imidazole column buffer; bound PapX-DNA complexes were eluted using 500 mm imidazole elution buffer. DNA, isolated from the eluted PapX-DNA complexes by phenol/chloroform extraction, was PCR-amplified using primers specific for the paired end adapters to ensure that only the library, and not contaminating DNA, was amplified. The enriched library DNA was quantified with real-time PCR using primers specific to the adapters (but internal to the amplification primers) and then applied to a new Ni2+-NTA column containing bound PapX-His6. The entire process of binding the library to Ni2+-NTA-bound PapX-His6, amplifying bound members, and reapplying the enriched library to Ni2+-NTA-bound PapX-His6 was repeated for four iterations. The amounts of PapX-His6 and DNA used were empirically determined to be the lowest quantities that still allowed for low cycle (i.e. unbiased) amplification (23) of each cycle's DNA output to regenerate sufficient template for the next cycle of SELEX.
TABLE 1.
Recombinant PapX purity by LC-MS/MS
| Protein | Ontology | emPAIa | Coverage |
|---|---|---|---|
| % | |||
| PapX | Transcriptional regulator | 28.21 | 60 |
| l-Glutamine:d-fructose-6-phosphate aminotransferase | Metabolism | 0.69 | 44 |
| cAMP receptor protein | Transcriptional regulator | 2.27 | 53 |
| FKBP-type peptidyl-prolyl cis-trans-isomerase | Protein folding | 0.55 | 17 |
| Aconitate hydrase B | Metabolism | 0.03 | 1 |
| 30 S ribosomal subunit S3 | Translation | 0.13 | 5 |
a Exponentially modified protein abundance index, an estimate of the prevalence of a species in a sample analyzed by LC-MS/MS.
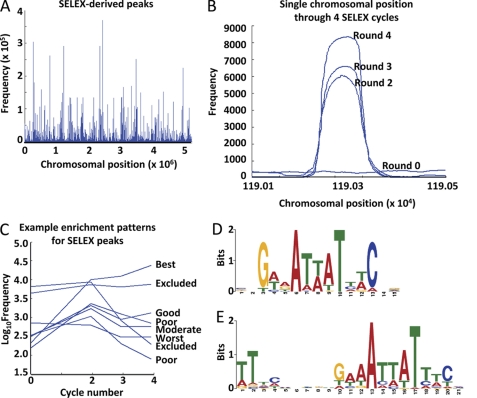
The initial CFT073 sheared genome library, as well as the second, third, and fourth libraries enriched by serial rounds of SELEX, were sequenced using a high throughput Illumina sequencing. On average, 24 ± 2.8 × 106 80-bp reads were obtained for each SELEX round, representing 400-fold coverage of the CFT073 genome. Sequences for each round were aligned to the CFT073 genome using a local BLAST (Fig. 7A), showing a clear pattern of site-specific enrichment. A dot plot of the frequencies of hits at each position in the chromosome is shown for a single representative peak across all sequenced rounds of SELEX (Fig. 7B). The width of peaks (160–200 bp) corresponds to roughly twice the size of an average member of the sheared genomic library, suggesting that PapX-His6 interacts with the CFT073 genome in a strongly sequence-specific manner. Peak positions generated from the library of the final round of SELEX were overlaid onto each sequenced round of enrichment, and the frequency of occurrence at that chromosomal position was plotted against the round of enrichment (Fig. 7C). Peaks were graded based on their rate of enrichment across rounds (Table 2), with heavy weight given to positions that enriched under the most stringent environment (i.e. the final rounds of enrichment), and discarded if they were too heavily represented in the control library at greater than 2 S.D. values above the mean positional frequency across the entire genome of the unselected initial library. Sufficient selective enrichment occurred, as indicated by the background representation of library members dropping to zero between peaks (Fig. 7B). These data were sufficiently rich to generate bell-shaped curves of predicted width (160–200 bp) and appropriate distribution from dot plots, indicating that the protocol was successful at amplifying only regions that contained short, PapX-specific binding sequences. Of the sequences generated from the fourth round of SELEX, frequencies at 94% of the chromosome were below the mean of the unselected library, and the frequency at 64% of chromosomal positions was 0. This demonstrates that the majority of the chromosome was successfully counterselected and did not interact with PapX as assessed by SELEX.
FIGURE 7.
Analysis of SELEX Illumina sequencing results. A, the fourth enrichment of SELEX was sequenced, and results were aligned to the genomic sequence of strain CFT073 using a local BLAST. Frequencies of a sequence read at every chromosomal position were plotted using a dot plot. B, dot plot of frequencies across SELEX rounds zero, two, three, and four in the chromosomal vicinity of a prototypical low frequency best grade peak. C, representative example of chromosomal peak positions and their frequencies plotted against SELEX rounds zero, two, three, and four. Grades of peaks are based on their pattern of frequencies across rounds and appear to the right (defined in the footnotes to Table 2). The MEME suite of sequence analysis tools was employed to determine if motifs were present in the list of 366 best grade peaks, generating two significant motifs. D, position weight matrix present in 362 of the 366 best grade peaks, E value 4.5 × 10−142. E, position weight matrix present in 269 of the 366 best grade peaks, E value 4.7 × 10−140.
TABLE 2.
Positional weight matrix motif derivative prevalence in graded peaks and randomly selected sequences
| Motif | Derivatives of GWWAWWWTWWC |
||||||
|---|---|---|---|---|---|---|---|
| All peaksa | Bestb | Goodc | Moderated | Poore | Worstf | Controlg | |
| % | % | % | % | % | % | % | |
| AAAAT | 50 | 62 | 49 | 41 | 49 | 46 | 18 |
| AAATT | 34 | 48 | 34 | 23 | 34 | 23 | 11 |
| AAATTT | 7 | 10 | 6 | 3 | 7 | 7 | 2 |
| AATTTC | 12 | 22 | 12 | 5 | 11 | 7 | 3 |
| ATTTC | 35 | 52 | 39 | 32 | 32 | 23 | 12 |
| TTATT | 47 | 69 | 48 | 38 | 45 | 30 | 12 |
a 4,407 peaks.
b “Best” quality peaks are the 366 peaks that increased in frequency in each round of SELEX.
c “Good” quality peaks are the 932 peaks that increased in frequency in the final round of SELEX and had a fourth round frequency greater than 2,800.
d “Moderate” quality peaks are the 269 peaks that increased in frequency in the final round of SELEX and had a fourth round frequency less than 2,800.
e “Poor” quality peaks are the 2,827 peaks that increased in frequency only in the first round of SELEX.
f “Worst” quality peaks are the 13 peaks that decreased in frequency in the first round of SELEX.
g Control peaks are six pooled groups of 366 randomly selected positions in the chromosome of strain CFT073.
Of 4407 total peaks in the final round of enrichment, 366 satisfied the “best” criteria for peaks (Table 2). Sequence flanking each best peak by 30 bases both up- and downstream in the chromosome was analyzed using the online MEME suite of tools (24). 362 of the 366 (99%) best peaks contained a single core motif represented by positional weight matrix (Fig. 7D), with an E value of 4.1 × 10−142. (E value is an estimate of the expected number of a particular motif one would find in a similar, random set of sequences; thus, an E value of 4.1 × 10−142 indicates a 4.1 × 10−142 probability that the motif arose by chance.) A second, modified version of this motif was found present in 269 of the 366 (73%) best peak sequences and represented by positional weight matrix with an E value of 4.7 × 10−140 (Fig. 7E). Because motifs were calculated presuming positional independence, palindromes were more difficult to identify using the bits scoring system. Within the 269 sequences, the second motif appeared as a palindrome, described using the International Union of Pure and Applied Chemistry (IUPAC) nomenclature as TT(7n)GWWAWWWTWWC(7n)AA (where W is A or T, and n is A, T, G, or C). Database analysis using Microsoft Access verified that the palindromic form of the second, modified core motif appeared significantly more often than if by chance within the 269 sequences in which the second motif was present (Table 2). Altogether, these represented the motifs with the lowest E values within the set; the next lowest value motif was represented by fewer than four members with E values greater than 10−5.
To verify that these motifs were not emerging at random, two methods of validation were employed. The input sequences were shuffled randomly and analyzed for motifs, which produced no motifs with E values lower than 1,000 (i.e. motifs generated from the shuffled set were very likely to have arisen by chance). Additionally, six groups of 366 random peaks were generated using Matlab and analyzed in the same manner as the best peaks. The rates of the predicted motif (Fig. 7D) and several derivatives were significantly less common in the random peaks using Microsoft Access (Table 2), and MEME could detect no significantly represented motifs from the random peak sets. Together, these data suggested that PapX binds to the CFT073 chromosome in a sequence-dependent manner at a motif 11–29 bases long.
PapX Interacts with a Short Sequence within the flhDC Promoter of CFT073
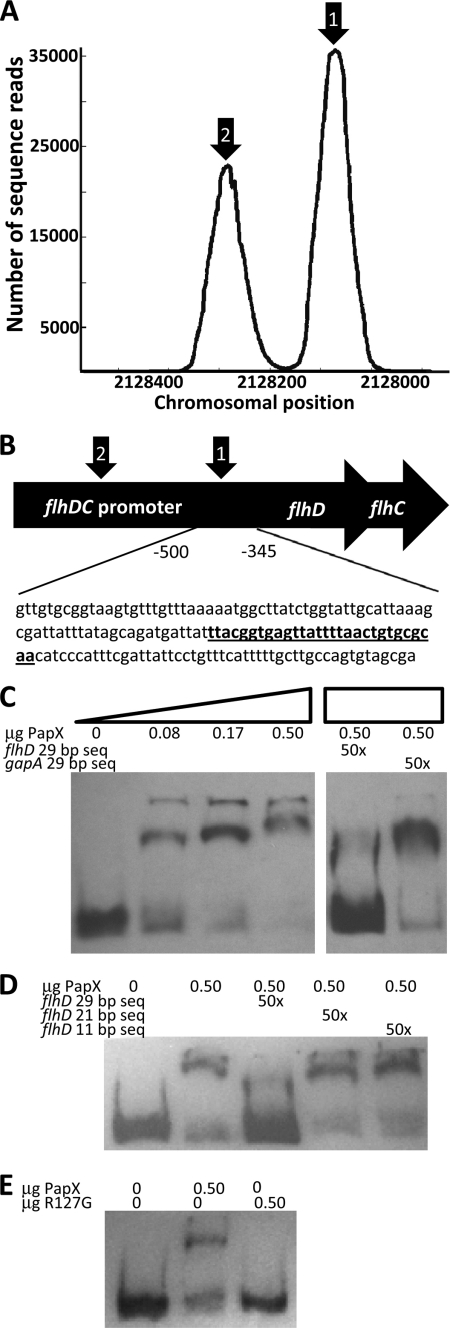
Two best quality peaks appear within the flhDC promoter (Fig. 8A). The larger of the two peaks, centered 410 bp upstream of the flhDC translational start site, centers around a perfect match to both predicted motifs, matching 11 of 11 and 29 of 29 bases (Fig. 8B). The second, smaller peak at −624 bases upstream of the flhDC translational start site is a degenerate version of the motif, matching only 9 of 11 and 25 of 29 bases. To validate our findings, we used EMSA to shift the 154-bp fragment, −501 to −346 bp upstream of the flhD translational start site, containing the best version of the predicted motif (Fig. 8C). The motility of the fragment through a 5% acrylamide gel was retarded in the presence of recombinant PapX-His6. This shift was dose-dependent with increasing concentrations of PapX. In a competition assay, we competed the DNA-binding reaction with unlabeled short sequences. A 29-bp sequence (Fig. 8B, boldface type, underlined) found within the 154-bp target fragment (representing a 29 of 29 nucleotide match with the second, modified core motif (TT(7n)GWWAWWWTWWC(7n)AA)) was added in 50-fold excess by mass (250-fold by molarity) to the DNA binding reaction between PapX and the 154-bp fragment and was able to compete away the shift. A second 29-bp fragment derived from gapA was unable to reverse the shift under the same conditions (Fig. 8C). The 29-bp fragment found in the flhDC promoter appears only once in the E. coli strain CFT073 genome, and no similar sequences appear in the TRANSFAC data base by TfBlast.
FIGURE 8.
PapX binds to the flhDC promoter. A, dot plot of chromosomal frequencies by SELEX revealed two significant peaks within the flhDC promoter (arrows 1 and 2). The peak under arrow 1 includes the predicted PapX binding site. B, schematic of the flhDC promoter, indicating the region −346 to −501 bases upstream of the flhD translational start used in subsequent gel shifts. A perfectly identical (29 of 29) predicted PapX binding site is indicated in underlined boldface type and is centered 410 bp upstream of the flhD translational start site. C, increasing amounts of PapX protein were added to 3.2 ng of DIG-UTP-end-labeled 154-base pair fragments derived from the flhDC promoter, the sequence shown in B. Unlabeled competitor DNA at 50-fold excess by mass was added to the protein-DNA binding reaction for each of the last two lanes. flhD 29 bp seq, sequence indicated in underlined boldface type in B. gapA 29 bp seq, sequence ACTCCACTCACGGCCGTTTCGACGGTACCG from gapA. D, the competition assay performed in C was repeated using only the 0.50-μg amount of PapX-His6 protein. Additional sequences derived from the 29-bp competitor (TTACGGTGAGTTATTTTAAC, indicated by flhD 21 bp seq, and GTTATTTTAAC, indicated by flhD 11 bp seq), determined by sequence analysis in Fig. 7, D and E, were also used in 50-fold excess by mass. E, gel shifts were repeated with the same 154-bp fragment as in C and with 0.50-μg amounts of wild-type PapX-His6 protein and PapX-His6 containing the R127G mutation.
We also tested the ability of the motifs predicted by SELEX (Fig. 7, D and E) to bind PapX using the same competition assay approach. Oligonucleotides matching the 11-bp sequence and 21-bp sequence were assayed for their ability to compete with DNA binding to the 154-bp flhD promoter fragment. All competing fragments were added in 50-fold excess by mass. Results indicated that only the 29-bp fragment was able to bind PapX, demonstrated by its unique ability among competitor DNA fragments to reverse the shift observed with wild-type recombinant purified PapX and the 154-bp flhD promoter fragment (Fig. 8D).
As mentioned earlier, structural modeling predictions based on I-TASSER and alignments suggested that Arg127 in PapX could represent a key residue required for DNA binding (33–36, 38). To test this hypothesis, recombinant purified PapX-His6 containing the R127G point mutation was used in an EMSA against the 154-bp fragment of the flhD promoter (Fig. 8E). Wild-type recombinant PapX-His6 was able to shift the fragment, indicating binding, whereas the binding of the R127G mutant was ablated (Fig. 8E).
Taken together, these results suggest that PapX binds to the flhDC promoter in a sequence-specific manner at a novel, unique motif predicted by SELEX, at a position consistent with the findings of our flhDC promoter fusion experiment.
DISCUSSION
Previously, we have shown that PapX regulates motility via transcriptional regulation of flhD and flhC in UPEC (16). The current study expands upon this finding, defining the mechanism by which PapX, a 43-kDa non-structural dimeric protein encoded by an operon that encodes P fimbria, mediates its effect. Our structural and functional results indicate that PapX is a dimeric, helix-turn-helix DNA-binding protein of the 17-kDa family, a MarR homolog that relies on Lys54 and Arg127 for full activity. Our novel use of SELEX in conjunction with high throughput sequencing in bacteria indicates that PapX exerts its effect by binding to a unique, specific, previously unknown sequence, TTACGGTGAGTTATTTTAACTGTGCGCAA, within the flhDC promoter, centered at a position 410 bp upstream of the flhD translational start site. Because PapX homologs are so closely related (>93% amino acid sequence identity), these findings have implications across a broad range of extraintestinal pathogenic E. coli strains and their likely regulation of motility and adherence.
Novel Application of SELEX with High Throughput Sequencing in Bacteria
When SELEX experiments were first performed, the limitations of analysis on the final, most enriched library (frequently involving subcloning and sequencing of fewer than 50 members of the library) (see Ref. 41 and references therein) typically meant that SELEX had to be performed so that the vast majority of the final library consisted of a single or very few species. However, doing so runs the risk of eliminating physiologically relevant but less strongly binding DNA targets. Using high throughput sequencing with SELEX, it is possible to identify the cycle at which all non-binding species have been selected against (when the background frequency drops below that of the initial library) and to halt iterative cycles before potentially biologically important species have been eliminated from the pool. In this way, SELEX combined with high throughput sequencing permits the accumulation of many more potential target species than SELEX as it has been classically performed, enriching the final data set and providing a potentially far more accurate picture of protein-DNA interaction.
Our modification of SELEX also included several unique improvements over past incarnations. Instead of using an aptameric library (39), our library was constructed directly from sheared CFT073 genomic DNA, generating paired end-ligated 80–100-bp fragments. A typical SELEX aptameric library contains 1013 to 1015 members of 10–20 nucleotides in length (42), whereas our library contained only 5 × 106 unique members (and 106 of each unique member per cycle of SELEX), each 80–100 bp in length, resulting in a high stringency selection throughout. Because we had substantial evidence that PapX exerts its effect on genes within the CFT073 genome (16) (Fig. 3), the best binding site was, of course, likely to be present in this library. Additionally, because aptameric libraries are usually generated at random and aptamers may be shorter than a complete binding site, a problem that SELEX encounters is questionable physiologic relevance of a determined motif (43). Therefore, a maximally bound motif may not actually be present in any organism. Our approach circumvented this problem. In addition, some physiologically relevant consensus sequences rarely appear as perfect matches; for example, the Shine-Dalgarno sequence, a ubiquitous ribosomal binding site found in E. coli, frequently appears as an imperfect 4 of 6 or 5 of 6 match yet still remains functional. In fact, varying the degree to which a given Shine-Dalgarno site matches the consensus is a means of regulating translation (44). Indeed, any positive hits from our library are, by definition, present in the CFT073 genome and are thus more likely to be physiologically relevant.
Additionally, genomic SELEX shares many of the advantages of ChIP-seq in terms of data analysis; in fact, the approach is quite similar (40), although SELEX does not require the generation of antibodies or the use of complex alignment parsing algorithms, such as MACS (45). We were able to analyze peaks because, as the 80–100-bp fragments overlap less and less over PapX binding sites, bell-curved shaped drop-offs in binding affinity can be seen, as represented by chromosomal positional frequency declining at sites that include only few or none of the maximal binding site sequence. Analysis of these curves produced by the alignment of hits using local BLAST allows for more precise motif analysis than using Hamming distances in aptameric libraries, which of necessity rely on similarity of aptamers rather than their position in a genomic context. Another advantage of our approach, because we use 80–100-bp probes, is that if PapX frequently binds in the proximity of other associated chromosomal target regions of other factors, those sites may be revealed and provide insight into novel interactions between PapX and other species. It is more difficult to precisely contextualize aptamers in an organism's genome because their short length reduces the strength of an alignment.
To identify target sequences using SELEX and high throughput sequencing, a sufficient coverage of the target genome must be used in the initial library. For each cycle of SELEX, roughly 3 × 1012 80-bp fragments (or 2.4 × 1014 nucleotides) were exposed to immobilized protein. This suggests that this technique could be adapted to eukaryotic genomes and still preserve 105-fold representation at each nucleotide position in the probed sample. The 2.5 × 107 reads per lane generated in high throughput sequencing limits the use of the control library in establishing quality scores for peak enrichment. However, the peak scoring algorithm applies between enriched rounds as well as between the control library and the first enrichment, so a few rounds of SELEX should be sufficient to generate initial peaks from which to base subsequent scoring in a eukaryotic schema.
Evaluating the Interaction of PapX and flhDC Promoter
MarR family proteins tend to bind motifs between 21 and 45 bases long (37). In addition, we demonstrated that neither the 21-bp motif (Fig. 7E) nor the 11-bp motif (Fig. 7D) is alone sufficient to bind PapX (Fig. 8D). The 29-bp palindromic form of the 21-bp motif, however, was sufficient to bind PapX, whereas a 29-bp sequence that did not contain either the 21- or 11-bp motifs generated from gapA was unable to bind PapX (Fig. 8C). These data indicate that both the specific sequence content and the length of the sequence are together necessary to mediate PapX binding to DNA. This may explain why the palindromic form of the 21-bp motif did not appear in the best grade peaks; the software used to identify motifs, MEME (24), analyzes sequences for conserved nucleotides at given positions but cannot predict important regions that are necessary for topological reasons or that contain degenerate sequence. Both motifs (Fig. 7, D and E) appear to be necessary and specific sequences, as indicated experimentally (Fig. 8, C and D). This is supported as well in silico by their comparably low E values, which were by far lower than any other motif identified. This suggests that SELEX with high throughput sequencing was successful at identifying necessary binding elements for PapX.
Interestingly, a degenerate version of the 11-bp motif exists at the 5′-end of the 360-bp fragment used in the lacZ fusions (Fig. 6). This may have produced a small degree of PapX interaction with the fragment and may explain the elevated variance seen between replicate assays with the 360-bp fragment. The variance appeared to be biological in origin, persisting across three repeats of the experiment, each using biological triplicates and technical duplicates. It is also possible that PapX acts by antagonizing an activator, so a PapX binding site may be present in the 360-bp fragment, but reduced upstream activator binding may explain the difference in β-galactosidase activity.
Over half of the 362 best peaks fall within predicted promoter regions for genes. As mentioned, overexpression and deletion of papX has a transcriptionally specific effect (16), with transcriptional regulation confined to a small list of genes or gene classes. There are several possible explanations for this mismatch phenomenon. First, SELEX is a molecular technique that helps to make inferences about binding energy between a ligand and a nucleic acid sequence. When aptameric libraries are used, limited inferences about the biological relevance of the strength of in silico ligand-protein interactions can be drawn. SELEX is best used to identify target motifs, but follow-up, such as by EMSA or competition assay, is necessary to demonstrate biological significance.
We have confirmed a direct interaction of PapX with a specific sequence within the flhDC promoter and demonstrated a transcriptional effect on the downstream genes. By investigating a binding site known to produce an effect, we may discover additional clues to the mechanism whereby PapX regulates gene expression. When theoretical sites are determined using aptameric libraries, there must be follow-up to confirm biological relevance because the fact of DNA-protein interaction by SELEX is not sufficient to guarantee physiologically relevant action. Our studies have successfully demonstrated the utility of SELEX to identify a short, specific sequence within which a protein acts to modulate transcriptional regulation.
Pathogen Specificity of PapX-flhDC Promoter Interaction
Our multiplex results (Fig. 2) and motility assays (Fig. 3) indicated that the effect of PapX is pathogen-specific. However, the binding site identified by SELEX (Fig. 8B, boldface type, underlined) appears as a 29 of 29 nucleotide match in the flhDC promoter of strain K12. In this and previous work, control of motility in UPEC by PapX is mediated by flhD and flhC transcriptional repression. Taken together, this suggests that there are additional factors present in strain CFT073 and absent in strain K12 that affect the activity of PapX on flhD and flhC transcription and that the mere presence of a nucleotide sequence with likely PapX affinity is insufficient to regulate downstream transcription in the commensal strain. For example, PapX may be regulated by other factors (such as small molecules, as with salicylate derivatives and MarR (34)) that locally affect PapX activity at the site of regulation. This may help to explain the discrepancy between the transcriptional effect of PapX described previously (16), which is limited to 42 differentially regulated genes, and the list of hundreds of peaks by PapX with SELEX. It is possible that PapX binds most or all of the peak regions identified but that other factors are necessary to produce a differential transcriptional effect. The 29-bp fragment found in the flhDC promoter appears only once in the E. coli CFT073 and E. coli K12 genomes, offering further explanation for the specificity of transcriptional effect by PapX.
Functional Characterization of PapX
Our data suggest that Arg127 in PapX is crucial for DNA binding (Fig. 8E). Taken together with the loss of wild-type PapX phenotype of the R127G mutant seen by motility assay (Figs. 3 and 4) and loss of transcriptional repression (Fig. 5), this suggests that it is essential for PapX to bind DNA in order to exert its motility-repressing phenotype.
In addition, within the PapX homologs, Ala to Thr substitutions are found commonly (Fig. 1), occurring at residue positions 52, 82, 114, and 160. Ala to Thr substitutions have been shown to affect protein activity in other proteins (46). Our A21T mutation had full PapX activity by motility assay, suggesting that at least at some positions, this substitution is inconsequential for PapX activity. It would stand to reason that substitutions that have a small or no impact on protein function will emerge by chance and not remain conserved, offering a potential explanation of why the Ala-Thr substitutions are common among the homologs.
The combination of SELEX and Illumina high throughput sequencing provided a high resolution map of putative PapX binding sites. A single unique sequence within the genome of E. coli CFT073, a prototype of UPEC strains, was identified within the flhDC promoter, which controls flagellar gene expression. Direct binding of the target sequence by PapX was demonstrated. Repression of motility was observed in the pathogenic strain but not a non-pathogenic strain, suggesting the presence of additional factors required for this regulation. Such accessory factors will be the target of future studies, which will also aim to address the discrepancy between the specific list of papX differentially regulated genes seen previously (16) and the more extensive number of peaks determined by SELEX.
Supplementary Material
Acknowledgments
We thank Oliver He, Allen Xiang, and Tristan Trutna for assistance with genome-wide BLAST and Matlab instruction, and we thank Ann Stapelton, Thomas Hooten, and James Johnson for strains described and analyzed previously (28, 29).
This work was supported, in whole or in part, by National Institutes of Health Public Health Service Grants AI43363, AI59722, AI007528, and T32 GM07863.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–3.
- UTI
- urinary tract infection
- UPEC
- uropathogenic E. coli
- SELEX
- systematic evolution of ligands by exponential enrichment
- IPTG
- isopropyl β-d-thiogalactopyranoside
- SOC
- Super Optimal broth with catabolite repression.
REFERENCES
- 1. Litwin M. S., Saigal C. S., Beerbohm E. M. (2005) J. Urol. 173, 1065–1066 [DOI] [PubMed] [Google Scholar]
- 2. DeFrances C. J., Lucas C. A., Buie V. C., Golosinskiy A. (2008) Natl. Health Stat. Report, 1–20 [PubMed] [Google Scholar]
- 3. Svanborg C., Godaly G. (1997) Infect. Dis. Clin. North Am. 11, 513–529 [DOI] [PubMed] [Google Scholar]
- 4. Connell I., Agace W., Klemm P., Schembri M., Mrild S., Svanborg C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9827–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson J. R. (1991) Clin. Microbiol. Rev. 4, 80–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leffler H., Svanborg-Edén C. (1981) Infect. Immun. 34, 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lund B., Lindberg F., Marklund B. I., Normark S. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5898–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korhonen T. K., Väisänen V., Saxén H., Hultberg H., Svenson S. B. (1982) Infect. Immun. 37, 286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mulholland S. G., Mooreville M., Parsons C. L. (1984) Urology 24, 232–235 [DOI] [PubMed] [Google Scholar]
- 10. Mobley H. L., Jarvis K. G., Elwood J. P., Whittle D. I., Lockatell C. V., Russell R. G., Johnson D. E., Donnenberg M. S., Warren J. W. (1993) Mol. Microbiol. 10, 143–155 [DOI] [PubMed] [Google Scholar]
- 11. Smith T. G., Hoover T. R. (2009) Adv. Appl. Microbiol. 67, 257–295 [DOI] [PubMed] [Google Scholar]
- 12. Lane M. C., Lockatell V., Monterosso G., Lamphier D., Weinert J., Hebel J. R., Johnson D. E., Mobley H. L. (2005) Infect. Immun. 73, 7644–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwan W. R. (2008) Int. J. Med. Microbiol. 298, 441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pearson M. M., Mobley H. L. (2008) Mol. Microbiol. 69, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lane M. C., Alteri C. J., Smith S. N., Mobley H. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simms A. N., Mobley H. L. (2008) Infect. Immun. 76, 4833–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soutourina O. A., Bertin P. N. (2003) FEMS Microbiol. Rev. 27, 505–523 [DOI] [PubMed] [Google Scholar]
- 18. Welch R. A., Burland V., Plunkett G., 3rd, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., Stroud D., Mayhew G. F., Rose D. J., Zhou S., Schwartz D. C., Perna N. T., Mobley H. L., Donnenberg M. S., Blattner F. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mobley H. L., Green D. M., Trifillis A. L., Johnson D. E., Chippendale G. R., Lockatell C. V., Jones B. D., Warren J. W. (1990) Infect. Immun. 58, 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clarke M. B., Sperandio V. (2005) Mol. Microbiol. 57, 1734–1749 [DOI] [PubMed] [Google Scholar]
- 21. Zhang X., Bremer H. (1995) J. Biol. Chem. 270, 11181–11189 [DOI] [PubMed] [Google Scholar]
- 22. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozarewa I., Ning Z., Quail M. A., Sanders M. J., Berriman M., Turner D. J. (2009) Nat. Methods 6, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey T. L., Elkan C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 25. Kelley L. A., Sternberg M. J. E. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 26. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y. (2007) Proteins 69, Suppl. 8, 108–117 [DOI] [PubMed] [Google Scholar]
- 28. Vigil P. D., Stapleton A. E., Johnson J. R., Hooton T. M., Hodges A. P., He Y., Mobley H. L. (2011) MBio 2, e00066–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spurbeck R. R., Stapleton A. E., Johnson J. R., Walk S. T., Hooton T. M., Mobley H. L. (2011) Infect. Immun. 79, 4753–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagan E. C., Mobley H. L. (2009) Mol. Microbiol. 71, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ott M., Hacker J., Schmoll T., Jarchau T., Korhonen T. K., Goebel W. (1986) Infect. Immun. 54, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simms A. N., Mobley H. L. (2008) J. Bacteriol. 190, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nichols C. E., Sainsbury S., Ren J., Walter T. S., Verma A., Stammers D. K., Saunders N. J., Owens R. J. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. (2001) Nat. Struct. Biol. 8, 710–714 [DOI] [PubMed] [Google Scholar]
- 35. Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005) Mol. Cell 20, 131–141 [DOI] [PubMed] [Google Scholar]
- 36. Kumarevel T., Tanaka T., Nishio M., Gopinath S. C., Takio K., Shinkai A., Kumar P. K., Yokoyama S. (2008) J. Struct. Biol. 161, 9–17 [DOI] [PubMed] [Google Scholar]
- 37. Wilkinson S. P., Grove A. (2006) Curr. Issues Mol. Biol. 8, 51–62 [PubMed] [Google Scholar]
- 38. Saito K., Akama H., Yoshihara E., Nakae T. (2003) J. Bacteriol. 185, 6195–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Djordjevic M. (2007) Biomol. Eng. 24, 179–189 [DOI] [PubMed] [Google Scholar]
- 40. Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., Taipale M., Vaquerizas J. M., Yan J., Sillanpää M. J., Bonke M., Palin K., Talukder S., Hughes T. R., Luscombe N. M., Ukkonen E., Taipale J. (2010) Genome Res. 20, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klug S. J., Famulok M. (1994) Mol. Biol. Rep. 20, 97–107 [DOI] [PubMed] [Google Scholar]
- 42. Sampson T. (2003) World Patent Information 25, 123 [Google Scholar]
- 43. Zimmermann B., Bilusic I., Lorenz C., Schroeder R. (2010) Methods 52, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McClure W. R. (1985) Annu. Rev. Biochem. 54, 171–204 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imai M., Shimada H., Watanabe Y., Matsushima-Hibiya Y., Makino R., Koga H., Horiuchi T., Ishimura Y. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7823–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.