Abstract
Laminarinases hydrolyzing the β-1,3-linkage of glucans play essential roles in microbial saccharide degradation. Here we report the crystal structures at 1.65–1.82 Å resolution of the catalytic domain of laminarinase from the thermophile Thermotoga maritima with various space groups in the ligand-free form or in the presence of inhibitors gluconolactone and cetyltrimethylammonium. Ligands were bound at the cleft of the active site near an enclosure formed by Trp-232 and a flexible GASIG loop. A closed configuration at the active site cleft was observed in some molecules. The loop flexibility in the enzyme may contribute to the regulation of endo- or exo-activity of the enzyme and a preference to release laminaritrioses in long chain carbohydrate hydrolysis. Glu-137 and Glu-132 are proposed to serve as the proton donor and nucleophile, respectively, in the retaining catalysis of hydrolyzation. Calcium ions in the crystallization media are found to accelerate crystal growth. Comparison of laminarinase and endoglucanase structures revealed the subtle difference of key residues in the active site for the selection of β-1,3-glucan and β-1,4-glucan substrates, respectively. Arg-85 may be pivotal to β-1,3-glucan substrate selection. The similarity of the structures between the laminarinase catalytic domain and its carbohydrate-binding modules may have evolutionary relevance because of the similarities in their folds.
Keywords: Glycosidases, Hydrolases, Polysaccharide, Structural Biology, X-ray Crystallography
Introduction
Thermotoga maritima is a hyperthermophilic, anaerobic, and fermentive saccharolytic bacterium, catabolizing sugars and its polymers to make energy. Laminarinase (3-β-d-glucan glucanohydrolase; EC 3.2.1.39, Lam),3 an endoglucosidase, hydrolyzes internal β-1,3-glucosyl linkages in β-d-glucans and is therefore crucial in carbohydrate degradation for nutrient uptake and energy production in bacteria. According to the sequenced genome of T. maritima MSB8 (1), the laminarinase gene encodes the enzyme composed of a catalytic domain and two carbohydrate-binding modules (CBMs) connected by a linker region on each terminus. The structure of TmLamCBM2, located on the C terminus, has previously been determined by Boraston et al. (2). The coexistence of CBM with catalytic domains is widespread in many modular bacterial polysaccharide hydrolases that contain separately folding modules (3). The prevalent role of CBMs is to facilitate the association of substrates with the catalytic module; moreover, they sometime boost the reaction efficiency of the catalytic domain (4, 5). The modularity in biological macromolecules draws scientists' attention in biocatalyst designs (6).
Because of the substrate diversity among glycosyl hydrolases (GHs), it is not easy to classify these enzymes according to their substrate specificity. Henrissat and Bairoch (7) developed a sequence similarity-based classification to categorize GH enzymes as an alternative to the traditional enzyme classification system. Except for a laminaripentaose-producing β-1,3-glucanase from Streptomyces matensis, which belongs to GH-64 (8), most of the bacterial laminarinases have been classified in GH-16, which share a β-jelly roll fold and catalyze the glycosyl hydrolysis reaction in a retaining mechanism. At the active site, a glutamate residue acts as a nucleophile to attack the C1 atom in the absence of water molecules, and then another glutamate serves as a proton donor to complete the double displacement mechanism.
Bacterial laminarinase crystal structures have been analyzed from alkaliphilic Nocardiopsis sp. strain F96 (9) and the hyperthermophile Pyrococcus furiosus (10). Although the latter report modeled the existence of laminarin trisaccharide in the protein catalytic cleft, the bacterial GH-16 laminarinase-sugar complex structure has not been reported so far. Here we report the catalytic domain structures of laminarinase from T. maritima (TmLamCD), and a loop controlling the opening of the gate in the active site of laminarinase is described for the first time. Structures of the enzyme in complex with a gluconolactone or an inhibitor were also determined to reveal the relevant residues in enzyme-substrate interactions. Residues that may be pivotal to selection of saccharosyl substrates are also elucidated and defined. We also discovered the structural similarity between catalytic and carbohydrate-binding modules of TmLam acting on 1,3-β-d-glucan, suggesting a possible evolutionary relationship between them.
EXPERIMENTAL PROCEDURES
Enzymes, Chemicals, and Bacterial Strains
The Escherichia coli strains used in this study were DH5α, BL-21 (DE3) (Novagen), and XL1-Blue (Stratagene) cells. The enzymes for DNA manipulation were purchased from New England Biolabs. Gluconolactone and cetyltrimethylammonium bromide (ctab) were purchased from Sigma-Aldrich.
Protein Expression and Purification
The full-length laminarinase gene was amplified from the genomic DNA of T. maritima MSB8 (protein identifier NP_227840.1). The gene fragment encoding residues 204–466 of TmLamCD was amplified by PCR using primers with NdeI and SalI sites and then subcloned into the NdeI/XhoI sites of pET-21a. The resulting plasmid pET-TmLamCD encoding the catalytic domain, fused to a C-terminal His6 tag, was confirmed by nucleotide sequencing. The numbering of TmLamCD amino acid residues mentioned in this study starts with residue Glu-204 in full-length protein as the second residue in the catalytic domain. The correct construct was then transformed in E. coli BL-21 (DE3), where the protein expression was induced by isopropyl β-thiogalactopyranoside in a final concentration of 0.5 mm. The protein was then purified by FPLC using a nickel-nitrilotriacetic acid column in 20 mm Tris-HCl buffer (pH 7.5) containing 400 mm NaCl and 10–300 mm imidazole. The purified His6-tagged larminarinase protein was concentrated and changed to enzyme storage buffer (50 mm Tris-HCl, 100 mm NaCl, 10% glycerol, pH 8.0).
Crystallization, Data Collection, and Processing
The TmLamCD protein crystals were grown by vapor diffusion in sitting drops from two kinds of reservoir solution at room temperature. Solution formula I contained 10–15% (w/v) PEG 8000 (Fluka), 0.25–0.45 m KH2PO4 (J.T. Baker), 20% glycerol (J.T. Baker), and 10 mm CaCl2 (Sigma), whereas formula II contained 41–46% (w/v) polypropylenglycol P400 (Fluka), 0.05–0.2 m (NH4)2SO4 (Hampton). The crystal, belonging to space group C2, was achieved in reservoir solution formula I with 5 mm DTT at a protein concentration of 75–100 mg/ml, and crystals belonging to other space groups were achieved in reservoir solution formula II at a protein concentration of 60–90 mg/ml in 50 mm Tris-HCl buffer (pH 8.0) containing 100 mm NaCl and 10% glycerol, pH 8.0. The P212121 TmLamCD structure in complex with gluconolactone was obtained using native crystals (in P43 space group) soaked in the reservoir solution with 50 mm gluconolactone for 2 min before data collection. The ctab was cocrystallized with TmLamCD in the same reservoir. X-ray diffraction data were collected at cryogenic temperatures using synchrotron radiation at SPring-8 in Japan and NSRRC in Taiwan. The diffraction images were processed using HKL2000 (11).
Structure Determination, Refinement, and Validation
All of the crystallographic computations were carried out using programs from the CCP4 suite (12). The structure was solved by molecular replacement with MolRep (13) using the structure of the PDB entry of 2HYK (endo-β-1,3-glucanase from Nocardiopsis sp. strain F96) as the search model. Automatic model building was performed with ARP/wARP (14) and Buccaneer (15). Model completion and refinement were performed with Refmac (16) and Coot (17). A subset of 5% randomly selected reflections was excluded from computational refinement to calculate the Rfree factor throughout the refinement (18). All of the final refinements were carried out using Refmac with TLS group tensor and anisotropic B factor without NCS restrains. The stereochemistry and structure of the final models were analyzed by RAMPAGE (19) and SFCHECK (20) of the CCP4 program suite. Data collection and refinement statistics are summarized in Table 1. The C2 structure was refined to a final Rwork of 19.8% and Rfree 24.8%. The C2 model was then used to solve the structures of two other crystal forms, space groups P43 and P212121. The P43 structure was refined to a final Rwork of 16.3% and Rfree of 19.2%, whereas Rwork of 17.0% and Rfree of 20.0% exist in the P212121 structure.
TABLE 1.
Data collection and refinement statistics of laminarinase from Thermotoga maritima
| Crystals | Gluconolactone | ctaba | Closed | ||
|---|---|---|---|---|---|
| Protein Data Bank code | 3AZX | 3AZY | 3AZZ | 3B00 | 3B01 |
| Data collection | |||||
| Radiation source | NSRRC BL13B1 | NSRRC BL13B1 | NSRRC BL13B1 | SPring-8 BL44XU | NSRRC BL13C1 |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | 0.90000 | 0.97622 |
| Space group | C2 | P43 | P212121 | P43 | P43 |
| Unit cell parameter | |||||
| a (Å) | 100.57 | 120.42 | 106.99 | 120.32 | 120.31 |
| b (Å) | 56.15 | 120.42 | 120.23 | 120.32 | 120.31 |
| c (Å) | 126.43 | 107.20 | 121.00 | 107.01 | 107.15 |
| β (o) | 105.26 | ||||
| ZA | 2 | 4 | 4 | 4 | 4 |
| Resolution (Å)b | 30-1.80 (1.86-1.80) | 30-1.65 (1.71-1.65) | 30-1.82 (1.89-1.82) | 30-1.74 (1.80-1.74) | 30-1.87 (1.94-1.87) |
| Number of reflections | 62313 (6005) | 183216 (18165) | 140042 (13940) | 155608 (15513) | 125562 (12561) |
| Completeness (%) | 98.8 (95.8) | 99.8 (100) | 99.6 (99.9) | 99.9 (100) | 99.7 (100) |
| Redundancy | 3.9 (3.7) | 5.6 (5.6) | 6.6 (6.8) | 7.4 (7.4) | 6.2 (6.1) |
| Rmerge (%) | 8.6 (49.6) | 5.7 (44.1) | 6.0 (57.6) | 14.3 (48.8) | 6.5 (51.7) |
| I/σ(I) | 16.4 (2.4) | 26.8 (3.2) | 29.2 (3.2) | 13.1 (5.5) | 27.4 (3.9) |
| Overall Wilson B factor (Å2) | 39.7 | 33.4 | 41.8 | 31.6 | 39.0 |
| Refinementc | |||||
| Reflections (work) | 59130 (8207) | 173849 (25165) | 132697 (18670) | 147427 (21357) | 119132 (17334) |
| Reflections (free) | 3167 (392) | 9197 (1338) | 7013 (1001) | 7787 (1147) | 6298 (908) |
| Rwork (%) | 19.8 (28.8) | 16.3 (19.4) | 17.0 (17.6) | 15.3 (16.7) | 15.2 (17.0) |
| Rfree (%) | 24.8 (36.6) | 19.2 (24.5) | 20.0 (23.3) | 18.4 (22.5) | 18.8 (23.0) |
| Geometry deviations | |||||
| Bond length (Å) | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 |
| Bond angles (o) | 1.4 | 1.4 | 1.4 | 1.4 | 1.4 |
| Mean B-values (Å2)/No. | |||||
| Protein atoms | 25.8/4080 | 20.8/8176 | 29.1/8127 | 20.5/8200 | 27.4/8226 |
| Water molecules | 41.3/359 | 35.3/809 | 42.0/585 | 34.9/779 | 39.1/664 |
| Ion atoms | 24.5/2 | 63.5/64 | 63.3/44 | 19.4/4 | 56.8/47 |
| Ligand atoms | 37.9/36 | 59.2/80 | |||
| Ramachandran plot (%)d | |||||
| Favored | 97.4 | 96.6 | 97.0 | 97.0 | 96.3 |
| Allowed | 2.4 | 3.4 | 3.0 | 3.0 | 3.5 |
| Disallowed | 0.2 | 0 | 0 | 0 | 0.2 |
a ctab, cetyltrimethylammonium bromide.
b The values in the parentheses are for the highest resolution shells.
c All positive reflections are used in the refinements.
d Categories were defined by RAMPAGE.
Multiple sequence alignments with secondary structure depiction as shown in Figs. 1B and 4B were adapted from ESPript (21). Root mean square deviation (RMSD) values were calculated by using the Structure Similarity service on the PDBeFold website (22) to assess structural difference among similar structures deposited in the data bank. Molecular graphics were produced using PyMOL (23). The Ligplot program was used to generate schematic diagrams of protein-ligand interactions (24). Based on the top five templates with the highest scores (3K4Z_A, 1CX1_A, 3P6B_A, 1GU3_A, and 1GUI_A), the structural model of the N-terminal CBM (TmLamCBM1, residues 38–180) was generated using the HHpred (25), a website of homology detection and structure prediction by Hidden Markov Models (HMM-HMM) comparison. Structural comparison presented in Fig. 4A was produced using the secondary structure matching algorithm of the PDBeFold server with default settings of multiple three-dimensional alignment.
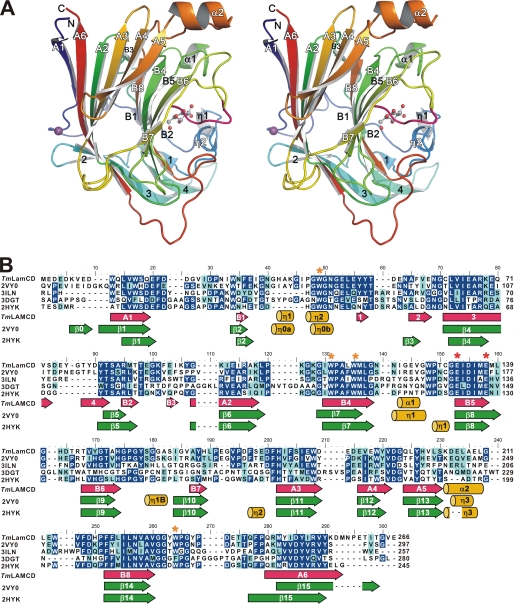
FIGURE 1.
Structural information of the catalytic domain of TmLam. A, chain A (residues 8–257) of TmLamCD in P212121 space group is shown as a cartoon and spectrum-colored from blue (N terminus) to red (C terminus). Calcium ions are shown as purple balls, and gluconolactone molecules are in ball-and-stick form. B, the sequence alignment of relative laminarinases. Identical and similar residues are highlighted in blue and cyan, respectively. TmLamCD, the catalytic domain of laminarinase from T. maritima MSB8; 2VY0, LamA from P. furiosus; 3ILN, Lam from R. marinus; 2HYK, β-1,3-glucanase from Nocardiopsis sp. strain F96; 3DGT, endo-1,3-β-glucanase from Streptomyces sioyaensis. Secondary structures depicted under the sequences are TmLamCD (in hot pink and yellow) according to our refined TmLamCD structures, 2VY0 and 2HYK (in green and yellow) followed by a previous annotation (8, 9). The catalytic Glu and conservative Trp residues are marked with red and orange asterisks, respectively.
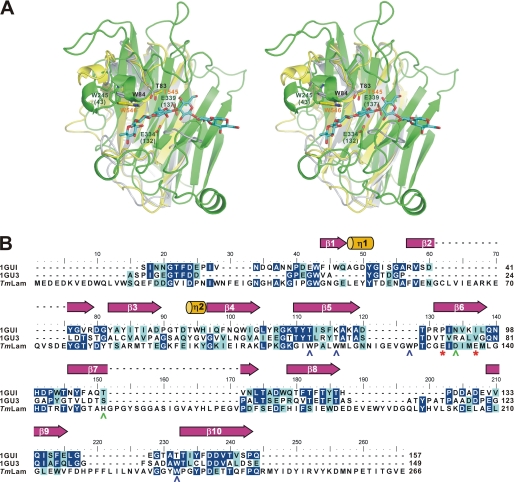
FIGURE 4.
Comparison of the carbohydrate-binding modules and the catalytic domain of TmLam. A, TmLamCD (chain C of 3AZZ, in green), TmLamCBM2 (1GUI, in yellow), and the modeled TmLamCBM1 (in gray) were superposed using PDBeFold server. The modeled TmLamCD-laminarihexaose molecule was then aligned with the superposed TmLamCD using PyMOL. The laminarihexaose is shown as cyan sticks. Amino acid residues are labeled in accordance with the overall TmLam protein numbering and with TmLamCD in parentheses. Residues belonging to TmLamCBM1 are labeled in black, those for TmLamCD are in dark green, and those for TmLamCBM2 are in orange. B, ESPript produced figure using the aligned sequence by ClustalW. 1GU3, CBM4–1 from Cellulomonas fimi; TmLam, the catalytic domain of TmLam. The catalytic Glu residues are denoted as red asterisks below the TmLam sequence and shown as green sticks in A. The secondary structure of the 1GUI is assigned above. Amino acids of TmLamCD with polar side chains around the active site are labeled in green, whereas aromatic amino acids are in blue.
To visualize the existence of laminarin in TmLamCD, a laminaritriose was modeled by superposing the pyranose annotated as 1003 in PDB 3N9K with gluconolactone in PDB 3AZZ. Then a laminaripentaose was modeled in TmLamCD by overlapping the pyranose 1001 of a laminaritriose with the previously built pyranose 1003 of the modeled laminaritriose to extend the following 1004 and 1005 pyranoses at the reducing end. Because of the stereohindrance with TmLamCD protein structure, bond angles between subsites −1 and +1, and +1 and +2 were adjusted, and the overall structure was optimized by energy minimization using the Refmac program in ccp4 suite.
Thin Layer Chromatography Analysis
The hydrolytic products of laminarin after treatment with TmLamCD were analyzed by TLC. Five micrograms of purified TmLamCD and 2.4% (w/v) laminarin were incubated in 25 mm sodium phosphate buffer (pH 7.0) at 45 °C for various time intervals. Approximately 1 μl of the reaction product was spotted on a silica plate (silica gel 60; Merck), developed with ethyl acetate/acetic acid/methanol/formic acid/water (8:4:1:1:1, by volume), and visualized by 4-methyoxybenzaldehyde (26).
PDB Accession Codes
The atomic coordinates and experimental structure factors for the catalytic domain of laminarinase from T. maritima have been deposited in the Protein Data Bank under codes 3AZX, 3AZY, and 3B01. The TmLamCD in complex with gluconolactone or ctab have been deposited in the PDB under codes 3AZZ and 3B00, respectively.
RESULTS
TmLamCD Structures in Three Crystal Packings
Three different packing forms were determined to 1.65, 1.80, and 1.82 Å resolution as shown in Table 1. All exhibited the same β-jelly roll fold shown in Fig. 1A. Protein sequence alignment analysis of the TmLamCD using Blast program (27) shows an identity of 48% (118 of 250 residues) with β-1,3-glucanase from Nocardiopsis sp. (PDB code 2HYK) (9), 61% (152 of 250 residues) with Lam from P. furiosus (PDB code 2VY0) (10), and 44% (112 of 260 residues) with Lam from Rhodothermus marinus (PDB code 3ILN). The superposition of Cα atoms of the TmLamCD structures in three different crystal packing forms reveals similar structures with the RMSDs in the range of 0.19–0.77 Å (supplemental Table S1). The structure in the C2 space group consists of two protein molecules (designated as chains A and B) with each bound with a calcium ion in the asymmetric unit, whereas both crystals with space groups P43 and P212121 contain four protein molecules (designated as chains A, B, C, and D), with each bound with a calcium ion in the asymmetric unit. In the structure with a P212121 space group, a gluconolactone molecule was located at each active site, except in chain D. Refinement was performed with Refmac followed by anisotropic thermal displacement factors using CCP4. Final data statistics are listed in Table 1.
Overall Structure
TmLamCD has a classical sandwich-like β-jelly roll fold, composed of two antiparallel β-sheets (Fig. 1 and supplemental Fig. S1) packed against each other. These β-sheets twist somewhat to form a concavity where the substrates are located. According to the ESPript analysis, the secondary structure of TmLamCD consists of 18 β-strands (two parallel β-sheets designated as A1–A6, B1–B8, and other β-strands numbered from 1 to 4), two α-helices, and two 310 helices (supplemental Fig. S1A). Interestingly, double occupancy was observed in the loop Β6-Β7 (158–162; sequence GASIG) when refining the chain A of TmLamCD structures in both space groups, P43 and P212121, regardless of whether the gluconolactone molecule was present or not. The GASIG fragment is thus a flexible loop, adopting two conformers, an open form and a closed form, with the farthest separation between corresponding Cα positions being 8.11 Å at Ile-161 (supplemental Fig. S2). This flexible loop is located at a similar location to the 310 helix (η1B) of LamA from P. furiosus (PDB accession code 2VY0). Around the B7 strand region of the TmLamCD structure, no helix structure was found, which is quite different from those observed in 2VY0 and 2HYK (Fig. 1B). In comparison with catalytic domains from other four laminarinases belonging to GH-16, TmLamCD folds similarly to most of them, including helix and β-strand regions (supplemental Fig. S3).
To assess the structural difference between TmLamCD and other similar structures deposited in the data bank, RMSD values were calculated by PDBeFold. The calcium-bound chain A of the TmLamCD structure is similar to those of P. furiosus Lam (2VY0, RMSDs ranged from 0.68 to 0.76 Å over 251 Cα atoms), Nocardiopsis sp. strain F96 Lam (2HYK, RMSD = 0.92 Å over 223 Cα atoms), and R. marinus Lam (3ILN, RMSDs ranged from 1.19 to 1.36 Å over 216–222 Cα atoms). Together, the basic β-jelly roll structure is well preserved among laminarinases belonging to class GH-16.
Active Site of the Catalytic Domain
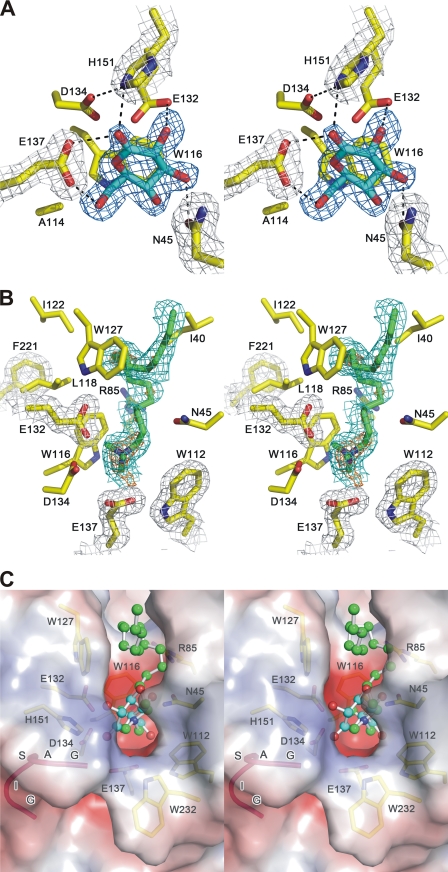
To confirm the residues important in enzyme catalysis, gluconolactone, a flattened hexose-like ring resembling the transition state geometry (28), was soaked into the crystal of TmLamCD. In the P212121 structure, a gluconolactone was found to bind at each active site of chains A-C, but not of chain D. Among residues around the active site, Asn-45, Glu-132, Glu-137, and His-151 formed direct hydrogen bonds with gluconolactone (Fig. 2A). The Oδ2 atom of Asp-134 interacting with Nϵ2 of His-151 by hydrogen bonding is also involved in stabilizing the ligand. Residues Trp-116 and Ala-114 contribute to hydrophobic interactions according to the Ligplot analysis (supplemental Fig. S4). On the basis of the known catalytic mechanism of Lam, the Glu-132 residue is assumed to be the nucleophile directly attacking C1 of the sugar ring. On the other hand, Glu-137 serves as the proton donor to complete the retaining catalysis. These two active carboxyl groups are in close proximity at a distance of 6.67 Å (Cδ to Cδ), which supports the space requirement for retaining enzymes (29).
FIGURE 2.
Stereo view of the active site of laminarinase from T. maritima. A, the hydrogen bond network between the active site residues (in yellow) and gluconolactone (in cyan). Hydrogen bonds are shown as dotted lines. B, the inhibitor ctab (in green sticks) is bound in the active site of chain A. In A and B, meshes in orange, blue, and green color represent the difference Fourier map (mFo − DFc) contoured at 4, 3, and 1.5 σ levels, respectively. Gray mesh represents the 2mFo − DFc map contoured at the 1 σ level. C, superposition of TmLamCD structures containing gluconolactone (carbons in cyan) and ctab (carbons in green). Both ligands are shown in ball-and-stick. The molecular surface of the TmLamCD structure is colored by electrostatic potential. Positive and negative values are represented in blue and red, respectively. TmLamCD residues are shown as yellow sticks.
To simulate the location of long chain substrates in the enzyme, ctab, a surfactant molecule with a polar head and a 16-carbon tail was cocrystallized. This structure was determined at 1.74 Å resolution. The ctab molecule, previously found to be an inhibitor of Lam (30), was located at the cleft of the active site, as shown in Fig. 2 (B and C). Residues Ile-40, Trp-43, Asn-45, Arg-85, Ala-114, Trp-116, Trp-127, Glu-132, and Glu-137 involved in hydrophobic interactions with the ctab molecule are shown in Fig. 2B, as Ligplot revealed (supplemental Fig. S4D). By superposition of the two TmLamCD structures in complex with gluconolactone and ctab, these two inhibitor molecules appeared to be located at the same position in the active site near the gate controlled by the GASIG fragment (Fig. 2C). In a distal view, there is a large catalytic groove crossing the TmLamCD structure, a reasonable pathway for the bent long chain β-1,3-glucan substrates (Fig. 3A). However, an opposite orientation of the long chain sugar is actually observed according to our modeled extended β-1,3-glucans based on the superposition of the laminaritriose structure solved in the PDB code 3N9K with the gluconolactone in our 3AZZ structure (supplemental Fig. S5A). With forward extension of β-1,3-glucans by slightly modifying bond angles to make the sugar chain penetrate the enclosure with energy minimization of the docked model, the spatial arrangement at the controlling gate just allows the glucan to pass by (supplemental Fig. S5B). Similar catalytic grooves have also been found in β-1,4-endoglucanases, as shown in Fig. 3B (31).
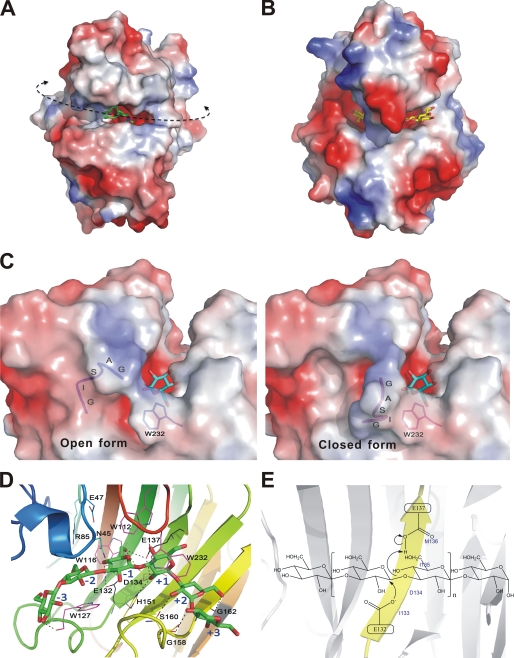
FIGURE 3.
Enzyme-carbohydrate interactions at the TmLamCD active site. A and B are electrostatic surface views of TmLamCD and TmCel12A, respectively. The ctab ligand is shown as green sticks, and the cellotetraose molecule is show with yellow sticks. The curved dashed line indicates the catalytic groove of TmLamCD, whereas the arrowheads show possible directions of a long chain substrate, the laminarin. C, electrostatic surface representation of the TmLamCD active site in complex with gluconolactone. The GASIG fragment located in a closed form is shown in magenta, and the open form is in blue. One of the double occupancies of Trp-232 was shown around the flexible loop, in which occupancy has no conflict over both open and closed forms. D, residues interacting with the modeled laminarihexaose by hydrophobic interaction are shown in magenta, whereas those forming hydrogen bonds are in yellow. Some nearby potential residues to interact with ligands are in cyan. The two catalytic Glu residues are drawn in black. Subsites of the pyranosides in the active site are labeled with blue text. E, depiction of the proposed mechanism for the retaining catalysis in TmLam. Both of the catalytic Glu residues are located on the B5 strand (in yellow).
Open and Closed Forms of the Catalytic Domain in Complex with Gluconolactone
A surface view around the active site reveals that the GASIG fragment plays the role of guarding the gate of the catalytic cleft (Fig. 3C). All Lam structures determined till now adopt the open form in crystal where the open gateway may allow the free passage of products or substrates. In the present work, a closed conformation caused by this loop was observed in some TmLamCD subunits. Residues Ala-159, Ser-160, and Ile-161 in this GASIG fragment are found to interact with the conserved Trp-232 residue. These hydrophobic interactions make the GASIG fragment move into a closed form and shrink the cavity. In some chains of our determined structures of TmLamCD, the side chain of Trp-232 has double occupancy, one of which collides with Ile-161 in the closed form. We hypothesize that Trp-232 is a key residue in bringing about closing of the gate through hydrophobic interaction. When the catalytic cleft of the active site is closed, the flexible loop filling part of the passageway may block the following hydrolysis reaction and help the expulsion of products. This closed conformation was also observed in another P43 crystal structure without the presence of gluconolactone (PDB code 3B01).
In TmLamCD, the distance between oxygen atoms on C6 and C2 of gluconolactone is 6.11 Å, which is similar to that in glucose, the basic component of laminarinase substrates. The width of the groove across the surface at the catalytic site is ∼10 Å, which constricts the substrates for proper catalysis. Regarding charge distribution in this region, the electronegatively charged residues (Glu-137, Asp-134, and Glu-132) in the same B5 strand are located at the catalytic cavity (Fig. 3D). In a proximal view of the TmLamCD with a modeled laminarihexaose, hydrophilic residues facing the substrate-binding sites, such as Arg-85, Glu-47, Asp-134, His-151, and Asn-45, may form direct or water-mediated hydrogen bonds in protein-laminarin binding to position substrates correctly in the catalytic groove (Fig. 3D). In the schematic view shown in Fig. 3E, Glu-132 is the base/nucleophile to attack the C1 atom of the sugar ring to promote the cleavage of β-1,3-linkage between sugars. Glu-137 serves as the acid to receive the electron and then transfer it back to adjacent water molecule to attack the same C1 atom in the β-position, and the Glu-132 base and product will then be released. These residues important in hydrolyzing β-1,3-glycosyl linkage are highly conserved among GH-16 laminarinases (Fig. 1B).
Calcium-binding Site
Calcium ion has been reported to increase thermal stability in bacterial hybrid glucanases (32). In our study, all TmLamCD x-ray structures contained one calcium ion in each molecule when calcium chloride was added in the crystallization buffer. The calcium ion was located on the convex side of the protein, whereas the substrate analog bound to the cleft on the concave side (Fig. 1A). The calcium coordinated to the backbone carbonyl oxygen atoms of Gly-61, Glu-17, and Asp-249, the carboxyl side chain oxygen of Asp-19 and Asp-249, and two water molecules (supplemental Fig. S6). The calcium ion was far away from the active site of TmLamCD, implying no direct relation with catalytic function. However, probably because of the stabilizing effect of the calcium, it helped the growth of TmLamCD crystals, making it about four times faster, if it was added in the crystallizing buffer.
Comparison between the Catalytic and Carbohydrate-binding Domains of TmLam
At first glance, the TmLamCD structures have a structural fold similar to the TmLamCBM2 structure, sharing the β-jelly roll fold and having a substrate-binding groove across the surface, although superposition of them indicates that β-strands bend and distribute differently from each other (Fig. 4A). They align with an overall RMSD of 2.7 Å (123 aligned Cα atoms), measured by PDBeFold. According to the sequence alignment shown in Fig. 4B, some polar amino acid residues interacting with sugars in the binding subsites of TmLamCBM2 are similar to those in the catalytic domain, but aromatic amino acid residues are not equivalent. However, both three-dimensional structures exhibit plenty of Trp residues along the substrate-binding groove, and those aromatic amino acid side chains are omnipresent for carbohydrate recognition (33).
Hydrophobic interactions are also important for substrate binding in the catalytic groove. Near the active site, Trp residues are highly concentrated around the substrate-binding site, and this phenomenon is also observed in glycan chain-binding CBMs (33). These Trp residues are expected to be involved in the protein-carbohydrate contacts by hydrophobic stacking interactions as those of CBMs, in which the orientation of aromatic side chains may determine the specificity of ligand binding (34). Viewing the identity of Trp residues among catalytic laminarinases, Trp-112, Trp-116, Trp-127, and Trp-232 of the TmLamCD are highly conserved (Fig. 1B), revealing the importance of hydrophobic residues in stabilizing substrates.
Comparing the structures of TmLamCD and TmLamCBM2 in a surface view reveals that they both have a substrate-binding groove on the solvent-accessible surface (supplemental Fig. S7A). An obvious difference is that the CBM2 binds glucan chains in a catalytic-like open cleft, whereas the catalytic domain has an enclosure ahead of the cleavage site, which probably contributes to holding the glucan chain in place for efficient catalysis.
To explore further a structural evolutionary relationship between the catalytic and carbohydrate-binding domains of TmLam, the structure of TmLamCBM1, which shares 27% identity in protein sequence with TmLamCBM2, was predicted and superposed with TmLamCD (3AZZ, chain C) and TmLamCBM2 (1GUI) (Fig. 4A). Both CBMs fold similarly and have a short 310 helix (η2 in TmLamCBM2) near the active site. On this 310 helix, the side chains of Thr-83/545 and Trp-84/546 residues can interact with sugar chains to help substrate binding. However, the catalytic domain developed a longer loop containing η1, η2 helices, and six interseptal Gly residues. This increases the flexibility in substrate binding and moving toward the catalytic site. The nucleophile (Glu-132) and proton donor (Glu-137) in the catalytic domain are not found in either of the TmLamCBM domains, which explains the resulting difference that the TmLamCBMs target and bind substrates to deposit enzymes on the surface of substrates to facilitate the catalytic domain to hydrolyze the β-1,3 glucosidic linkage.
Comparison of β-1,3- and β-1,4-Endoglucanases in T. maritima
Because only subtle differences of stereochemistry exist in carbohydrates, it is interesting to investigate the key residues required by β-glucanases to select their substrate specificity. The crystal structure of TmCel12A, a β-1,4-glucanase, has been determined by Cheng et al. (31) recently (Fig. 3B and supplemental Fig. S7B). To compare the structural difference between TmLamCD and TmCel12A, these two structures were superposed by residues 123–140 of TmLamCD with residues 126–143 of TmCel12A based on PDBeFold analysis (supplemental Fig. S8A). In the active site as shown in Fig. 5, there is a subsite shift for cleavage. In TmLamCD, Trp-112 and Trp-116 on B4 strand contribute in aromatic stacking interaction with pyranose at −1 subsite of β-1,3-glucan; also, Asp-134 on B5 strand, Asn-225 on B8 strand, and Asn-45 followed by η2 interact with the same d-glucosyl residue by hydrophilic interactions. As to the following subsite −2 of β-1,3-glucan, Arg-85 on B2 strand fits the curvature of β-1,3 glucan (Fig. 5) and is highly conserved among GH-16 laminarinases (Fig. 1B). If a β-1,4-glucan is located at the active site with one of the pyranose fixed on subsite −1 of laminarinase, a serious conflict with Arg-85 will happen at subsite −2 of laminarinase, suggesting its importance in determining substrate specificity. Leu-118 and Ile-223 are supposed to interact hydrophobically with pyranose carbons on subsites −3 and −2, respectively. Most of the essential TmLamCD residues mentioned above are located on B4, B5, or B8 strands, the highly conservative regions in GH-16 laminarinases, showing their indispensability in substrate selection. Another difference is that the TmCel12A holds its β-1,4-glucan substrate at the cleavage subsite −1, but TmLamCD is enclosed at subsite +1 (supplemental Fig. S8, B and C). The former clutches the −1 sugar ring to cleave the glucosidic bond efficiently, but the latter may contribute to dual roles in both holding substrates and expelling products cooperating with the coexistence of open and closed forms.
FIGURE 5.
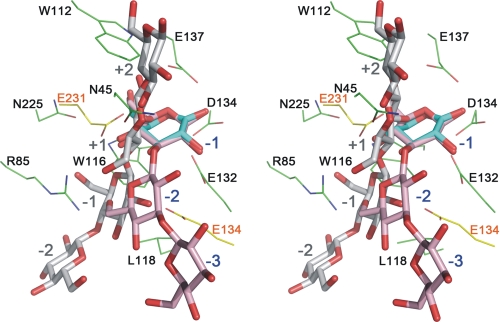
Overlap showing of the ligands bound in TmLamCD and TmCel12A. The gluconolactone molecule is shown with cyan stick models, laminaritriose is shown with pink ball-and-stick models (labeled from −1 to −3 in blue), and cellotetraose is shown with gray ball-and-stick models (labeled from +2 to −2 in gray). The laminaritriose in the active site was modeled based on the orientation of the gluconolactone molecule we determined in PDB code 3AZZ. The amino acid residues of TmLamCD and TmCel12A are denoted in green and yellow lines, respectively.
DISCUSSION
Among glucanases, including β-glucosidases, endo- and exo-glucanases, proteins fold in different shapes to bind substrates around the active site. In many endoglucanases, such as cellulases 12A from T. maritima (supplemental Fig. S7B) (31), 44A from Clostridium thermocellum (35), and endoglucanase V from Humicola insolens (36), long substrate-binding clefts with ends open at both sides have been observed. In most exoglucanases, blockers bumped into the nonreducing end of the carbohydrate polymeric chain to stop substrates moving forward (supplemental Fig. S7C). In β-glucosidases, only sufficient space for placing a disaccharide molecule in the active site has been observed (supplemental Fig. S7D) (37). The spatial restraint of binding sites not only accommodates properly shaped sugars but also confines the cleavage type of substrates. The TmLamCD we report herein is supposed to be an endo-acting glycosyl hydrolase that performs endo-cleavage of β-1,3-glucans (38). However, if the active site adopts the closed configuration caused by the GASIG loop, the cavity suspiciously becomes suitable for exo-cleavage.
To identify this, the hydrolyzed products of laminarin by TmLamCD were analyzed by TLC. As expected, TmLamCD performs its endoglucanase activity with a preference to produce laminaritriose at the beginning of laminarin hydrolysis (supplemental Fig. S9). The trioses were then hydrolyzed to bioses and glucoses, showing that the TmLamCD also involves exoglucanase and β-glucosidase activity. Similar results in the activity analysis of endo-β-1,3-glucanase from Thermotoga petrophila were also observed (39).
In some exoglucanases, such as chitinase B from Serratia marcescens (40) and cellobiohydrolase II (Cel6A) from Trichoderma reesei (41), the enzymes bind long chain substrates in an endo mode but catalyze them as an exocellulase or exochitinase. This structural feature is thought to be advantageous to substrate ends accessing the active site (42). The closed configuration we observed at the catalytic site of TmLamCD may have functional significance in switching the three types of activity.
Being an endoglucanase, the TmLamCD shows a preference in starting a cleavage on laminarin to produce laminaritrioses, as shown in supplemental Fig. S9. In our modeling of extended β-1,3-glucans based on the laminaritriose structure in PDB 3N9K, the GASIG loop in the open form is found to be interactive with both +3 and +2 subsites of laminarin (supplemental Fig. S10). The long chain sugar may be pulled forward by the flexible loop when it changes from the closed form to an open form through van der Waals interaction. This may explain why TmLamCD produces most laminaritriose in the catalysis. Sugars extended further toward the reducing end, such as subsite +4, may protrude away from the protein and be surrounded by solvent. No evident dragging force toward the extra protruding pyranose can be observed.
An enclosure at the TmLamCD catalytic cavity is formed by Gly-158 and Trp-232, which is conservative in 2VY0, 3ILN, and 2HYK. Two kinds of side chain orientation have been observed in this conserved Trp (supplemental Fig. S11A). This agrees with our finding of dual occupancy of the Trp-232 side chain in some of our determined structures. However, the electron density map of Trp-232 side chain in the direction similar to the one of 2HYK and 3ILN overlaps with the side chain of Ile-161 in the closed form of the GASIG loop (supplemental Fig. S11B). Therefore, we only built the coordinate without structural hindrance. Interestingly, closely similar to our observation, this conserved Trp in chain A of 2VY0 was also built in dual occupancy in the PDB structure deposited by Ilari et al. (10). This Trp (numbered as 270 in 2VY0) also forms a closure with a nearby loop as TmLamCD in the open form, different from the catalytic cavity structures in 2HYK and 3ILN (supplemental Fig. S11, C–E). Further mutational studies on the conserved Trp-232 of TmLamCD and the relative flexible GASIG loop will be more informative to identify the real role of the structural enclosure.
Microbial carbohydrate-active enzymes are found to be modular enzymes in many cases. The coexistence of two CBMs with the catalytic domain contributes to efficient catalysis in TmLam (2). Unlike the fitting of the curvature of the β-1,3 linked ligand with the orientation of aromatic ring planes in TmLamCBM2 (2), the stacking between Trp side chains and glucosyl residues is more variable in TmLamCD and TmCel12A. This may correlate with their dynamic requirements during catalysis, including signaling right positions in substrate binding, linkage cleavage, and product release. In the era of energy deficiency, solar energy stored in plants and marine algae may be a good substitute for fossil fuel. Carbohydrate-degrading enzymes with efficient catalysis appear to be pivotal. Modular enzymes with domains having a synergistic effect to facilitate catalysis have potential utility in biocatalyst design. The structural investigation of the catalytic domain of TmLam in combination with its CBMs can be expected to have a practical application in biomass degradation.
Supplementary Material
Acknowledgments
We thank Hui-Lin Shr of the Core Facility for protein production and x-ray structural analysis supported by NRPGM for crystal screening and Dr. Yan-Chung Lo for help in TLC analysis. We are grateful to NSRRC in Taiwan and SPring-8 in Japan for beam time allocations and data collection assistance.
This work was supported by Academia Sinica and National Science Council (Taiwan) Grants NSC96-3114-P-001-004, NSC97-3114-P-001-001, and NSC99-3113-B-001-001 (to A. H.-J. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S11.
- Lam
- laminarinase
- CBM
- carbohydrate-binding module
- ctab
- cetyltrimethylammonium bromide
- GH
- glycosyl hydrolase
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- TmCel12A
- cellulose 12A from T. maritima
- TmLam
- laminarinase from T. maritima
- TmLamCBM
- CBM of TmLam
- TmLamCD
- catalytic domain of TmLam.
REFERENCES
- 1. Nelson K. E., Clayton R. A., Gill S. R., Gwinn M. L., Dodson R. J., Haft D. H., Hickey E. K., Peterson J. D., Nelson W. C., Ketchum K. A., McDonald L., Utterback T. R., Malek J. A., Linher K. D., Garrett M. M., Stewart A. M., Cotton M. D., Pratt M. S., Phillips C. A., Richardson D., Heidelberg J., Sutton G. G., Fleischmann R. D., Eisen J. A., White O., Salzberg S. L., Smith H. O., Venter J. C., Fraser C. M. (1999) Nature 399, 323–329 [DOI] [PubMed] [Google Scholar]
- 2. Boraston A. B., Nurizzo D., Notenboom V., Ducros V., Rose D. R., Kilburn D. G., Davies G. J. (2002) J. Mol. Biol. 319, 1143–1156 [DOI] [PubMed] [Google Scholar]
- 3. Zverlov V. V., Volkov I. Y., Velikodvorskaya G. A., Schwarz W. H. (2001) Microbiology 147, 621–629 [DOI] [PubMed] [Google Scholar]
- 4. Burstein T., Shulman M., Jindou S., Petkun S., Frolow F., Shoham Y., Bayer E. A., Lamed R. (2009) FEBS Lett. 583, 879–884 [DOI] [PubMed] [Google Scholar]
- 5. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khosla C., Harbury P. B. (2001) Nature 409, 247–252 [DOI] [PubMed] [Google Scholar]
- 7. Henrissat B., Bairoch A. (1996) Biochem. J. 316, 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu H. M., Liu S. W., Hsu M. T., Hung C. L., Lai C. C., Cheng W. C., Wang H. J., Li Y. K., Wang W. C. (2009) J. Biol. Chem. 284, 26708–26715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fibriansah G., Masuda S., Koizumi N., Nakamura S., Kumasaka T. (2007) Proteins 69, 683–690 [DOI] [PubMed] [Google Scholar]
- 10. Ilari A., Fiorillo A., Angelaccio S., Florio R., Chiaraluce R., van der Oost J., Consalvi V. (2009) FEBS J. 276, 1048–1058 [DOI] [PubMed] [Google Scholar]
- 11. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 12. Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 13. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 14. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Brünger A. T. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 24–36 [DOI] [PubMed] [Google Scholar]
- 19. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 20. Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 21. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 22. Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 23. DeLano W. L. (2002) The PyMol Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 24. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 25. Söding J., Biegert A., Lupas A. N. (2005) Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fried B., Sherma J. (1982) Thin-layer Chromatography: Techniques and Applications, Marcel Dekker, New York [Google Scholar]
- 27. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 28. Vasur J., Kawai R., Andersson E., Igarashi K., Sandgren M., Samejima M., Ståhlberg J. (2009) FEBS J. 276, 3858–3869 [DOI] [PubMed] [Google Scholar]
- 29. Davies G., Henrissat B. (1995) Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 30. Jirk V., Kraxnerová B., Krumphanzl V. (1980) Folia Microbiol. 25, 24–31 [DOI] [PubMed] [Google Scholar]
- 31. Cheng Y. S., Ko T. P., Wu T. H., Ma Y., Huang C. H., Lai H. L., Wang A. H., Liu J. R., Guo R. T. (2011) Proteins 79, 1193–1204 [DOI] [PubMed] [Google Scholar]
- 32. Welfle K., Misselwitz R., Welfle H., Politz O., Borriss R. (1995) Eur. J. Biochem. 229, 726–735 [DOI] [PubMed] [Google Scholar]
- 33. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simpson P. J., Xie H., Bolam D. N., Gilbert H. J., Williamson M. P. (2000) J. Biol. Chem. 275, 41137–41142 [DOI] [PubMed] [Google Scholar]
- 35. Kitago Y., Karita S., Watanabe N., Kamiya M., Aizawa T., Sakka K., Tanaka I. (2007) J. Biol. Chem. 282, 35703–35711 [DOI] [PubMed] [Google Scholar]
- 36. Davies G. J., Tolley S. P., Henrissat B., Hjort C., Schülein M. (1995) Biochemistry 34, 16210–16220 [DOI] [PubMed] [Google Scholar]
- 37. Jeng W. Y., Wang N. C., Lin M. H., Lin C. T., Liaw Y. C., Chang W. J., Liu C. I., Liang P. H., Wang A. H. (2011) J. Struct. Biol. 173, 46–56 [DOI] [PubMed] [Google Scholar]
- 38. Chhabra S. R., Shockley K. R., Ward D. E., Kelly R. M. (2002) Appl. Environ. Microbiol. 68, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cota J., Alvarez T. M., Citadini A. P., Santos C. R., de Oliveira Neto M., Oliveira R. R., Pastore G. M., Ruller R., Prade R. A., Murakami M. T., Squina F. M. (2011) Biochem. Biophys. Res. Commun. 406, 590–594 [DOI] [PubMed] [Google Scholar]
- 40. Sikorski P., Sørbotten A., Horn S. J., Eijsink V. G., Vårum K. M. (2006) Biochemistry 45, 9566–9574 [DOI] [PubMed] [Google Scholar]
- 41. Boisset C., Fraschini C., Schülein M., Henrissat B., Chanzy H. (2000) Appl. Environ. Microbiol. 66, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., Eijsink V. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.