Background: The active site iron of [NiFe] hydrogenases is equipped with a carbonyl ligand of undetermined origin.
Results: The carbonyl ligand derives exclusively from the cellular metabolism, and the CO scavenger PdCl2 mediates severe retardation of hydrogenase-driven growth.
Conclusion: The data indicate multiple, growth mode-dependent biosynthetic pathways for the carbonyl ligand.
Significance: Understanding the intricate cofactor assembly of [NiFe] hydrogenase is crucial for hydrogen-based biotechnology.
Keywords: Bioenergetics, Biophysics, Energy Metabolism, Enzyme Processing, Metalloenzymes, Carbon Monoxide, Enzyme Maturation, FTIR Spectroscopy
Abstract
The O2-tolerant [NiFe] hydrogenases of Ralstonia eutropha are capable of H2 conversion in the presence of ambient O2. Oxygen represents not only a challenge for catalysis but also for the complex assembling process of the [NiFe] active site. Apart from nickel and iron, the catalytic center contains unusual diatomic ligands, namely two cyanides (CN−) and one carbon monoxide (CO), which are coordinated to the iron. One of the open questions of the maturation process concerns the origin and biosynthesis of the CO group. Isotope labeling in combination with infrared spectroscopy revealed that externally supplied gaseous 13CO serves as precursor of the carbonyl group of the regulatory [NiFe] hydrogenase in R. eutropha. Corresponding 13CO titration experiments showed that a concentration 130-fold higher than ambient CO (0.1 ppmv) caused a 50% labeling of the carbonyl ligand in the [NiFe] hydrogenase, leading to the conclusion that the carbonyl ligand originates from an intracellular metabolite. A novel setup allowed us to the study effects of CO depletion on maturation in vivo. Upon induction of CO depletion by addition of the CO scavenger PdCl2, cells cultivated on H2, CO2, and O2 showed severe growth retardation at low cell concentrations, which was on the basis of partially arrested hydrogenase maturation, leading to reduced hydrogenase activity. This suggests gaseous CO as a metabolic precursor under these conditions. The addition of PdCl2 to cells cultivated heterotrophically on organic substrates had no effect on hydrogenase maturation. These results indicate at least two different pathways for biosynthesis of the CO ligand of [NiFe] hydrogenase.
Introduction
Hydrogenases are metalloenzymes catalyzing the reversible heterolytic cleavage of dihydrogen into two protons and two electrons (1, 2). According to the metal content of the active site, these enzymes are classified into three groups of [NiFe] and [FeFe] hydrogenases and [Fe] hydrogenases, all of which emerged from convergent lines of evolution (3). Despite the different evolutionary origin, the catalytic centers of hydrogenases share common features. They contain at least one iron atom coordinated by one or two cysteine-stemming thiols. Additional ligation of the iron by the diatomic ligands carbon monoxide (CO) and cyanide (CN−) is suggested to maintain the low-spin iron (II) state (3). In case of the [Fe] hydrogenase, the CN− ligand is replaced by a pyridinol group that probably fulfills a similar function (4). Biosynthesis and incorporation of these non-proteinogenic ligands requires a precisely operating maturation machinery to ensure correct stoichiometry and arrangement of the ligands.
In contrast to iron-only hydrogenases, which are assembled under completely anaerobic conditions (3, 5, 6), a number of [NiFe] hydrogenases is synthesized and catalytically active under aerobic conditions (7–9). Hence, biosynthesis of the metal cofactors needs to be protected against detrimental effects of oxygen. [NiFe] hydrogenase maturation proceeds via entirely different and considerably more complex routes than that of [FeFe] hydrogenases (3, 9, 10). This is partly due to the fact that the active site contains two different metals. The nickel atom is coordinated by four cysteine-stemming thiols, two of which serve as bridging ligands to the iron. Furthermore, two cyanides and one carbonyl group are ligated to the iron. At least six specific accessory proteins, the so-called Hyp proteins, are involved in the synthesis of the NiFe(CN)2(CO) cofactor (for recent reviews see Refs. 3, 7, 10).
Hydrogenase-3 from Escherichia coli, which has been intensively investigated by Böck et al. (3), still represents the best established system for [NiFe] hydrogenase maturation. The HypF and HypE proteins of E. coli were identified to synthesize CN− from carbamoyl phosphate as the metabolic precursor (11–16). The CN− groups are then transferred to a transient HypC-HypD complex, which is discussed as a scaffold for the formation of the Fe(CN)2(CO) fragment of the active site (17). However, it is currently unknown whether CO is already a constituent of the complex at this stage. It is anticipated that the resulting cofactor intermediate is subsequently transferred to the apo-hydrogenase (13, 18–20). In a process assisted by the HypA and HypB proteins, nickel is inserted (21), and maturation of the active site-containing large subunit is completed through specific endoproteolytic cleavage of a C-terminal amino acid extension that triggers protein folding and the final assembly with the electron-transferring small subunit (22, 23).
While biosynthesis of the CN− ligands is fairly established, the origin of the CO ligand in [NiFe] hydrogenases remains open. Previous studies, conducted with Allochromatium vinosum, indicated that the 13C atom of the carboxyl group of acetate is preferentially incorporated into the CO ligand of [NiFe] hydrogenase in this anaerobic phototroph (16). For hydrogenase-2 of E. coli as well as for the regulatory hydrogenase of Ralstonia eutropha, it has been shown that carbamoyl phosphate serves as a precursor of the CN− ligands, but not for CO. Furthermore, infrared (IR)3 spectroscopy revealed that 13CO, externally supplied to hydrogenase-synthesizing cell cultures, is efficiently incorporated into the active site of the enzymes (12, 14). Taking an average CO concentration of 0.1 ppmv (24) in the northern hemisphere into account, it is theoretically conceivable that atmospheric CO serves as the source of the carbonyl ligand.
We have addressed the question of the origin of the CO ligand by employing the aerobic H2 -oxidizing proteobacterium R. eutropha H16 as a model. R. eutropha harbors at least three well characterized [NiFe] hydrogenases that sustain growth on H2 as the energy source and O2 as the terminal electron acceptor. The energy collected from this process can be used for autotrophic CO2 fixation (25). All three hydrogenases of R. eutropha are catalytically active in the presence of O2 and, hence, rely on an O2-tolerant maturation process (7). The RH of R. eutropha functions as an H2 sensor and controls H2-dependent synthesis of the two energy-converting [NiFe] hydrogenases (25).
The RH was selected for the present investigations for three reasons: 1) Previous IR analyses uncovered well resolved distinct absorption bands characteristic for CN− and CO (14, 26, 27); 2) the RH does not display multiple redox changes as commonly observed with [NiFe] hydrogenases (27); and 3) maturation of the RH requires fewer steps than that of the membrane-bound counterpart (27). In this study, we have screened a number of 13C-labeled compounds for their potential to act as direct or indirect precursors of the CO ligand in [NiFe] hydrogenase. The physiological, biochemical and spectroscopic data obtained in this study unambiguously show that the CO ligand is derived from the cellular metabolism. Experiments with palladium chloride as an efficient CO-specific scavenger revealed that CO gas formed within the cells serves as a crucial intermediate for carbonyl ligand biosynthesis under certain growth conditions.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
For experiments involving the regulatory [NiFe] hydrogenase of R. eutropha, a transconjugant of R. eutropha HF574 (28) harboring the expression plasmid pGE567 (henceforth designated pRH) was used for overproduction of the heterodimeric RHstop protein that carries a Strep®Tag II affinity tag instead of the last 55 amino acid residues of the C terminus of the small subunit (29).
For experiments with 13C-labeled fructose, glycerol, and acetate, R. eutropha cells were grown heterotrophically under hydrogenase-derepressing conditions under air in mineral salt medium containing a mixture of 0.2% (w/v) fructose and 0.2% (w/v) glycerol and tetracycline at a final concentration of 10 μg/ml, for plasmid maintenance (30). The cultures are incubated at 30 °C under continuous shaking at 100 rpm. In the case of CO limitation experiments, cells were cultivated in desiccators filled initially with a defined gas atmosphere. For heterotrophic cultures (in fructose-glycerol medium), the headspace gas composition was 85% N2, 10% CO2, and 5% O2 (all gas concentrations are given in v/v). Alternatively, the cells were grown lithoautotrophically in the same mineral medium used for heterotrophic cultures but without organic carbon sources. Instead, H2 and CO2 served as the energy and carbon sources and were provided in a gas mixture of 10% H2, 10% CO2, 5% O2, and 75% N2.
CO limitation was realized through addition of 1 mm PdCl2 (relative to the total culture volume) that was deposited in dialysis capsules (Carl Roth GmbH, Karlsruhe, Germany) sealed with a gas-permeable polytetrafluoroethylene (PTFE) membrane of 0.125 μm thickness (H. Saur Laborbedarf, Reutlingen, Germany).
13C Labeling
For labeling experiments with fructose-glycerol, the non-labeled substrates were replaced by their counterparts containing 13C atoms at different positions, i.e. D-[13C6]fructose, [13C3]glycerol, [1,3-13C2]glycerol, and [2-13C]glycerol. [1-13C] and [2-13C]acetate (99%) were added directly to the fructose-glycerol medium at concentrations indicated in the text. Labeling experiments with gaseous 13CO (< 2% 18O) were performed in rubber-sealed 1100-ml serum bottles containing 200 ml fructose-glycerol medium. CO was added at concentrations indicated in the text. In this setup, the cells were grown for 72 h under an atmosphere of 70% N2 and 30% O2. All isotopes were obtained from EURISO-TOP GmbH (Saarbrücken, Germany) with a purity greater than 99%.
Protein Purification
Strep-tagged RH was purified from 3- to 5-g cells (wet weight). The cell pellet was resuspended in buffer W (100 mm Tris-HCl (pH 8.0), 150 mm NaCl; 1 ml buffer W per g of cells), and cells were disrupted by two passages with 1.1 × 108 Pa through a French pressure cell (G. Heinemann, Schwäbisch Gmünd, Germany). Cell debris was removed by ultracentrifugation for 45 min at 90,000 × g and 4 °C. RH purification from the soluble cell extract was performed with Strep-Tactin® spin columns (IBA, Göttingen, Germany) following the protocol of the manufacturer. Portions of 700 μl of soluble extract were applied to the column, which was subsequently centrifuged at 700 × g and 4 °C. The spin columns were washed four times with 100 μl of buffer W. Protein elution took place by adding 6 × 150 μl of buffer BE (buffer W containing 5 mm d-Biotin). RH-containing fractions were pooled, and the buffer was exchanged by buffer S (10 mm Tris-HCl (pH 8.0), 5% (w/v) glycerol) using Amicon Ultra-15 and Microcon filtration devices (Millipore). Finally, the samples were concentrated to a final volume of 20 μl with a protein concentration ranging between 8.9 and 31.5 mg/ml.
Infrared Spectroscopy
Infrared spectra were recorded on a Bruker Tensor 27 spectrometer equipped with a liquid nitrogen-cooled Mercury-cadmium-telluride detector at a spectral resolution of 2 cm−1. The sample compartment was purged with dry air, and the sample was held in a temperature-controlled (1.0 °C) gas-tight liquid cell (volume 10 μl, optical path length = 50 μm) with CaF2 windows at 10 °C. Spectra were corrected by using a spline function implemented within OPUS 6.5 software supplied by Bruker.
Western Immunoblot Analysis
Proteins were separated using denaturing polyacrylamide gel electrophoresis (31). Western immunoblot analysis was performed according to a standard protocol (32). For immunological detection of hydrogenase subunits, HoxG and HoxH antisera were applied at a dilution of 1:10,000.
Determination of Hydrogenase Activity in Whole Cells
The activity of the NAD+-reducing soluble hydrogenase (EC 1.12.1.2) was determined with a previously described whole-cell assay (33), except that 20 μl of 48 mm NAD+ were added per ml of reaction volume.
RESULTS
The Carbonyl Ligand of the [NiFe] active Site Originates from the Cellular Metabolism
For investigations on the origin of the CO ligand in [NiFe] hydrogenase, we used a truncated form of the RH (RHstop). This RH variant is still active in H2 oxidation but no longer able to interact with its cognate histidine protein kinase (34). The RHstop protein is encoded as a Strep-tagged version on the overexpression plasmid, pRH, under control of the strong PSH promoter of the soluble, NAD-reducing hydrogenase of R. eutropha. Plasmid pRH was transferred to the hydrogenase knockout derivative R. eutropha HF574, which carries in-frame deletions in the large subunit genes of both energy-converting hydrogenases and the RH structural genes hoxBC. For RH overproduction, the transconjugant strain was grown heterotrophically in a minimal medium supplemented with fructose and glycerol as the energy and carbon sources. Under these conditions, R. eutropha displays classical diauxic growth. Upon exhaustion of the preferentially utilized fructose, the cells switch to the poor substrate glycerol. The resulting decrease of the growth rate because of energy limitation provides the signal for enhanced hydrogenase gene expression (30).
To investigate whether the CO ligand is derived from the carbon/energy sources, cells were cultivated with all combinations of unlabeled and uniformly 13C-labeled fructose and glycerol. Furthermore, differentially labeled 12C/13C-glycerol was used in combination with 12C6-fructose. In all experiments, the RH was purified from cells, harvested in the glycerol phase of growth, and the protein was subjected to IR spectroscopic analysis.
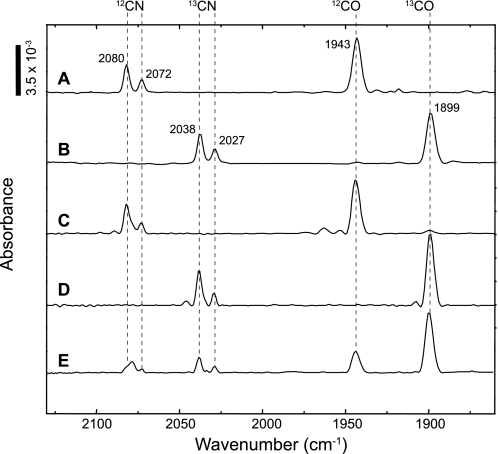
A typical IR spectrum of the unlabeled RHstop protein in the oxidized state (27) is shown in Fig. 1A. The two absorptions at wavenumbers of 2080 cm−1 and 2072 cm−1 correspond to the 12C-cyanide ligands, whereas the 12C-carbonyl ligand shows a CO stretching vibration at 1943 cm−1. The RHstop protein isolated from cells cultivated on uniformly labeled 13C6-fructose and 13C3-glycerol showed a completely different signal pattern. All three bands were shifted to lower frequencies (Fig. 1B, νCN = 2038/2027 cm−1, νCO = 1899 cm−1). This characteristic shift of 42–45 wavenumbers is related to the heavier 13C isotope and proves that both the cyanides and the CO ligand originate from metabolic precursors that are synthesized from the growth substrates.
FIGURE 1.
IR spectroscopic analysis of the regulatory [NiFe] hydrogenase purified from cells grown heterotrophically on fructose-glycerol. The following substrates were employed: A, 12C6-fructose and 12C3-glycerol. B, 13C6- fructose and 13C3-glycerol. C, 13C6- fructose and 12C3-glycerol. D, 12C6-fructose and 13C3-glycerol. E, 12C6-fructose and 1,3-13C2-glycerol. Hatched lines indicate the ν(CN) and ν(CO) vibrational modes. Bands at wavenumbers 2080 cm−1 and 2072 cm−1 are attributed to the 12CN− ligands and the absorption at 1943 cm−1 to the 12CO ligand. Incorporation of the 13C atom leads to band shifts to 2038 cm−1 and 2027 cm−1 for the CN stretchings and to 1899 cm−1 for the CO stretching, respectively.
In the next series of experiments we analyzed the IR pattern of RHstop proteins isolated from cultures containing either 13C6-fructose/12C3-glycerol (Fig. 1C) or 12C6-fructose/13C3-glycerol (Fig. 1D). Noticeable, band shifts were exclusively observed with protein prepared from 13C3-glycerol-grown cells. This result supports the assumption that synthesis of the [NiFe] cofactor, including its diatomic ligands, proceeds only during growth on glycerol.
The fact that glycerol contains three carbon atoms, two of which are stereochemically identical, raised the question whether they are equally involved in CO ligand formation. To explore this question, we grew the cells on 12C-fructose and glycerol carrying 13C atoms only at the positions C1 and C3. The IR pattern of the corresponding RHstop protein revealed a ν(12CO): ν(13CO) stretching ratio of 1:4.1 ± 0.3 (Fig. 1E) instead of the initially expected ratio of 1:2. Although all three glycerol-derived carbons occur as CO ligands, the terminal carbon atoms appear to be preferentially incorporated into the cofactor. This observation is supported by a control experiment using [2-13C]glycerol as the substrate (supplemental Fig. S1).
Acetate Does Not Serve as a Precursor of the CO Ligand
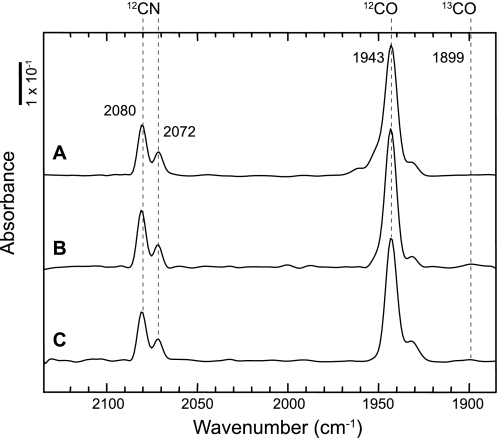
To examine if acetate serves as the precursor of the CO ligand in the RH of R. eutropha, cells were grown on fructose and glycerol. After the transition from fructose to glycerol growth, at an A436 nm of approximately 6.5, 13CH3COOH or CH313COOH, were added to the medium at a final concentration of 5 mm. As R. eutropha metabolizes acetate as a carbon and energy source (35), labeled acetate (2.5 mm, final concentration) was again added after 5 h, and cultivation was continued for another 19 h. The cells were harvested and used for purification of the RH. IR analysis did not show the slightest shift of neither the CO- nor CN−-related absorption bands (Fig. 2), supporting the notion that acetate is not the metabolic precursor of the carbonyl ligand of the R. eutropha [NiFe] hydrogenase.
FIGURE 2.
IR spectra of 13C-acetate-labeled RH purified from R. eutropha. RH-synthesizing cells were grown heterotrophically in fructose-glycerol medium supplemented with 5 mm (final concentration) of 12C-acetate (A), 1-13C-acetate (B), or 2-13C-acetate (C) at an A436 of 6.5 just at the beginning of hydrogenases synthesis. After 5 h of continuous cultivation, a further portion of acetate corresponding to 2.5 mm (final concentration) was added.
Gaseous Carbon Monoxide Serves as Precursor of the Carbonyl Ligand in Cells Grown on H2, CO2, and O2
It has been shown previously that externally added CO is incorporated as carbonyl ligand into E. coli hydrogenase-2 and recombinant RH (12, 14). This observation suggests that CO may be released from an internal metabolite prior to insertion into the [Ni-Fe] cofactor. If this is the case, trapping of CO from the culture medium should have an effect on active hydrogenase synthesis. To eliminate CO selectively, a novel approach was designed that exploits the specific CO-mediated turnover of palladium(II)chloride to metallic palladium,
which is a well established forensic method for specific detection of CO intoxication in human blood (36). As palladium(II)chloride is toxic to the cells, the PdCl2 solution was deposited in a dialysis capsule sealed with a gas-permeable membrane. In a first experiment, PdCl2-loaded capsules were added to R. eutropha cells grown heterotrophically on fructose-glycerol medium, which represented the condition under which the labeling of the RH by externally added 13CO has been observed previously (14). Palladium(II)chloride had no effect on growth (Fig. 3A), which was expected because heterotrophic growth is independent of hydrogenases, although these enzymes are synthesized at high levels under these conditions (30, 35). However, this result underlines that the membrane-sealed capsules indeed prevented leakage of toxic palladium into the medium. To test whether the energy-generating hydrogenases of R. eutropha were affected by the CO scavenger, the H2-dependent NAD+ reduction activity of the soluble [NiFe] hydrogenase (SH) of R. eutropha was measured. The SH activity determined in cells grown in the presence of PdCl2 was almost the same as in control cells grown without a CO scavenger (Fig. 3B). A complementary immunological analysis conducted at the end of the growth phase on fructose revealed no significant amount of hydrogenase (Fig. 3, C and D, lanes 1 and 4). After 35 h, however, during growth on glycerol, the large subunits of SH (HoxH) and membrane-bound hydrogenase (HoxG) occurred predominantly in the processed, mature form (Fig. 3, C and D, lanes 2 and 5), irrespective of the presence of PdCl2. These results indicate that hydrogenase maturation proceeded normally and was not affected by the limitation of the metabolic precursor of the carbonyl ligand. The reaction of PdCl2 with CO leads to the formation of metallic Pd that accumulates on the membranes of the capsules as silver-gray precipitate. Notably, no significant amounts of Pd precipitate were formed in the palladium capsules deposited in heterotrophically grown cell cultures (supplemental Fig. S2). These data strongly indicate that no significant amounts of gaseous CO are released by the cells and, therefore, may not serve as a precursor in carbonyl ligand synthesis under these conditions.
FIGURE 3.
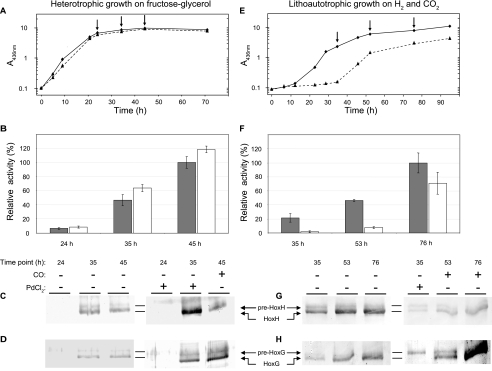
Effect of CO-limitation in R. eutropha H16. A and E, growth of cells (measured as A at 436 nm) cultivated under lithoautotrophic (H2, CO2, O2) and heterotrophic (fructose-glycerol, O2) conditions, respectively, with (dashed lines) and without (solid lines) CO limitation. CO limitation was induced at inoculation through addition of gas-permeable capsules containing PdCl2. After 35 h of cultivation, the capsules were removed, and 5000 ppmv of CO was added to the culture headspace. Samples were withdrawn at individual time points (arrows). Activity of the soluble, NAD+-reducing hydrogenase was assayed for lithoautotrophically (B) and heterotrophically cultivated (F) cells. Bars represent the activities measured in cultures with (white) and without (gray) initial CO limitation. An activity of 100% corresponds to 3.7 ± 0.4 Units/mg and 1.3 ± 0.1 Units/mg in B and F, respectively. Error bars represent the S.D. of at least three independent experiments. C, D, G, and H, corresponding immunological analysis of the large subunits, HoxH and HoxG, of the soluble and membrane-bound hydrogenase, respectively. Proteins were separated on denaturing 12% (w/v) SDS gels. The premature forms (68.8 kDa for pre-HoxG and 54.9 kDa for pre-HoxH) are slightly larger than the fully matured forms (67.1 kDa for HoxG and 52.3 kDa for HoxH).
A completely different picture was obtained when R. eutropha cells were grown lithoautotrophically in minimal medium with H2, CO2, and O2 in the presence or absence of the PdCl2-loaded capsules. The growth curve (Fig. 3E) clearly demonstrates that PdCl2 delayed growth of the cells by more than 24 h.
The retardation of H2-driven growth of PdCl2-treated cells correlated well with a diminished hydrogenase activity, which was measured as H2-dependent NAD+ reduction by the SH of R. eutropha (Fig. 3F). At low cell densities (i.e. after 35 h of cultivation), the PdCl2-treated cells showed an approximately 20-fold lower specific activity than cells grown in the absence of the PdCl2 capsules. Furthermore, a significant deposition of metallic palladium on the gas-permeable membrane was observed (supplemental Fig. S2), indicating that the cells had released a substantial amount of CO. The removal of the PdCl2 capsules and a concomitant addition of 5000 ppmv CO led to growth recovery and a significant increase in hydrogenase activity. After 76 h of cultivation, the hydrogenase activity of the formerly CO-depleted cells was only slightly lower than that of the untreated cells (Fig. 3F).
Parallel immunological analysis revealed that the low hydrogenase activity correlated with the occurrence of the unprocessed precursor form of the SH large subunit, HoxH. At 35 h, just before removal of the PdCl2 capsule, the processed and unprocessed forms appeared in almost equal amounts (Fig. 3G, lane 4), whereas in PdCl2-free cell cultures, most of the HoxH protein occurred in the processed form (lane 1). This effect became even more evident when the large subunit of MBH, HoxG, was examined (Fig. 3H, lane 4). The accumulation of the precursor forms of HoxH and HoxG clearly indicates incomplete assembly of the [NiFe] active site. We interpret this observation as limited availability of free CO in the formation of the carbonyl ligand of the iron caused by the CO-scavenging activity of PdCl2.
Titration of the Active Site Carbonyl Group in the Regulatory Hydrogenase with 13CO
The CO scavenging experiments described above revealed that free CO does not serve as the direct precursor of the carbonyl ligand during hydrogenase synthesis in cells grown on fructose-glycerol. However, previous studies showed that the metabolic precursor can be displaced by externally supplied gaseous CO (12, 14). This prompted us to conduct a titration experiment with 13C-labeled gaseous CO to monitor the incorporation of the carbonyl ligand into the [NiFe] active site. For this purpose, cells of R. eutropha HF574 harboring pRH were grown in 200 ml fructose-glycerol minimal medium in 1100-ml rubber-sealed serum flasks. The headspace contained 70% N2, 30% O2 (all gas concentrations are given in v/v) and varying concentrations of gaseous 13CO. RHstop was purified from glycerol-grown cells at an optical density of A436 nm = 10.0 and subsequently subjected to IR spectroscopy.
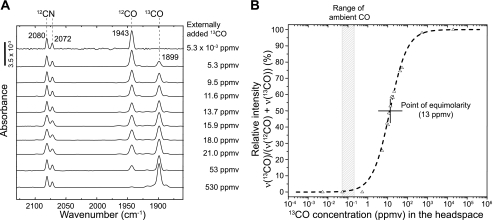
Upon addition of 530 ppmv 13CO, the CO stretching was completely shifted toward 1899 cm−1, indicating that externally added 13CO was entirely incorporated as carbonyl ligand at the catalytic center of the RH (Fig. 4A). A successive decrease of the 13CO concentration led to reappearance of the 12CO peak at 1943 cm−1, finally yielding the typical spectrum of oxidized, 12CO-carrying RHstop protein. The 13CO band intensities relative to the 12CO band heights were plotted against the CO concentration, and the resulting data points were fitted using a logistic function (Fig. 4B).
FIGURE 4.
Incorporation of gaseous 13CO into the RH of R. eutropha. A, IR spectroscopic analysis of RH purified from cells grown heterotrophically on fructose and glycerol at varying concentrations of gaseous 13CO added to the head space. The spectra were normalized on the CN stretchings. B, the peak height ratio (13CO peak height compared with the sum of 12CO and 13CO peak heights) as a function of 13CO concentration in the headspace, was fitted with a logistic, sigmoidal function: y(x0) = (Ai-Af/1+(x/x0)ρ)+Af. Fitting parameters: Ai (initial y value) = 0; Af (final y value) = 100; x0 center value: y(x0) = (Ai+Af)/2) = 13.0186 and ρ (rate) = 1.0135.
The point of equimolarity, which represents the external 13CO concentration that leads to a 50% labeling of the active site CO ligand, was found to be 13 ppmv. According to Henry's law and the van't Hoff equation, the amount of 13 ppmv corresponds to a CO concentration of 10.5 nmol/l in solution under the conditions used. This value is about 130-fold higher than the ambient CO concentration, which usually ranges between 0.05 and 0.2 ppmv, with an average of 0.1 ppmv in the northern hemisphere (24). Saturation with 13CO is met at concentrations approximately 5300 times higher than the ambient CO concentration. These data provide further evidence that the carbonyl ligand of the [NiFe] active site is of metabolic origin, suggesting a cellular concentration of its direct precursor in the nanomolar range.
DISCUSSION
[NiFe] active site assembly is a highly complex process involving at least six dedicated auxiliary Hyp proteins, whose function is rather well understood because of the elegant work carried out with E. coli (reviewed in Ref. 3). These studies shed light on the biosynthesis of the cyanide ligands (11–16), incorporation of the nickel ion into the active site (37–42), and folding of the active site pocket by proteolytic processing (22, 23). Two major questions remained open: the origin of the carbonyl ligand and the mechanism of its attachment to the iron atom.
In this study, we focused our attention on the origin of the CO ligand. Using the regulatory [NiFe] hydrogenase of R. eutropha as a model and IR spectroscopy as a powerful analytic tool, we show that the carbonyl ligand has a metabolic origin and that its cellular precursor occurs in nanomolar concentrations during heterotrophic growth on fructose-glycerol. Furthermore, application of the specific CO scavenger PdCl2 revealed that gaseous CO plays a major role in biosynthesis of RH during growth of R. eutropha on H2, CO2 and O2.
Glycine, serine, formate, N10-formyl-THF, acetate/acetyl-CoA, and CO2 have been discussed as possible precursors of the CO ligand in [NiFe] hydrogenases. Tyrosine appears to serve as the direct precursor for both the CO and CN− ligands in [FeFe] hydrogenases (5, 43–45). In this case, the conserved auxiliary protein HydG, which belongs to the class of radical S-adenosylmethionine proteins, mediates biosynthesis of the diatomic ligands. Although only little information is available on [Fe] hydrogenase maturation, it probably also involves radical chemistry (6). However, there is currently no indication that radical S-adenosylmethionine proteins are involved in maturation of [NiFe] hydrogenases, and so far only CO2 and acetate were tested experimentally as possible precursors for the CO ligand. Although CO2 could be excluded (14), Roseboom et al. (16) showed by IR spectroscopy that 20% of the active site carbonyl groups from the A. vinosum hydrogenase derive from the carboxyl group of 13C-labeled acetate, when the cells were grown photoautotrophically in the presence of 3 mm CH313COOH and 58 mm Na212CO3. Acetate represents a good carbon and energy source for R. eutropha (35), and, therefore, should be taken up rapidly by the organism. Thus, we cannot completely exclude that the provided 13C-acetate is metabolized before delivering components for the active site. However, even with twice the amount of labeled acetate, which had been used by Roseboom et al. (16), a 13CO-derived absorption band at 1899 cm−1 was not detected. Hence, our data indicate that acetate does not serve as direct precursor of the CO ligand for R. eutropha [NiFe] hydrogenases and, therefore, cannot be considered as universal.
Results from previous studies suggested that gaseous CO competes with a cellular precursor in the formation of the carbonyl ligand, allowing the possibility that even ambient CO may fulfill this function (12, 14). The carbon monoxide titration experiments provided an estimate of the concentration of the native precursor on the basis of its displacement by externally added 13CO. An excess of 130-fold 13CO relative to the ambient CO concentration of 0.1 ppmv is necessary to achieve a 50% labeling of the carbonyl ligand in the RH. This excludes the possibility that atmospheric CO contributes significantly to the synthesis of the carbonyl ligand. This conclusion has been confirmed by the quantitative labeling of both the CN− and CO ligands of the RH in cells grown with 13C-glycerol as the carbon and energy source. The 13CO titration experiments, performed with heterotrophically grown cells, revealed a rather low carbon monoxide concentration of 10.5 nmol/liter to be sufficient to outcompete the “natural” CO precursor. This is clearly below the cellular concentrations of most metabolites that have been determined for E. coli as a reference system (46) and can be interpreted as a so far unknown metabolic side reaction. Nonetheless, this low concentration fits to the hydrogenase content in the cell that has been proposed to be in the nanomolar range (12). Although a quantitative displacement of the cellular precursor of the carbonyl ligand by externally added CO is possible, it is unlikely that gaseous CO serves as the natural precursor under heterotrophic conditions. Addition of the CO scavenger PdCl2 did neither alter hydrogenase maturation nor activity under these conditions. The absence of metallic palladium in the capsules after cultivation indicates that no significant amounts of CO gas were formed. This also supports that CO ligand synthesis of [NiFe] hydrogenases is different from that of [FeFe] hydrogenases. In the latter case, both the CN− and CO ligands are synthesized anaerobically from tyrosine (44, 45). The accessory radical AdoMet protein HydG forms gaseous CO as determined spectroscopically using deoxyhemoglobin as a CO-sensitive reporter (45).
In contrast to observations with R. eutropha cells grown on fructose-glycerol medium, the application of CO limitation to cells cultivated with H2, CO2, and O2, led to a dramatic effect. Severe growth retardation was observed in the presence of the PdCl2-containing capsules, strongly indicating that gaseous CO is required for normal growth under these conditions. An obvious interpretation is that CO serves as the direct precursor for the carbonyl group of the [NiFe] active site, which is supported by the accumulation of unprocessed precursors of the large subunits of SH and membrane-bound hydrogenase upon CO limitation. Indeed, cells exposed to an excess of CO gas resumed growth at rates comparable with non-treated cells. These results show that CO plays an important role in maturation of the R. eutropha hydrogenases synthesized under lithoautotrophic growth conditions.
Besides CO formation of from tyrosine in the course of [FeFe] hydrogenase maturation, the results from this study revealed two different additional biosynthetic pathways for synthesis of the carbonyl ligand in the case of [NiFe] hydrogenases of R. eutropha. Future investigations in our lab will aim at the elucidation of the biosynthetic pathway that provides gaseous CO required for [NiFe] hydrogenase during lithoautotrophic growth of R. eutropha on H2 and CO2.
Supplementary Material
Acknowledgments
We thank Angelika Strack, Josta Hamann, and Janna Schoknecht for skillful assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) via the Cluster of Excellence “Unifying Concept in Catalysis” and the Sfb 498.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- IR
- infrared
- RH
- regulatory hydrogenase
- SH
- soluble hydrogenase.
REFERENCES
- 1. Lubitz W., Reijerse E. J., Messinger J. (2008) Energy Environ. Sci. 1, 15–31 [Google Scholar]
- 2. Fontecilla-Camps J. C., Volbeda A., Cavazza. C., Nicolet Y. (2007) Chem. Rev. 107, 4273–4303 [DOI] [PubMed] [Google Scholar]
- 3. Böck A., King P. W., Blokesch M., Posewitz M. C. (2006) Adv. Microb. Physiol. 51, 1–71 [DOI] [PubMed] [Google Scholar]
- 4. Shima S., Pilak O., Vogt S., Schick M., Stagni M. S., Meyer-Klaucke W., Warkentin E., Thauer R. K., Ermler U. (2008) Science 321, 572–575 [DOI] [PubMed] [Google Scholar]
- 5. Mulder D. W., Shepard E. M., Meuser J. E., Joshi N., King P. W., Posewitz M. C., Broderick J. B., Peters J. W. (2011) Structure 19, 1038–1052 [DOI] [PubMed] [Google Scholar]
- 6. Thauer R. K., Kaster A. K., Goenrich M., Schick M., Hiromoto T., Shima S. (2010) Annu. Rev. Biochem. 79, 507–536 [DOI] [PubMed] [Google Scholar]
- 7. Lenz O., Ludwig M., Schubert T., Bürstel I., Ganskow S., Goris T., Schwarze A., Friedrich B. (2010) Chemphyschem 11, 1107–1119 [DOI] [PubMed] [Google Scholar]
- 8. Schwartz E., Friedrich B. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., eds) Vol. 2, 3rd Ed., pp. 496–563, Springer, New York [Google Scholar]
- 9. Soboh B., Krüger S., Kuhns M., Pinske C., Lehmann A., Sawers R. G. (2010) FEBS Lett. 584, 4109–4114 [DOI] [PubMed] [Google Scholar]
- 10. Forzi L., Sawers R. G. (2007) Biometals 20, 565–578 [DOI] [PubMed] [Google Scholar]
- 11. Blokesch M., Paschos A., Bauer A., Reissmann S., Drapal N., Böck A. (2004) Eur. J. Biochem. 271, 3428–3436 [DOI] [PubMed] [Google Scholar]
- 12. Forzi L., Hellwig P., Thauer R. K., Sawers R. G. (2007) FEBS Lett. 581, 3317–3321 [DOI] [PubMed] [Google Scholar]
- 13. Jones A. K., Lenz O., Strack A., Buhrke T., Friedrich B. (2004) Biochemistry 43, 13467–13477 [DOI] [PubMed] [Google Scholar]
- 14. Lenz O., Zebger I., Hamann J., Hildebrandt P., Friedrich B. (2007) FEBS Lett. 581, 3322–3326 [DOI] [PubMed] [Google Scholar]
- 15. Reissmann S., Hochleitner E., Wang H., Paschos A., Lottspeich F., Glass R. S., Böck A. (2003) Science 299, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 16. Roseboom W., Blokesch M., Böck A., Albracht S. P. (2005) FEBS Lett. 579, 469–472 [DOI] [PubMed] [Google Scholar]
- 17. Blokesch M., Albracht S. P., Matzanke B. F., Drapal N. M., Jacobi A., Böck A. (2004) J. Mol. Biol. 344, 155–167 [DOI] [PubMed] [Google Scholar]
- 18. Blokesch M., Böck A. (2002) J. Mol. Biol. 324, 287–296 [DOI] [PubMed] [Google Scholar]
- 19. Blokesch M., Böck A. (2006) FEBS Lett. 580, 4065–4068 [DOI] [PubMed] [Google Scholar]
- 20. Watanabe S., Matsumi R., Arai T., Atomi H., Imanaka T., Miki K. (2007) Mol. Cell 27, 29–40 [DOI] [PubMed] [Google Scholar]
- 21. Zhang J. W., Butland G., Greenblatt J. F., Emili A., Zamble D. B. (2005) J. Biol. Chem. 280, 4360–4366 [DOI] [PubMed] [Google Scholar]
- 22. Bernhard M., Benelli B., Hochkoeppler A., Zannoni D., Friedrich B. (1997) Eur. J. Biochem. 248, 179–186 [DOI] [PubMed] [Google Scholar]
- 23. Thiemermann S., Dernedde J., Bernhard M., Schroeder W., Massanz C., Friedrich B. (1996) J. Bacteriol. 178, 2368–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conrad R. (1996) Microbiol. Rev. 60, 609–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgdorf T., Lenz O., Buhrke T., van der Linden E., Jones A. K., Albracht S. P., Friedrich B. (2005) J. Mol. Microbiol. Biotechnol. 10, 181–196 [DOI] [PubMed] [Google Scholar]
- 26. Pierik A. J., Schmelz M., Lenz O., Friedrich B., Albracht S. P. (1998) FEBS Lett. 438, 231–235 [DOI] [PubMed] [Google Scholar]
- 27. Bernhard M., Buhrke T., Bleijlevens B., De Lacey A. L., Fernandez V. M., Albracht S. P., Friedrich B. (2001) J. Biol. Chem. 276, 15592–15597 [DOI] [PubMed] [Google Scholar]
- 28. Buhrke T., Brecht M., Lubitz W., Friedrich B. (2002) J. Biol. Inorg. Chem. 7, 897–908 [DOI] [PubMed] [Google Scholar]
- 29. Buhrke T., Lenz O., Krauss N., Friedrich B. (2005) J. Biol. Chem. 280, 23791–23796 [DOI] [PubMed] [Google Scholar]
- 30. Schwartz E., Gerischer U., Friedrich B. (1998) J. Bacteriol. 180, 3197–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Friedrich B., Heine E., Finck A., Friedrich C. G. (1981) J. Bacteriol. 145, 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buhrke T., Löscher S., Lenz O., Schlodder E., Zebger I., Andersen L. K., Hildebrandt P., Meyer-Klaucke W., Dau H., Friedrich B., Haumann M. (2005) J. Biol. Chem. 280, 19488–19495 [DOI] [PubMed] [Google Scholar]
- 35. Friedrich C. G., Friedrich B., Bowien B. (1981) J. Gen. Microbiol. 122, 69–78 [Google Scholar]
- 36. Reimann W., Prokop O. (1980) Gerichtsmedizin, VEB Verlag Volk und Gesundheit, Berlin [Google Scholar]
- 37. Atanassova A., Zamble D. B. (2005) J. Bacteriol. 187, 4689–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blokesch M., Rohrmoser M., Rode S., Böck A. (2004) J. Bacteriol. 186, 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hube M., Blokesch M., Böck A. (2002) J. Bacteriol. 184, 3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jacobi A., Rossmann R., Böck A. (1992) Arch. Microbiol. 158, 444–451 [DOI] [PubMed] [Google Scholar]
- 41. Leach M. R., Sandal S., Sun H., Zamble D. B. (2005) Biochemistry 44, 12229–12238 [DOI] [PubMed] [Google Scholar]
- 42. Waugh R., Boxer D. H. (1986) Biochimie 68, 157–166 [DOI] [PubMed] [Google Scholar]
- 43. McGlynn S. E., Boyd E. S., Shepard E. M., Lange R. K., Gerlach R., Broderick J. B., Peters J. W. (2010) J. Bacteriol. 192, 595–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Driesener R. C., Challand M. R., McGlynn S. E., Shepard E. M., Boyd E. S., Broderick J. B., Peters J. W., Roach P. L. (2010) Angew. Chem. Int. Ed. Engl. 49, 1687–1690 [DOI] [PubMed] [Google Scholar]
- 45. Shepard E. M., Duffus B. R., George S. J., McGlynn S. E., Challand M. R., Swanson K. D., Roach P. L., Cramer S. P., Peters J. W., Broderick J. B. (2010) J. Am. Chem. Soc. 132, 9247–9249 [DOI] [PubMed] [Google Scholar]
- 46. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.