Background: IFN activates JAK-STAT signaling, where STAT1 phosphorylation is crucial for ISG induction and expression of IFIT2 to limit West Nile virus infection.
Results: IKKϵ mediates STAT1 serine 708 phosphorylation exclusive of tyrosine phosphorylation but dependent on nuclear export and ISG synthesis.
Conclusion: IKKϵ-mediated STAT1 S708 phosphorylation is crucial for IFIT2 expression to control WNV.
Significance: We define a novel anti-WNV innate immune effector pathway.
Keywords: Flaviviruses, Immunology, Innate Immunity, Interferon, Signal Transduction, STAT Transcription Factor, Viral Immunology, West Nile Virus
Abstract
West Nile virus is an emerging virus whose virulence is dependent upon viral evasion of IFN and innate immune defenses. The actions of IFN-stimulated genes (ISGs) impart control of virus infection, but the specific ISGs and regulatory pathways that restrict West Nile virus (WNV) are not defined. Here we show that inhibitor of κB kinase ϵ (IKKϵ) phosphorylation of STAT1 at serine 708 (Ser-708) drives IFIT2 expression to mediate anti-WNV effector function of IFN. WNV infection was enhanced in cells from IKKϵ−/− or IFIT2−/− mice. In IKKϵ−/− cells, the loss of IFN-induced IFIT2 expression was linked to lack of STAT1 phosphorylation on Ser-708 but not Tyr-701 nor Ser-727. STAT1 Ser-708 phosphorylation occurs independently of IRF-3 but requires signaling through the IFN-α/β receptor as a late event in the IFN-induced innate immune response that coincides with IKKϵ-responsive ISGs expression. Biochemical analyses show that STAT1 tyrosine dephosphorylation and CRM1-mediated STAT1 nuclear-cytoplasmic shuttling are required for STAT1 Ser-708 phosphorylation. When compared with WT mice, WNV-infected IKKϵ−/− mice exhibit enhanced kinetics of virus dissemination and increased pathogenesis concomitant with loss of STAT1 Ser-708 phosphorylation and IFIT2 expression. Our results define an IFN-induced IKKϵ signaling pathway of specific STAT1 phosphorylation and IFIT2 expression that imparts innate antiviral immunity to restrict WNV infection and control viral pathogenesis.
Introduction
West Nile virus (WNV)2 is an emerging flavivirus that has recently spread into the Western hemisphere from points of origin within Asia, Africa, or the Middle East (1, 2). Infection by WNV is now a leading cause of arboviral encephalitis and imparts 4% overall case fatality frequency in the United States. Typically maintained within avian reservoirs, WNV is spread to other vertebrates, including humans as dead-end hosts, through mosquito bite. WNV circulates as two major lineages and minor clades, with specific clades of lineage 1 representing the emergent and virulent strain in North America and elsewhere, whereas lineage 2 strains are typically endemic to Africa and Asia and are not known to cause disease in humans (1, 3–6). WNV infection is controlled in part through type I IFN immune defenses (7, 8). IFN actions comprise a major component of the innate immune response to virus infection, which serves to restrict virus replication and spread in part through the actions of interferon-stimulated genes (ISGs). WNV suppression of IFN signaling is linked to virus dissemination, neuroinvasion, and virulence of emergent lineage 1 strains (8, 9).
WNV acutely induces IFN-β expression from infected cells upon engagement of RNA products, including viral RNA by the RIG-I-like receptors, RIG-I and MDA5 (10–12). The RIG-I-like receptors signal the downstream activation of interferon regulatory factor (IRF) 3 and NF-κB transcription factors through the actions of the IKK-related kinases (Tank binding kinase 1 (TBK1) and inhibitor of κB kinase ϵ (IKKϵ)). As a result, IRF-3 and NF-κB activation drive the expression of IFN-β, other proinflammatory cytokines, chemokines, and direct IRF-3-target genes that confer antiviral and immune-activating functions (13, 14). Secreted IFN-β drives the innate immune response characterized by ISG expression. This process is triggered upon IFN-β binding to the interferon receptor (IFNAR) to induce receptor dimerization, autophosphorylation of receptor-associated kinases Tyk2 and JAK1 leading to tyrosine phosphorylation of signal transducer and activator of transcription (STAT)1 at residue Tyr-701, phosphorylation of STAT2, and assembly of the STAT1-STAT2-IRF9 ISGF3 complex (15–18). ISGF3 function is further augmented or sustained by STAT1 serine phosphorylation at residues Ser-727 and Ser-708 (19–21). ISGF3 translocates to the nucleus and triggers the transcription of hundreds of ISGs (13, 15). ISG products serve as immunomodulators and restriction factors against virus infection (22–24).
Among the ISGs, IFN-induced protein with tetratricopeptide repeats (IFIT)2, also known as ISG54, has been identified as an ISG restriction factor of WNV (23). IFIT2 belongs to the IFIT gene family whose members function to restrict virus infection through alteration of cellular protein synthesis (reviewed in Ref. 25), and IFIT2 mediates these action by inhibiting eIF3 function in translation initiation (26–28). Our recent study revealed that in the absence of IFN, ectopic IFIT2 expression can impose a blockade that ultimately restricts WNV replication. However, emergent WNV can evade IFIT2 restriction through 2′-O modification of the 5′ non-translated region of the viral RNA mediated by the methyltransferase activity of the viral NS5 protein (23). These observations define IFIT2 as a critical host factor of IFN action and WNV restriction, and underscore the IFIT2/WNV interaction as a critical virus/host interface governing innate antiviral immunity and infection outcome.
IFIT2 is expressed after virus infection directly upon IRF-3 activation as well as upon IFN signaling because of the presence of both IRF-3 and ISGF3 binding sites in the Ifit2 promoter (24, 29–31). However, STAT1−/− mice failed to induce IFIT2 expression in the CNS following Lymphocytic Choriomeningitis virus (LCMV) and WNV-infection in vivo (32). Importantly, IFN-induced expression of murine IFIT2 is dependent upon STAT1 Ser-708 phosphorylation, as recently described by tenOever et al. (21). In this respect, Ifit2 is among a set of ISGs, including Adar1, Mx1, and Oas1b, and others, whose promoters lack a purine-rich region upstream of their ISRE, which otherwise serves as STAT2 binding sites, wherein STAT1 Ser-708 phosphorylation is thought to increase the affinity for STAT1 binding sites to confer gene expression by ISGF3 in the absence of the STAT2 binding site (21). Notably, STAT1 Ser-708 phosphorylation is induced after IFN treatment of cells. However, the temporal relationship of Ser-708 phosphorylation to other STAT1 phosphorylation sites during IFN-stimulation, innate immune responses, or WNV infection is not known. We therefore conducted the current study to define the cell signaling pathway and STAT1 phosphorylation interactions that drive IFN-induced IFIT2 expression for the restriction of WNV infection. Our results reveal an IKKϵ-dependent pathway of STAT1 Ser-708 phosphorylation whose activation requires IFN signaling for ISG expression and that plays a key role in the temporal regulation of STAT1 phosphorylation and the expression of IFIT2 crucial to the control of WNV infection and immunity.
EXPERIMENTAL PROCEDURES
Cell Culture, Interferons, Viruses
Parental WT 2fTGH fibrosarcoma cells, U3A (2fTGH-derived mutant cells, deficient in STAT1) and U5A (IFRNAR-deficient, provided by Dr. George Stark, Cleveland Clinic), immortalized human PH5CH8 hepatocytes (provided by Dr. Nobuyuki Kato, Okayama University, Japan), Baby Hamster Kidney (BHK) cells, and HEK293 cells were grown in DMEM supplemented with 10% FBS, 2 mm L-glutamine, 1 mm sodium pyruvate, antibiotic-antimycotic solution, and 1× nonessential amino acids (complete DMEM). Primary mouse embryonic fibroblasts (MEFs) were isolated from IKKϵ−/−, IRF-3−/−, IFNAR−/−, IFIT2−/−, and age-matched WT control mice as described previously (7, 33), and grown in DMEM. Human IFNα-2a, human IFN-β, and murine IFNβ (PBL InterferonSource) were used at a concentration of 100 IU/ml, whereas human IFN-γ and IFN-λ1 (PBL InterferonSource) were used at concentrations of 50 ng/ml and 100 ng/ml, respectively. The Madagascar-AnMg798 strain of WNV (WNV-MAD) was obtained from the World Reference Center of Emerging Viruses and Arboviruses and passaged in Vero cells as described previously (8). The Cantell strain of Sendai virus (SenV) was obtained from the Charles River Laboratory. Where indicated, cells were mock-treated, treated with IFN, or infected with either WNV-MAD at an m.o.i. of 1 or SenV at 100 HA units/ml for the indicated times before harvesting for immunoblot assay (8, 34). Cycloheximide (CHX, Sigma) and leptomycin B (LMB, Sigma) was used at a concentration of 50 μg/ml and 100 nm, respectively. Pervanadate was prepared by mixing equal volumes of 50 mm H2O2 and 50 mm sodium orthovanadate to make 50 mm pervanadate before adding it into growth media for a final concentration of 50 μm.
Transfection and Promoter-Luciferase Analyses
pFLAG-IKKϵ, pFLAG-STAT1 WT, pFLAG-STAT1 Y701F, and pFLAG-STAT1 S727A expression plasmids were gifts from Dr. Curt Horvath (Northwestern University). pFLAG-STAT1 mutant constructs were generated by site-directed mutagenesis using the QuikChange XL site-directed mutagenesis kit (Stratagene) and the following primers: 5′-CAAGACTGAGTTGATTGCTGTGTCTGAAGTTC-3′ (forward) and 5′-GAACTTCAGACACAGCAATCAACTCAGTCTTG-3′ (reverse) for S708A; 5′-CAAGACTGAGTTGATTGATGTGTCTGAAGTTCACCCTTCTAGAC-3′ (forward) and 5′-GTCTAGAAGGGTGAACTTCAGACACATCAATCAACTCAGTCTTG-3′ (reverse) for S708D; and 5′-GATGGCCCTAAAGGAACTGGAGAGATCAAGACTGAGTTG-3′ (forward) and 5′-CAACTCAGTCTTGATCTCTCCAGTTCCTTTAGGGCCATC-3′ (reverse) for Y701E.
pIFIT2-Luc was a gift from Kineta, Inc. (Seattle). pISG15-Luc and pIFN-β-Luc were described previously (34). pADAR1-CAT was a gift from Dr. Charles Samuel (University of California-San Diego), and pADAR1-Luc was generated by subcloning into the pGL3 Luciferase reporter vector. Transfections were carried out for 16 h using FuGENE 6 transfection reagent (Roche) and following the manufacturer's instructions. For luciferase analyses, cells were cotransfected with either the IFN-β-Luciferase, ISG15-Luciferase, or ADAR1-Luciferase construct; CMV-Renilla; and the indicated cDNA expression plasmids. Cell extracts were collected 24 h post-infection or post-treatment and analyzed for dual luciferase activity (Promega).
In Vivo Mouse Infection
C57BL/6 (Bl6) WT and IKKϵ−/− mice were purchased from The Jackson Laboratory (21). Mice were bred in the animal facility at the University of Washington under specific pathogen-free (SPF) conditions. Experiments using these animals were completed within the approval and guidelines by the University of Washington Institutional Animal Care and Use Committee. IKKϵ−/− and age-matched WT control mice were inoculated subcutaneously in the left rear footpad with 103 or 104 pfu of WNV-MAD as described previously (12). Mice were monitored daily for morbidity and mortality. Clinical symptoms were numerically scored: 1, ruffled fur/lethargic/hunched/no paresis; 2, very mild to mild paresis; 3, frank paresis in at least one hind limb or mild paresis in two hind limbs; 4, severe paresis, still retains feeling and possibly limbic; 5, true paralysis; 6, moribund; 7, dead (12). For the in vivo viral burden analysis, infected mice were bled and perfused with 20 ml of PBS following euthanasia. Spleens were collected and homogenized for immunoblot analysis as described previously (12).
Immunoblot Analysis
Protein extracts were prepared by lysing cells in radioimmune precipitation assay buffer (50 mm Tris HCl (pH 7.5), 150 mm NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 1 mm EDTA, and 0.1% sodium dodecyl sulfate) supplemented with 1 μm okadaic acid, 1 μm phosphatase inhibitor mixture II (Calbiochem), and 10 μm protease inhibitor (Sigma), followed by 4 °C centrifugation at 16,000 × g for 10 min to clarify the lysate. Equivalent protein amounts were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting. Affinity-purified rabbit polyclonal anti-STAT1 Ser-708 antibody was generated by repeat-immunization with the STAT1 Ser-708 phospho-specific peptide (YIKTELI{pS}VSEVHP, amino acids 701–714) (GenScript). The following primary antibodies were used for immunoblot analyses: α-ADAR1 (Abnova); α-IRF-3 (M. David, UCSD); α-IFIT1, α-murine IFIT2 and α-murine IFIT3 (G. Sen, Cleveland Clinic); α-ISG15 (A. Haas, Louisiana State University); α-WNV (Centers for Disease Control and Prevention); α-p-STAT1 Tyr-701, α-p-STAT1 Ser-727, α-STAT1, α-p-IRF-3 (Cell Signaling Technology, Inc.); α-murine IRF-3 (Invitrogen); α-IKKϵ (Imgenex); α-PKR (Santa Cruz Biotechnology, Inc.); α-SenV (Biodesign International); α-FLAG (M2), and α-Tubulin (Sigma). HRP-conjugated goat anti-rabbit, goat anti-mouse, and donkey anti-goat (Jackson ImmunoResearch Laboratories, Inc.) were used as secondary antibodies.
Immunoprecipitation
Following FLAG-STAT1 reconstitution of U3A cells and IFN-β treatment, cell extracts were immunoprecipitated using α-FLAG (M2)-conjugated agarose beads (Sigma) for 2 h at 4 °C. Samples were then washed three times with radioimmune precipitation assay buffer before elution. The eluate was heated for 5 min and analyzed by SDS gel electrophoresis and immunoblot analysis. Subsequently, 20 μl of 50% slurry of protein G-agarose beads (Calbiochem) was added and incubated for 2 h at 4 °C. Beads were washed and eluted as described above. Clean-Blot HRP-conjugated immunoprecipitation detection reagent (Thermo Scientific) was used as a secondary antibody for the immunoblot assay.
RESULTS
IKKϵ and IFIT2 Impose Restriction of WNV Infection
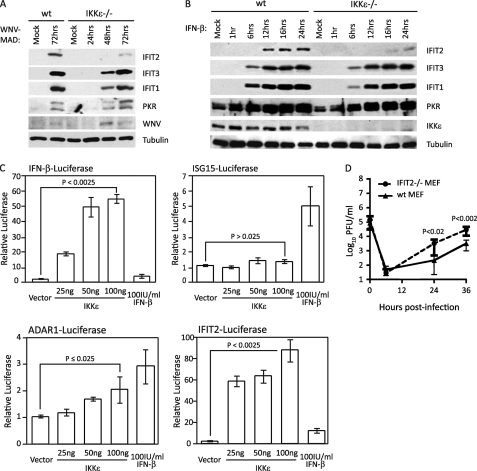
To determine the role of IKKϵ in IFIT2 expression during WNV infection, we evaluated the IFIT2 abundance and virus replication in primary mouse embryonic fibroblasts (MEF) isolated from WT or IKKϵ−/− mice. For these studies we utilized the avirulent lineage 2 Madagascar strain of WNV (WNV-MAD). As opposed to virulent lineage 1 strains, WNV-MAD lacks the ability to block IFN signaling and is highly sensitive to the innate immune antiviral actions of IFN (8), thus allowing studies of IFN actions against WNV without confounding influences of viral antagonism of the IFN response. As shown in Fig. 1A, WNV infection induced the expression and accumulation of IFIT2 in a manner dependent on IKKϵ, but IFIT1, IFIT3, and PKR were induced by WNV regardless of IKKϵ. Following WNV infection of wild-type MEFs, IFIT2 was expressed 48 h post-infection, whereas IFIT1 and IFIT3 expression was first observed by 24 h (supplemental Fig. S1). However, virus-induced expression of IFIT2 was severely attenuated in IKKϵ−/− MEF infected with WNV, whereas induction of IFIT1 and IFIT3 expression remained comparable with WT controls (Fig. 1A). Furthermore, in absence of IKKϵ, we observed delayed and impaired IFIT2 expression following IFN-β stimulation, demonstrating that IKKϵ was important for IFIT2 induction in MEFs. In contrast, IFN-β stimulation efficiently induced the expression of IFIT1, IFIT3, and PKR regardless of IKKϵ expression (Fig. 1B). To further assess the specific role of IKKϵ in regulating ISG expression, we evaluated the ISG promoter induction in HEK293 cells ectopically expressing IKKϵ. Ectopic overexpression of IKKϵ has been shown to induce its multimerization through the coiled-coil domain, causing its trans-autoactivation. Trans-autoactivation of IKKϵ results in its signaling to activate downstream substrates (such as IRF-3, IκBα, and AKT) and induce the expression of target genes (35–37). We found that ectopic expression of IKKϵ induced activation of IFN-β-promoter (which contains IRF-3 but not the ISGF3 binding site) occurred in a dose-dependent manner, whereas treatment of cells with exogenous IFN-β did not induce promoter expression, as expected (Fig. 1C, p < 0.0025, top left panel). Importantly, we observed a dose-dependent promoter activation of ADAR1 and IFIT2 (Fig. 1C, p ≤ 0.025, bottom panels) but not ISG15 upon IKKϵ ectopic expression (C, p > 0.025, top right panel), further demonstrating the role of IKKϵ in induction of an ISG subset recently shown to be sensitive to IKKϵ and STAT1 Ser-708 phosphorylation (21). Unlike ADAR1, whose promoter contains ISRE but not IRF-3 binding sites, the IFIT2 promoter contains both sites, each of which are IKKϵ responsive. Our results show that IKKϵ is essential for both virus-induced and IFN-induced IFIT2 expression, demonstrating dual roles of IKKϵ to induce innate effector ISG expression.
FIGURE 1.
IKKϵ and IFIT2 impose restriction on WNV infection. WT or IKKϵ−/− MEFs were mock-infected or infected with WNV-MAD at an m.o.i. of 1 (A) and mock-stimulated or stimulated with 100 IU/ml IFN-β (B). Protein lysate was collected at the indicated times and immunoblotted using IFIT2, IFIT3, IFIT1, PKR, and IKKϵ antibodies. Tubulin was used as loading control. C, HEK293 cells were cotransfected with pCMV-Renilla and either pIFN-β-Luciferase (top left panel), pISG15-Luciferase (top right panel), pADAR1-Luciferase (bottom left panel), or pIFIT2-Luciferase (bottom right panel). 16 h later, cells were either mock-stimulated (vector cotransfection); stimulated by transfection with 25 ng, 50 ng, or 100 ng of an IKKϵ expression plasmid; or treated with 100 IU/ml IFN-β. Cells were harvested 48 h post-transfection, and luciferase expression was measured and normalized to Renilla. The relative luciferase value was calculated as fold induction over induction of the vector that was set to 1. Statistical analysis was performed with Student's t test. D, WT or IFIT2−/− MEFs were infected with WNV-MAD at an m.o.i. of 5. At the indicated time points post-infection, culture supernatants were collected, and virus titers were determined by plaque assay on BHK cells. C and D, data are mean ± S.D. of three independent experiments performed in triplicate. p values were calculated using Student's t test to determine statistical significance.
Because IFIT2 expression is induced by WNV infection, we assessed the antiviral actions of IFIT2 in restricting WNV-MAD infection in MEFs from WT and IFIT2−/− mice. Culture supernatants were collected 0, 6, 24, and 48 h post-infection, where time 0 represents the input virus. Analysis of single-step virus growth revealed that WNV-MAD replication was enhanced in cells lacking IFIT2 compared with WT cells, thus validating IFIT2 function as a WNV restriction factor (Fig. 1D) (23). These results define IFIT2 as an IKKϵ-dependent ISG whose expression is induced by WNV infection and IFN-β to restrict WNV growth in primary cells.
Virus Infection Induces Delayed STAT1 Ser-708 Phosphorylation
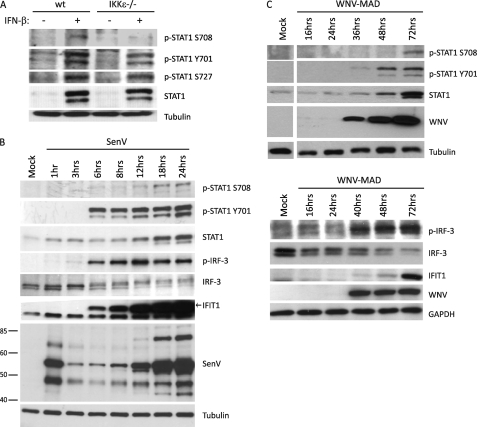
Because IFIT2 is induced by WNV infection in a manner dependent on IKKϵ, we sought to characterize the IKKϵ and STAT1 Ser-708 phosphorylation kinetics in response to cell treatment with IFN-β. IKKϵ has been shown previously to directly phosphorylate STAT1 Ser-708 in vitro (21). We therefore generated novel phospho-specific, affinity-purified polyclonal antibody against a phosphorylated peptide representing phospho-STAT1 Ser-708 (α-p-STAT1 Ser-708, supplemental Fig. S2, A–C). We used this antibody to assess STAT1 Ser-708 phosphorylation status after IFN-β-treatment, confirming that IKKϵ−/− MEFs were deficient in IFN-β-induced Ser-708 STAT1 phosphorylation. In contrast, IFN-β-induced STAT1 Tyr-701 and Ser-727 phosphorylation occurred independently of IKKϵ (Fig. 2A). IKKϵ-mediated STAT1 Ser-708 phosphorylation is independent of its role in IRF-3 activation, as activated IRF-3 efficiently translocated into the nucleus following SenV infection in the absence of IKKϵ (supplemental Fig. S3). These results demonstrate the specificity of the α-p-STAT1 Ser-708 antibody and confirm that IFN-induced STAT1 Ser-708 phosphorylation is dependent on IKKϵ expression.
FIGURE 2.
Virus infection induces delayed STAT1 Ser-708 phosphorylation. A, WT or IKKϵ−/− MEFs were mock-stimulated or stimulated with 100IU/ml IFN-β. Protein lysate was collected 16 h post-IFN stimulation and immunoblotted using p-STAT1 Ser-708, p-STAT1 Ser-727, p-STAT1 Tyr-701, and total STAT1 antibodies. B and C, Sendai and WNV virus infections induce STAT1 Ser-708 phosphorylation. HEK293 cells were infected with 100 HA units/ml SenV (B) or West Nile virus strain Madagascar (WNV-MAD) (C) at an m.o.i. of 1. At the indicated times following infection, protein lysates were collected and immunoblotted for p-STAT1 Ser-708, p-STAT1 Tyr-701, total STAT1, p-IRF-3, total IRF-3, IFIT1, and SenV or WNV. Tubulin and GAPDH were used as loading controls.
To further determine the kinetics of STAT1 Ser-708 phosphorylation in response to RNA virus infection, we analyzed p-STAT1 Ser-708 abundance during infection of HEK293 cells by the prototypic Paramyxovirus, SenV, or WNV-MAD (Fig. 2, B and C). Immunoblot analysis revealed that phosphorylated STAT1 (p-STAT1) Ser-708 occurred at later time points during the infection cycle of either virus, happening between 18 to 24 h post-infection with SenV and 72 h post-infection with WNV-MAD. In both SenV and WNV infection models, phosphorylation of Ser-708 appeared to be occurring much later than the canonical phosphorylation of STAT1 Tyr-701, which began at 6 h post-SenV infection and as early as 36 h post-WNV infection. Moreover, phosphorylation of STAT Tyr-701 was preceded by detectable levels of phosphorylated/activated p-(IRF-3), which is consistent with endogenous IFN being expressed to drive IFNAR signaling of STAT1 phosphorylation (Fig. 2B). Together, these results demonstrate that both SenV and WNV-MAD infections stimulate STAT1 Ser-708 phosphorylation late in the virus replication cycle and that that IFIT2 expression associates with IFN-induced accumulation of p-STAT1 Ser-708.
Type I, Type II, and Type III IFNs Induce STAT1 Ser-708 Phosphorylation
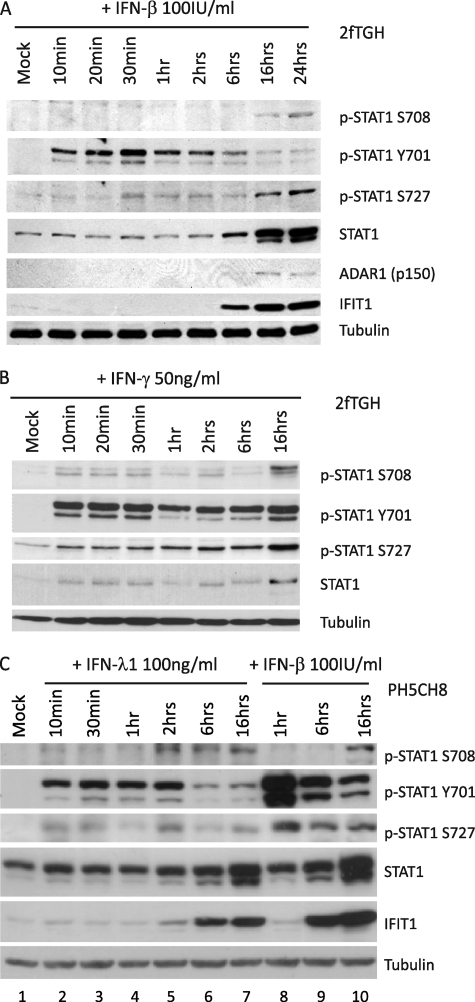
To determine whether different classes of IFNs stimulate STAT1 Ser-708 phosphorylation, we compared the ability of type I, II, and III IFNs to induce Ser-708 phosphorylation in 2fTGH human fibrosarcoma cells after treatment with IFN-β (type I IFN), IFN-γ (type II IFN), or IFN-λ (type III IFN). We found that IFN-β and IFN-γ treatment of cells induced STAT1 S708 phosphorylation after the onset of Tyr-701 and Ser-727 phosphorylation at a late time post-treatment and similar to the kinetics of p-STAT1 Ser-708 accumulation during WNV and SenV infection (Fig. 3, A and B). In IFN-β-treated cells, STAT1 was immediately phosphorylated at Tyr-701 and Ser-727 within 10 and 30 min after treatment, respectively. In contrast, STAT1 Ser-708 phosphorylation was first detectable at 16 h and peaked at 24 h post-treatment, which coincides with diminishing level of Tyr-701 phosphorylation and expression of ADAR1 (like IFIT2, an IKKϵ-dependent ISG). Likewise, when treated with IFN-γ, 2fTGH cells induced STAT1 Tyr-701 and Ser-727 phosphorylation within 10 min and through 16 h after treatment, whereas STAT1 Ser-708 phosphorylation occurred at 16 h post-treatment. To evaluate the ability of type III IFN to stimulate STAT1 Ser-708 phosphorylation, we examined the phosphorylation status of Ser-708 in lysates of PH5CH8 cells, an immortalized hepatocyte cell line that expresses endogenously the type III IFN receptor. Interestingly, we found that PH5CH8 cells treated with IFN-λ1 exhibited faster STAT1 Ser-708 phosphorylation kinetics compared with IFN-β treatment (Fig. 3C, lanes 5–7 and 10), with p-STAT1 Ser-708 accumulating within 2 h after IFN-λ1 treatment. In addition, we observed that p-STAT1 Ser-727 was weakly phosphorylated following IFN-λ1 treatment compared with IFN-β. Furthermore, STAT1 Tyr-701 phosphorylation was no longer sustained after 6 h of IFN-λ1 treatment, a finding which contrasted with the phosphorylation kinetics observed in IFN-β-treated cells. In all cases, we found that IFN treatment induces phosphorylation of STAT1 at the Tyr-701 residue before phosphorylation is detected on Ser-708 (Fig. 3, A–C). Together, these observations demonstrate that type I, II, and III IFN are all able to induce STAT1 phosphorylation at residue Ser-708, albeit with different kinetics.
FIGURE 3.
Type I, type II, and type III IFNs induce STAT1 Ser-708 phosphorylation. 2fTGH cells were mock-treated or treated with 100 IU/ml IFN-β (A) or 50 ng/ml IFN-γ (B). C, PH5CH8 cells were mock-treated (lane 1), treated with 100 ng/ml IFN-λ1 (lanes 2-6), or 100 IU/ml IFN-β (lanes 7-9). Protein lysate was collected at respective time points following IFN treatment and immunoblotted for p-STAT1 Ser-708, p-STAT1 Tyr-701, p-STAT1 Ser-727, total STAT1, ADAR1, and IFIT1.
Signaling through IFNAR Is Required for STAT1 Ser-708 Phosphorylation Following Type I IFN Treatment or Virus Infection
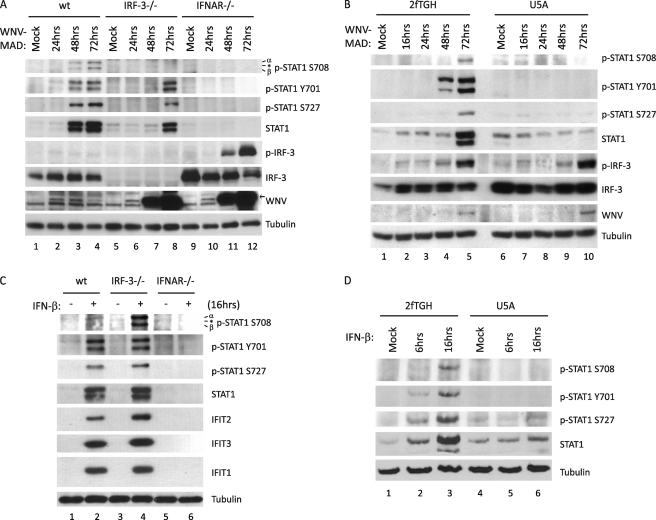
To evaluate the signaling requirements for WNV-MAD-induced STAT1 Ser-708 phosphorylation, we infected WT, IRF-3−/−, and IFNAR−/− MEFs with WNV-MAD (m.o.i. = 1) and analyzed STAT1 tyrosine and serine phosphorylation by immunoblot assay. As expected, phosphorylation of STAT1 at Tyr-701 and Ser-727 were ablated in the absence of IFNAR. Phosphorylation at Ser-708 was similarly blocked despite the high abundance of phospho/active IRF-3 and IFN-β secretion of IFNAR−/− MEFs, indicating that active signaling through IFNAR is also required for STAT1 Ser-708 phosphorylation during WNV-MAD infection (Fig. 4A, lanes 9–12, supplemental Fig. S4). Similarly, WNV-infected U5A human fibrosarcoma cells, which lack IFNAR2, also failed to induce STAT1 Ser-708 phosphorylation compared with parental 2fTGH cells (Fig. 4B) (38). Next, we evaluate the IRF-3-signaling requirement for WNV-MAD-induced STAT1 Ser-708 phosphorylation. In IRF-3−/− MEFs, there was a lack of STAT1 Ser-708 phosphorylation during WNV-MAD infection (Fig. 4A, lanes 5–8), a finding that might be explained by the cellular defect in IFN-β production in the absence of IRF-3 (supplemental Fig. S4) (39). Thus, to assess the possible outcome because of loss of IFN-β production and its impact on p-STAT1 Ser-708 accumulation in these cells, we compared p-STAT1 Ser-708 abundance in WT, IRF-3−/−, and IFNAR−/− MEFs after treatment with 100 IU/ml IFN-β. p-STAT1 Ser-708 levels in IRF-3−/− MEFs were similar or greater to the level found in WT MEFs after IFN-β treatment (Fig. 4C, lanes 3–4). However, IFN-β failed to induce STAT1 phosphorylation in IFNAR−/− MEFs and U5A cells (Fig. 4C, lanes 5–6, and Fig. 4D). Thus, IFNAR signaling but not IRF-3 signaling is required for STAT1 Ser-708 phosphorylation.
FIGURE 4.
Signaling through IFNAR is required for STAT1 Ser-708 phosphorylation following type I IFN treatment or virus infection. WT, IRF-3−/−, or IFNAR−/− (A) and parental 2fTGH cells or their derivative U5A cells (which lack IFNAR) (B) were infected with WNV-MAD at an m.o.i. of 1. Protein lysates were collected at the indicated time points and immunoblotted for p-STAT1 Ser-708, p-STAT1 Tyr-701, p-STAT1 Ser-727, total STAT1, p-IRF-3, total IRF-3, and WNV. C and D, the same cells were also mock-stimulated or stimulated with 100 IU/ml IFN-β for 6 or 16 h. Protein lysates were collected at the indicated time points and immunoblotted for p-STAT1 Ser-708, p-STAT1 Tyr-701, p-STAT1 Ser-727, total STAT1, p-IRF-3, total IRF-3, IFIT2, IFIT3, and IFIT1. Asterisk, nonspecific band.
STAT1 Ser-708 Phosphorylation Requires de Novo Protein Synthesis
Because STAT1 Ser-708 phosphorylation first requires IFN signaling, we sought to determine whether an IFN-responsive factor(s) might be required to induce p-STAT1 Ser-708 accumulation and the subsequent expression of IFIT2. To test this notion, we assessed p-STAT1 Ser-708 abundance in 2fTGH cells that were either mock-treated or treated with CHX for 30 min prior to a 16-h IFN-β treatment time course. We observed that IFN-β-induced STAT1 phosphorylation at Ser-708, but not Tyr-701, is abrogated when de novo protein synthesis is blocked (Fig. 5A, lanes 6–10, supplemental Fig. S5A, lane 3). We found that when cells were pretreated with CHX for 16 h and then subsequently treated with IFN-β for 1 h, p-STAT1 Tyr-701 still accumulated to high levels, revealing that available STAT1 molecules can be readily phosphorylated at Tyr-701 during long-term protein synthesis inhibition, and that IFN receptor signaling remains intact under these conditions. Thus, STAT1 phosphorylation on Tyr-701 does not require de novo protein synthesis (Fig. 5A, lane 11). These observations also demonstrate that the CHX-treated cells were viable and responsive following 16 hours of protein synthesis inhibition (supplemental Fig. S5B). Similar to treatment with IFN-β, IFN-γ-induced accumulation of p-STAT1 Ser-708, but neither Tyr-701 nor Ser-727 phosphorylation, was also blocked in the presence of CHX (data not shown). Taken together, these data show that de novo protein synthesis is required for STAT1 Ser-708 phosphorylation but not Tyr-701 or Ser-727 phosphorylation following treatment of cells with IFN-β or IFN-γ. Thus, STAT1 Ser-708 phosphorylation is induced and regulated though the actions of ISG product(s) whose expression precedes p-STAT1 Ser-708 accumulation. Furthermore, we found that STAT1 Ser-708 phosphorylation only occurred when CHX was added more than 9 h after the addition of IFN-β (Fig. 5A, lanes 7–9), suggesting that the ISG product(s) required for STAT1 Ser-708 phosphorylation is synthesized between 9 and 12 hours after the initiation of IFN-β treatment. In contrast, IFN-β stimulation of STAT1 Tyr-701 phosphorylation occurred regardless of CHX treatment, indicating that the requirement for a de novo synthesized IFN-responsive gene product is specific to Ser-708 phosphorylation (Fig. 5A, lanes 6-11). Because STAT1 itself is an ISG, we assessed whether or not de novo STAT1 expression is required for Ser-708 phosphorylation. We found that ectopic overexpression of STAT1 does not induce its phosphorylation at Ser-708. Moreover, in the presence of IFN-β, we did not observe an acceleration of STAT1 Ser-708 phosphorylation kinetics when compared with vector-transfected control cells (data not shown), demonstrating that de novo STAT1 expression does not immediately result in Ser-708 phosphorylation. Thus, an IFN-responsive factor(s) but not STAT1 itself is the primary ISG product(s) driving STAT1 Ser-708 phosphorylation and the IFN-induced expression IFIT2.
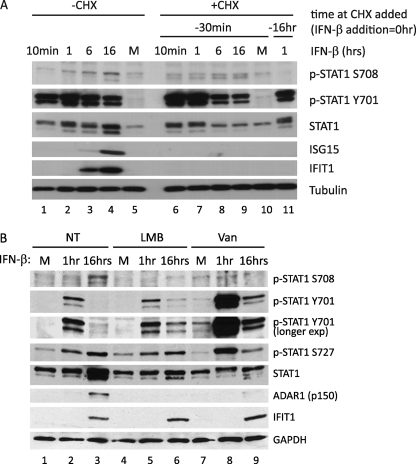
FIGURE 5.
STAT1 Ser-708 phosphorylation requires de novo protein synthesis, STAT1 tyrosine dephosphorylation, and nuclear export. A, 2fTGH cells were mock-treated (-CHX, lanes 1-5) or treated with CHX (+CHX, lanes 6-11) to block protein synthesis. At 30 min (lanes 6-10) or 16 h (lane 11) following CHX treatment, cells were mock-stimulated (M) or stimulated with IFN-β. Cells were harvested at 10 min as well as 1, 6, and 16 h post-IFN stimulation and immunoblotted to detect p-STAT1 Ser-708, p-STAT1 Tyr-701, total STAT1, ISG15, and IFIT1. B, 2fTGH cells were not treated (NT, lanes 1-3), pretreated with 100 nm LMB (lanes 4-6), or 50 mm pervanadate (Van, lanes 7-9) 1 h before mock stimulation or stimulation with 100 IU/ml IFN-β. Cells were harvested at 1 and 16 h post-stimulation. Immunoblot analysis was performed using p-STAT1 Ser-708, p-STAT1 Tyr-701, p-STAT1 Ser-727, total STAT1, ADAR1, and IFIT1 antibodies.
STAT1 Ser-708 Phosphorylation Requires STAT1 Tyrosine Dephosphorylation and Nuclear Export
Given the different kinetics of STAT1 phosphorylation at various phospho-residues following IFN-β treatment and the requirement for ISG expression to drive p-STAT1 Ser-708 accumulation, we investigated whether the phosphorylation of STAT1 Tyr-701 and Ser-708 are linked. We assessed the impact of STAT1 Tyr-701 phosphorylation on the accumulation of p-STAT1 Ser-708 by pretreatment of cells to sustain STAT1 Tyr-701 phosphorylation upon subsequent treatment with IFN-β. In the absence of inhibitor, IFN-β stimulation resulted in early induction of p-STAT1 Tyr-701. However, p-STAT1 Tyr-701 levels diminished after 16 h of IFN-β stimulation despite increased total STAT1 abundance (Figs. 5B, lanes 1-3, and 3A). Pretreatment of 2fTGH cells with CRM1 nuclear export inhibitor LMB or protein tyrosine-phosphatase (PTP) inhibitor pervanadate for 1 h before the start of IFN treatment resulted in the sustained accumulation of p-STAT1 Tyr-701 within IFN-treated cells (Fig. 5B, lanes 4-9, and Refs. 40–42). In agreement with a previous report, pervanadate treatment of cells induced low level STAT1 activation in the absence of IFN-stimulation and further enhanced STAT1 tyrosine phosphorylation following IFN stimulation (Fig. 5B, lanes 7-9, and Ref. 42). Importantly, there was an absence of p-STAT1 Ser-708 concomitant with the lack of IFN-induced ADAR1 expression (like IFIT2, an IKKϵ- and p-STAT1 Ser-708-dependent ISG (21)) in cells pretreated with either LMB (Fig. 5B, lanes 4–6) or pervanadate (Fig. 5B, lanes 7-9). However, STAT1 Ser-727 phosphorylation and non-IKKϵ-dependent ISG expression were effectively induced upon IFN-β treatment of these cells. This observation suggests that STAT1 phosphorylation at the Tyr-701 and Ser-708 residues are mutually exclusive and that removal of Y701 phosphorylation and subsequent STAT1 nuclear export are prerequisites the phosphorylation of STAT1 on Ser-708.
To further assess the relationship of p-STAT1 Tyr-701 and Ser-708, we evaluated STAT1 site-specific phosphorylation in STAT1-negative U3A cells reconstituted with transfected FLAG-tagged constructs containing either WT FLAG-STAT1, FLAG-STAT1 Y701E phosphomimetic, FLAG-Y701F phosphomutant, FLAG-S708A phosphomutant, FLAG-S708D phosphomimetic, FLAG-S727A phosphomutant, or a FLAG vector control. We assessed STAT1 phosphorylation at each site after cells were treated with IFN-β for 16 h. We found that although STAT1 Tyr-701 phosphorylation was absent in IFN-treated cells reconstituted with FLAG-STAT1 S708D, it was present in cells reconstituted with STAT1 S708A (supplemental Fig. S6, lane 6). Furthermore, we detected STAT1 Ser-708 phosphorylation only in cells reconstituted with the FLAG-STAT1 Y701F or FLAG-STAT1 S708D, the latter observation defining the FLAG-STAT1 S708D construct as a direct phosphomimetic recognized by our anti-phospho STAT1 Ser-708 antibody. STAT1 Ser-708 phosphorylation was not detected in cells expressing FLAG-STAT1 WT or FLAG-STAT1 S727A, both of which were phosphorylated on Tyr-701 (supplemental Fig. S6, lanes 2 and 7). In fact, we found that each of these constructs becomes immediately phosphorylated at Tyr-701 upon IFN treatment and are sustained as such throughout the course of IFN stimulation (data not shown). Cells reconstituted with FLAG-STAT1 Y701E failed to display Ser-708 phosphorylation (supplemental Fig. S6, lane 3). Thus, p-STAT1 Ser-708 likely takes place only after Tyr-701 dephosphorylation and nuclear export, which occurs approximately at 16 h post IFN-β stimulation.
IKKϵ Mediates IFIT2 Expression and Protection against WNV Pathogenesis in Vivo
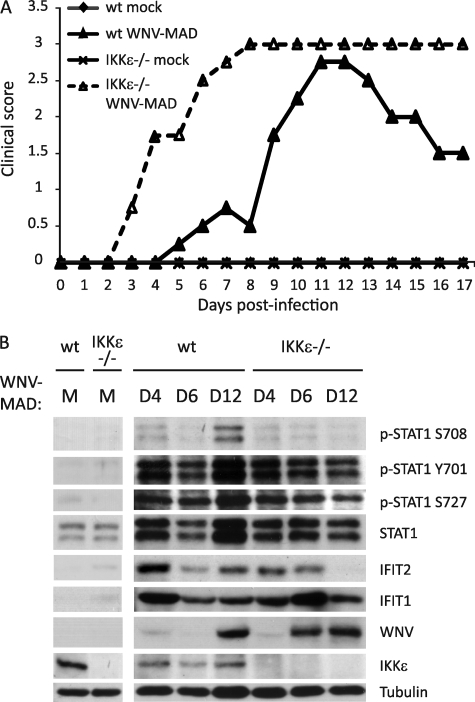
To determine the role of IKKϵ and STAT1 Ser-708 phosphorylation in IFIT2 expression and protection against WNV infection in vivo, we examined the response of WT and IKKϵ−/− mice to WNV challenge. WT and IKKϵ−/− mice were challenged with 103 pfu WNV-MAD by subcutaneous injection into the footpad. Clinical symptoms were monitored daily during the course of infection to observe the occurrence of disease and neurovirulence (12). When compared with the WT controls, IKKϵ−/− mice displayed earlier neurological symptoms, a higher degree of neurovirulence, and a failure to recover from acute WNV-MAD infection (Fig. 6A). Furthermore, we observed lack of sustained IFIT2 expression in the spleen of IKKϵ−/− mice during infection, and this associated with earlier virus entry to the spleen compared with WT controls (Fig. 6B). For comparison, we also challenged WT and IKKϵ−/− mice with the virulent/emergent lineage 1 Texas 02 strain of WNV (WNV-TX). This strain mediates a robust block to IFN signaling while evading the antiviral actions of IFIT2 (8, 23). As expected, we observed a similar but more rapid pathology defined by neurovirulence among WT and IKKϵ−/− mice infected with WNV-TX (data not shown). These observations reveal a dependence of IKKϵ for IFIT2 expression during WNV infection in vivo and demonstrate that IKKϵ plays a role in programming the innate immune response for the expression of IFIT2 and the control WNV infection. Our data also underscore the pathogenic outcome of WNV infection linked to viral evasion of IFN defenses.
FIGURE 6.
IKKϵ mediates IFIT2 expression and protection against WNV pathogenesis in vivo. WT Bl6 and IKKϵ−/− mice were mock-infected (PBS only) or infected with 103 pfu of WNV-MAD subcutaneously through footpad injection. A, mice were monitored and scored daily for clinical symptoms over 17 days. Clinical scores from four representative mice per group were graphed. B, spleens from WT or IKKϵ−/− mice, mock-infected or infected with WNV-MAD, were collected at days 4, 6, and 12 post-infection. Protein lysates were extracted by homogenizing spleens with radioimmune precipitation assay buffer and immunoblotted using p-STAT1 Ser-708, p-STAT1 Tyr-701, p-STAT1 Ser-727, total STAT1, IFIT2, IFIT1, WNV, and IKKϵ antibodies. The immunoblot analysis panel is a representative from four mice per infection group.
DISCUSSION
Our study identifies IFIT2 as an innate immune effector gene that can restrict WNV replication and defines the IKKϵ-mediated signaling pathway of IFN action that drives the expression of IFIT2 and a subset of ISGs through phosphorylation of STAT1 Ser-708. Furthermore, we reveal that this IKKϵ pathway is dependent on an ISG product(s) to stimulate the IKKϵ-directed STAT1 Ser-708 phosphorylation at late times in the IFN response. Recent studies have demonstrated the importance of a variety of ISGs in controlling WNV infection, such as Viperin, IFITM2, IFITM3, ISG20, PKR, and IFIT2 (23, 43). Moreover, the IFIT family members have been shown to suppress protein synthesis, thus restricting replication of Alphavirus, Papillomavirus, and hepatitis C virus (28, 44, 45). Our results now show that IFIT2 can restrict WNV growth in vitro and demonstrate that its expression within an IKKϵ-dependent innate immune effector pathway associates with the control of virus spread and pathogenesis in vivo. Although the magnitude of the increase of WNV-MAD replication in IFIT2−/− cells was only 10-fold (one log), it is notable that this difference was statistically significant and caused by loss of a single ISG of several hundred known ISGs, many of which might restrict WNV infection (43). These observations indicate the importance of IFIT2 in controlling the growth of WNV and indeed may in part explain the immune protection and lack of pathogencity after infection by low virulence WNV strains such as WNV-MAD and others in animals (8, 46, 47).
IKKϵ has multiple roles in activating the innate immune response to virus infection, including the phosphorylation and activation of IRF-3, which leads to IFN-β production and phosphorylation of STAT1 at residue Ser-708 following IFNAR signaling (21, 48, 49). The role of IKKϵ in STAT1 phosphorylation is independent of its role in IRF-3 activation, a role which is redundant with the related kinase TBK1. MEFs lacking TBK1 are deficient in IRF-3 phosphorylation, indicating that TBK1 and not IKKϵ is the dominant kinase for IRF-3 activation, at least in MEFs (50). We conclude that IKKϵ functions to induce STAT1 Ser-708 phosphorylation and a specific ISG expression signature that includes IFIT2 in WNV-infected cells. This conclusion is supported by our findings that ectopic overexpression of IKKϵ alone in human cells specifically stimulated IFIT2 and ADAR1 promoter induction, whereas IFN-induced IFIT2 expression is strictly linked to STAT1 Ser-708 phosphorylation and is dependent on IKKϵ in vitro and in vivo (see Figs. 1, 2, and 6) (21). Ectopic overexpression of a kinase such as IKKϵ induces its trans-autoactivation facilitated by its multimerization, therefore bypassing the requirement for upstream signaling (35, 36). In agreement with previous reports, IKKϵ overexpression also induces IFN-β promoter activation (Fig. 1C and Ref. 36), suggesting that general activation of IKKϵ target genes occurs upon its overexpression. However, IKKϵ expression alone did not induce the ISG15 promoter, an ISG that can be induced through canonical ISGF3 function, which instead required IFN treatment (see Fig. 1). These data suggest that the expression of a specific ISG subset that includes IFIT2, ADAR1, and others directly depends on IKKϵ, thus supporting the novel role for IKKϵ in antiviral immunity (21).
Our data now implicate this IKKϵ-dependent pathway and its specific expression of IFIT2 as important components of the innate immune response to WNV infection and indicate that this response is governed by IKKϵ phosphorylation of STAT1 Ser-708. IKKϵ−/− MEFs, which are deficient in STAT1 Ser-708 phosphorylation, showed defects in IFN-induced IFIT2 expression but not in other related ISGs, including IFIT3 and IFIT1. We found that STAT1 Ser-708 phosphorylation was induced by type I, II, and III IFNs in addition to being induced during infection by WNV or SenV. Moreover, the kinetics of IFN-induced STAT1 Ser-708 phosphorylation varied from phosphorylation at Tyr-701 and Ser-727. Although STAT1 Tyr-701 and Ser-727 phosphorylation occurred immediately following type I and type II IFN stimulation as known previously (reviewed in Ref. 15), Ser-708 phosphorylation occurred later, ∼16 h post-IFN treatment. In comparison, type III IFN-induced phosphorylation of STAT1 Ser-708 occurred more rapidly. This observation agrees with a previous report that demonstrated the differential kinetics and duration of JAK-STAT signaling activity induced by type I and III IFN (51) and suggests that antiviral immune actions of these IFNs are each mediated in part through STAT1 Ser-708-responsive ISGs. Similarly, STAT1 Ser-708 phosphorylation was induced at later times following RNA virus infection, with delayed kinetics associated with WNV-MAD compared with SenV infection, and likely because of the slower growth rate and IFN induction of the former. These observations indicate that WNV and likely RNA virus infections in general indirectly stimulate STAT1 Ser-708 phosphorylation via viral induction of IFN production from the infected cell, which then stimulates STAT1 Ser-708 phosphorylation through the actions of IKKϵ.
We found that active signaling through the type I IFN receptor was required for IFN-β- and virus-induced STAT1 Ser-708 phosphorylation. IFN-induced STAT1 Ser-708 phosphorylation, however, did not require IRF-3 expression. These observations are consistent with further data that IKKϵ-dependent IFIT2 induction can occur independently of IRF-3 (see Fig. 4C). However, during the course of virus infection, STAT1 Ser-708 phosphorylation failed to take place in the absence of IRF-3 because of a lack of IFN-β induction, secretion, and signaling. Indeed, de novo protein synthesis downstream of IFN signaling was required for STAT1 Ser-708 phosphorylation, suggesting that one or more ISG product signals IKKϵ to catalyze STAT1 Ser-708 phosphorylation. This requirement of de novo IFN-induced factor synthesis for STAT1 Ser-708-responsive ISG expression can be bypassed by IKKϵ overexpression, which induces its autoactivation, suggesting that an IKKϵ activator ISG would function upstream of IKKϵ (see Fig. 1C). Although we have yet to determine the IFN-inducible factor that promotes Ser-708 phosphorylation, on the basis of CHX pulse-chase experiments, it appears to be synthesized between 9 and 12 h after IFN-β stimulation. More detailed time course-dependent transcriptome profiling experiments may narrow down a list of candidate ISGs that directly or indirectly activate the kinase activity of IKKϵ that is responsible for Ser-708 phosphorylation. Potential candidates could include the IFN-induced protein kinase PKR, which interacts with STAT1 without directly phosphorylating the Tyr-701 residue (52) and restricts WNV infection in cells and in vivo (53, 54), and p38, which has been implicated in ISRE activation following type I IFN stimulation but is not required for IFN-dependent STAT1 Tyr-701 or Ser-727 phosphorylation (55–57). Alternatively, protein phosphatases or non-enzymatic ISG products might be involved in modulating IKKϵ action and STAT1 Ser-708 phosphorylation either through regulation of a signaling network of IKKϵ control or through direct binding to signaling factors of IKKϵ relevance or IKKϵ.
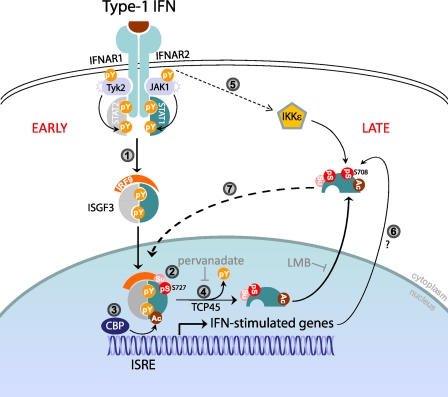
Our studies suggest that the order of STAT1 phosphorylation during the course of IFN stimulation could be an important contributor to the kinetics of ISG expression as STAT1 Tyr-701 phosphorylation temporally precedes Ser-708 phosphorylation and the induction of IFIT2 expression. Moreover, we observed minimal Ser-708 phosphorylation in IFN-treated cells under conditions of pervanadate or LMB treatment, which blocks STAT1 tyrosine dephosphorylation and nuclear export, respectively (see Fig. 5B). Additionally, these treatments result in sustained Tyr-701 phosphorylation of STAT1. These observations suggest that Tyr-701 and Ser-708 could be mutually exclusive on the same molecule. Consistent with this, STAT1 molecules that are phosphorylated at Tyr-701 following IFN-β treatment are not phosphorylated on Ser-708 (supplemental Fig. S6). These observations suggest one of two possible scenarios of STAT1 phosphorylation kinetics in which 1) Tyr-701 and Ser-708 phosphorylation cannot occur simultaneously in a single STAT1 molecule, whereas Ser-727, which is more distantly located, can; or 2) STAT1 molecules phosphorylated at Tyr-701 cannot dimerize with STAT1 molecules phosphorylated at residue Ser-708. We favor the former hypothesis because of the close proximity of the Tyr-701 and Ser-708 residues, as phosphorylation at Tyr-701 may result in steric hindrance of Ser-708 phosphorylation. Importantly, we note that Tyr-701 phosphorylation has been shown to diminish at later time points after IFN stimulation or virus infection because of nuclear STAT1 acetylation and dephosphorylation of Tyr-701 by tyrosine phosphatase TCP45, which has been reported previously (40, 58, 59). Additionally, chromatin-bound STAT1 can be phosphorylated at Ser-727, resulting in its sumoylation by UBC9 (60, 61). As unphosphorylated STAT1 cycles back to the cytoplasm via CRM1-mediated nuclear export, acetylation and sumoylation results in STAT1 latency by inhibiting IFN-induced STAT1 Tyr-701 phosphorylation, which should then permit Ser-708 phosphorylation (58, 59, 61). These studies concluded that non-tyrosine phosphorylated STAT1 is the “unphosphorylated STAT1,” which functions to sustain expression of some ISGs (62, 63). Our findings now suggest that the actual nature of these unphosphorylated STAT1 entities may be STAT1 phosphorylated at Ser-708, thus promoting the expression of a specific subset of ISGs whose expression occurs later after IFN treatment, such as IFIT2, thus “sustaining” the IFN response. We therefore propose a model of early and late type I IFN response programs (Fig. 7). Early after type I IFN stimulation, STAT1 is phosphorylated at Tyr-701, translocates to the nucleus, and induces expression of IKKϵ-independent ISGs. Following tyrosine dephosphorylation, STAT1 molecules are exported back to the cytoplasm. At a later time, as yet undetermined IFN-inducible factor(s) activate the IKKϵ-mediated STAT1 phosphorylation at the Ser-708 residue, which results in sustained expression of IKKϵ-dependent ISGs, including IFIT2. Similar to the actions of IFIT2 against WNV infection, we propose that IKKϵ-dependent ISGs include genes whose products direct antiviral and immune-modulatory actions to mediate innate immunity. Defining the nature of these ISGs within the innate immune response to WNV infection will be an important contribution toward identifying therapeutic targets for enhancement of immune protection against WNV and other flaviviruses. Therefore, a genomics-based assessment of the response to WNV infection in WT and IKKϵ−/− mice, as well as a targeted chromatin immunoprecipitation assay and analyses to define p-STAT1 Ser-708-responsive genes, is warranted for future studies aimed at characterizing this novel IKKϵ-dependent pathway of innate immunity.
FIGURE 7.
A model illustrating that early and late ISGs induction is regulated by multiple STAT1 posttranslational modifications. 1, the canonical JAK-STAT signaling is activated following type I IFN binding to its receptor, which results in STAT1 Tyr-701 phosphorylation, ISGF3 formation, and its nuclear translocation. ISGF3 binding to the ISRE element induces transcription of ISGs. 2, chromatin-bound STAT1 can be phosphorylated by MAPK at residue Ser-727, which induces its sumoylation. 3 and 4, nuclear STAT1 is also acetylated by histone acetyltransferase (HAT) CREB-binding protein (CBP), resulting in recruitment of TCP1, which catalyzes STAT1 tyrosine dephosphorylation. Sumoylated-acetylated STAT1 cycles back to the cytoplasm, and both modifications render STAT1 unable to be further tyrosine-phosphorylated. 5 and 6, type I IFN signaling and unknown IFN-stimulated factor(s) activate IKKϵ phosphorylation of STAT1 Ser-708. 7, STAT1 molecules phosphorylated at Ser-708 can enter the nucleus and induce expression of a specific ISG subset. pY, tyrosine phosphorylation; pS, serine phosphorylation; Ac, acetylation; Su, sumoylation.
The importance of IFIT2 and the IKKϵ pathway of ISG induction is underscored by the observation that virulent WNV effectively suppresses IFIT2 function to ensure efficient virus replication (23). Moreover, we have shown that a WNV strain that specifically lacks the ability to modulate the effect of IFIT genes is attenuated in WT mice (23, 24, 26). These observations, coupled with the present study showing that MEFs lacking IFIT2 support greater replication of WNV-MAD, a strain of WNV that only inefficiently antagonizes the antiviral effects of IFN (8), define IFIT2 as a innate immune effector gene that restricts WNV replication. Our studies also examined the significance of STAT1 Ser-708 phosphorylation during the course of infection in vivo, through infection of IKKϵ−/− mice with WNV-MAD or WNV-TX, the latter being the emergent strain of WNV that is highly virulent and now circulates in North America (8). Although the WNV-MAD strain does not cause neurovirulence in adult WT C57BL/6 mice, the WNV-TX strain is highly neurovirulent (8, 64). Importantly, in isogenic IKKϵ−/− mice, WNV-MAD disseminated to the spleen at earlier times and caused increased clinical disease, although none of the animals succumbed to lethal infection over the study time course. Furthermore, IFIT2 expression was not sustained in the spleen of these animals at later time during WNV-MAD infection. However, early IFIT2 induction likely resulted from IRF-3 activation, as IFIT2 is responsive to both IRF-3 and IFN-stimulation, which can be differentially regulated in different cell types of the spleen. These studies confirmed the IKKϵ dependence for STAT1 Ser-708 phosphorylation and sustained IFIT2 expression during virus infection to demonstrate a role for the IKKϵ pathway of IFIT2 expression in vivo during WNV infection (see Fig. 6). Viral suppression of IFIT2 or IKKϵ signaling may therefore impart replication fitness for the support of viral spread and tissue dissemination and therefore represents a virulence determinant among strains of WNV and possibly other pathogenic viruses. The IKKϵ pathway could therefore prove attractive for therapeutic strategies aimed at limiting virus replication and enhancing innate antiviral immunity. Further work to define this pathway will reveal the nature of IKKϵ signaling control during the response to IFN.
Supplementary Material
Acknowledgments
We thank Drs. Stacy Horner, Yueh Ming-Loo, Courtney Wilkins, Helene Liu, and Renee Ireton for critical reading of the manuscript and Arjun Rustagi for the generation of IRF-3 antibody.
This work was supported, in whole or in part, by National Institutes of Health grants R01AI074973 and U19AI083019.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- WNV
- West Nile virus
- ADAR
- double-stranded RNA-specific adenosine deaminase
- IFNAR
- interferon-α/β receptor
- ISG
- interferon-stimulated gene
- IRF
- interferon regulatory factor
- IKK
- IκB kinase
- SenV
- Sendai virus
- m.o.i.
- multiplicity of infection
- CHX
- cycloheximide
- LMB
- leptomycin B
- MEF
- mouse embryonic fibroblast
- ISRE
- interferon- stimulated regulatory element
- PKR
- protein kinase R.
REFERENCES
- 1. Lanciotti R. S., Roehrig J. T., Deubel V., Smith J., Parker M., Steele K., Crise B., Volpe K. E., Crabtree M. B., Scherret J. H., Hall R. A., MacKenzie J. S., Cropp C. B., Panigrahy B., Ostlund E., Schmitt B., Malkinson M., Banet C., Weissman J., Komar N., Savage H. M., Stone W., McNamara T., Gubler D. J. (1999) Science 286, 2333–2337 [DOI] [PubMed] [Google Scholar]
- 2. Smithburn K. C., Hughes T. P., Burke A. W., Paul J. H. (1940) J. Trop. Med. Hyg. 20, 471–492 [Google Scholar]
- 3. Berthet F. X., Zeller H. G., Drouet M. T., Rauzier J., Digoutte J. P., Deubel V. (1997) J. Gen. Virol. 78, 2293–2297 [DOI] [PubMed] [Google Scholar]
- 4. Lanciotti R. S., Ebel G. D., Deubel V., Kerst A. J., Murri S., Meyer R., Bowen M., McKinney N., Morrill W. E., Crabtree M. B., Kramer L. D., Roehrig J. T. (2002) Virology 298, 96–105 [DOI] [PubMed] [Google Scholar]
- 5. Lvov D. K., Butenko A. M., Gromashevsky V. L., Kovtunov A. I., Prilipov A. G., Kinney R., Aristova V. A., Dzharkenov A. F., Samokhvalov E. I., Savage H. M., Shchelkanov M. Y., Galkina I. V., Deryabin P. G., Gubler D. J., Kulikova L. N., Alkhovsky S. K., Moskvina T. M., Zlobina L. V., Sadykova G. K., Shatalov A. G., Lvov D. N., Usachev V. E., Voronina A. G. (2004) Arch. Virol. Suppl. 85–96 [DOI] [PubMed] [Google Scholar]
- 6. Jia X. Y., Briese T., Jordan I., Rambaut A., Chi H. C., Mackenzie J. S., Hall R. A., Scherret J., Lipkin W. I. (1999) Lancet 354, 1971–1972 [DOI] [PubMed] [Google Scholar]
- 7. Daffis S., Samuel M. A., Suthar M. S., Keller B. C., Gale M., Jr., Diamond M. S. (2008) J. Virol. 82, 8465–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keller B. C., Fredericksen B. L., Samuel M. A., Mock R. E., Mason P. W., Diamond M. S., Gale M., Jr. (2006) J. Virol. 80, 9424–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keller B. C., Johnson C. L., Erickson A. K., Gale M., Jr. (2007) Cytokine Growth Factor Rev. 18, 535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fredericksen B. L., Keller B. C., Fornek J., Katze M. G., Gale M., Jr. (2008) J. Virol. 82, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., Gale M., Jr. (2008) J. Virol. 82, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suthar M. S., Ma D. Y., Thomas S., Lund J. M., Zhang N., Daffis S., Rudensky A. Y., Bevan M. J., Clark E. A., Kaja M. K., Diamond M. S., Gale M., Jr. (2010) PLoS Pathog. 6, e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gale M., Jr., Foy E. M. (2005) Nature 436, 939–945 [DOI] [PubMed] [Google Scholar]
- 14. Saito T., Gale M., Jr. (2007) Curr. Opin. Immunol. 19, 17–23 [DOI] [PubMed] [Google Scholar]
- 15. Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 16. Hengel H., Koszinowski U. H., Conzelmann K. K. (2005) Trends Immunol. 26, 396–401 [DOI] [PubMed] [Google Scholar]
- 17. Levy D. E., García-Sastre A. (2001) Cytokine Growth Factor Rev. 12, 143–156 [DOI] [PubMed] [Google Scholar]
- 18. Sen G. C. (2001) Annu. Rev. Microbiol. 55, 255–281 [DOI] [PubMed] [Google Scholar]
- 19. Uddin S., Sassano A., Deb D. K., Verma A., Majchrzak B., Rahman A., Malik A. B., Fish E. N., Platanias L. C. (2002) J. Biol. Chem. 277, 14408–14416 [DOI] [PubMed] [Google Scholar]
- 20. Varinou L., Ramsauer K., Karaghiosoff M., Kolbe T., Pfeffer K., Müller M., Decker T. (2003) Immunity. 19, 793–802 [DOI] [PubMed] [Google Scholar]
- 21. Tenoever B. R., Ng S. L., Chua M. A., McWhirter S. M., García-Sastre A., Maniatis T. (2007) Science 315, 1274–1278 [DOI] [PubMed] [Google Scholar]
- 22. Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 23. Daffis S., Szretter K. J., Schriewer J., Li J., Youn S., Errett J., Lin T. Y., Schneller S., Zust R., Dong H., Thiel V., Sen G. C., Fensterl V., Klimstra W. B., Pierson T. C., Buller R. M., Gale M., Jr., Shi P. Y., Diamond M. S. (2010) Nature 468, 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fensterl V., Sen G. C. (2011) J. Interferon Cytokine Res. 31, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarkar S. N., Sen G. C. (2004) Pharmacol. Ther. 103, 245–259 [DOI] [PubMed] [Google Scholar]
- 26. Terenzi F., Hui D. J., Merrick W. C., Sen G. C. (2006) J. Biol. Chem. 281, 34064–34071 [DOI] [PubMed] [Google Scholar]
- 27. Hui D. J., Terenzi F., Merrick W. C., Sen G. C. (2005) J. Biol. Chem. 280, 3433–3440 [DOI] [PubMed] [Google Scholar]
- 28. Wang C., Pflugheber J., Sumpter R., Jr., Sodora D. L., Hui D., Sen G. C., Gale M., Jr. (2003) J. Virol. 77, 3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daffis S., Samuel M. A., Keller B. C., Gale M., Jr., Diamond M. S. (2007) PLoS Pathog. 3, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bluyssen H. A., Vlietstra R. J., Faber P. W., Smit E. M., Hagemeijer A., Trapman J. (1994) Genomics 24, 137–148 [DOI] [PubMed] [Google Scholar]
- 31. Bandyopadhyay S. K., Leonard G. T., Jr., Bandyopadhyay T., Stark G. R., Sen G. C. (1995) J. Biol. Chem. 270, 19624–19629 [DOI] [PubMed] [Google Scholar]
- 32. Wacher C., Müller M., Hofer M. J., Getts D. R., Zabaras R., Ousman S. S., Terenzi F., Sen G. C., King N. J., Campbell I. L. (2007) J. Virol. 81, 860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., Akira S. (2004) J. Exp. Med. 199, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Foy E., Li K., Wang C., Sumpter R., Jr., Ikeda M., Lemon S. M., Gale M., Jr. (2003) Science 300, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 35. Peters R. T., Liao S. M., Maniatis T. (2000) Mol. Cell 5, 513–522 [DOI] [PubMed] [Google Scholar]
- 36. Breiman A., Grandvaux N., Lin R., Ottone C., Akira S., Yoneyama M., Fujita T., Hiscott J., Meurs E. F. (2005) J. Virol. 79, 3969–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie X., Zhang D., Zhao B., Lu M. K., You M., Condorelli G., Wang C. Y., Guan K. L. (2011) Proc. Nat. Acad. Sci. U.S.A. 108, 6474–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pellegrini S., John J., Shearer M., Kerr I. M., Stark G. R. (1989) Mol. Cell. Biol. 9, 4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Immunity. 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 40. Haspel R. L., Salditt-Georgieff M., Darnell J. E., Jr. (1996) EMBO J. 15, 6262–6268 [PMC free article] [PubMed] [Google Scholar]
- 41. McBride K. M., Reich N. C. (2003) Sci. STKE 2003, RE13. [DOI] [PubMed] [Google Scholar]
- 42. Mowen K., David M. (2000) Mol. Cell. Biol. 20, 7273–7281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang D., Weidner J. M., Qing M., Pan X. B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P. Y., Block T. M., Guo J. T. (2010) J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y., Burke C. W., Ryman K. D., Klimstra W. B. (2007) J. Virol. 81, 11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Terenzi F., Saikia P., Sen G. C. (2008) EMBO J. 27, 3311–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Daffis S., Suthar M. S., Gale M., Jr., Diamond M. S. (2009) J. Innate Immun. 1, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daffis S., Lazear H. M., Liu W. J., Audsley M., Engle M., Khromykh A. A., Diamond M. S. (2011) J. Virol. 85, 5664–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 49. tenOever B. R., Sharma S., Zou W., Sun Q., Grandvaux N., Julkunen I., Hemmi H., Yamamoto M., Akira S., Yeh W. C., Lin R., Hiscott J. (2004) J. Virol. 78, 10636–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McWhirter S. M., Fitzgerald K. A., Rosains J., Rowe D. C., Golenbock D. T., Maniatis T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maher S. G., Sheikh F., Scarzello A. J., Romero-Weaver A. L., Baker D. P., Donnelly R. P., Gamero A. M. (2008) Cancer Biol. Ther. 7, 1109–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wong A. H., Tam N. W., Yang Y. L., Cuddihy A. R., Li S., Kirchhoff S., Hauser H., Decker T., Koromilas A. E. (1997) EMBO J. 16, 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Samuel M. A., Whitby K., Keller B. C., Marri A., Barchet W., Williams B. R., Silverman R. H., Gale M., Jr., Diamond M. S. (2006) J. Virol. 80, 7009–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gilfoy F. D., Mason P. W. (2007) J. Virol. 81, 11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Katsoulidis E., Li Y., Mears H., Platanias L. C. (2005) J. Interferon Cytokine Res. 25, 749–756 [DOI] [PubMed] [Google Scholar]
- 56. Uddin S., Lekmine F., Sharma N., Majchrzak B., Mayer I., Young P. R., Bokoch G. M., Fish E. N., Platanias L. C. (2000) J. Biol. Chem. 275, 27634–27640 [DOI] [PubMed] [Google Scholar]
- 57. Uddin S., Majchrzak B., Woodson J., Arunkumar P., Alsayed Y., Pine R., Young P. R., Fish E. N., Platanias L. C. (1999) J. Biol. Chem. 274, 30127–30131 [DOI] [PubMed] [Google Scholar]
- 58. Krämer O. H., Knauer S. K., Greiner G., Jandt E., Reichardt S., Gührs K. H., Stauber R. H., Böhmer F. D., Heinzel T. (2009) Genes Dev. 23, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krämer O. H., Heinzel T. (2010) Mol. Cell. Endocrinol. 315, 40–48 [DOI] [PubMed] [Google Scholar]
- 60. Vanhatupa S., Ungureanu D., Paakkunainen M., Silvennoinen O. (2008) Biochem. J. 409, 179–185 [DOI] [PubMed] [Google Scholar]
- 61. Zimnik S., Gaestel M., Niedenthal R. (2009) Nucleic Acids Res. 37, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cheon H., Stark G. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang J., Stark G. R. (2008) Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 64. Beasley D. W., Li L., Suderman M. T., Barrett A. D. (2002) Virology 296, 17–23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.