Background: The precise role of the hemopexin domain of matrix metalloproteinase-1 (MMP-1) in collagenolysis is unknown.
Results: The hemopexin domain collagen binding site is on β-propeller blades 1 and 2, and includes a Phe that is buried in the interface with the catalytic domain in the MMP-1 crystal structure.
Conclusion: Domain dislocation is required for exosite exposure.
Significance: MMP-1 may undergo significant domain rearrangements during collagenolysis.
Keywords: Collagen, Extracellular Matrix Proteins, Matrix Metalloproteinase (MMP), Nuclear Magnetic Resonance, Protein Structure, Site-directed Mutagenesis, Surface Plasmon Resonance (SPR), X-ray Scattering
Abstract
Matrix metalloproteinase-1 (MMP-1) is an instigator of collagenolysis, the catabolism of triple helical collagen. Previous studies have implicated its hemopexin (HPX) domain in binding and possibly destabilizing the collagen substrate in preparation for hydrolysis of the polypeptide backbone by the catalytic (CAT) domain. Here, we use biophysical methods to study the complex formed between the MMP-1 HPX domain and a synthetic triple helical peptide (THP) that encompasses the MMP-1 cleavage site of the collagen α1(I) chain. The two components interact with 1:1 stoichiometry and micromolar affinity via a binding site within blades 1 and 2 of the four-bladed HPX domain propeller. Subsequent site-directed mutagenesis and assay implicates blade 1 residues Phe301, Val319, and Asp338 in collagen binding. Intriguingly, Phe301 is partially masked by the CAT domain in the crystal structure of full-length MMP-1 implying that transient separation of the domains is important in collagen recognition. However, mutation of this residue in the intact enzyme disrupts the CAT-HPX interface resulting in a drastic decrease in binding activity. Thus, a balanced equilibrium between these compact and dislocated states may be an essential feature of MMP-1 collagenase activity.
Introduction
The regulated proteolysis of collagen (known as collagenolysis) in the extracellular matrix is an important process in normal physiological events such as wound healing and connective tissue remodeling (1). In contrast, loss of collagenolytic control is associated with conditions such as cancer, arthritis, and cardiovascular disease (1).
The fibrillar collagens (types I, II, III, V, and XI) are highly resistant to degradation by nonspecific proteases, a property that is imparted by the tightly wound, triple helical conformation in individual α-chain trimers, and the ordered lateral association of these trimers into microfibrils. Thus, in higher eukaryotes, specific collagenase enzymes are present to perform this function. The majority of collagenases are members of the matrix metalloproteinase (MMP)4 family of zinc-dependent endopeptidases (2). Those MMPs with significant collagenolytic activity are MMP-1, -8, -13, -14, and, to a lesser extent, MMP-2 (also known as collagenases 1, 2, and 3, membrane type 1 MMP, and gelatinase A, respectively). These enzymes cleave type I collagen (the most abundant fibrillar form) between Gly775-Ile776 of α1(I) chains and Gly775-Leu776 of the α2(I) chain generating characteristic ¾- and ¼-length fragments that exhibit conformational instability and thus undergo further enzymatic processing by the gelatinases, MMP-2 and -9 (2).
MMPs are multidomain enzymes, each expressed as an inactive zymogen in which the inhibitory N-terminal propeptide (“PRO”) domain docks into the active site cleft of the catalytic (“CAT”) domain thus blocking substrate entry and so maintaining the latency of the enzyme. In so doing, a conserved PRO domain Cys residue coordinates the active site Zn2+ ion that is held in place by three His residues in the conserved CAT domain sequence motif HEXXHXXGXXH (2, 3). For many MMPs (including each of the collagenases), cleavage of a protease-sensitive “bait” region in the PRO domain initiates a chain of other proteolytic events that ultimately result in its complete removal and concomitant activation of the zymogen (4–6). All collagenolytic (and most other) MMPs also contain a C-terminal hemopexin (“HPX”) domain, which is linked to the CAT domain by an extended proline-rich linker.
MMP-1, the prototypic collagenase, has been the subject of intense study and is thus the best characterized in terms of its structure, function, and dynamics (7–10). A comparison of the crystal structures of pro-MMP-1 and MMP-1 shows that not only does the PRO domain sterically block the CAT domain active site, but it also draws the CAT and HPX domains together by interacting with the C-D loop in blade 1 of the HPX β-propeller (8, 9, 11). These structures suggest that, upon activation, a groove opens between the CAT and HPX domains; this groove has been proposed to form the collagen binding site (8, 9), a hypothesis that would explain the apparent lack of binding of pro-MMP-1 to collagen (12, 13), whereas MMP-1 exhibits micromolar affinity (13, 14).
The success of collagenolysis relies on determinants in both the collagen substrate and the collagenase enzyme. In fibrillar collagens, the region surrounding the collagenase cleavage site contains numerous nonpolar residues (which are otherwise rare) and has a low imino acid content that together may make the local triple helical conformation relatively unstable at physiological temperatures (15, 16). Furthermore, Ile776 in the leading strand is more conformationally labile than the equivalent residue in the middle and lagging strands, and thus may serve as a signal for collagenase cleavage (17). In turn, the collagenolytic MMPs contain numerous “exosites,” regions that, although lacking in any hydrolytic apparatus (this is limited to the active site of the CAT domain), are required for recognition and cleavage of the natural substrate (reviewed by Ref. 18). The presence of such exosites may, in part, explain the mystery that has surrounded collagenases since the early crystal structures were solved: how do these enzymes cleave collagen when the active site cleft is only one-third as wide as the collagen triple helix? The exosites are believed to impart the collagenases with helicase activity, the ability to bind and partially “unwind” the triple helical collagen substrate, so allowing a single α-chain to enter the active site cleft of the CAT domain (14, 19).
Numerous studies have identified determinants of helicase/collagenase activity in the CAT domain, the HPX domain, and the inter-domain linker of MMP-1. In the CAT domain, both the active site itself and the loop connecting the fifth β-strand and second α-helix have been shown to be involved (20, 21). In addition, mutation of a specific Gly residue in the interdomain linker abrogates collagenolysis possibly by restricting interdomain flexibility (22). Such flexibility has been inferred from nuclear magnetic resonance (NMR) spectroscopic and small angle x-ray scattering (SAXS) studies of MMP-1 in solution (10).
The presence of one or more exosites in the HPX domain is evident from the lack of collagenolytic activity in HPX deletion mutants of MMP-1, -8, -13, and -14 (23–28). However, the precise location of any exosite(s) has, until recently, remained largely elusive; unlike the CAT domain, the four-bladed β-propeller of the HPX domain lacks any obvious binding clefts or pockets that could accommodate a polypeptide (8, 9, 11). A recent study examining the interaction between an active site mutant (E219A) of MMP-1 and a synthetic triple helical peptide (THP) using hydrogen/deuterium exchange mass spectrometry implicated blades 1 and 4 of the HPX domain in collagen binding (29). Subsequently, using mutagenesis and assay residues, Ile290 and Arg291 in the A-B loop of blade 1 were identified as an exosite for collagenolysis (29).
Here, we present a biophysical study of the interaction between the MMP-1 HPX domain (hereafter referred to as HPX-1) and a synthetic THP, which encompasses the MMP-1 cleavage site of the α1(I) chain and is analogous to that used recently by Lauer-Fields and co-workers (29). Using analytical ultracentrifugation (AUC) and solution NMR spectroscopy, we establish that HPX-1 binds the homotrimeric THP with a 1:1 stoichiometry that involves an extensive convex surface of HPX-1 covering much of β-propeller blades 1 and 2. Subsequently, through a program of mutagenesis and assay using surface plasmon resonance (SPR), we highlight residues in blade 1 as having a significant role in collagen binding. Intriguingly, the most vital of these, Phe301, is buried in the interface between the CAT and HPX domains in the crystal structure of MMP-1(E219A). However, using SAXS we confirm previous results that, in solution, the domains undergo a transient separation thus exposing residues, such as Phe301, which are concealed between the domains. Furthermore, in the full-length enzyme, mutation of Phe301 causes a complete dislocation of the domains and a drastic drop in collagen binding activity, implying that both the dislocated and compact domain arrangements are important for the recognition and/or unwinding of the triple helix.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression
The hydrolytically inactive E219A mutants of pro-MMP-1 and MMP-1 (hereafter abbreviated as pro-MMP-1* and MMP-1*, respectively) were produced as previously described (14) but with a modified refolding protocol (10). Unlabeled wild-type (WT) human HPX-1 (corresponding to residues Pro272-Asn469 of pro-MMP-1 with an additional N-terminal Met) previously cloned into a pET-3a plasmid (Merck) was expressed from Escherichia coli BL21(DE3)RIPL cells as described previously (6). Briefly, the recombinant protein was refolded from chaotrope-solubilized inclusion bodies in the presence of a redox pair (5 mm β-mercaptoethanol, 1 mm 2-hydroxyethyl disulfide) to enable formation of the intramolecular disulfide bond connecting Cys278 and Cys466. The refolded protein was then purified to homogeneity (as judged by SDS-PAGE) by cation-exchange chromatography on a Mono S HR5/5 column (GE Healthcare). The identity of the purified product was confirmed by mass spectrometry (BMS Mass Spectrometry and Proteomics Facility, University of St Andrews). HPX-1 proteins uniformly labeled with 15N or 13C/15N were produced by growth and induction of the cells in M9 minimal media containing (15NH4)2SO4 or [13C]glucose/(15NH4)2SO4, respectively. Prior to undertaking experiments, protein samples were exchanged into the appropriate buffer by dialysis using a 3.5-kDa cut-off membrane (Spectra/Por) and concentrated by centrifugal ultrafiltration using 5-kDa cut-off spin filters (Amicon). The protein solutions were then quantified by UV absorbance at 280 nm using theoretical molar extinction coefficients.
Synthetic α1(I)772–787 Triple Helical Peptide
Two α1(I)772–787 THPs, either with or without an N-terminal amino group, were produced by solid-phase synthesis as described previously (30). Each THP had the sequence: C6-(GPO)4 GPQ G∼IA GQR GVV GLP GPO (GPO)3-NH2 in which “O” denotes 4-hydroxyproline, the underlined sequence is identical to residues Gly772-Gly787 of the collagen α1(I) chain, and the tilde (∼) marks the MMP-1 cleavage site. MMP-1 cleaves the α1(I)772–787 THP at the analogous position to type I collagen (30). Previous work has shown that the N-terminal hexanoic moiety (designated C6) and the GPO repeats induce a triple helical conformation at room temperature (30). The fidelity of synthesis and homogeneity of each THP was confirmed by mass spectrometry. The THP with a free N-terminal amino group was utilized in SPR experiments (see later); the C6-THP lacking this moiety was used in all other experiments. Prior to each experiment, the THP monomers were annealed into a triple helix at a high concentration (>1 mm) by heating for 15 min at 65 °C followed by slow cooling from 65 to 4 °C over 2 h. Because of their lack of aromatic residues, these α1(I)772–787 THP solutions were quantitated spectrophotometrically according to the formula: [THP] (mg/ml) = ((A215 − A225) × 0.144)/e; where e is the path length in cm (31).
Site-directed Mutagenesis
The solvent-accessible surface area of candidate HPX-1 residues for site-directed mutagenesis was determined using a 1.4-Å diameter rolling ball in the program MOLMOL (32). Mutagenesis was performed using the QuikChange XL II Kit (Stratagene). The sense and antisense strand oligonucleotide primers for the chosen mutations are given in supplemental Table S1. In each case, the mutated codon was flanked by 10–20 bases of complementary sequence. The DNA sequence of each mutant pET-3a:HPX-1 construct was confirmed by sequencing (Beckman Coulter Genomics). Those HPX mutants that successfully expressed in BL21(DE3)RIPL cells were refolded and purified as described above for the WT protein.
Analytical Ultracentrifugation
Sedimentation equilibrium (SE) AUC experiments were performed on a Beckman Coulter Optima XL-A analytical ultracentrifuge using absorbance optics and an AN-Ti-50 rotor at speeds selected based on the anticipated apparent Mr values (33). All studies were performed at 25 °C in 150 mm NaCl, 10 mm CaCl2, 20 mm NaOAc, pH 4.8. SE studies on HPX-1 in isolation were performed at 15,000 and 18,000 rpm at protein concentrations of 0.5, 1.0, and 5.0 mg/ml (20.9, 41.7, and 208 μm, respectively). SE analysis of the complex formed between HPX-1 and the α1(I)772–787 THP was performed at 15,000 rpm with a constant HPX-1 concentration of 50 μm and α1(I)772–787 THP:HPX-1 molar ratios of 0:1, 1:1, 2:1, 5:1, 10:1, and 20:1. AUC detection was performed at 303 nm, higher than the absorbance maximum of HPX-1 (280 nm) to allow for higher protein concentrations to be analyzed using the absorbance optics (at 200 μm, the A303 for HPX-1 is 0.7). The α1(I)772–787 THP lacks aromatic residues and, therefore, has negligible absorbance at this wavelength. Thus, a single species model could be used to simplify interpretation of the apparent Mr data. Data analysis was performed using the Origin-based DC/DT software (Beckman Coulter).
NMR Data Collection and Chemical Shift Assignments
For sequential assignment of HPX-1, a suite of standard through-bond, triple-resonance experiments (e.g. HNCA, HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH) were recorded on the 14.1 T (600 MHz for 1H) Varian Inova spectrometer at the University of Portsmouth. These were supplemented by a through-space 50-ms mixing time 1H,15N-NOESY-HSQC spectrum recorded on a 18.8 tesla (800 MHz for 1H) Bruker Avance spectrometer at the National Institute for Medical Research (NIMR). All NMR spectra were acquired at 25 °C on 0.5 mm samples of HPX-1 dissolved in 10% (v/v) D2O, 10 mm CaCl2, 0.02% (w/v) NaN3, 20 mm NaOAc at pH 4.8. Spectral processing was performed using the program NMRPipe (34) and assignment was undertaken using the program analysis2 in the CCPN software suite (35). Sequential assignment of WT HPX-1 1H, 13C, and 15N resonances was performed predominantly using a main chain-directed strategy. Any cases of ambiguity were resolved through identifying backbone-backbone HN-HN NOEs that were consistent with the crystal structures of MMP-1* and pro-MMP-1* (8, 9). The structural integrity of HPX domain mutants that showed perturbed collagen binding activity (see below) was confirmed through comparison of a 1H15N-HSQC spectrum of the uniformly 15N-labeled mutant with that of the WT protein.
NMR-monitored Ligand Titrations
Ligand-induced resonance broadening on HPX-1 was measured following serial additions of small volume aliquots of 4 mm unlabeled α1(I)772–787 THP into a 0.2 mm sample of 15N-labeled HPX-1 up to a THP:HPX-1 molar ratio of 5:1. For each HPX-1 residue, the degree of line broadening was inferred from the loss of backbone NH-HN cross-peak intensity. No attempt was made to measure the line widths of the individual 1HN and 15NH resonances explicitly.
Surface Plasmon Resonance
SPR experiments were performed on a BIACore T100 instrument (GE Healthcare). The N-terminal aminated α1(I)772–787 THP was immobilized to the dextran surface of a CM4 sensorchip (GE Healthcare) flow cell using amine coupling in 10 mm HEPES pH 7.4. The more commonly used CM5 sensorchip (which has a greater degree of dextran carboxylation) was found to exhibit unacceptably high levels of electrostatic binding to WT HPX-1 (which has a pI of 9.45). The running buffer for all SPR binding assays was HBSCP+ (10 mm HEPES, 150 mm NaCl, 10 mm CaCl2, 0.05% (v/v) polysorbate-20), pH 7.4. Due to the extremely rapid dissociation rates of WT HPX-1 from immobilized α1(I)772–787 THP seen in preliminary experiments, the dissociation constants for the wild-type and each mutant were determined from the SPR response at equilibrium. Thus, long-duration (10-min contact time) triplicate injections were performed with a suitable range of analyte concentrations (e.g. ∼40 nm to ∼40 μm for WT HPX-1) at a flow rate of 1 μl/min. In each case, the equilibrium point was taken to be 5 s from the end of the analyte injection. Data analysis was performed using BIAevaluation (GE Healthcare) by nonlinear least squares fitting using Equation 1: Req = Rmax × c/(c + Kd), where Req is the equilibrium response at a given concentration (c) of the WT or mutant protein, Rmax is the maximum response possible if the immobilized THP ligand were saturated with analyte, and Kd is the dissociation constant.
Small Angle x-ray Scattering
SAXS experiments were conducted on the I22 beamline at the Diamond Light Source with a wavelength (λ) of 1.0 Å and an energy of 12.4 keV, and equipped with a photon counting detector at a distance of 3.25 m from the sample cell. The scattering patterns were measured for several protein concentrations in the range from 0.84 to 8.25 mg/ml, each with multiple successive 90-s exposures (to check for radiation-induced damage and aggregation), and processed using PRIMUS (36) and GNOM (37) within the ATSAS software package (38). The data were normalized to the intensity of the incident beam, and the scattering of the buffer was subtracted using an in-house program. The Guinier approximation was used to interpret the data, at low angle, the isotropic scattering intensities, I(Q) (where Q is the scattering vector 4πsinθ/λ and 2θ is the scattering angle), can be described by I(Q) = I(0) exp(−Rg2Q2/3), where Rg is the radius of gyration. Subsequently, the I(Q) data were transformed to particle distribution functions, P(r), which were in turn used to estimate the particle maximum dimensions Dmax and calculate the real space Rg. Simulated small-angle scattering curves were generated from the atomic resolution crystal structures of pro-MMP-1* and MMP-1* using the program CRYSOL (39). Ten independent ab initio low resolution atomic reconstructions were generated from the P(r) data by simulated annealing using the DAMMIF program with default parameters (40, 41), and then aligned, averaged, and filtered using DAMAVER (40). The use of 20 structures did not significantly alter the average model. Overlay of the SAXS models with those from x-ray crystallography was performed with the program SUPCOMB (42), in which a low normalized spatial discrepancy (NSD) value indicates an acceptable fit.
Notes
Numbering for MMP-1 and HPX-1 corresponds to that in the crystal structure of pro-MMP-1 (PDB accession code 1SU3), which has Phe20 as the N-terminal residue. The numbering for collagen chain α1(I) (accession number P02452) is relative to the estimated start of the helical region. Theoretical molar extinction coefficients and molecular weights were calculated using the ProtParam tool on the Expasy website. Molecular structure figures were prepared using PyMOL (43). NMR chemical shift assignments for WT HPX-1 at pH 4.8 and 25 °C are given in supplemental Table S2 and have been deposited in the BioMagResBank.
RESULTS
HPX-1 Binds the α1(I)772–787 THP with 1:1 Stoichiometry
Initially, SE AUC studies were performed to assess the solution ideality and oligomeric state of HPX-1 in solution. The residuals measure the difference between the actual and theoretical absorbance at a given radius for a particle of a given Mr. An upward or downward curve on these residuals would be evidence of aggregation or nonideality, respectively. However, the residuals for each study were evenly distributed about the x axis suggesting that HPX-1 is monodisperse in solution under the conditions studied (see supplemental Fig. S1). The apparent Mr of the HPX domain is largely independent of concentration over the range of 0.5–5.0 mg/ml (21–210 μm). The average apparent molecular mass (± S.D.) over the six measurements is 23.5 ± 2.1 kDa, which is in close agreement with the theoretical molecular mass for monomeric HPX-1 (23.6 kDa) calculated from the amino acid sequence. Thus, at concentrations up to ∼200 μm, the HPX-1 is monomeric in solution.
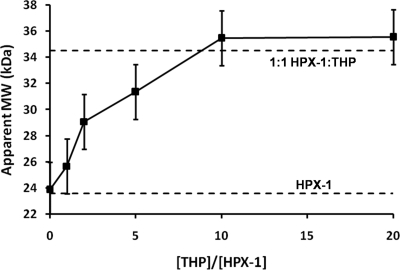
SE AUC analysis of the complex formed between HPX-1 and the α1(I)772–787 THP reveals a dose-dependent increase in apparent molecular mass up to a maximal apparent molecular mass of 35.5 (±0.04) kDa at α1(I)772–787 THP:HPX-1 molar ratios of 10:1 and 20:1 (Fig. 1). As with the analysis of isolated HPX-1 above, the even distribution of the residuals indicates that the complex is monodisperse in solution (see supplemental Fig. S2).
FIGURE 1.
Determination of the binding stoichiometry between HPX-1 and the α1(I)772–787 THP by sedimentation equilibrium AUC. The horizontal dashed lines indicate the theoretical Mr for the free monomeric HPX-1 (23.6 kDa) and a 1:1 complex of HPX-1 with the α1(I)772–787 THP (34.5 kDa). The error in apparent molecular mass (2.1 kDa) for each point is estimated as the S.D. of the apparent molecular mass from the AUC studies on the free HPX-1 (see supplemental Fig. S1).
Given that the theoretical molecular mass of the THP is 10.9 kDa, the maximal apparent molecular mass of 35.5 kDa for the complex is strongly indicative of a 1:1 stoichiometry (i.e. Langmuir binding). In contrast, if the THP:HPX-1 interaction stoichiometry were 2:1 or 1:2 then one would expect significantly different maximal molecular mass values of ∼45.4 or 58.1 kDa, respectively.
α1(I)772–787 THP Binds to Blades 1 and 2 of the HPX-1 β-Propellor
NMR has the capability of providing site-specific information on protein-ligand interactions, but to identify the ligand binding site, it is first necessary to sequentially assign the protein resonances. HPX-1 had previously been assigned at pH 7.2 (10) but, due to the elevated rate of amide proton exchange at near neutral pH, 16 of the backbone 1HN and 15NH resonances could not be assigned. Most of these residues are solvent-exposed and thus may be involved in collagen binding. In this study we aimed to achieve near complete backbone resonance assignments of HPX-1 under mildly acidic conditions (at pH 4.8), with a view to providing complete coverage of any potential HPX-1 collagen-binding surface(s) in subsequent NMR-monitored ligand titrations. The reduced amide proton exchange rate at pH 4.8 allowed sequential assignment of all HPX-1 backbone 1HN and 15NH resonances, with the exception of Ala350. This residue was one of those unassigned in the previous study (10).
During the NMR-monitored titration of 15N-labeled HPX-1 with unlabeled α1(I)772–787 THP, no significant changes in HPX-1 chemical shift were observed, nor did any cross-peaks appear that could be attributed to the complex, even at the highest THP:HPX-1 molar ratio of 5:1. Instead, certain HPX-1 HSQC cross-peaks were seen to extensively broaden and reduce in intensity indicative of an intermediate exchange regime (see supplemental Fig. S3). All of the backbone HSQC cross-peaks show some degree of intensity loss during the course of the titration (Fig. 2), which we attribute to the increasing Mr upon complex formation and hence slower tumbling of the HPX-1 in solution. This slower tumbling is exacerbated by the increasing viscosity that accompanies increasing protein concentration, and results in shorter T1 relaxation times and thus broader line widths. Nevertheless, a subset of HSQC cross-peaks show further loss of intensity, which we attribute to site-specific binding of the α1(I)772–787 THP ligand on an intermediate (millisecond to microsecond) exchange time scale. However, a detailed analysis of the biophysical basis of the resonance line broadening is beyond the scope of this report.
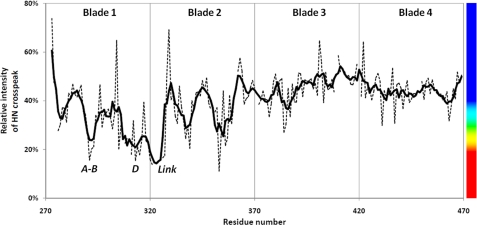
FIGURE 2.
HPX-1 line broadening upon addition of α1(I)772–787 THP. The sequence dependence of the HPX-1 backbone 1HN-15NH cross-peak broadening following an equimolar addition of THP is shown. The values show the intensity of cross-peaks at a 1:1 molar ratio of THP:HPX-1 relative to the sample of HPX-1 alone. The dashed lines show the raw data, whereas the thick solid line depicts a 5-residue roaming average of this data. The vertical lines show the approximate boundaries between the four blades of the HPX-1 β-propeller. The vertical bar on the right-hand side shows the rainbow color scheme used for the molecular representations in Fig. 3.
The majority of HPX-1 residues whose backbone amide resonances are most affected by THP binding are found in blades 1 and 2 of the β-propeller (Figs. 2 and 3A). Specifically, in blade 1, extensive line broadening is observed in the A-B loop (Arg291-Glu293), the C-D loop (Phe308-Glu311), β-strand D (Glu313-Phe316), the α-helical turn (Ser318-Val319), and the following unstructured region that connects blade 1 to blade 2 (Phe320-Gly328). Extensive line broadening is less widespread in blade 2 where only the C-D loop (Gly353-Val356) is significantly affected.
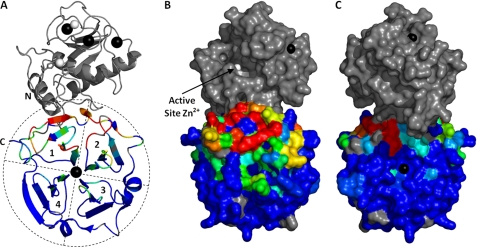
FIGURE 3.
α1(I)772–787 THP recognition surface on HPX-1 as identified by NMR. A, ribbon representation of the crystal structure of MMP-1 (9) with HPX domain residues colored according to the line broadening data in Fig. 2. The colors range from blue through cyan, green, yellow, and red over the relative intensity range of 40–20%. Proline residues of HPX-1 and the unassigned Ala350 are colored gray. For clarity, the four blades of the β-propeller are outlined and labeled 1–4. B and C, surface representations of the HPX-1 domain using the same color scheme as in A. In C, the HPX-1 has been rotated through 180° about the y axis. Note that, although shown here in gray, the CAT domain is absent from the protein fragment assayed in this experiment.
When mapped onto the solvent-accessible surface of the MMP-1 crystal structure, the affected residues form a putative convex binding site that extends across both “faces” of the HPX domain β-propeller (Fig. 3, B and C). The majority of affected residues map to the “front” face, i.e. the side that is oriented in the same direction as the catalytic cleft in the MMP-1 crystal structures (Fig. 3B). The “rear” facing portion of the putative binding site is comprised of the solvent-exposed residues in the linker between blades 1 and 2 (Fig. 3C).
Collagen Binding Involves Hydrophobic Residues on Blade 1 of HPX-1
It should be noted that line broadening of a particular residue in backbone 1HN and/or 15NH resonance does not necessarily imply that its side chain is directly involved in ligand binding. Instead, the line width perturbation may arise from an interaction on the millisecond to microsecond time scale that involves a neighboring side chain. Thus, we used site-directed mutagenesis and assay to confirm or refute the involvement of the residues implicated by the NMR-monitored ligand titration in collagen binding. The criteria for choosing which residues to mutate were 3-fold: 1) close proximity of side chain (less than approximately 10 Å) to one or more extensively broadened backbone 1HN/15NH resonances; 2) a side chain solvent exposure of >50% in the HPX domain; and 3) not Gly or Ala. Thus, 15 residues were chosen for mutation to alanine: Asp299, Arg300, Phe301, Phe308, Tyr309, Phe316, Ser318, Val319, Pro325, Arg337, Asp338, Gln352, Gln354, His358, and Pro361.
Each of these mutant pET3a:HPX-1 constructs was successfully produced by site-directed mutagenesis as confirmed by DNA sequencing. Furthermore, when transformed into BL21(DE3)RIPL cells, each was found to express recombinant protein at levels approaching or even exceeding that of the WT HPX-1. However, during the refolding stage that follows inclusion body isolation and solubilization, four of the mutant HPX-1 domains (D299A, F308A, F316A, and Q352A) precipitated almost quantitatively, leaving insufficient material for functional analysis. Those 11 HPX-1 mutants that remained soluble during the refolding stage (R300A, F301A, Y309A, S318A, V319A, P325A, R337A, D338A, Q354A, H358A, and P361A) were found to elute from the cation exchange column at an identical NaCl concentration to that of the WT HPX-1, suggesting that each possesses a native-like structure, i.e. is capable of binding a Ca2+ ion at the center of the β-propeller.
In preliminary SPR experiments, WT HPX-1 was found to exhibit fast dissociation kinetics from immobilized α1(I)772–787 THP (kd = ∼1 s−1) that is somewhat beyond the dissociation rate detection limit of 0.5 s−1 for the Biacore T100 (Biacore T100 manual). Therefore, because point mutants of HPX-1 may increase this off-rate still further, we decided to characterize the binding affinity of the WT and mutants for the α1(I)772–787 THP using equilibrium analysis rather than kinetic analysis. Because the problem of mass transfer limitation does not apply to equilibrium analysis, we used relatively high immobilization levels of ∼500 response units for the α1(I)772–787 THP, to increase the sensitivity of subsequent ligand binding experiments.
In the equilibrium SPR assays, the WT HPX-1 and each of the mutants tested exhibited dose-dependent binding to the immobilized α1(I)772–787 THP (Fig. 4, A and B). Binding activities were found to be reproducible in this assay with triplicate Kd measurements typically differing by less than 10% (Fig. 4C). Repeated assays of the WT protein, at both the beginning and end of the series of analyses, revealed that there was little or no degradation of the HPX-1 binding sensorchip capacity. Thus, the regeneration conditions did not damage the immobilized THP in any way.
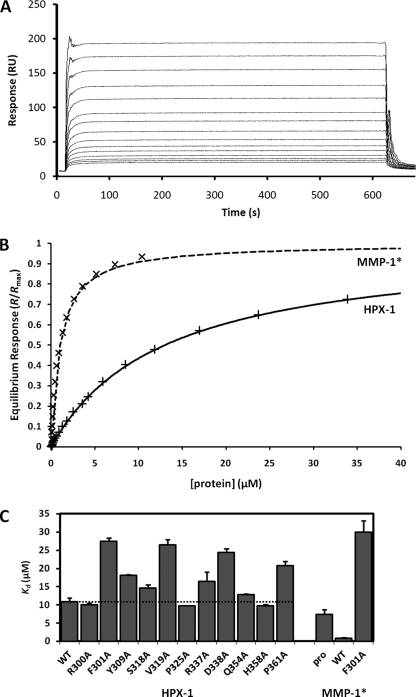
FIGURE 4.
SPR assay of HPX-1 collagen binding activity. A, overlaid SPR sensorgrams showing the dose-dependent binding of WT HPX-1 to immobilized α1(I)772–787 THP. The WT HPX-1 concentration ranges from 0.2 to 34 μm. B, hyperbolic binding curves for WT HPX-1 and WT MMP-1* from the equilibrium SPR data. The fitted curves are of the form R/Rmax = c/(c + Kd), where c is the analyte protein concentration. C, equilibrium dissociation constants (Kd) for the binding of wild-type and mutant proteins to immobilized THP. Error bars indicate the S.D. from triplicate measurements. The horizontal dashed line emphasizes the Kd of the WT HPX-1 protein for comparative purposes.
The Kd for the interaction of WT HPX-1 to the immobilized THP was found to be 10.9 ± 1.0 μm. This is an order of magnitude higher than that for MMP-1*, which in this assay binds with a Kd of 0.92 ± 0.09 μm (Fig. 4, B and C). Thus, it is clear that the CAT domain and/or CAT-HPX linker help to stabilize the MMP-THP complex in the intact enzyme. In contrast to previous reports (12, 13, 24), collagen-binding activity was detected for the pro-MMP-1* zymogen, which bound the α1(I)772–787 THP with a Kd of 7.37 ± 1.34 μm.
As seen in Fig. 4C, the binding activity of HPX-1 mutants R300A, S318A, P325A, Q354A, and H358A were very similar to that of the WT protein implying that they play no direct role in contacting the collagen substrate. Moderate increases in Kd were observed for mutants Y309A, R337A, and P361A. However, mutants F301A, V319A, and D338A show a more than 2-fold increase in Kd relative to WT HPX-1 (i.e. have less than 50% of the binding activity) suggesting that these residues are important contributors to collagen recognition, i.e. they comprise an exosite. Fig. 5 shows the location of these residues in the crystal structure of MMP-1*. Surprisingly, the introduction of the highest impact HPX-1 mutation, F301A, into the full length MMP-1* resulted in a drastic (30-fold) decrease in binding activity (Fig. 4C).
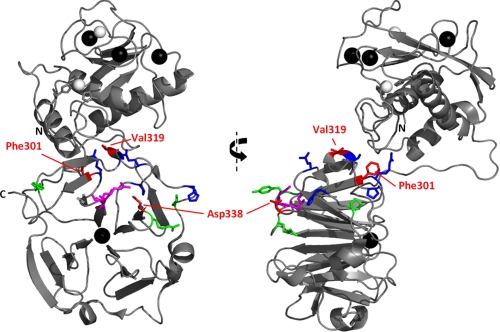
FIGURE 5.
Residues of HPX-1 implicated in collagen binding by mutagenesis and assay. Orthogonal views of the crystal structure of MMP-1* are shown as a ribbon diagram with the left-hand side in the same orientation as that in Fig. 3A. Mutated HPX-1 residues are shown as sticks and colored according to the collagen binding activity of the mutant, i.e. Phe301, Val319, and Asp338 in red (Kd ≥ 24 μm); Tyr309, Arg337, and Pro361 in green (16 μm < Kd < 24 μm); and Arg300, Ser318, Pro325, Gln354, and His358 in blue (Kd ≤ 16 μm). Residues Ile290 and Arg291, previously identified as an exosite (29), are shown as magenta sticks.
The possibility exists that the reduced binding of any HPX-1 point mutant is due to a global disruption of the three-dimensional structure of the domain rather than the selective removal of a critically important side chain. Thus, we analyzed the two HPX-1 point mutants with the lowest collagen binding activity, F301A and V319A, by heteronuclear NMR spectroscopy to investigate this possibility. In each case, the 1H,15N-HSQC of the 15N-labeled HPX-1 mutant was very similar to that of the WT protein with excellent chemical shift dispersion in each dimension (see supplemental Fig. S4). In neither case were any extensive changes in 1H or 15N chemical shifts observed that would indicate a substantial change in the HPX-1 structure. Furthermore, no significant line broadening was observed in the HPX-1 mutants that could indicate a conformational exchange and thus destabilization of the domain. Therefore, we are confident that the reduced binding activity of these HPX-1 mutants is a direct result of the loss of their collagen-interacting Phe301 or Val319 side chains rather than any perturbation to the structure and/or dynamics of the domain.
Disrupting the Interface between CAT and HPX Domains Impairs Collagen Binding
The structural basis for the dramatic reduction in binding activity of MMP-1*(F301A) compared with MMP-1* was investigated using SAXS (supplemental Fig. S5), a solution state method that has previously been applied to conformational studies of MMPs-1, -9, and -12 (10, 44, 45). In addition, we examined pro-MMP-1* to investigate whether the compact arrangement of domains in the crystal structure is representative of the conformation in solution. For each protein, the distance distribution function (P(r) plot) was found to be asymmetric and thus characteristic of an elongated protein (Fig. 6A). That for MMP-1* has a distinct shoulder indicative of a discrete domain structure (46). In the MMP-1*(F301A) mutant, this shoulder is exaggerated to the point where it becomes a separate peak that implies that the CAT and HPX domains have become more separated.
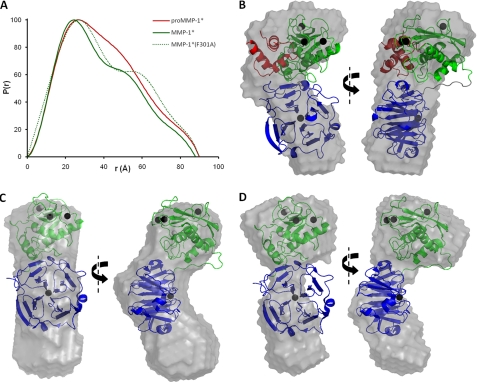
FIGURE 6.
SAXS analysis of pro-MMP-1*, MMP-1*, and MMP-1*(F301A). A, the particle distribution functions, P(r), are shown for each protein. Note the increased Dmax (x intercept) and enhanced bilobal form of the F301A mutant relative to MMP-1*. B-D, orthogonal views of the average low resolution solution state models of pro-MMP-1* (B), MMP-1* (C), and MMP-1*(F301A) (D). In B, the SAXS model was overlaid automatically over the crystal structure of pro-MMP-1* (with an NSD of 1.65). In C and D, a manual overlay was performed over the crystal structure of MMP-1* as automated overlays gave unacceptably poor fits (NSD values of 2.07 and 2.11, respectively). In the crystal structures, the PRO domain is colored red, the CAT domain green, and the HPX domain blue. Note that, in C and D, unique orientations of the MMP-1* crystal structure with respect to the ab initio low resolution structures could not be identified due to the similar sizes of the CAT and HPX domains. Thus, their relative positioning here is arbitrary and only serves to highlight the discrepancy between solution-state and solid-state observations.
Each of the P(r) plots was used to calculate a real space Rg value. The Rg of 28.5 ± 0.2 Å for MMP-1* agrees with the previous report of 29 ± 1 Å (10), and is significantly less than the 32.8 ± 0.2 Å determined here for pro-MMP-1*. This reduction is accounted for by the loss of the ∼9-kDa PRO domain upon activation, which also results in a slight decrease in the Dmax from 90 to 88 Å (Fig. 6A). However, MMP-1*(F301A) has an Rg of 33.6 ± 0.4 Å, 18% higher than that for MMP-1*. Indeed, the MMP-1*(F301A) mutant has comparable Rg and Dmax values to pro-MMP-1* despite the absence of the PRO domain in the former. This is further evidence of a disruption to the CAT-HPX domain interface in the F301A mutant.
Ab initio modeling of the pro-MMP-1* conformation from the SAXS data results in a low resolution model that largely agrees with the compact arrangement of domains in the crystal structure (Fig. 6B), as shown by the reasonably low NSD value of 1.65. The discrepancies that do exist between the two may be due to the fact that a significant proportion of the electron density map for pro-MMP-1* is missing from the crystal structure, namely that for the N-terminal residues (Phe20-Val31), the bait region (Asn46-Ser57), and the PRO-CAT linker (Gln99-Asn106) (8).
In contrast, ab initio modeling from the MMP-1* SAXS data produces a molecular envelope that is considerably larger than expected from its compact crystal structure, and a superposition of these low and high resolution structures results in an considerably higher NSD of 2.07 and leaves an entire lobe of the solution-state model unaccounted for (Fig. 6C). To ensure that this discrepancy is not due to a fault in the modeling protocol, we generated models based on artificial scattering data synthesized from the crystal coordinates of MMP-1* and pro-MMP-1*. These control low resolution structures were in very close agreement with the solid state coordinates (supplemental Fig. S5, D and E) with NSD values of 1.19 and 1.24, respectively. Thus, the elongated low-resolution solution structure of MMP-1* is indicative of an equilibrium between the compact state seen in crystallographic analyses and a state in which the interface between CAT and HPX is disrupted. This is in agreement with the previous study by Bertini and co-workers (10).
This apparent transient dislocation of the CAT and HPX domains in mature MMP-1* becomes more permanent in the F301A mutant, the low resolution ab initio structure that exhibits two discrete lobes (Fig. 6D), which we attribute to the separate CAT and HPX domains. Thus, the P(r) distance distribution functions, the Rg values, and the low resolution models all indicate that the F301A mutation has disrupted the dynamic inter-domain interaction that is a feature of the WT enzyme and, from the SPR data presented here, is an important contributor to collagen binding.
DISCUSSION
The AUC study described here has shown that HPX-1 remains monomeric in solution at protein concentrations up to 0.2 mm. This implies that the HPX domain-mediated dimerization seen in MMP-14 that is crucial for collagenolysis and pro-MMP-2 activation (47–49) is not a common feature of all MMPs.
The finding that HPX-1 binds to the α1(I)772–787 THP with 1:1 stoichiometry has important consequences. It implies that (a) HPX-1 binds specifically to only one of the three staggered α-chains in the pseudo-symmetric THP; or (b) HPX-1 binding to any one of the three α-chains inhibits binding of a second HPX-1 molecule to either of the other two α-chains by steric hindrance; or (c) the HPX-1 binds to two or three α-chains simultaneously. The last interpretation is perhaps the most intriguing in terms of the mechanism of collagenolysis. In intact MMP-1, the binding of two α-chains simultaneously by the HPX domain may leave the third free to enter the catalytic cleft of the CAT domain that, at 5 Å across, is too narrow to accommodate the 15-Å diameter intact triple helix. Indeed, it is well established that the α2 chain of α1(I)2α2(I) heterotrimers is cleaved preferentially by the MMP-1 CAT domain (14), suggesting that the HPX domain may favor binding to the two α1 chains, thus releasing the α2 chain for hydrolysis.
Our SPR-based assay has revealed the affinities (Kd values) for binding to an immobilized homotrimeric collagen THP. We are mindful that in nondiseased adult tissue, the majority of type I collagen exists as α1(I)2α2(I) heterotrimers (50). Indeed, α1(I)3 homotrimers only exist in significant quantities in vivo in fetal tissues (51), carcinomas (50, 52–55), fibrotic tissues (56–58), and in genetic disorders in which the α2(I) chain is deficient (59–61). Thus, the homotrimeric THP used in this study is not strictly representative of the homeostatic state. However, a recent study has demonstrated that the affinity of MMP-1* for homotrimeric and heterotrimeric collagen is effectively identical, with Kd = 1.31 ± 0.2 and 1.27 ± 0.3 μm, respectively (19). Therefore, the use of the homotrimeric THP is justified when studying collagen binding by the HPX-1 domain. Furthermore, the affinity observed here by SPR between MMP-1* and the immobilized THP (Kd = 0.92 ± 0.09 μm) is in keeping with the above values.
Previously, Ottl and co-workers (62) saw no interaction between HPX-1 and a heterotrimeric THP using a kinetic SPR analysis. Clearly, this contradicts the data presented here. However, each chain of the α1(I)2α2(I) THP used in the previous study lacked Leu785, the end of the hydrophobic stretch that is believed to contribute to MMP-1 recognition (15, 16). Indeed, it is noticeable that the Kd of 3.7 ± 1.1 μm observed for intact MMP-1* binding to their THP (62) is ∼4-fold higher than observed here. Additionally, in the previous study, the inclusion of Ca2+-chelating EDTA in the dissociation phase buffer (62) may have contributed to the lack of observed HPX-1 binding; extraction of the Ca2+ ion from the β-propeller of HPX-1 is likely to destabilize the domain thus accelerating its dissociation rate from the immobilized THP and so increasing the Kd.
For the most part, the data shown here is in agreement with the recent results of Lauer-Fields and co-workers (29), who used hydrogen/deuterium exchange mass spectrometry to probe the collagen binding site of MMP-1. Both studies have implicated β-propeller blade 1 in collagen binding. Furthermore, using site-directed mutagenesis and assay, the previous study identified two specific residues, Ile290 and Arg291, as playing an important role in collagenolysis. These residues lie on the A-B loop of blade 1 (Fig. 5) and are among those implicated by the NMR-monitored titration in this study (Figs. 2 and 3A). In addition, in the crystal structures of both pro-MMP-1 and MMP-1, Arg291 is salt-bridged to Asp338, the mutation of which significantly reduces the collagen binding activity of HPX-1 in our SPR-based assay (Fig. 4C). The previous study also implicated β-propeller blade 4 in collagen binding (29), but here we find no strong evidence for its involvement. However, it is important to note that the hydrogen/deuterium exchange mass spectrometry experiments did not detect direct contact to blade 4 per se, but rather changes in its dynamics upon complex formation. Therefore, we believe that THP binding to blades 1 and 2 stabilizes blade 4, thus restricting its dynamic behavior and reducing its hydrogen/deuterium exchange rates.
In addition to Asp338, we have identified residues Phe301 and Val319 as important contributors to collagen binding activity. Residue Phe301 lies at the end of β-strand D in HPX-1 blade 1 with its aromatic ring oriented away from the core of the domain but entirely buried in an interface with the CAT domain (Fig. 5). Val319 is located within the α-helical turn of blade 1 with its side chain solvent exposed on the “equator” of the oblate HPX domain (Fig. 5). Unlike Phe301, Val319 does not itself contact the CAT domain, but it is in close proximity to the CAT domain loop connecting the fifth β-strand and second α-helix (sequence Arg202-Tyr210), a region previously shown to be required for collagenolytic activity of intact MMP-1 (63).
Together with Phe316 and Phe320, residues Phe301 and Val319 form a hydrophobic strip along the HPX-1 surface in the cleft formed with the CAT domain, which is proximal to the Arg291-Asp338 salt bridge (Fig. 5). This hydrophobic strip may form the interaction surface for the nonpolar residues that surround the scissile bond in each α chain of type I collagen. We note that although expression of the F316A HPX-1 mutant was successful, its refolding and purification failed to produce sufficient quantities of soluble protein for assay. Thus, at this time, we cannot ascertain whether Phe316 (or indeed Phe320) has any involvement in collagen recognition.
At first sight, the positioning of Phe301 in the CAT-HPX domain interface would appear to preclude its involvement in collagen binding in the intact enzyme. It forms part of a “ball and socket” joint between the domains, in which the cluster of Arg300, Phe301, and Phe316 side chains (the “ball”) in the HPX domain docks into a hydrated cavity encapsulated by residues Gly233-Gln247 (the “socket”) of the loop connecting the second and third helices in the CAT domain (Fig. 7). The importance of this interaction is seen in the MMP-1*(F301A) mutant that, from the SAXS data, appears to completely dislocate the domains (Fig. 6, A and C) and results in a ∼30-fold reduction in collagen binding activity relative to MMP-1* (Fig. 4C).
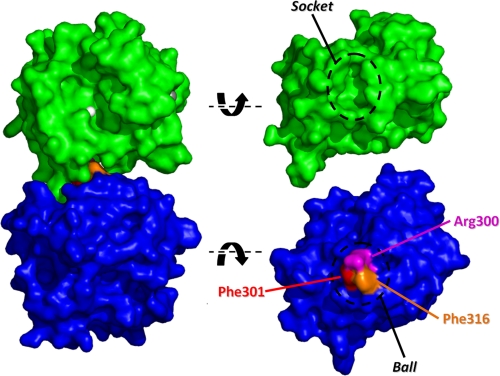
FIGURE 7.
The ball and socket joint in the CAT-HPX interface of MMP-1*. A space-filling model of the crystal structure of MMP-1* is shown looking into the active site cleft with the Zn2+ ion colored white. Dislocation of the ball and socket joint and rotation of the CAT (green) and HPX (blue) domains reveals the interacting surfaces. The three residues comprising the ball are highlighted.
However, this static picture of MMP-1 is not the full story. A recent combined study using NMR and SAXS demonstrated that the compact arrangement of domains in the crystal is not fully representative of the conformation in solution (10). Instead, for approximately one-third of the time, the enzyme exists as a more extended arrangement of the CAT and HPX domains. This discrepancy between the solid and solution states is further highlighted here in the ab initio modeling of MMP-1* from SAXS data (Fig. 6C). Furthermore, the previous study identified residues involved in the domain-domain interface whose high levels of exposure to a paramagnetic probe were inconsistent with the compact arrangement of domains in the crystal structure. Among these were residues Asp245-Gln247 (10), which form part of the above socket. Therefore, the domain separation most probably involves dislocation of the ball and socket joint with concomitant exposure of the collagen binding exosite involving Phe301 (Fig. 7).
The results presented here support the presence of a collagen binding exosite in blade 1 of the HPX domain of MMP-1 that requires dislocation from the CAT domain for full exposure to the substrate. However, whether this domain separation is necessary for the initial recognition of the collagen, or for its local unwinding, or both, remains to be seen. Furthermore, we have demonstrated that the compact conformation of MMP-1* observed by x-ray crystallography is also biologically relevant, as complete disruption of the CAT-HPX interface drastically reduces collagen binding activity. Thus, MMP-1 collagenase activity most probably depends on a subtle equilibrium between these compact and dislocated states. In the future, disruption of this equilibrium by small molecules that block the CAT-HPX interaction may prove to be effective inhibitors of collagenase activity.
Supplementary Material
Acknowledgments
We are extremely grateful to Hideaki Nagase and Robert Visse for providing the original pET-3a plasmid constructs for expressing pro-MMP-1* and HPX-1. We thank Tom Frenkiel, Geoff Kelly, and Alain Oregioni for help at the MRC Biomedical NMR Centre, Mill Hill, UK. We are also indebted to Helen Vincent, Anastasia Callaghan, and Marc Malfois for help in the acquisition and processing of SAXS data at the Diamond Light Source. Funding for the University of Portsmouth NMR facility was provided by the Wellcome Trust.
This work was supported, in whole or in part, by National Institutes of Health Grant CA098799 (to G. B. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
- MMP
- matrix metalloproteinase
- AUC
- analytical ultracentrifugation
- CAT
- catalytic
- HPX
- hemopexin
- HPX-1
- HPX domain of MMP-1
- PRO
- pro-peptide
- SAXS
- small angle X-ray scattering
- SPR
- surface plasmon resonance
- THP
- triple helical peptide
- SE
- sedimentation equilibrium
- NSD
- normalized spatial discrepancy
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Hadler-Olsen E., Fadnes B., Sylte I., Uhlin-Hansen L., Winberg J. O. (2011) FEBS J. 278, 28–45 [DOI] [PubMed] [Google Scholar]
- 2. Nagase H., Visse R., Murphy G. (2006) Cardiovasc. Res. 69, 562–573 [DOI] [PubMed] [Google Scholar]
- 3. Van Wart H. E., Birkedal-Hansen H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stricklin G. P., Jeffrey J. J., Roswit W. T., Eisen A. Z. (1983) Biochemistry 22, 61–68 [DOI] [PubMed] [Google Scholar]
- 5. Nagase H., Enghild J. J., Suzuki K., Salvesen G. (1990) Biochemistry 29, 5783–5789 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki K., Enghild J. J., Morodomi T., Salvesen G., Nagase H. (1990) Biochemistry 29, 10261–10270 [DOI] [PubMed] [Google Scholar]
- 7. Pardo A., Selman M. (2005) Int. J. Biochem. Cell Biol. 37, 283–288 [DOI] [PubMed] [Google Scholar]
- 8. Jozic D., Bourenkov G., Lim N. H., Visse R., Nagase H., Bode W., Maskos K. (2005) J. Biol. Chem. 280, 9578–9585 [DOI] [PubMed] [Google Scholar]
- 9. Iyer S., Visse R., Nagase H., Acharya K. R. (2006) J. Mol. Biol. 362, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertini I., Fragai M., Luchinat C., Melikian M., Mylonas E., Sarti N., Svergun D. I. (2009) J. Biol. Chem. 284, 12821–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., Brick P., O'Hare M. C., Skarzynski T., Lloyd L. F., Curry V. A., Clark I. M., Bigg H. F., Hazleman B. L., Cawston T. E., et al. (1995) Structure 3, 541–549 [DOI] [PubMed] [Google Scholar]
- 12. Allan J. A., Hembry R. M., Angal S., Reynolds J. J., Murphy G. (1991) J. Cell Sci. 99, 789–795 [DOI] [PubMed] [Google Scholar]
- 13. Welgus H. G., Jeffrey J. J., Eisen A. Z., Roswit W. T., Stricklin G. P. (1985) Coll. Relat. Res. 5, 167–179 [DOI] [PubMed] [Google Scholar]
- 14. Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., Nagase H. (2004) EMBO J. 23, 3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leikina E., Mertts M. V., Kuznetsova N., Leikin S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1314–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fields G. B. (1991) J. Theor. Biol. 153, 585–602 [DOI] [PubMed] [Google Scholar]
- 17. Xiao J., Addabbo R. M., Lauer J. L., Fields G. B., Baum J. (2010) J. Biol. Chem. 285, 34181–34190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Overall C. M. (2002) Mol. Biotechnol. 22, 51–86 [DOI] [PubMed] [Google Scholar]
- 19. Han S., Makareeva E., Kuznetsova N. V., DeRidder A. M., Sutter M. B., Losert W., Phillips C. L., Visse R., Nagase H., Leikin S. (2010) J. Biol. Chem. 285, 22276–22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knäuper V., Patterson M. L., Gomis-Rüth F. X., Smith B., Lyons A., Docherty A. J., Murphy G. (2001) Eur. J. Biochem. 268, 1888–1896 [PubMed] [Google Scholar]
- 21. O'Farrell T. J., Guo R., Hasegawa H., Pourmotabbed T. (2006) Biochemistry 45, 15411–15418 [DOI] [PubMed] [Google Scholar]
- 22. Tsukada H., Pourmotabbed T. (2002) J. Biol. Chem. 277, 27378–27384 [DOI] [PubMed] [Google Scholar]
- 23. Clark I. M., Cawston T. E. (1989) Biochem. J. 263, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphy G., Allan J. A., Willenbrock F., Cockett M. I., O'Connell J. P., Docherty A. J. (1992) J. Biol. Chem. 267, 9612–9618 [PubMed] [Google Scholar]
- 25. Knäuper V., Osthues A., DeClerck Y. A., Langley K. E., Bläser J., Tschesche H. (1993) Biochem. J. 291, 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knäuper V., Cowell S., Smith B., López-Otin C., O'Shea M., Morris H., Zardi L., Murphy G. (1997) J. Biol. Chem. 272, 7608–7616 [DOI] [PubMed] [Google Scholar]
- 27. Ohuchi E., Imai K., Fujii Y., Sato H., Seiki M., Okada Y. (1997) J. Biol. Chem. 272, 2446–2451 [DOI] [PubMed] [Google Scholar]
- 28. Hurst D. R., Schwartz M. A., Ghaffari M. A., Jin Y., Tschesche H., Fields G. B., Sang Q. X. (2004) Biochem. J. 377, 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lauer-Fields J. L., Chalmers M. J., Busby S. A., Minond D., Griffin P. R., Fields G. B. (2009) J. Biol. Chem. 284, 24017–24024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lauer-Fields J. L., Nagase H., Fields G. B. (2000) J. Chromatogr. A 890, 117–125 [DOI] [PubMed] [Google Scholar]
- 31. Sober H. A., Chemical Rubber Co., Cleveland, (1968) Handbook of Biochemistry: Selected Data for Molecular Biology, Chemical Rubber Co., Cleveland, OH [Google Scholar]
- 32. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–5, 29–32 [DOI] [PubMed] [Google Scholar]
- 33. Chervenka C. H. (1970) Anal. Biochem. 34, 24–29 [DOI] [PubMed] [Google Scholar]
- 34. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 35. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 36. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 37. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 38. Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Svergun D. I., Richard S., Koch M. H., Sayers Z., Kuprin S., Zaccai G. (1998) Proc. Natl. Acad Sci. U.S.A. 95, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Svergun D. I. (1999) Biophys. J. 77, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozin M. B., Svergun D. I. (2001) J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 43. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 44. Rosenblum G., Van den Steen P. E., Cohen S. R., Grossmann J. G., Frenkel J., Sertchook R., Slack N., Strange R. W., Opdenakker G., Sagi I. (2007) Structure 15, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 45. Bertini I., Calderone V., Fragai M., Jaiswal R., Luchinat C., Melikian M., Mylonas E., Svergun D. I. (2008) J. Am. Chem. Soc. 130, 7011–7021 [DOI] [PubMed] [Google Scholar]
- 46. Glatter O. (1979) J. Appl. Crystallogr. 12, 166–175 [Google Scholar]
- 47. Itoh Y., Takamura A., Ito N., Maru Y., Sato H., Suenaga N., Aoki T., Seiki M. (2001) EMBO J. 20, 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Itoh Y., Ito N., Nagase H., Evans R. D., Bird S. A., Seiki M. (2006) Mol. Biol. Cell 17, 5390–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tochowicz A., Goettig P., Evans R., Visse R., Shitomi Y., Palmisano R., Ito N., Richter K., Maskos K., Franke D., Svergun D., Nagase H., Bode W., Itoh Y. (2011) J. Biol. Chem. 286, 7587–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makareeva E., Han S., Vera J. C., Sackett D. L., Holmbeck K., Phillips C. L., Visse R., Nagase H., Leikin S. (2010) Cancer Res. 70, 4366–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jimenez S. A., Bashey R. I., Benditt M., Yankowski R. (1977) Biochem. Biophys. Res. Commun. 78, 1354–1361 [DOI] [PubMed] [Google Scholar]
- 52. Moro L., Smith B. D. (1977) Arch. Biochem. Biophys. 182, 33–41 [DOI] [PubMed] [Google Scholar]
- 53. Shapiro F. D., Eyre D. R. (1982) J. Natl. Cancer Inst. 69, 1009–1016 [PubMed] [Google Scholar]
- 54. Yamagata S., Yamagata T. (1984) J. Biochem. 96, 17–26 [DOI] [PubMed] [Google Scholar]
- 55. Asokan R., Puvanakrishnan R., Ravichandran L. V., Kokila V., Reddy G. K., Dhar S. C. (1993) Mol. Cell. Biochem. 121, 99–107 [DOI] [PubMed] [Google Scholar]
- 56. Rojkind M., Giambrone M. A., Biempica L. (1979) Gastroenterology 76, 710–719 [PubMed] [Google Scholar]
- 57. Narayanan A. S., Page R. C., Meyers D. F. (1980) Biochemistry 19, 5037–5043 [DOI] [PubMed] [Google Scholar]
- 58. Ehrlich H. P., Brown H., White B. S. (1982) Biochem. Med. 28, 273–284 [DOI] [PubMed] [Google Scholar]
- 59. Schwarze U., Hata R., McKusick V. A., Shinkai H., Hoyme H. E., Pyeritz R. E., Byers P. H. (2004) Am. J. Hum. Genet. 74, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Malfait F., Symoens S., Coucke P., Nunes L., De Almeida S., De Paepe A. (2006) J. Med. Genet. 43, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pace J. M., Wiese M., Drenguis A. S., Kuznetsova N., Leikin S., Schwarze U., Chen D., Mooney S. H., Unger S., Byers P. H. (2008) J. Biol. Chem. 283, 16061–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ottl J., Gabriel D., Murphy G., Knäuper V., Tominaga Y., Nagase H., Kröger M., Tschesche H., Bode W., Moroder L. (2000) Chem. Biol. 7, 119–132 [DOI] [PubMed] [Google Scholar]
- 63. Chung L., Shimokawa K., Dinakarpandian D., Grams F., Fields G. B., Nagase H. (2000) J. Biol. Chem. 275, 29610–29617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.