Background: Signaling from RTKs is regulated at multiple levels; however, the role played by dephosphorylation in this process remains poorly understood.
Results: We show that loss of PTP1B activity leads to abrogated receptor trafficking via the endocytic pathway.
Conclusion: Dephosphorylation of the vesicle fusion machinery component NSF by PTP1B is required for RTK trafficking.
Significance: This identifies a novel mechanism through which PTP1B can modulate RTK signaling.
Keywords: Epidermal Growth Factor Receptor (EGFR), Phosphatase, Protein Phosphatase, Receptor Endocytosis, Receptor Tyrosine Kinase, Signaling
Abstract
The endoplasmic reticulum-localized non-receptor protein-tyrosine phosphatase 1B (PTP1B) is associated with oncogenic, metabolic, and cytokine-related signaling and functionally targets multiple receptor tyrosine kinases (RTKs) for dephosphorylation. Loss of PTP1B activity leads to enhanced ligand-dependent biological activity of the Met RTK among others. Here, we demonstrate that knockdown of PTP1B or expression of a PTP1B trapping aspartic acid-to-alanine substitution (D/A) mutant delayed ligand-induced degradation of the Met and EGF RTKs. Loss of PTP1B function abrogated trafficking of Met and EGF receptor to Rab5- and phosphatidylinositol 3-phosphate (Pl3P)-positive early endosomes and subsequent trafficking through the degradative pathway. Under these conditions, internalization of the Met and EGF receptors was unaltered, suggesting a block at the level of early endosome formation. We show that the N-ethylmaleimide-sensitive factor (NSF), an essential component of the vesicle fusion machinery, was hyperphosphorylated in PTP1B knockdown or PTP1B D/A-expressing cells and was a target for PTP1B. NSF knockdown phenocopied PTP1B knockdown, demonstrating a mechanism through which PTP1B regulates endocytic trafficking. Finally, we show that PTP1B dephosphorylated NSF and that this interaction was required for physiological RTK trafficking and appropriate attenuation of downstream signaling.
Introduction
Signaling via receptor tyrosine kinases (RTKs)3 is regulated at multiple levels, including ligand binding affinity, ligand concentration, post-translational modification, receptor internalization, trafficking, localization, and degradation. Following ligand-dependent activation, RTKs are removed from the plasma membrane via internalization and subsequently either recycled back to the cell surface or targeted for lysosomal degradation (1). Defects in the mechanisms involved in RTK down-regulation can result in inappropriate activation of receptor-responsive downstream signaling cascades, which have been linked to cancer development and progression (2, 3). Hence, under normal physiological conditions, modulation of receptor signaling is finely regulated, and different RTKs are often subject to coordinate control by multiple mechanisms.
The hepatocyte growth factor (HGF) receptor Met and the epidermal growth factor receptor (EGFR) undergo ligand-dependent internalization and continue to signal from the endosomal compartment (4–8). Following internalization, these receptors can recycle back to the cell surface or continue down the degradative pathway in which the internalized RTKs enter multivesicular bodies (MVBs) and are subsequently degraded as MVBs fuse with lysosomes (1). Once they are internalized into MVBs, their cytoplasmic tails are no longer exposed to signaling molecules (4).
Internalization of receptors into MVBs is regulated in part by receptor ubiquitination, which provides a recognition motif for proteins of the endosomal sorting complex required for transport machinery, such as hepatocyte growth factor substrate 1 (Hrs1). Ubiquitination of Met and EGFR is required for efficient receptor degradation (6, 9), and uncoupling receptors from ubiquitination leads to sustained signaling and cell transformation (3, 6, 10). Notably, naturally occurring Met mutants identified in lung cancers were found to uncouple the receptor from ubiquitination-mediated lysosomal degradation and promote sustained activation of the MAPK pathway (11), underscoring the importance of receptor endocytosis in regulating signaling.
Although internalization and degradation are now accepted mechanisms for down-regulation of RTKs, little is known regarding the role played by dephosphorylation in the attenuation of RTK-dependent signaling. Mathematical modeling studies suggest that tyrosine-specific phosphatases regulate the duration but not the amplitude of the RTK-dependent signal (12, 13). Evidence exists supporting a role for endoplasmic reticulum-localized tyrosine phosphatases, such as PTP1B, and perinuclearly localized SHP-1 in processing the c-kit, FLT-3, PDGFβ, and Ros receptors to their mature form (14). Additionally, ligand binding-induced production of reactive oxygen species can inhibit tyrosine phosphatase activity, resulting in an increase in the phosphorylation of non-ligand-bound receptors at the cell membrane (15–17). Multiple studies have also implicated protein-tyrosine phosphatases in the regulation of RTKs following growth factor activation. The insulin receptor is a substrate for PTP1B (18–20), T-cell protein-tyrosine phosphatase (21, 22), and PTPϵ (23) among others. In vitro studies suggest that the EGFR is a substrate for PTP1B, T-cell protein-tyrosine phosphatase, receptor-type protein-tyrosine phosphatase, leukocyte antigen related protein tyrosine phosphatase, and SHP-1 (24–27), and PTP1B has been proposed to regulate EGFR signaling from endosomes (28, 29). Met is a substrate for TCPTP and PTP1B (30) as well as DEP-1 (31) and leukocyte antigen related protein tyrosine phosphatase (32). Although PTPs have been shown to modulate signaling downstream of RTKs and selectively dephosphorylate specific tyrosine residues on RTKs, the physiological relevance of these interactions in vivo is still unclear.
PTP1B is a ubiquitously expressed non-receptor tyrosine phosphatase, which is localized to the cytoplasmic face of the endoplasmic reticulum (33, 34). PTP1B activity is required to maintain receptors in an inactive state as they undergo post-translational modifications following initial synthesis. Biochemical and in vivo studies have also revealed a role for PTP1B in the attenuation of multiple receptor-mediated signaling pathways, including those downstream from receptors that are inactivated and recycled back to the cell surface (e.g. the insulin and IGF-1 receptors) as well as those that more readily undergo degradation after activation (e.g. the Met, EGF, and PDGFβ receptors) (35). The complexity of the mode of action of PTP1B is underscored by the observation that its loss results in hyperphosphorylation of its RTK substrates and an increased susceptibility to B-cell lymphoma in a p53-null background on one hand (36) and delayed tumor formation in an ErbB2-induced mammary cancer mouse model on the other hand (37), suggesting a pleiotropic role for PTP1B in the maintenance of cellular homeostasis.
Previous studies have shown that internalization of the Met, EGF, and PDGF receptors is required for their interaction with PTP1B, suggesting a potential relationship between the interaction of these receptors with PTP1B and their trafficking (30, 38, 39). Although increased receptor-mediated signaling has been observed as a consequence of the loss of PTP1B, the mechanism through which this occurs is still unclear. One potential mechanism involves regulation of the components of the endocytic machinery by PTP1B.
Upon internalization, endocytic vesicles are incorporated into an endosomal compartment, requiring the action of the vesicle docking proteins v-SNAREs and t-SNAREs. After vesicle fusion, the SNARE complex, which includes α-soluble N-ethylmaleimide-sensitive factor (NSF)-associated protein (α-SNAP), must be disassembled via ATP hydrolysis of its α-SNAP subunit by the NSF; inefficient disassembly of the SNARE complex halts further vesicle fusion. The activity of NSF is regulated via its phosphorylation status; the kinases Fes and Ca2+/calmodulin-dependent protein kinase II (40) and the phosphatases PTP-MEG2 (41) and PTP1B (42) have been shown to control NSF phosphorylation. PTP-MEG2 is located on the cytoplasmic face of secretory vesicles where it regulates vesicle size by promoting homotypic vesicle fusion via tyrosine dephosphorylation of NSF (41). Recently, PTP1B has been identified as a PTP that dephosphorylates NSF in acrosomes (42). Additionally, PTP1B has been implicated in the dephosphorylation and inactivation of internalized RTKs, including Met and EGFR (26, 30), and in MVB maturation in response to EGF (39). We therefore investigated whether PTP1B plays a role in the control of RTK endocytosis at points other than MVB formation. Here, we show that loss of PTP1B delayed growth factor-induced Met and EGFR degradation and that this occurred via disruption of the vesicle fusion machinery specifically caused by the inability of NSF to undergo dephosphorylation.
EXPERIMENTAL PROCEDURES
DNA Constructs and siRNA
PTP1B WT and aspartic acid-to-alanine substitution (D/A) were inserted into the BglII and EcoRI sites of the pEGFPC2 vector (Clontech) as described previously (30). Cherry-PTP1B WT and D/A were constructed in a pEGFPC2 vector by replacing enhanced GFP from the constructs above with a cherry sequence. The cherry construct was a gift from Roger Tsien (University of California, Los Angeles, CA). GFP-FYVE was a gift from Harald Stenmark (University of Oslo, Oslo, Norway), and GFP-Rab5 was a gift from John Presley (McGill University, Montreal, Quebec). NSF was a gift from William Trimble (University of Toronto, Toronto, Ontario). siRNA duplexes against the PTP1B target sequence CACGTGGGTATTTAATAAGAA and the NSF target sequence UGGAAAUGCUUAACGCUUU were obtained from Qiagen (Toronto, Ontario, Canada) and transfected into HeLa cells for 48 h using HiPerFect (Qiagen) according to the manufacturer's instructions.
Cell Culture and DNA Transfections
HeLa cell lines were maintained in DMEM containing 10% FBS and antibiotics. For expression of cDNAs, 1 × 105 HeLa cells were seeded 18 h prior to transient transfection using Lipofectamine Plus (Invitrogen) and carried out according to the manufacturer's instructions. Medium was replaced 3 h post-transfection, and cells were lysed 24 h post-transfection in lysis buffer (50 mm HEPES, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 1% Triton X-100, and 10% glycerol) lacking sodium vanadate unless otherwise indicated in figure legends. 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride (PMSF) were added to the lysis buffer prior to use.
Antibodies and Reagents
HGF was a kind gift from Genentech, Inc. (San Francisco, CA). A rabbit polyclonal antibody raised against a peptide from the carboxyl terminus of human Met (43) was used for immunoprecipitation and Western blotting of Met (44). Alexa Fluor 555-EGF, Alexa Fluor 647-EGF, Alexa Fluor 555-transferrin, and all secondary antibodies used for immunofluorescence were purchased from Molecular Probes/Invitrogen; anti-early endosomal antigen 1 (EEA1), anti-NSF, and anti-GFP antibodies were purchased from Upstate Biotechnologies Inc. (Lake Placid, NY); and anti-actin antibody was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). An antibody against the extracellular region of Met (anti-Met AF276) used for immunofluorescence was purchased from R&D Systems (Minneapolis, MN).
Immunoprecipitations and Western Blotting
500 μg of HeLa cell lysates were used for each immunoprecipitation. The antibody was allowed to bind for 1 h at 4 °C, and 10 μl of Protein A-Sepharose beads were then added to collect immune complexes. Beads were washed twice in lysis buffer, then resuspended in 20 μl of Laemmli sample buffer, and boiled for 10 min. Supernatants were loaded onto 10% SDS-polyacrylamide gels and separated by electrophoresis. Proteins were transferred onto nitrocellulose membranes (Hybond-C, GE Healthcare), which were blocked with 3% BSA and probed with the appropriate antibody as indicated in the figure legends. Horseradish peroxidase-linked secondary antibodies (Amersham Biosciences) indicating the positions of target proteins were visualized by enhanced chemiluminescence (Amersham Biosciences). Results were quantified using NIH ImageJ software (Version 1.43u).
Confocal Immunofluorescence Microscopy
2 × 104 HeLa cells were seeded on glass coverslips (Bellco Glass Inc., Vineland, NJ) in 24-well plates (Nalgene NUNC, Rochester, NY) and transfected with the indicated DNA constructs using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. 24 hours later, cells were serum-starved for 2 h with cycloheximide (0.1 mg/ml) prior to HGF treatment (1.5 nm) as indicated. Coverslips were washed once with PBS and then fixed in 2% paraformaldehyde (Fisher Scientific) in PBS for 20 min. Coverslips were washed four times in PBS, and residual paraformaldehyde was removed by three 5-min washes with 100 mm glycine in PBS. Cells were permeabilized using 0.3% Triton X-100 in PBS and blocked for 30 min with blocking buffer (5% bovine serum albumin, 0.2% Triton X-100, and 0.05% Tween 20 in PBS). Coverslips were incubated with primary and secondary antibodies diluted in blocking buffer for 1 h and 40 min, respectively, at room temperature and washed four times in IF buffer (0.5% bovine serum albumin, 0.2% Triton X-100, and 0.05% Tween 20 in PBS) between primary and secondary and after secondary antibody incubations. Coverslips were mounted with Immu-Mount (Thermo-Shandon, Pittsburgh, PA). Confocal images were taken using a Zeiss 510 Meta laser-scanning confocal microscope (Carl Zeiss, Canada Ltd., Toronto, Ontario, Canada) with a 100× objective. Image analysis was carried out using the LSM 5 image browser software (Empix Imaging, Mississauga, Ontario, Canada). To define the degree of co-localization, Nikon NIS Analysis software was used to determine the Pearson co-localization coefficient (a value of 1 indicates complete co-localization, and −1 indicates complete exclusion). This analysis was performed on ≥20 cells for each siRNA condition and a minimum of n = 5 for overexpression studies. Significance was assessed using a two-tailed heteroscedastic t test with a significance threshold of p < 0.05.
Transferrin Assay
1 × 105 HeLa cells were seeded on 6-well dishes containing four glass coverslips (Bellco Glass Inc.) in each well and transfected with the indicated cDNA constructs. 24 h post-transfection, cells were loaded with transferrin conjugated to Alexa Fluor 555 and either HGF or Alexa Fluor 647-labeled EGF for 3 min after which cells were washed with cold PBS for 1 min on ice, and medium was replaced with ligand-free 37 °C medium. Cells were fixed at the times indicated and processed for microscopy as described above.
RESULTS
Loss of PTP1B Activity Abrogates Met Degradation and Leads to Sustained Downstream MEK1/2 Activation
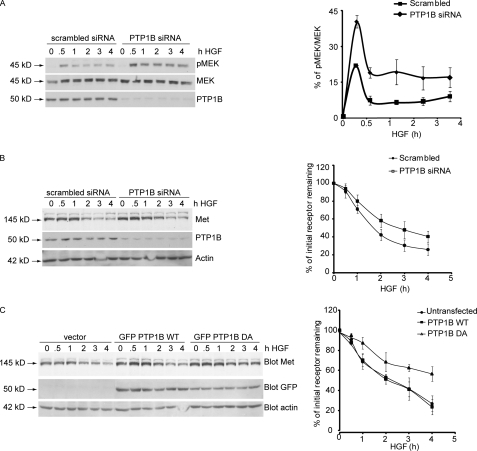
We have shown previously that depletion of PTP1B leads to Met receptor hyperphosphorylation and increased cell invasiveness in response to HGF, indicating enhanced biological activity of the receptor (30). To determine whether this increase in the invasive potential of Met in the absence of PTP1B involves deregulation of Met processing, we examined the effect of PTP1B ablation on HGF-induced degradation of the Met receptor. For this purpose, HeLa cells transfected with either PTP1B-specific or scrambled (nonspecific) siRNA were stimulated with HGF for the times indicated, lysed, and probed for Met protein by Western blotting. We found that depletion of PTP1B resulted in a 2-fold higher phosphorylation of MEK 30 min post-HGF stimulation (Fig. 1A). Because changes in pMEK activation can be attributed to an alteration in Met down-regulation (6), we investigated the effect of PTP1B depletion on Met receptor levels and observed delayed degradation of Met (Fig. 1B). In cells treated with PTP1B-specific siRNA, ∼60% of endogenous receptor was present 2 h post-HGF stimulation compared with ∼40% in scrambled siRNA-transfected cells. We then tested whether the catalytic activity of PTP1B was required for normal Met degradation. For this purpose, we utilized the PTP1B D/A mutant. In this mutant, the Asp-181 residue responsible for protonating the tyrosyl leaving group of the substrate, which is followed by bond cleavage and substrate release, is replaced by a catalytically inactive alanine, which cannot mediate substrate release. Thus, the D/A mutant forms a complex with PTP1B substrates, thereby “trapping” them. In cells overexpressing either GFP vector or GFP-PTP1B WT, receptor degradation began 2 h after stimulation, and 70% of the receptor was degraded 4 h post-HGF stimulation (Fig. 1C). However, in GFP-PTP1B D/A-overexpressing cells, Met levels remained elevated for a longer period, and only 40% of the receptor was degraded after 4 h (Fig. 1C). These results demonstrate that ablation of PTP1B activity either by overexpression of the PTP1B D/A mutant or by siRNA-mediated depletion of endogenous PTP1B results in delayed Met degradation and affects signaling downstream of the receptor and suggest that PTP1B is an important novel determinant of Met receptor degradation.
FIGURE 1.
Loss of function of PTP1B leads to protracted Met signaling and delayed Met dephosphorylation and degradation following HGF stimulation but does not affect Met ubiquitination. A, HeLa cells were transfected either with scrambled siRNA or siRNA against PTP1B and stimulated with HGF for the times indicated. Protein lysates from these cells were subjected to immunoblotting with antibody against MEK, pMEK, or PTP1B. The graph on the right depicts the percentage of phosphorylated MEK. B, HeLa cells were transfected either with scrambled siRNA or siRNA against PTP1B and stimulated with HGF for the times indicated. Protein lysates from these cells were subjected to immunoblotting with antibody against Met, PTP1B, or actin (loading control). The graph on the right depicts the percentage of initial receptor remaining. C, HeLa cells were transfected with either GFP-PTP1B WT or the mutant GFP-PTP1B D/A and stimulated with HGF in the presence of cycloheximide for the times indicated. Protein lysates from these cells were subjected to immunoblotting with antibody against Met, GFP, or actin (loading control). The graph on the right depicts the percentage of initial receptor remaining.
Previous studies from our and other laboratories have reported that degradation of the Met receptor requires its ubiquitination (4–6, 10, 45). To ascertain whether Met ubiquitination was altered by the expression of the dominant negative PTP1B D/A allele, HeLa cells transfected with either GFP-PTP1B WT or D/A were stimulated with HGF, and receptor ubiquitination was evaluated. No obvious difference in Met receptor ubiquitination was observed between cells expressing PTP1B WT or D/A (supplemental Fig. S1A). Furthermore, HGF-stimulated HeLa cells transfected with scrambled or PTP1B-specific siRNA did not exhibit differences in Met receptor ubiquitination status (supplemental Fig. S1B). These data suggest that the mechanism through which PTP1B regulates Met receptor degradation does not involve changes in receptor ubiquitination.
PTP1B Is Required for Progression of Met Receptor from Internalized Vesicle to Early Endosome
Having demonstrated that PTP1B is required for attenuation of Met-mediated signaling, we investigated the mechanisms involved. Upon ligand binding, the Met RTK is activated and internalized (4–6, 10, 45). Met then traffics along the endocytic pathway during which time it continues to signal until it is eventually degraded (6). Although overexpression of the dominant negative variant of PTP1B (D/A) or siRNA-mediated PTP1B knockdown did not affect Met receptor ubiquitination (supplemental Fig. S1), both led to delayed degradation of the receptor (Fig. 1, B and C). We therefore hypothesized that the effect of PTP1B on receptor stability may be mediated via control of receptor trafficking.
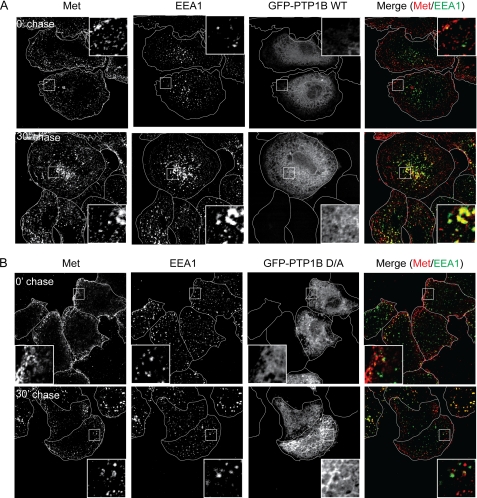
To investigate Met endocytosis in HeLa cells, we stimulated cells with HGF at 4 °C (cold load), which leads to receptor activation but prevents its internalization, and then monitored Met trafficking by exposing cells to 37 °C for the indicated time points (chase). Receptor endocytosis was tracked using the established early endosomal markers EEA1 and Rab5, which have been shown previously to co-localize with Met during trafficking (6). In cells transfected with a GFP vector or GFP-PTP1B WT, Met was internalized and entered EEA1-positive early endosomes within 15 min postchase as demonstrated previously (data not shown and Ref. 6). 30 min postchase, Met-positive EEA1 endosomes began to cluster in a perinuclear compartment (Fig. 2A). Similarly, expression of PTP1B did not affect co-localization of Met with Rab5 (supplemental Fig. S2A). However, in cells expressing GFP-PTP1B D/A, co-localization of Met with EEA1 or Rab5 30 min poststimulation was significantly abrogated when compared with PTP1B WT-expressing cells (Fig. 2B; Pearson's co-localization coefficient, 0.17 ± 0.003 versus 0.61 ± 0.02 in WT cells; supplemental Fig. S2B; Pearson's coefficient, 0.10 ± 0.04 versus 0.62 ± 0.05 in WT cells). Strikingly, in the presence of the D/A mutant, Met-positive puncta remained present at the cell periphery and even at later time points did not localize to a perinuclear compartment (Fig. 2B, supplemental Fig. S2B, and data not shown), suggesting that the PTP1B D/A dominant negative mutant induces a block in Met trafficking following internalization. Moreover, the observed defect in Met trafficking through the endocytic pathway is consistent with the delayed Met receptor degradation observed in the presence of the PTP1B D/A mutant (Fig. 1C) and suggests that the catalytic activity of PTP1B is required for efficient endocytic progression of Met. Importantly, overexpression of PTP1B D/A did not disrupt the formation of EEA1- or Rab5-positive endosomes, which were comparable in size and number with those observed in control and PTP1B WT-expressing cells (Fig. 2 and supplemental Fig. S2).
FIGURE 2.
Loss of function of PTP1B delays Met trafficking following HGF stimulation. A and B, HeLa cells were seeded on glass coverslips and transfected with either GFP-PTP1B WT (A) or D/A (B) 16 h postseeding. 24 h later, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with HGF at 4 °C for 1 h. Cells were chased with warm medium without HGF and fixed at the times indicated followed by staining with anti-Met and anti-EEA1 antibodies. The merge of Met (red) and EEA1 (green) is shown in GFP-expressing cells. Pearson's coefficient of co-localization, 0.17 ± 0.01 (0 min (0′)) and 0.61 ± 0.02 (30 min (30′)) for PTP1B WT-transfected cells and 0.13 ± 0.03 (0 min) and 0.16 ± 0.03 (30 min) for PTP1B D/A-transfected cells. p values, 0.2550 (0 min) and 0.0002 (30 min).
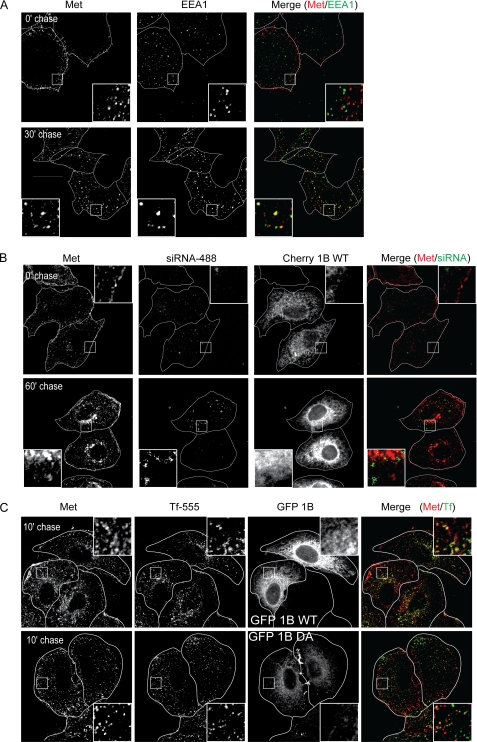
To address the possibility that the defect in Met trafficking may be an artifact caused by PTP1B D/A overexpression, we examined the effect on Met trafficking of PTP1B depletion, which also resulted in delayed Met degradation (Fig. 1B). Interestingly, significantly unlike what was seen in PTP1B D/A-expressing cells, Met co-localized with EEA1 and Rab5 in PTP1B siRNA-treated cells (Fig. 3A; Pearson's coefficient, 0.11 ± 0.01 at 0 min and 0.46 ± 0.03 at 30 min in PTP1B siRNA-treated cells versus 0.05 ± 0.02 at 0 min and 0.45 ± 0.04 at 30 min in scrambled siRNA-treated cells (supplemental Fig. S3)) but failed to reach a perinuclear compartment, providing an explanation for the delay in Met degradation seen under these conditions (Figs. 3A and 1B and supplemental Fig. S7A). These data suggest that in the absence of PTP1B Met is internalized and can reach an early endosome but is not efficiently targeted to a perinuclear compartment for subsequent degradation. Importantly, expression of an siRNA-resistant PTP1B WT construct was able to rescue the defect in Met trafficking, stability, and MEK activation in cells treated with siRNA against endogenous PTP1B (Fig. 3B and supplemental Fig. S7A). The ability of Met to reach an EEA1-positive compartment in PTP1B siRNA-treated cells but not in PTP1B D/A-expressing cells may be due to the presence of low levels of functional endogenous PTP1B in the siRNA-treated cells. Taken together, our data confirm that PTP1B is required for the Met receptor to traffic into a late endosomal compartment. These data are in agreement with a recent observation that PTP1B is required for the sequestration of the EGFR into MVBs (39).
FIGURE 3.
Loss of PTP1B delays Met trafficking but does not affect transferrin recycling following HGF stimulation. HeLa cells were seeded on coverslips and transfected with siRNA directed against PTP1B (A). siRNA-treated cells were rescued by overexpression of cherry-PTP1B (Cherry 1B) WT 24 h post-siRNA transfection (B). 48 h post-siRNA transfection, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with HGF at 4 °C for 1 h. Cells were chased with warm medium without HGF and fixed at the times indicated followed by staining with anti-Met and anti-EEA1 antibodies. A, PTP1B siRNA-treated cells. The merge of Met (red) and EEA1 (green) is shown. Pearson's coefficient of co-localization for PTP1B siRNA transfected cells, 0.11 ± 0.01 (0 min (0′)) and 0.46 ± 0.03 (30 min (30′)). p values, 0.0697 (0 min) and 0.7663 (30 min). B, Alexa Fluor 488-labeled PTP1B siRNA-treated cells overexpressing cherry-PTP1B WT. Trafficking of Met to the perinuclear compartment in cells overexpressing siRNA-resistant cherry-PTP1B WT is shown. C, HeLa cells were seeded on coverslips and transfected with GFP-PTP1B WT. Cells were loaded with HGF and Alexa Fluor 555-transferrin (Tf-555), allowed to internalize, and chased for the times indicated. Cells were fixed and stained for extracellular Met. Pearson's coefficient of co-localization, 0.49 ± 0.01 (10 min) for PTP1B WT-transfected cells and 0.28 ± 0.05 (10 min) for PTP1B D/A-transfected cells. p values, 0.5669 (0 min) and 0.1493 (30 min).
PTP1B Regulates RTK Trafficking but Not That of Constitutively Internalizing Transferrin Receptor
To further address the importance and specificity of PTP1B in trafficking, we investigated whether transport of the constitutively recycling transferrin receptor, which also traffics to early endosomes (46), is altered when PTP1B activity is ablated. Labeled transferrin was loaded onto HeLa cells and allowed to internalize (for 3 min), and then excess ligand in the medium was removed. Cells were subsequently fixed at the indicated time points, and transferrin recycling was followed. Recycling is indicated by the loss of labeled transferrin within the cell and its release back into the medium. Importantly, we found that expression of the PTP1B D/A mutant did not alter trafficking of the transferrin receptor as the clearance of labeled transferrin from cells after a 20-min chase was the same in cells expressing either the PTP1B WT or PTP1B D/A mutant (Fig. 3C). Moreover, in cells also stimulated with HGF, 10 min after chase, far less transferrin co-localized with Met in PTP1B D/A-expressing cells as compared with PTP1B WT-expressing cells (Fig. 3C). This is in agreement with our previous observation that Met was present in a compartment near the membrane in the presence of PTP1B D/A (Fig. 2B). We therefore conclude that PTP1B regulates receptor trafficking in a cargo-specific manner and that the point at which it acts is downstream from ligand-induced receptor internalization.
PTP1B Is Required for EGFR Trafficking through Endocytic Pathway
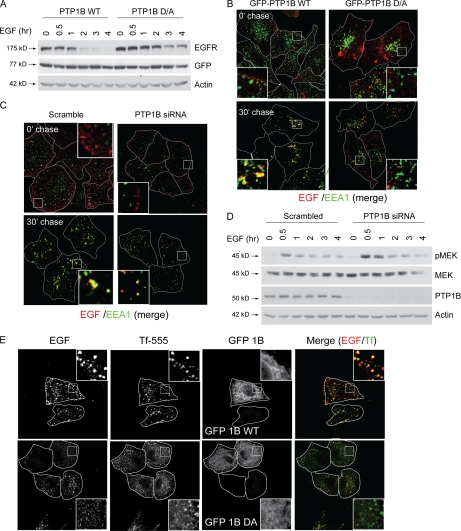
Our results show that PTP1B is required for the trafficking of the Met receptor toward the perinuclear compartment of the cell, a stage that is associated with lysosomal degradation of cargo proteins (3). Because PTP1B was not required for transferrin receptor trafficking, we investigated whether the requirement for PTP1B in Met receptor trafficking was unique to the Met receptor or whether this mechanism was common to other RTKs. The trafficking and down-regulation of the EGFR has been extensively studied, and this receptor is also a substrate for PTP1B (26). Recently, Eden et al. (39) have demonstrated that PTP1B is required for MVB maturation and lysosomal degradation of the EGFR. Hence, we asked whether PTP1B also regulates EGFR down-regulation earlier in the endocytic pathway as was seen for the Met receptor. Upon expression of the PTP1B D/A mutant or siRNA-mediated depletion of the endogenous protein from HeLa cells, we observed a similar delay in EGF-induced EGFR degradation as was demonstrated previously for the Met receptor (Fig. 4A and supplemental Fig. S4). Furthermore, using fluorescently labeled EGF to follow EGFR trafficking, we also observed a severe delay in EGFR localization to the perinuclear compartment in cells expressing the D/A trapping mutant or depleted of PTP1B (Fig. 4B). Similar to our results with the Met receptor, in cells expressing cherry-PTP1B D/A, we observed significantly decreased co-localization of the EGFR with EEA1 (Pearson's coefficient, 0.22 ± 0.04 versus 0.46 ± 0.02 at 30 min postchase) or GFP-Rab5 (Pearson's coefficient, 0.20 ± 0.01 versus 0.52 ± 0.07 at 60 min postchase), confirming that the EGFR also requires PTP1B to reach an early endosomal compartment (Fig. 4B and supplemental Fig. S5). However, as seen with the Met receptor, labeled EGF co-localized with EEA1 in cells depleted of PTP1B compared with cells transfected with scrambled siRNA (Fig. 4C). Importantly, the inability of the EGFR to reach the perinuclear compartment and be degraded in cells depleted of endogenous PTP1B using siRNA was accompanied by more robust phosphorylation of MEK in response to EGF (Fig. 4D). In addition, the sustained MEK phosphorylation caused by PTP1B depletion was reverted by expression of an siRNA-resistant GFP-PTP1B construct (supplemental Fig. S7B). Although transferrin receptor recycling was unaffected in PTP1B D/A-expressing cells (Figs. 3C and 4E and supplemental Fig. S6), as with Met, we observed reduced EGFR co-localization with transferrin in D/A-expressing cells compared with cells expressing PTP1B WT (Pearson's coefficient, 0.49 ± 0.02 versus 0.35 ± 0.03 at 10 min postchase). Taken together, our data demonstrate that PTP1B regulates the trafficking of at least two RTKs but not of the transferrin receptor.
FIGURE 4.
Loss of function of PTP1B leads to delayed EGFR degradation and protracted EGFR signaling following EGF stimulation. A, HeLa cells were transfected with either GFP-PTP1B WT or the mutant GFP-PTP1B D/A and stimulated with EGF in the presence of cycloheximide for the times indicated. Protein lysates from these cells were subjected to immunoblotting with antibodies against EGFR, GFP (transfection control), and actin (loading control). B, HeLa cells were seeded on glass coverslips and transfected with either GFP-PTP1B WT or D/A 16 h postseeding. 24 h later, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with Alexa Fluor 555-EGF at 4 °C for 1 h. Cells were chased with warm medium without EGF and fixed at the times indicated followed by staining with an anti-EEA1 antibody. The merge of EGF (red) and EEA1 (green) is shown. Pearson's coefficient of co-localization, 0.13 ± 0.02 (0 min (0′)) and 0.46 ± 0.02 (30 min (30′)) for PTP1B WT-transfected cells and 0.14 ± 0.02 (0 min) and 0.22 ± 0.04 (30 min) for PTP1B D/A-transfected cells. p values, 0.6702 (0 min) and 0.0322 (30 min). C, HeLa cells were seeded on glass coverslips and transfected with either scrambled or PTP1B-specific siRNA 16 h postseeding. 24 h later, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with Alexa Fluor 555-EGF at 4 °C for 1 h. Cells were chased with warm medium without EGF and fixed at the times indicated followed by staining with an anti-EEA1 antibody. The merge of EGF (red) and EEA1 (green) is shown. Pearson's coefficient of co-localization, 0.14 ± 0.02 (0 min) and 0.37 ± 0.03 (30 min) for scrambled siRNA-transfected cells and 0.08 ± 0.04 (0 min) and 0.49 ± 0.03 (30 min) for PTP1B siRNA-transfected cells. p values, 0.2766 (0 min) and 0.0427 (30 min). D, HeLa cells were transfected either with scrambled siRNA or siRNA against PTP1B and stimulated with EGF for the times indicated. Protein lysates from these cells were subjected to immunoblotting with antibodies against pMEK, MEK, PTP1B, and actin (loading control). E, HeLa cells were seeded on coverslips and transfected with GFP-PTP1B WT. Cells were loaded with Alexa Fluor 647-EGF and Alexa Fluor 555-transferrin (Tf-555), allowed to internalize, chased for the times indicated, and fixed. Pearson's coefficient of co-localization, 0.49 ± 0.02 (10 min) for PTP1B WT-transfected cells and 0.35 ± 0.03 (10 min) for PTP1B D/A-transfected cells. p values, 0.8252 (0 min) and 0.0255 (30 min).
PTP1B Is Required for Progression of Met Receptor from Internalized Vesicle to PI3P-rich Endosomal Compartment
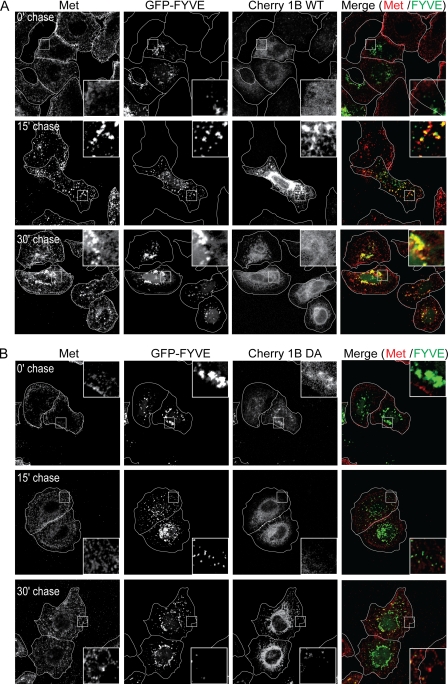
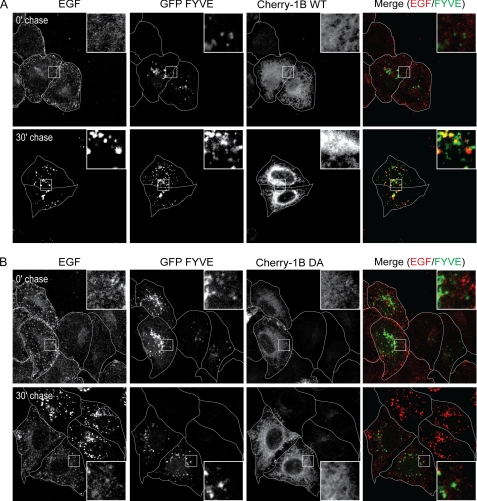
Formation of classical early endosomes as defined by the recruitment of Rab5 and EEA1 requires the conversion of the phosphatidylinositol 3,4,5-trisphosphate- and phosphatidylinositol 4,5-bisphosphate-rich plasma membrane envelope to a PI3P-rich endosomal membrane. Because this conversion cannot be directly observed, we utilized a fluorescent probe containing GFP fused to the FYVE domain of EEA1, which selectively binds PI3P, to mark PI3P-rich vesicles. In cells expressing PTP1B WT, 30 min postchase almost all PI3P-rich vesicles contained Met in HGF-stimulated cells and EGF in EGF-stimulated cells (Figs. 5A and 6A). However, at the same time point, in cells expressing PTP1B D/A, GFP-FYVE-containing endosomes showed significantly reduced association with Met or EGF, indicating that the catalytic activity of PTP1B is required for these RTKs to reach PI3P-rich early endosomes (Figs. 5B and 6B). These results suggest that PTP1B activity is essential for the maturation of a nascent plasma membrane-derived vesicle containing the EGF or Met RTK into a PI3P-rich early endosome. However, the activity of PTP1B is not required for the constitutive formation of these compartments as such structures were able to form in cells expressing the dominant negative D/A variant of PTP1B and trafficking of the transferrin receptor was not abrogated (Figs. 2B, 3A, 4B, 4C, 5B, and 6B).
FIGURE 5.
Met does not reach FYVE-positive compartment in HeLa cells overexpressing PTP1B D/A. HeLa cells were seeded on coverslips and at 16 h postseeding transfected with cherry-PTP1B (Cherry 1B) WT (A) or D/A (B) and GFP-FYVE. 20 h post-transfection, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with HGF at 4 °C for 1 h. Cells were chased with warm medium without HGF and fixed at the times indicated followed by staining with an antibody directed against the extracellular region of Met. A, GFP-FYVE- and cherry-PTP1B WT-overexpressing cells. The merge of Met (red) and FYVE (green) is shown. B, GFP-FYVE- and cherry-PTP1B D/A-overexpressing cells. The merge of Met (red) and FYVE (green) is shown. Pearson's coefficient of co-localization, 0.07 ± 0.02 (0 min (0′)), 0.47 ± 0.08 (15 min (15′)), and 0.35 ± 0.02 (30 min (30′)) for PTP1B WT-transfected cells and 0.11 ± 0.02 (0 min), 0.18 ± 0.02 (15 min), and 0.16 ± 0.04 (30 min) for PTP1B D/A-transfected cells. p values, 0.2145 (0 min), 0.0259 (15 min), and 0.0232 (30 min).
FIGURE 6.
EGF does not reach FYVE-positive compartment in HeLa cells overexpressing PTP1B D/A. HeLa cells were seeded on coverslips and at 16 h postseeding transfected with cherry-PTP1B (Cherry-1B) WT (A) or D/A (B) and GFP-FYVE. 16 h post-transfection, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with Alexa Fluor 647-EGF at 4 °C for 1 h. Cells were chased with warm medium without EGF. A, GFP-FYVE- and cherry-PTP1B WT-overexpressing cells. The merge of EGF (red) and FYVE (green) is shown. B, GFP-FYVE- and cherry-PTP1B D/A-overexpressing cells. The merge of EGF (red) and FYVE (green) is shown. Pearson's coefficient of co-localization, 0.07 ± 0.02 (0 min (0′)) and 0.44 ± 0.06 (30 min (30′)) for PTP1B WT-transfected cells and 0.05 ± 0.02 (0 min) and 0.18 ± 0.01 (30 min) for PTP1B D/A-transfected cells. p values, 0.4359 (0 min) and 0.0132 (30 min).
Depletion of NSF, a Substrate of PTP1B, Phenocopies Effect of PTP1B Depletion on Met and EGFR Down-regulation
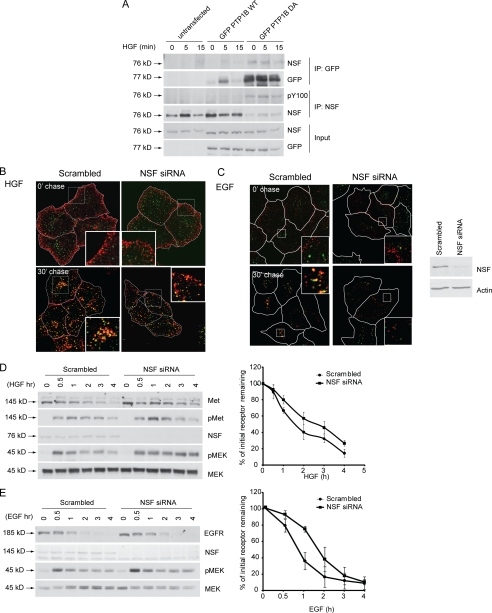
Our data thus far suggest that functional PTP1B is required for the Met and EGF receptors to progress from the small vesicles formed upon internalization of the RTKs to PI3P-rich, Rab5- and EEA1-positive compartments. Newly formed vesicles enter the endosomal network through fusion with acceptor vesicles (i.e. early endosomes), a process dependent on the action of vesicle docking proteins (v-SNAREs and t-SNAREs) (47). Following vesicle fusion, the SNARE complex must be disassembled via ATP hydrolysis of its α-SNAP subunit by NSF (48, 49), the localization of which is regulated via its phosphorylation status. We therefore hypothesized that the defect in Met and EGFR trafficking observed upon depletion or loss of function of PTP1B could be due to abrogation of NSF dephosphorylation. To address this possibility, we first confirmed that NSF was indeed a substrate for PTP1B in our system. Upon HGF stimulation, we observed that endogenous NSF associated with PTP1B WT (Fig. 7A). Moreover, this interaction was dramatically increased with the substrate trapping mutant (D/A) to the extent that the remaining pool of NSF isolated from sequential immunoprecipitation of NSF was reduced compared with that in PTP1B WT-transfected cells (Fig. 7A). Similarly, use of the trapping mutant resulted in increased phosphorylation of NSF, further supporting that NSF is a substrate for PTP1B (Fig. 7A).
FIGURE 7.
NSF is substrate of PTP1B, and knockdown of NSF delays RTK trafficking and protracts signaling. A, immunoblotting of PTP1B immunoprecipitates with the anti-NSF antibody shows that NSF is trapped by PTP1B. HeLa cells were transfected with either GFP-PTP1B WT or GFP-PTP1B D/A and stimulated with HGF in the presence of cycloheximide for the times indicated. Serial immunoprecipitations were performed with anti-GFP and anti-NSF antibodies, and immunoprecipitates and total protein lysates from these cells were subjected to immunoblotting with antibodies against NSF, GFP, and phosphotyrosine Tyr(P)-100 (pY100). Note that NSF is hyperphosphorylated in the absence of PTP1B and is unaffected by presence of the Met ligand, HGF. B, HeLa cells were seeded on coverslips and at 16 h postseeding transfected with either siRNA against NSF or a scrambled siRNA control. 48 h post-transfection, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with HGF at 4 °C for 1 h. Cells were chased with warm medium without HGF and fixed at the times indicated followed by staining with an antibody against the extracellular region of Met. The merge of EEA1 (green) and Met (red) is shown. Side panel, immunoblot depicting knockdown of NSF for this experiment. Pearson's coefficient of co-localization, 0.05 ± 0.02 (0 min (0′)) and 0.45 ± 0.02 (30 min (30′)) for scrambled siRNA-transfected cells and 0.13 ± 0.01 (0 min) and 0.44 ± 0.07 (30 min) for NSF1 siRNA-transfected cells. p values, 0.0330 (0 min) and 0.9188 (30 min). C, HeLa cells were seeded on coverslips and at 16 h postseeding transfected with either siRNA against NSF or a scrambled siRNA control. 48 h post-transfection, cells were serum-starved in the presence of cycloheximide for 2 h and loaded with Alexa Fluor 555-EGF at 4 °C for 1 h. Cells were chased with warm medium without HGF and fixed at the times indicated followed by staining with an antibody against the extracellular region of Met. The merge of EEA1 (green) and Met (red) is shown. Pearson's coefficient of co-localization, 0.09 ± 0.02 (0 min) and 0.51 ± 0.01 (30 min) for scrambled siRNA-transfected cells and 0.12 ± 0.04 (0 min) and 0.52 ± 0.03 (30 min) for NSF1 siRNA-transfected cells. p values, 0.5462 (0 min) and 0.7938 (30 min). D, HeLa cells transfected with either scrambled siRNA or siRNA against PTP1B were serum-starved overnight and stimulated with HGF in the presence of cycloheximide for the times indicated. Protein lysates from these samples were subjected to immunoblotting with antibodies against Met, phosphorylated Met (pMet), NSF, phosphorylated MEK, and total MEK. The graph on the right depicts the percentage of initial Met receptor remaining. E, HeLa cells transfected with either scrambled siRNA or siRNA against NSF were serum-starved overnight and stimulated with EGF in the presence of cycloheximide for the times indicated. Protein lysates from these samples were subjected to immunoblotting with antibodies against EGFR, NSF, phosphorylated MEK, and total MEK. Note that the loss of NSF results in delayed RTK degradation and sustained pMEK activation.
To determine whether loss of NSF resulted in altered RTK trafficking, we examined the localization of the Met and EGFR RTKs in cells treated with siRNA against NSF. Importantly, we found that depletion of NSF led to significant changes in localization, namely retention of Met and EGFR in small vesicles at the cell periphery following ligand stimulation. These vesicles were unable to progress to a perinuclear compartment (Fig. 7, B and C), mimicking the defect seen in the absence of PTP1B. This delay in RTK endocytosis also correlated with delayed degradation of Met or the EGFR (Fig. 7, D and E). Similar to results obtained using PTP1B siRNA, 40% of Met remained in cells treated with scrambled siRNA 2 h post-HGF stimulation compared with 60% in NSF-siRNA treated cells, a delay that was also seen for the EGFR (5% receptor remaining in control cells versus 35% in NSF-depleted cells at 2 h post-EGF stimulation; Fig. 7, D and E). In addition, loss of NSF protein also resulted in sustained activation of MEK1/2 downstream from both Met and the EGFR upon ligand stimulation (Fig. 7, D and E), demonstrating that perturbation of NSF function leads to sustained signaling downstream of these RTKs.
DISCUSSION
Following receptor activation, maintenance of cellular homeostasis requires that mechanisms exist to attenuate and eventually extinguish the signal once the stimulus is no longer present. In the case of receptor tyrosine kinases, multiple levels of regulation have been shown to exist, including receptor endocytosis and eventual degradation as well as dephosphorylation. However, the mechanisms through which specific phosphatases regulate receptor trafficking are still not fully understood. However, a role for the endocytic pathway in regulating receptor signaling has now been established, and it has been shown that deregulation of receptor endocytosis can lead to altered signaling and cellular transformation (3). We have demonstrated previously that like the EGFR the Met RTK continues to signal as it traverses the endocytic pathway (6). Ubiquitination-deficient variants of Met (Y1003F) or EGFR (Y1045F) become oncogenically active due to escape from lysosomal degradation; such receptors are retained in an endosomal compartment and induce prolonged activation of the MAPK pathway (6, 51). These results suggest that signaling downstream of receptors at least through the MAPK pathway is down-regulated by eventual receptor degradation rather than receptor dephosphorylation. Recently, Eden et al. (39) have demonstrated a role for PTP1B in sequestration of the EGFR into multivesicular bodies. Here, we further demonstrate that phosphatases can regulate RTK endocytosis at other levels of the endocytic pathway and show that the action of PTP1B was required for the degradation of the Met and EGF RTKs; loss of PTP1B catalytic activity resulted in delayed receptor degradation (Figs. 1, B and C, and 4A and supplemental Fig. S4) and subsequent prolonged activation of the MAPK signaling pathway (Figs. 1A and 4D). Our data are in agreement with those of Eden et al. (39) wherein loss of PTP1B results in a delay in EGFR degradation.
We and others have demonstrated previously that in the absence of PTP1B its RTK substrates are hyperphosphorylated (19, 52). In the case of the Met receptor, this leads to enhanced cell invasion in response to HGF stimulation (30). In this study, we demonstrate that depletion of PTP1B led to sustained activation of MEK1/2 downstream from both the Met and EGF receptors (Figs. 1A and 4D). Examination of Met and EGFR stability revealed that the sustained signaling downstream of these RTKs upon loss of PTP1B catalytic activity correlated with delayed degradation of these receptors (Figs. 1, B and C, and 4A and supplemental Fig. S4). Together, these data demonstrate that sustained signaling in the absence of PTP1B is due to a reduced rate of receptor degradation. We have demonstrated previously that uncoupling of Met from Cbl-mediated ubiquitination results in delayed degradation and sustained activation of the MAPK pathway (6). However, examination of Met ubiquitination when uncoupled from PTP1B catalytic activity demonstrated that the receptor was still efficiently ubiquitinated (Fig. S1), suggesting that an alternative mechanism is responsible for the observed delay in degradation.
To examine whether PTP1B was required for receptor internalization and/or trafficking, we examined the trafficking of the Met and EGF RTKs via confocal microscopy. Surprisingly, we found that in the absence of PTP1B catalytic activity trafficking of both receptors through the endocytic pathway was abrogated (Figs. 2, 3, and 4 and supplemental Figs. S2, S3, S5, and S6). Upon internalization, receptors are incorporated into small pinocytic vesicles, which eventually fuse with larger PI3P-rich early endosomes containing EEA1 and Rab5 (53). Our data show that the Met and EGF receptors are internalized in the presence of the catalytically inactive variant of PTP1B (D/A) but remain on small vesicles near the plasma membrane (Figs. 2B and 4B). Under these conditions, we did not observe co-localization of either receptor with the early endosome markers EEA1 and Rab5 (Figs. 2B and 4B and supplemental Figs. S2B, S3B, and S8). Moreover, using a GFP-FYVE probe, we demonstrated that the small receptor-containing vesicles lacked PI3P (Figs. 5 and 6). When PTP1B was depleted using a specific siRNA, we did observe co-localization of both receptors with the early endosomal marker EEA1 (Figs. 3A and 4C). However, these endosomes were always smaller in size and occurred at later time points following growth factor stimulation, and we did not observe the characteristic perinuclear accumulation of either receptor that occurs as early endosomes mature into late endosomal compartments (Figs. 3A and 4C). The ability of receptors to reach an early endosomal compartment when PTP1B-specific siRNA was transfected but not upon expression of the dominant negative PTP1B D/A mutant may be due to the small amount of remaining functional endogenous PTP1B. Altogether, these data demonstrate a requirement for PTP1B catalytic activity in the progression of RTKs through the endocytic pathway and suggest that PTP1B is involved in regulating endosome fusion. Importantly, re-expression of wild-type PTP1B in siRNA-treated cells rescued both the delayed degradation of Met and EGFR and the sustained downstream signaling observed (supplemental Fig. S7). This role of PTP1B is dependent on the cargo being internalized because recycling of the transferrin receptor was unaffected by the loss of PTP1B activity (Fig. 3C and 4E). Our data are supported by a recent study showing that EphA3 interacts with PTP1B at the cell surface and that overexpression of PTP1B D/A blocks trafficking of EphA3 but does not affect transferrin receptor trafficking (54). Our data also suggest that co-localization of both the Met and EGF receptors with transferrin is abrogated in the presence of a catalytically inactive mutant of PTP1B, suggesting that the Met and EGFR RTKs are unable to reach the recycling compartment and further strengthening our hypothesis that PTP1B is required early in the endocytic pathway.
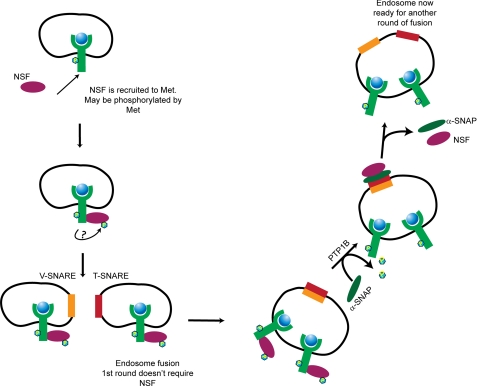
Recently, the ATPase NSF was identified as a substrate for PTP1B (42). NSF activity is tightly modulated by its phosphorylation status; dephosphorylation promotes its interaction with endosomal components and results in endosome fusion. We therefore hypothesized that the defect in RTK trafficking observed upon loss of PTP1B catalytic activity could be due to abrogation of NSF dephosphorylation and consequent disruption of endosome fusion. We demonstrated that HGF-induced tyrosine-phosphorylated NSF is a substrate for PTP1B (Fig. 7A) and importantly that depletion of NSF through specific siRNA phenocopied the loss of PTP1B catalytic activity, resulting in abrogation of Met trafficking, delayed Met degradation, and sustained activation of MEK1/2 (Fig. 7). Under these conditions, Met was retained on small vesicles at the periphery of the plasma membrane and did not accumulate in a perinuclear compartment as was observed with PTP1B siRNA (Fig. 7B). These data demonstrate that PTP1B-dependent regulation of RTK signaling occurs primarily through regulation of RTK trafficking via NSF-mediated endosome fusion rather than via direct dephosphorylation of RTKs. We therefore propose a model (Fig. 8) in which NSF is recruited to and potentially phosphorylated by the Met receptor followed by the fusion of endosomes mediated by the actions of v-SNAREs and t-SNAREs. Because the first round of endosome fusion does not require NSF (55), small vesicles containing Met are thus formed. We propose that PTP1B then dephosphorylates NSF, allowing the removal of α-SNAP and permitting the next round of endosome fusion to occur.
FIGURE 8.
Proposed model for regulation of PTP1B and NSF-mediated vesicle fusion during endocytosis of Met RTK.
Our study identifies a novel role for PTP1B whereby PTP1B can modulate RTK signaling through regulation of a component of the endocytic machinery. We demonstrate that for both the Met and EGF RTKs, loss of PTP1B or its substrate NSF resulted in abrogated receptor trafficking through the endocytic pathway, thus allowing receptors to escape degradation and permitting sustained activation of the MAPK pathway. Intriguingly, alterations in expression levels of α-SNAP have been implicated in the development of aggressive neuroendocrine tumors (50), indicating that disruption of NSF-mediated endosome fusion may be a common mechanism in cancer development. Until now, the role of PTP1B in regulating RTKs has mainly been attributed to direct dephosphorylation of RTK substrates. However, our data clearly demonstrate a new mechanism by which phosphatases have the potential to regulate a wide variety of RTKs, including those that are not direct substrates, by regulating a common pathway utilized by many receptors.
Supplementary Material
Acknowledgments
We thank members of the Park laboratory and Robert Annan for helpful comments on the manuscript and Genentech for the kind gift of HGF.
This work was supported in part by fellowships from the Cancer Research Society and the Terry Fox Foundation (to V. S.), United States Department of Defense Breast Cancer Research Initiative Grant DAMD17-99-1-9284 (to J. A.), and Canadian Institutes of Health Research Operating Grants MOP-62887 (to M. L. T.) and MOP-1154 (to M. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- RTK
- receptor tyrosine kinase
- PTP
- protein-tyrosine phosphatase
- NSF
- N-ethylmaleimide-sensitive factor
- D/A
- aspartic acid-to-alanine substitution
- HGF
- hepatocyte growth factor
- EGFR
- epidermal growth factor receptor
- MVB
- multivesicular body
- v-SNARE
- vesicle SNARE
- t-SNARE
- target SNARE
- α-SNAP
- α-soluble NSF-associated protein
- pMEK
- phosphorylated MEK
- EEA1
- early endosomal antigen 1
- PI3P
- phosphatidylinositol 3-phosphate.
REFERENCES
- 1. Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peschard P., Park M. (2003) Cancer Cell 3, 519–523 [DOI] [PubMed] [Google Scholar]
- 3. Abella J. V., Park M. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E973–E984 [DOI] [PubMed] [Google Scholar]
- 4. Hammond D. E., Urbé S., Vande Woude G. F., Clague M. J. (2001) Oncogene 20, 2761–2770 [DOI] [PubMed] [Google Scholar]
- 5. Hammond D. E., Carter S., McCullough J., Urbé S., Vande Woude G., Clague M. J. (2003) Mol. Biol. Cell 14, 1346–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abella J. V., Peschard P., Naujokas M. A., Lin T., Saucier C., Urbé S., Park M. (2005) Mol. Cell. Biol. 25, 9632–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorkin A., Goh L. K. (2009) Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 8. Vieira A. V., Lamaze C., Schmid S. L. (1996) Science 274, 2086–2089 [DOI] [PubMed] [Google Scholar]
- 9. Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. (1998) Genes Dev. 12, 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peschard P., Fournier T. M., Lamorte L., Naujokas M. A., Band H., Langdon W. Y., Park M. (2001) Mol. Cell 8, 995–1004 [DOI] [PubMed] [Google Scholar]
- 11. Kong-Beltran M., Seshagiri S., Zha J., Zhu W., Bhawe K., Mendoza N., Holcomb T., Pujara K., Stinson J., Fu L., Severin C., Rangell L., Schwall R., Amler L., Wickramasinghe D., Yauch R. (2006) Cancer Res. 66, 283–289 [DOI] [PubMed] [Google Scholar]
- 12. Heinrich R., Neel B. G., Rapoport T. A. (2002) Mol. Cell 9, 957–970 [DOI] [PubMed] [Google Scholar]
- 13. Hornberg J. J., Bruggeman F. J., Binder B., Geest C. R., de Vaate A. J., Lankelma J., Heinrich R., Westerhoff H. V. (2005) FEBS J. 272, 244–258 [DOI] [PubMed] [Google Scholar]
- 14. Schmidt-Arras D. E., Böhmer A., Markova B., Choudhary C., Serve H., Böhmer F. D. (2005) Mol. Cell. Biol. 25, 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reynolds A. R., Tischer C., Verveer P. J., Rocks O., Bastiaens P. I. (2003) Nat. Cell Biol. 5, 447–453 [DOI] [PubMed] [Google Scholar]
- 16. Meng T. C., Fukada T., Tonks N. K. (2002) Mol. Cell 9, 387–399 [DOI] [PubMed] [Google Scholar]
- 17. Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 18. Seely B. L., Staubs P. A., Reichart D. R., Berhanu P., Milarski K. L., Saltiel A. R., Kusari J., Olefsky J. M. (1996) Diabetes 45, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 19. Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., Ramachandran C., Gresser M. J., Tremblay M. L., Kennedy B. P. (1999) Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 20. Klaman L. D., Boss O., Peroni O. D., Kim J. K., Martino J. L., Zabolotny J. M., Moghal N., Lubkin M., Kim Y. B., Sharpe A. H., Stricker-Krongrad A., Shulman G. I., Neel B. G., Kahn B. B. (2000) Mol. Cell. Biol. 20, 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galic S., Hauser C., Kahn B. B., Haj F. G., Neel B. G., Tonks N. K., Tiganis T. (2005) Mol. Cell. Biol. 25, 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galic S., Klingler-Hoffmann M., Fodero-Tavoletti M. T., Puryer M. A., Meng T. C., Tonks N. K., Tiganis T. (2003) Mol. Cell. Biol. 23, 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakagawa Y., Aoki N., Aoyama K., Shimizu H., Shimano H., Yamada N., Miyazaki H. (2005) Zoolog. Sci. 22, 169–175 [DOI] [PubMed] [Google Scholar]
- 24. Suárez Pestana E., Tenev T., Gross S., Stoyanov B., Ogata M., Böhmer F. D. (1999) Oncogene 18, 4069–4079 [DOI] [PubMed] [Google Scholar]
- 25. Kulas D. T., Goldstein B. J., Mooney R. A. (1996) J. Biol. Chem. 271, 748–754 [DOI] [PubMed] [Google Scholar]
- 26. Flint A. J., Tiganis T., Barford D., Tonks N. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tenev T., Keilhack H., Tomic S., Stoyanov B., Stein-Gerlach M., Lammers R., Krivtsov A. V., Ullrich A., Böhmer F. D. (1997) J. Biol. Chem. 272, 5966–5973 [DOI] [PubMed] [Google Scholar]
- 28. Eden E. R., White I. J., Futter C. E. (2009) Biochem. Soc. Trans. 37, 173–177 [DOI] [PubMed] [Google Scholar]
- 29. Stuible M., Abella J. V., Feldhammer M., Nossov M., Sangwan V., Blagoev B., Park M., Tremblay M. L. (2010) J. Biol. Chem. 285, 23899–23907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sangwan V., Paliouras G. N., Abella J. V., Dubé N., Monast A., Tremblay M. L., Park M. (2008) J. Biol. Chem. 283, 34374–34383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palka H. L., Park M., Tonks N. K. (2003) J. Biol. Chem. 278, 5728–5735 [DOI] [PubMed] [Google Scholar]
- 32. Machide M., Hashigasako A., Matsumoto K., Nakamura T. (2006) J. Biol. Chem. 281, 8765–8772 [DOI] [PubMed] [Google Scholar]
- 33. Tonks N. K. (2003) FEBS Lett. 546, 140–148 [DOI] [PubMed] [Google Scholar]
- 34. Cheng A., Dubé N., Gu F., Tremblay M. L. (2002) Eur. J. Biochem. 269, 1050–1059 [DOI] [PubMed] [Google Scholar]
- 35. Stuible M., Doody K. M., Tremblay M. L. (2008) Cancer Metastasis Rev. 27, 215–230 [DOI] [PubMed] [Google Scholar]
- 36. Dubé N., Bourdeau A., Heinonen K. M., Cheng A., Loy A. L., Tremblay M. L. (2005) Cancer Res. 65, 10088–10095 [DOI] [PubMed] [Google Scholar]
- 37. Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 38. Haj F. G., Verveer P. J., Squire A., Neel B. G., Bastiaens P. I. (2002) Science 295, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 39. Eden E. R., White I. J., Tsapara A., Futter C. E. (2010) Nat. Cell Biol. 12, 267–272 [DOI] [PubMed] [Google Scholar]
- 40. Matveeva E. A., Whiteheart S. W., Vanaman T. C., Slevin J. T. (2001) J. Biol. Chem. 276, 12174–12181 [DOI] [PubMed] [Google Scholar]
- 41. Huynh H., Bottini N., Williams S., Cherepanov V., Musumeci L., Saito K., Bruckner S., Vachon E., Wang X., Kruger J., Chow C. W., Pellecchia M., Monosov E., Greer P. A., Trimble W., Downey G. P., Mustelin T. (2004) Nat. Cell Biol. 6, 831–839 [DOI] [PubMed] [Google Scholar]
- 42. Zarelli V. E., Ruete M. C., Roggero C. M., Mayorga L. S., Tomes C. N. (2009) J. Biol. Chem. 284, 10491–10503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodrigues G. A., Naujokas M. A., Park M. (1991) Mol. Cell Biol. 11, 2962–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X. M., Park M. (1993) Dev. Biol. 157, 308–320 [DOI] [PubMed] [Google Scholar]
- 45. Hammond D. E., Carter S., Clague M. J. (2004) Curr. Top. Microbiol. Immunol. 286, 21–44 [DOI] [PubMed] [Google Scholar]
- 46. Steele-Mortimer O., Méresse S., Gorvel J. P., Toh B. H., Finlay B. B. (1999) Cell. Microbiol. 1, 33–49 [DOI] [PubMed] [Google Scholar]
- 47. Söllner T. H., Rothman J. E. (1996) Experientia 52, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 48. Rodriguez L., Stirling C. J., Woodman P. G. (1994) Mol. Biol. Cell 5, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Block M. R., Glick B. S., Wilcox C. A., Wieland F. T., Rothman J. E. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 7852–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andreeva A. V., Kutuzov M. A., Voyno-Yasenetskaya T. A. (2006) Expert Opin. Ther. Targets 10, 723–733 [DOI] [PubMed] [Google Scholar]
- 51. Waterman H., Katz M., Rubin C., Shtiegman K., Lavi S., Elson A., Jovin T., Yarden Y. (2002) EMBO J. 21, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sangwan V., Paliouras G. N., Cheng A., Dubé N., Tremblay M. L., Park M. (2006) J. Biol. Chem. 281, 221–228 [DOI] [PubMed] [Google Scholar]
- 53. Mills I. G., Jones A. T., Clague M. J. (1999) Mol. Membr. Biol. 16, 73–79 [DOI] [PubMed] [Google Scholar]
- 54. Nievergall E., Janes P. W., Stegmayer C., Vail M. E., Haj F. G., Teng S. W., Neel B. G., Bastiaens P. I., Lackmann M. (2010) J. Cell Biol. 191, 1189–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Littleton J. T., Barnard R. J., Titus S. A., Slind J., Chapman E. R., Ganetzky B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12233–12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.