Abstract
Following organ transplantation, lifelong immunosuppressive therapy is required to prevent the host immune system from destroying the allograft. This can cause severe side effects and increased recipient morbidity and mortality. Complete cessation of immunosuppressive drugs has been successfully accomplished in selected transplant recipients, providing proof of principle that operational allograft tolerance is attainable in clinical transplantation. The intra-graft molecular pathways associated with successful drug withdrawal, however, are not well defined. In this study, we analyzed sequential blood and liver tissue samples collected from liver transplant recipients enrolled in a prospective multicenter immunosuppressive drug withdrawal clinical trial. Before initiation of drug withdrawal, operationally tolerant and non-tolerant recipients differed in the intra-graft expression of genes involved in the regulation of iron homeostasis. Furthermore, as compared with non-tolerant recipients, operationally tolerant patients exhibited higher serum levels of hepcidin and ferritin and increased hepatocyte iron deposition. Finally, liver tissue gene expression measurements accurately predicted the outcome of immunosuppressive withdrawal in an independent set of patients. These results point to a critical role for iron metabolism in the regulation of intra-graft alloimmune responses in humans and provide a set of biomarkers to conduct drug-weaning trials in liver transplantation.
Introduction
In human solid organ transplantation, long-term graft survival requires the administration of lifelong immunosuppressive therapy. This may result in severe side effects and increased recipient morbidity and mortality. Over the past two decades, studies primarily conducted in rodents have revealed that certain immunologic interventions induce transplantation tolerance, a state in which the allograft is specifically accepted without the need to administer chronic immunosuppression (1). Translation of these strategies into the clinic, however, has been remarkably difficult (1–3). On the other hand, human transplant recipients occasionally develop spontaneous operational tolerance, a phenomenon defined by the maintenance of stable graft function in the absence of harmful immune responses in recipients receiving no immunosuppressive therapy (4–9). In order to identify a putative signature of operational tolerance in blood, the immunologic characteristics of kidney and liver operationally tolerant recipients have been recently investigated in cross-sectional case-control studies (6, 10–12). The mechanistic interpretation of these studies, however, has been hampered by their retrospective design and by the lack of molecular analyses simultaneously conducted on allograft tissues. To unambiguously define the molecular pathways associated with human allograft operational tolerance, we collected sequential blood and liver tissue samples from liver recipients enrolled in a prospective multicenter immunosuppression withdrawal clinical trial. We sought to define the blood and intra-graft molecular patterns discriminating between recipients who can successfully discontinue immunosuppressive therapy (operationally tolerant) and those who reject when immunosuppressive drugs are discontinued (non-tolerant). Before the start of drug minimization, an expansion of NK cells and related transcripts was noted in blood samples collected from operationally tolerant recipients. In contrast, liver tissue samples obtained from operationally tolerant and non-tolerant recipients differed in the expression of a set of genes involved in the regulation of iron homeostasis. Among them, the gene encoding for hepcidin, the master regulator of systemic iron metabolism, was significantly overexpressed in operationally tolerant grafts. Furthermore, hepcidin serum levels and markers of systemic iron availability were higher in operationally tolerant than in non-tolerant recipients. These differences were not influenced by clinical parameters, nor by the type of immunosuppressive drugs administered, and in the case of liver tissue markers could accurately predict the outcome of the immunosuppressive withdrawal protocol. Our results reveal what we believe to be a novel role of iron metabolism in regulating the capacity of allografts to protect themselves against the cytotoxic effects of alloimmune responses, and provide mechanistic insight into the pathogenesis of operational tolerance in human liver transplantation.
Results
Clinical characteristics of the study population.
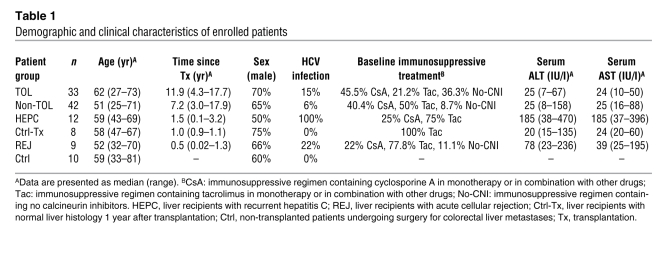
Of the 102 liver recipients included in the immunosuppressive drug withdrawal trial, 75 recipients from whom cryopreserved liver tissue samples had been obtained before the initiation of drug minimization and were available for transcriptional analyses were enrolled in the current study (Figure 1). Thirty-three recipients successfully discontinued all immunosuppressive drugs (operationally tolerant), while 42 rejected their allografts (non-tolerant). Clinical and demographic characteristics of enrolled patients before the initiation of drug minimization are summarized in Table 1. Operationally tolerant recipients had been transplanted for a longer period of time (P < 0.0001), were older (P < 0.0005), and were more likely to be receiving no calcineurin inhibitors (P < 0.014) than non-tolerant recipients. There were no other significant differences in clinical parameters between operationally tolerant and non-tolerant recipients. Similarly, no differences in liver function and graft histopathologic characteristics were noted between the two groups of patients at enrollment (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI59411DS1). Liver tissue specimens were collected before initiation of drug minimization in all patients, at the time of rejection in non-tolerant recipients, and 12 months after complete drug withdrawal in operationally tolerant patients. PBMC and serum samples were sequentially collected in all patients (Supplemental Table 2 contains a list of all biological specimens collected from enrolled recipients).
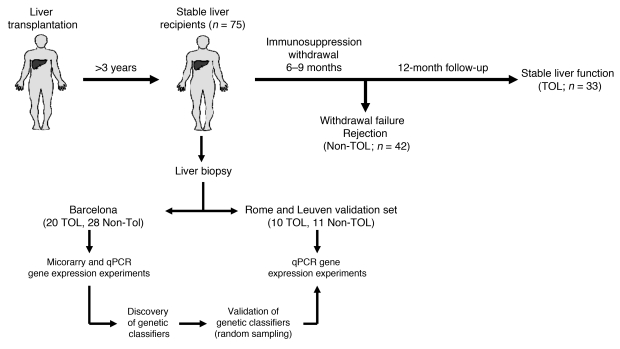
Figure 1. Study outline.
Stable liver transplant recipients with follow-up beyond 3 years after transplantation were enrolled in an immunosuppression withdrawal clinical trial. Drugs were gradually discontinued over a 6- to 9-month period, and patients were followed for an additional 12-month period. Protocol liver biopsies were obtained at study onset, at any time rejection was suspected, and at the end of the study in patients who did not reject. Patients who maintained stable graft function during the entire duration of the study and in whom no signs of rejection were noticed in protocol biopsies were considered operationally tolerant (TOL). Patients who underwent rejection at any time during the follow-up were labeled as non-tolerant (Non-TOL). Pre-weaning liver biopsy differential gene expression was assessed in a training set of TOL and Non-TOL recipients (all from Hospital Clinic Barcelona) employing whole-genome microarrays. A panel of selected genes was then analyzed by qPCR, followed by a search for predictive genetic classifiers. This was accomplished employing a random splitting strategy that incorporated an independent set of TOL and Non-TOL recipients (from University of Rome “Tor Vergata” and University Hospitals Leuven).
Table 1 .
Demographic and clinical characteristics of enrolled patients
Global intra-graft gene expression assessment of liver tissue samples.
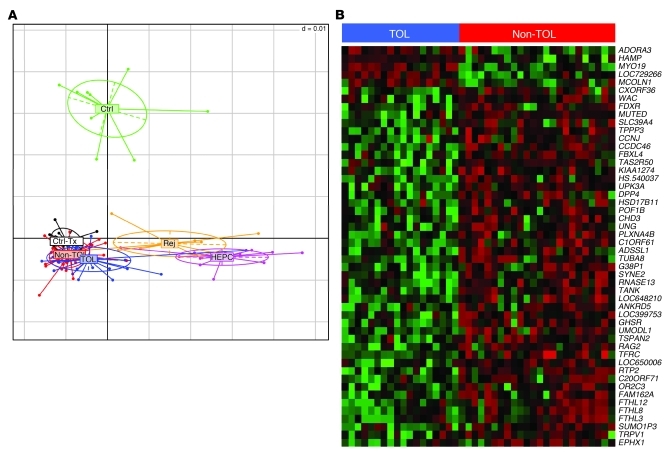
We conducted whole-genome transcriptional profiling experiments employing Illumina microarrays on pre-minimization liver samples collected from 20 operationally tolerant and 28 non-tolerant recipients and on rejecting samples obtained from 18 non-tolerant recipients (all from Hospital Clinic Barcelona). In addition, microarray experiments were also performed on liver tissue samples collected from the following control patient groups: (a) HCV-negative liver recipients under maintenance immunosuppression with normal liver function and normal liver graft histology 1 year after transplantation (n = 8); (b) liver transplant recipients under maintenance immunosuppression with chronic hepatitis due to recurrent HCV infection (n = 12); (c) immunosuppressed liver recipients undergoing mild acute cellular rejection during the first year after transplantation (n = 9); and (d) normal liver tissue from non-transplanted patients undergoing surgery for colorectal liver metastases (n = 10). To visualize the main differences in global intra-graft transcriptional patterns between these groups of samples, we performed a non-supervised correspondence analysis of the entire filtered probe list (Figure 2A). Marked differences in gene expression were noted between non-transplanted liver tissue samples and biopsy samples obtained from transplant recipients. Furthermore, among transplant recipients, HCV-associated expression patterns were easily discernible from those of rejecting grafts. In contrast, global gene expression patterns of samples collected from operationally tolerant and non-tolerant immunosuppressed liver transplant recipients before the initiation of drug minimization closely resembled each other regardless of whether immunosuppression could eventually be successfully withdrawn or resulted in graft rejection. These two groups also clustered with the group of control biopsies harvested from liver recipients with normal histology 1 year after transplantation. Thus, the overall differences in global gene expression among liver biopsies collected from transplant recipients closely paralleled the presence or absence of intra-graft inflammatory infiltrates (Supplemental Table 1), with fewer divergences being detectable between liver samples devoid of noticeable inflammatory infiltrates (i.e., operationally tolerant, non-tolerant, control liver recipients with normal histology).
Figure 2. Intra-graft microarray gene expression profiling of TOL and Non-TOL recipients before immunosuppression drug withdrawal.
(A) Non-supervised between-group analysis (BGA) conducted employing liver tissue expression data from the total filtered 33082 Illumina probeset obtained from the following groups of patients: TOL, Non-TOL, liver recipients with recurrent HCV infection (HEPC), liver recipients with acute cellular rejection (Rej), Ctrl-Tx, and Ctrl. Individuals (dots) and groups (ellipses) are positioned on the 2D plane on the basis of their multiple gene expression measurements. The areas delimited by the ellipses cluster 95% of the points belonging to the estimated binomial expression distribution of each of the groups analyzed. (B) Heatmap display of the 50 genes with the most significantly different expression between TOL and Non-TOL recipients. Rows represent genes, and columns represent samples. The intensity of each color denotes the standardized ratio between each value and the average expression of each gene across all samples. Red pixels correspond to an increased abundance of the transcript in the indicated sample, whereas green pixels indicate decreased transcript levels.
Before initiation of drug minimization, liver tissue samples from operationally tolerant and non-tolerant recipients differ in genes involved in the regulation of iron metabolism.
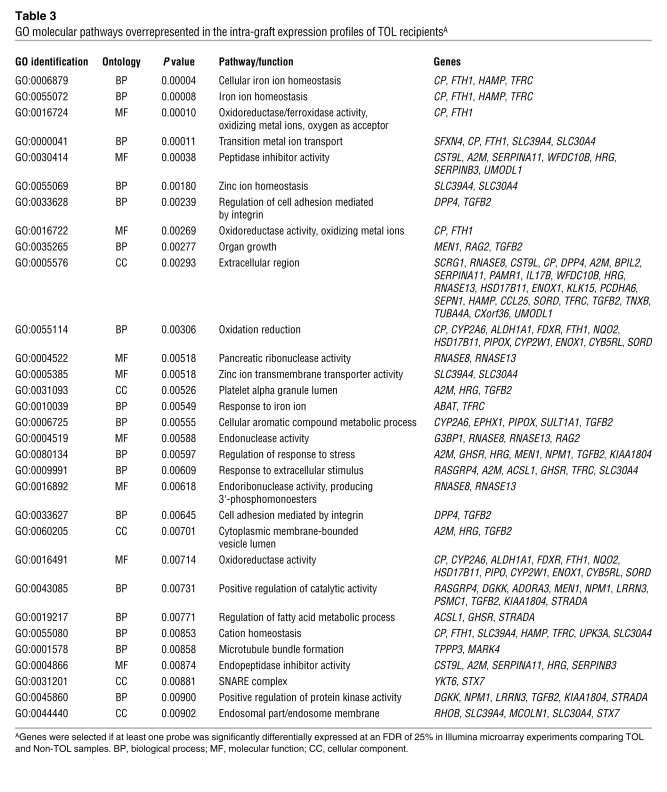
To explore in more detail the differences in gene expression profiles between liver tissue samples from operationally tolerant and non-tolerant recipients obtained before the initiation of drug minimization, we then performed a pairwise comparison between these two groups of samples employing significance analysis of microarrays (SAM) (ref. 13, Figure 2B, and Table 2). Given the marked impact of HCV infection on intra-graft expression patterns, the 3 samples from HCV-infected recipients were excluded from the analysis. Liver tissue samples from operationally tolerant and non-tolerant recipients differed in 8 transcripts at a false discovery rate (FDR) of 0 and in 176 transcripts at FDR less than 25% (see the complete dataset at http://bioinfo.ciberehd.org/asf2011/index.html). The genes with the most significant change in expression were HAMP and TFRC (FDR, 0; fold-change, 3.74 and –2.03, respectively), known to be critical for the regulation of iron homeostasis, and two ferritin-like pseudogenes, FTHL12 and FTHL8 (FDR, 0; fold-change, –1.41 and –1.43, respectively). Additional genes involved in iron metabolism among the set with FDR less than 25% were EPHX1, A2M, CP, FTHL3, FTHL11, ABAT, and SFXN4. The significance of iron-related genes was further confirmed by an unbiased functional analysis employing the Gene Ontology (GO) database. Iron ion homeostasis was the biological pathway most significantly overrepresented in the operational tolerance–associated expression profile (Table 3). Only a small number of genes associated with inflammation and immunopathogenesis (CD26, CD32, ADORA3, PTPN22) were differentially expressed between samples from operationally tolerant and non-tolerant recipients. In contrast, at the time of rejection non-tolerant recipients exhibited marked overexpression of various immune-related genes (e.g., STAT1, IL32, CXCL9, CXCL10, CD83, CD8A), none of which were detectable in the same group of patients before the initiation of drug minimization (Supplemental Table 3). We selected samples from 9 operationally tolerant and 10 non-tolerant recipients to conduct a cross-platform validation on whole-genome Affymetrix microarrays. In agreement with the Illumina experiments, only a small set of genes was differentially expressed between operationally tolerant and non-tolerant liver samples (Supplemental Figure 1A). Furthermore, a strong correlation between the two platforms was noted, with HAMP and TFRC being among the 10 genes whose expression differed the most between samples from operationally tolerant and non-tolerant recipients in the Affymetrix experiments as well (Supplemental Figure 1B).
Table 2 .
Comparison of liver tissue samples from TOL and Non-TOL recipients at baseline employing Illumina microarraysA
Table 3 .
GO molecular pathways overrepresented in the intra-graft expression profiles of TOL recipientsA
Microarray intra-graft gene expression differences can be validated by real-time quantitative PCR.
To validate the most informative microarray targets and to further investigate the role of previously identified immune tolerance–related genes, we performed quantitative PCR (qPCR) expression measurements for a set of 104 target genes on the same samples employed in the Illumina microarray experiments (Supplemental Table 4). Operationally tolerant and non-tolerant samples significantly differed in the expression of HAMP and TFRC. In addition, qPCR revealed differences in the expression of HMOX1, MIF, IFNG, SOCS1, CDHR2, DAB2, SLC5A12, and PEBP1 (Supplemental Figure 3). The differences in gene expression could not be attributed to differences in clinical parameters between operationally tolerant and non-tolerant recipients, since the association between expression markers and operational tolerance remained significant in most cases after adjustment for recipient’s age, sex, time since transplantation, and immunosuppressive therapy (Supplemental Table 5).
Intra-graft transcriptional biomarkers measured before initiation of immunosuppressive drug minimization accurately predict the outcome of drug withdrawal independent of any clinical parameter.
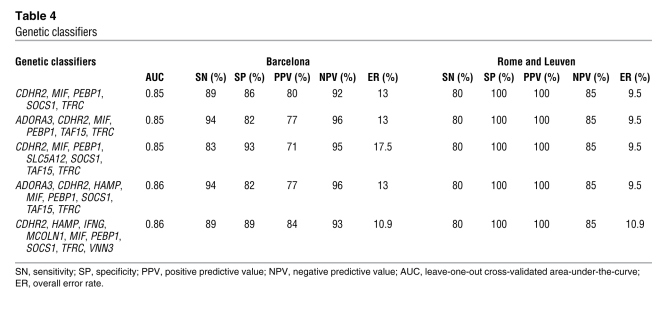
To determine whether biopsy-based gene expression classifiers could be employed to predict the success of immunosuppression withdrawal, we undertook an exhaustive search for gene predictive models on the qPCR-derived expression dataset. To conduct this analysis, we employed the same set of samples previously hybridized on Illumina microarrays (all of them from Barcelona), together with an independent group of biopsies from 10 operationally tolerant and 11 non-tolerant recipients (from Rome and Leuven), none of which had been included in the microarray experiments. To build the most robust and stable gene classifiers, we chose a repeated random sampling cross-validation strategy. These analyses resulted in the identification of 5 gene expression signatures capable of predicting the outcome of the immunosuppression weaning protocol, with less than a 17.5% overall error rate and very high sensitivity (SN), specificity (SP), negative predictive value (NPV), and positive predictive value (PPV), regardless of the center in which recipients had been enrolled (Table 4). The predictive accuracy of the gene classifiers was also independent of recipient age, sex, time from transplantation, and type of baseline immunosuppression (Supplemental Table 6) and was more powerful than any combination of clinical parameters (data not shown).
Table 4 .
Genetic classifiers
In operationally tolerant recipients, intra-graft transcriptional measurements are not influenced by the discontinuation of immunosuppressive therapy.
To investigate the impact of immunosuppressive drug withdrawal on liver tissue–derived expression patterns, we compared liver samples collected before initiation of drug minimization and 12 months after complete drug discontinuation. This was performed employing Affymetrix microarrays in a group of 5 operationally tolerant recipients. No major differences in gene expression were observed between the two time points, indicating that the presence or absence of pharmacological immunosuppression did not significantly alter the intra-graft gene expression profile of operationally tolerant recipients (only 6 probe sets were statistically significant with FDR <25%; Supplemental Figure 2). Furthermore, the 5 gene expression signatures defined above classified “pre-weaning” and “12 months after drug discontinuation” liver tissue samples from operationally tolerant recipients with similar accuracy (79%–88% in “pre-weaning” samples as compared with 77%–85% in the corresponding “12 months after drug discontinuation” samples; n = 13; data not shown).
Non-tolerant recipients exhibit decreased serum levels of hepcidin.
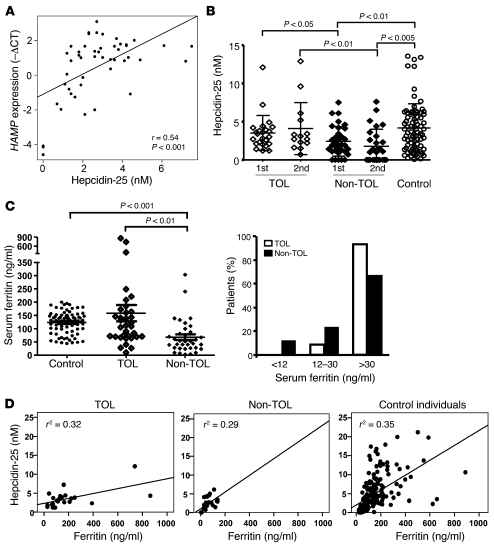
HAMP-encoded hepcidin is the central regulatory molecule of systemic iron homeostasis (14). We investigated whether operationally tolerant and non-tolerant recipients differed in hepcidin serum levels both before the initiation of drug minimization and 12 months after complete immunosuppressive drug withdrawal or rejection. A correlation between intra-graft hepcidin expression and hepcidin serum levels was noted (Figure 3A). In keeping with the gene expression results, non-tolerant recipients exhibited lower serum hepcidin levels than operationally tolerant patients both at baseline and at the end of the study. Hepcidin serum levels were also lower in non-tolerant recipients than in non-transplanted age- and sex-matched control individuals from the general population (Figure 3B). Furthermore, among transplant patients, undetectable hepcidin levels were only observed in non-tolerant recipients (<0.5 nM in 27.9% of non-tolerant individuals; Figure 3B). Hepcidin serum levels correlated with serum ferritin, a marker of body iron stores. Accordingly, ferritin serum levels were significantly higher in operationally tolerant than in non-tolerant recipients (Figure 3C), and absence of iron stores (serum ferritin <12 ng/ml, a highly specific indicator of iron deficiency; refs. 15–17) was exclusively observed among non-tolerant recipients (Figure 3C). The relationship between ferritin and hepcidin, however, differed between groups (Figure 3D). Thus, in operationally tolerant recipients, the increase in serum hepcidin per unit of serum ferritin was lower than in non-tolerant recipients (P = 0.044) and control individuals (P = 0.005), while it was similar in non-tolerant and control individuals (P = 0.488). The association between either hepcidin or ferritin and tolerance was not confounded by recipient age, sex, time from transplantation, or baseline immunosuppressive therapy, as demonstrated by their independent predictive value in a logistic regression multivariable analysis (Supplemental Table 7). Finally, operationally tolerant and non-tolerant recipients did not differ in blood hemoglobin, soluble transferrin receptor, serum iron, or C-reactive protein levels; transferrin saturation rate; incidence of ferropenic anemia; or HFE gene polymorphisms (Supplemental Table 8).
Figure 3. Non-TOL liver recipients exhibit lower hepcidin and ferritin serum levels than TOL patients.
(A) Correlation between HAMP expression in liver tissue samples collected before initiation of drug minimization as assessed by qPCR and hepcidin-25 serum levels. (B) Hepcidin-25 serum levels analyzed in TOL and Non-TOL recipients and in a group of 80 age- and sex-matched non-transplanted control individuals. Measurements were performed before the initiation of drug minimization in TOL and Non-TOL recipients (1st), and 12 months after complete drug withdrawal in TOL or 12 months after rejection in Non-TOL recipients (2nd). P values greater than 0.05 are not shown. (C) Ferritin serum levels in TOL and Non-TOL recipients before the initiation of drug minimization. The left panel shows the absolute values of ferritin serum levels; the right panel shows the proportion of TOL and Non-TOL recipients with ferritin levels less than 12 ng/ml (depleted iron stores); 12–30 ng/ml (reduced iron stores); and greater than 30 ng/ml (replete iron stores). (D) Relationship between ferritin and hepcidin serum levels in TOL, Non-TOL, and control individuals. The straight line represents the regression line. In TOL recipients, the increase in serum hepcidin per unit of serum ferritin was lower than in Non-TOL recipients (P = 0.044) and control individuals (P = 0.005), while it was similar in Non-TOL and control individuals (P = 0.488).
Before the initiation of drug minimization, liver grafts from operationally tolerant and non-tolerant recipients contain similar numbers of intra-hepatic lymphocytes but differ in hepatocyte iron accumulation.
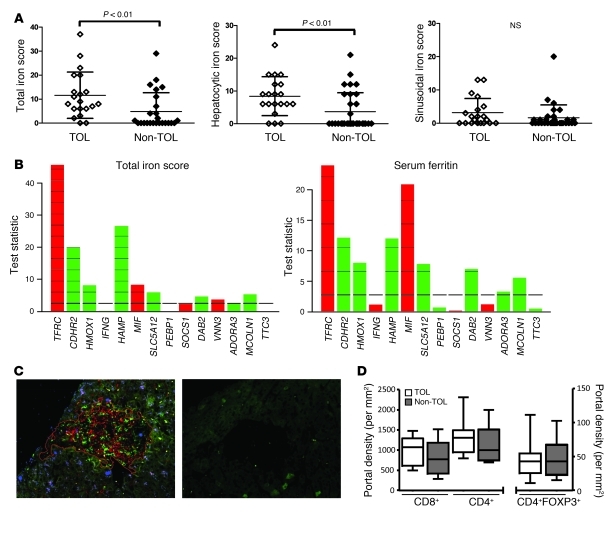
We assessed intra-graft iron accumulation by employing Perls staining in HCV-negative recipients. Mild periportal hepatocyte iron deposition was noted in most grafts from operationally tolerant recipients. In contrast, this was found in only a minority of liver biopsy samples from non-tolerant recipients. These differences were exclusively due to hepatocyte, rather than mesenchymal (endothelial and Kupffer cell), iron accumulation (Figure 4A). The periportal hepatocellular distribution of iron deposits noted in operationally tolerant recipients resembled findings previously reported in a minority of male healthy individuals with normal liver function (18). We then explored the relationship between iron stores and the differentially expressed gene set by deriving gene influence plots (Figure 4B). HAMP, TFRC, MIF, CDHR2, HMOX1, SLC5A12, DAB2, and MCOLN1 were significantly associated with either intra-hepatic iron stores or serum ferritin levels. In contrast, other gene expression markers, such as IFNG, PEBP1, SOCS1, VNN3, ADORA3, and TTC3, showed no or marginal association with iron stores, suggesting the potential involvement in the maintenance of operational tolerance of pathways other than iron metabolism. Similar results were observed when gene expression measurements were correlated with serum ferritin levels (Figure 4B). Immunofluorescence experiments were performed in parallel to investigate intra-graft effector and regulatory T cell subset infiltration. Lymphocyte infiltrates were very small in the portal tracts and almost absent in the lobules. Liver samples from operationally tolerant and non-tolerant recipients exhibited similar numbers of CD4+, CD8+, and CD4+FOXP3+ T cell subpopulations and CD4+FOXP3+/T cell ratios (Figure 4, C and D, and Supplemental Table 9). Among FOXP3+ lymphocytes, 93.7% were CD4+ and only 6.3% were potentially activated CD8+ cells. Thus, the immunophenotyping of liver biopsy samples did not distinguish operationally tolerant from non-tolerant patients.
Figure 4. In contrast to Non-TOL recipients, TOL patients exhibit mild hepatocyte stainable iron in their liver biopsies.
(A) Total iron score (TIS) values derived from TOL and Non-TOL liver biopsy samples collected at baseline (left panel). Significant differences between TOL and Non-TOL samples were only detected in hepatocytes (middle panel) and not in mesenchymal (Kupffer and endothelial) cells (right panel). (B) Plot showing the influence of individual gene expression measurements on iron stores as assessed by TIS (left panel) and on ferritin serum levels (right panel). Bars above the reference line indicate influential genes. Red indicates negative association; green indicates positive association. Reference line is the expected influence under the null hypothesis. (C) Visualization (original magnification, ×200) of CD4+, CD8+ and FoxP3+ infiltrating cells in a representative portal tract from a liver biopsy with the use of CD4- (red), CD8- (green), and FoxP3-specific (blue) monoclonal antibodies. The right panel shows the negative control employing the secondary fluorescent antibodies only. (D) Number of CD4+, CD8+, and FoxP3+ lymphocytes per portal tract area in biopsy samples obtained from TOL and Non-TOL individuals. All analyses were conducted on liver biopsy samples collected before immunosuppressive drugs were withdrawn. Box plots display medians for each category (center line), interquartile range (box upper and lower boundaries), and minimum and maximum (whiskers).
The transcriptional profiles of liver tissue samples from operationally tolerant and non-tolerant recipients differ from those observed before the initiation of drug minimization in PBMCs.
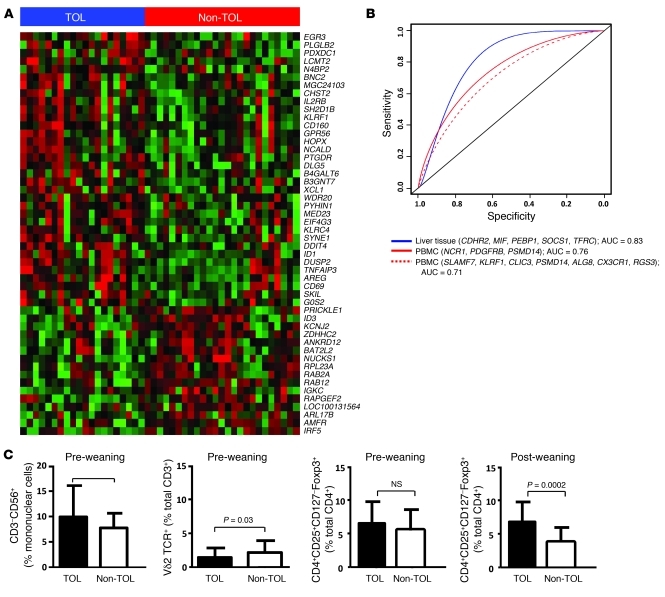
Our group previously reported the results of a retrospective cross-sectional study in which we observed that drug-free operationally tolerant liver recipients and liver recipients requiring maintenance immunosuppression differed in PBMC expression patterns (19). In order to investigate the similarities between PBMC and liver tissue expression patterns, we extracted RNA samples from PBMCs collected before the initiation of drug minimization from 20 operationally tolerant and 25 non-tolerant recipients (all of them from Hospital Clinic Barcelona) and performed gene expression experiments using both Affymetrix microarrays and qPCR (Supplemental Table 10). Microarray expression experiments conducted at baseline on PBMC samples revealed that the two groups of patients differed in the expression of 124 genes (FDR <5%, fold change ±1.32; Figure 5A and Supplemental Table 11), with NK-related gene sets being significantly overrepresented in the operational tolerance expression profile (Supplemental Table 12). None of these genes overlapped with the differentially expressed data set derived from liver tissue samples. Of the 94 targets tested by qPCR in the same set of PBMC samples, 11 were significantly different between operationally tolerant and non-tolerant recipients (Supplemental Table 13). Eight of these 11 genes (NCR1, KLRF1, IL2RB, GZMB, SH2D1B, CLIC3, KLRC4, NCAM1), all of them preferentially expressed by NK cells, had been previously described by our group as associated with liver operational tolerance (19). In order to compare the predictive performance of liver tissue and PBMC-derived gene expression signatures in the 45 recipients from whom baseline PBMC and liver tissue qPCR expression data were available, we computed receiver operating characteristic (ROC) curves for the following three predictive signatures: (a) CDHR2, MIF, PEBP1, SOCS1, and TFRC (representative liver tissue–based signature described in Table 4); (b) SLAMF7, KLRF1, CLIC3, PSMD14, ALG8, CX3CR1, and RGS3 (representative PBMC-based signature described in ref. 19); and (c) NCR1, PDGFRB, and PSMD14 (most performant PBMC-based logistic regression model derived from the set of 11 differentially expressed genes described above). The liver tissue–derived signature outperformed both the PBMC-based model identified in our previous work and the new model derived from the samples included in the current study (Figure 5B).
Figure 5. Before the initiation of immunosuppressive drug withdrawal, TOL and Non-TOL recipients differ in cellular and transcriptional peripheral blood markers.
(A) Heatmap displaying the 50 genes with the most significantly different expression when comparing PBMC samples collected from TOL and Non-TOL recipients (rows represent genes, and columns represent samples; the intensity of each color denotes the standardized ratio between each value and the average expression of each gene across all samples; red pixels correspond to an increased abundance of the transcript in the indicated samples, whereas green pixels indicated decreased transcript levels). (B) Overall diagnostic performance of liver tissue and PBMC-derived transcriptional signatures measured at patient enrollment in the prediction of successful drug withdrawal. (C) Differences between TOL and Non-TOL recipients in the proportion of PBMC subsets at enrollment and at the end of the study.
We next employed qPCR to measure in the liver tissue samples collected before the initiation of drug minimization the expression levels of the most informative genes identified in PBMCs (IL2RB, KLRF1, SLAMF7, KLRD1, CX3CR1, LINGO2, BNC2, NCR1, COL13A1, IGFBP7, NCAM1, KLRK1, KLRC1). Again, none of the genes significantly associated with operational tolerance in the liver tissue experiments overlapped with the set of genes differentially expressed in the PBMC samples (Supplemental Table 4).
Before the initiation of drug minimization, operationally tolerant recipients exhibit an expansion of NK cells in peripheral blood.
Before the start of drug minimization, operationally tolerant recipients exhibited an increased proportion of NK lymphocytes and a decreased proportion of Vδ2-TCR γδT cells as compared with non-tolerant patients. Furthermore, operationally tolerant recipients displayed a higher proportion of circulating CD4+CD25+CD127–Foxp3+ T cells than non-tolerant recipients. This was, however, only detectable 12 months after drug withdrawal or rejection and not at enrollment (Figure 5C).
Discussion
Liver cells are continually exposed to bacterial degradation products and food-derived antigens through the portal venous blood. This results in a microenvironment biased toward tolerance rather than immunity (20, 21) and likely contributes to the systemic liver tolerance effect observed in most animal models of liver transplantation and in selected human recipients who require no immunosuppressive drugs. The exact mechanisms (i.e., immunological tolerance, ignorance, immunodeficiency, or graft accommodation) responsible for the successful discontinuation of immunosuppressive drugs in human liver transplant recipients are, however, unknown. Furthermore, there are currently no tools to detect operationally tolerant liver recipients prior to drug withdrawal. To identify predictive biomarkers and investigate the pathogenesis of operational tolerance, we conducted a comprehensive molecular analysis of blood and allograft tissue specimens collected from liver recipients before immunosuppressive therapy was discontinued. Genetic classifiers were then derived and employed to predict the outcome of drug withdrawal. Liver tissue samples from operationally tolerant and non-tolerant patients differed in the expression of an unexpectedly small number of genes. Those with the most significantly different expression were genes involved in the regulation of iron homeostasis. These results were validated employing three different transcriptional platforms and further confirmed by the observation that intra-hepatic iron stores and hepcidin and ferritin serum levels were different in operationally tolerant and non-tolerant individuals. The association between iron metabolism markers and operational tolerance was not influenced by recipient characteristics or immunosuppressive therapy and was stable over time. Furthermore, intra-graft gene expression measurements accurately predicted the outcome of immunosuppression withdrawal independent of any clinical parameter.
Iron participates in most cell functions and is essential for cell survival. Hepcidin, mainly secreted by hepatocytes, acts as the main regulator of systemic iron metabolism by triggering the degradation of ferroportin and thus preventing the export of dietary and macrophage iron into the circulation (14, 22). Hepcidin expression is inversely related to body iron requirements, being increased when iron stores are sufficient and decreased when iron requirements are high. Furthermore, hepcidin expression is enhanced by inflammation and inhibited by hypoxia and erythropoietic signals (22). In our study, the lower hepcidin transcript and protein levels observed in non-tolerant recipients correlated with lower serum ferritin levels and with upregulation of TFRC expression (commonly seen in situations of decreased hepatic iron stores), while no differences in other pathways potentially influencing hepcidin expression were detected. Indeed, according to serum ferritin levels, more than one-third of non-tolerant recipients exhibited a state of non-anemic iron deficiency. In contrast, in operationally tolerant recipients, both ferritin and hepcidin serum levels were similar to those in healthy individuals. Operationally tolerant recipients, however, exhibited a lower increase in serum hepcidin per unit of serum ferritin than both non-tolerant and control individuals. This finding, suggestive of an inappropriate response to iron stores, is the hallmark of most diseases of iron metabolism resulting in iron overload. Consistently, as compared with non-tolerant recipients, operationally tolerant individuals displayed increased hepatocyte iron accumulation. Of note, these differences in liver iron loading were mild, clinically silent, and not associated with liver injury.
Changes in iron availability can influence microbial growth and affect the function of macrophages and other innate immune cells (23, 24). Furthermore, iron redistribution via modulation of hepcidin-ferroportin expression is a well-established antimicrobial strategy in mammals. This mechanism appears to have a major pathogenic role in liver infections by pathogens such as HCV and Plasmodium, where iron overload is associated with poor prognosis (25–29). Whether these effects are mainly due to pathogen growth inhibition, regulation of host immune responses, or both remains to be clarified. Similarly, the interactions among iron metabolism, adaptive immune responses, and allograft tolerance also need to be adequately investigated. In renal transplantation, several studies have linked post-transplant anemia with rejection and lower recipient and graft survival (30, 31). However, the prevalence and potential contribution of iron deficiency to these outcomes has not been systematically assessed. Similarly, the association between iron status and immunological outcomes in clinical liver transplantation remains to be explored. In an experimental rodent model of endotoxin-mediated liver inflammation, iron deficiency and decreased hepcidin levels resulted in exacerbated inflammatory responses, and this phenomenon was attenuated by exogenous hepcidin administration (32). In a similar model, hepcidin was shown to exert its intrinsic antiinflammatory effect through the phosphorylation of Stat3 (33). While this could be relevant to liver transplantation given the immunoregulatory function of Stat3 (34–37), the role of hepcidin and iron metabolism in models of immune-mediated liver diseases has yet to be investigated. Ours is therefore the first report to our knowledge indicating that modulation of hepcidin expression and redistribution of intra-graft iron stores could be involved in the capacity of the liver allograft to restrain alloreactive immune responses. Whether our findings constitute a cause or a consequence of the operationally tolerant state cannot be unambiguously established in the current study and will need to be formally demonstrated in future trials analyzing sequentially collected liver biopsy samples and addressing the immunologic outcome of iron metabolism therapeutic manipulations.
Regulation of iron homeostasis is unlikely to be the sole mechanism contributing to operational tolerance in liver recipients. Indeed, liver tissue samples from operationally tolerant and non-tolerant recipients also differed in the expression of a limited number of genes with cytoprotective and immunoregulatory function (CD26, ADORA3, DAB2, MIF, HMOX1, SOCS1). These genes were coregulated in a consistently antiinflammatory mode. Thus, as compared with non-tolerant grafts, operationally tolerant samples exhibited increased transcript levels of immune-inhibitory DAB2 and HMOX1 and decreased levels of immune-activatory CD26 and MIF. CD26 and ADORA3 are involved in the metabolism of adenosine, an immunosuppressive mediator responsible in part for the suppressive function of regulatory T cells (38). In experimental animal models, SOCS1 plays an important cytoprotective role by preventing harmful TLR-mediated cytokine responses and appears to be a critical negative regulator of LPS signaling following LPS restimulation (the so-called LPS tolerance effect) (39, 40). Furthermore, operationally tolerant and non-tolerant grafts also differed in the expression of CD32 and PTPN22, genes whose variants are associated with multiple autoimmune disorders (41). In contrast, prototypical immunoregulatory molecules identified in experimental rodent models of allograft tolerance, such as FoxP3, PD1/PDL1, CTLA4, IDO, IL-10, and TGF-β (1, 42–45), were not found to be associated with operational tolerance in our prospective clinical trial. Similarly, no differences were noted in the liver tissue expression of proinflammatory cytokines (other than IFNG, which is also crucial for regulatory T cell function in vivo; ref. 46) in the number of intra-graft effector and/or regulatory lymphocyte subsets, in the expression of proinflammatory mediators in PBMCs, or in serum C-reactive protein levels.
Exploratory experiments conducted to identify blood immunologic markers of successful drug withdrawal showed an association between NK cells and related transcripts and operational tolerance. This is consistent with our observations made in previous cross-sectional retrospective studies (19). Our current demonstration that NK-related transcriptional markers can be detected in PBMCs before immunosuppressive drugs are withdrawn indicates that they constitute a true biomarker of operational tolerance in liver transplantation. The availability of noninvasive biomarkers of operational tolerance has major clinical implications. The predictive performance of PBMC-derived transcriptional signatures, however, was less satisfactory than originally described (6, 10–12) and lower than that of liver tissue–based expression markers. Furthermore, clinical implementation of transcriptional tests requiring Ficoll-isolated PBMCs can be challenging. Further clinical validation specifically assessing reproducibility and clinical applicability will therefore be required before a blood-based in vitro diagnostic test of operational tolerance can be proposed.
Our findings suggest that the capacity of the liver allograft to resist immune-mediated injuries is likely to be a central mechanism in the maintenance of operational tolerance. This is supported by the demonstration that liver allografts from operationally tolerant and non-tolerant recipients differ in the expression of multiple genes potentially involved in the attenuation of intra-graft inflammatory responses, and by the absence of significant differences between the two groups of recipients in blood and liver tissue markers of adaptive immunity. A definitive answer to this question, however, will require analysis of donor-specific alloimmune responses and evaluation of the potential role of time, recipient age, and immunosenescence.
In short, we have identified several liver tissue–based prognostic expression models that can accurately identify operationally tolerant recipients before immunosuppressive drugs are discontinued. This could allow for the routine implementation of immunosuppression weaning strategies in liver transplantation. Furthermore, we have shown for the first time to our knowledge that graft acceptance in the absence of immunosuppression is strongly associated with the state of iron homeostasis. Our results suggest that in clinical transplantation, in which complete ablation of allospecific immune responses can rarely be accomplished, graft-specific mechanisms potentially capable of preventing inflammatory tissue destruction (such as the iron-hepcidin, adenosinergic, and SOCS1 immunomodulatory pathways, among others) are critical for the maintenance of operational tolerance. These mechanisms may be redundant for the prevention of graft rejection when nonspecific immunosuppressive drugs are administered but become essential as soon as immunosuppressive drugs are withdrawn. Our findings raise the prospect for therapeutic manipulations that could result in the widespread establishment of operational tolerance in liver transplantation.
Methods
Patient population and study design.
One hundred and two stable liver recipients at least 3 years after transplantation were enrolled in a prospective multicenter trial of immunosuppressive drug withdrawal ( ClinicalTrials.gov NCT00647283). Immunosuppressive drug doses were gradually decreased until complete discontinuation over 6–9 months. Patients were then followed up for 12 additional months. Protocol liver biopsy samples were obtained in all patients at baseline, 12 months after successful drug withdrawal (in operationally tolerant recipients), and at the time of rejection (in non-tolerant recipients). Patients who did not develop rejection were classified as operationally tolerant as long as immunosuppressive drug cessation was maintained for at least 12 months and no histopathologic evidence of acute and/or chronic rejection were observed. Rejection was diagnosed by the combination of allograft dysfunction and characteristic liver biopsy findings according to Banff criteria (47). Of the 102 recipients participating in the trial, 75 from whom baseline liver samples had been cryopreserved were included in the current study. Recipients were enrolled from Hospital Clinic Barcelona, University of Rome “Tor Vergata,” and University Hospitals Leuven. A description of the patient population and clinical outcome of the immunosuppression withdrawal clinical trial is provided in Supplemental Figure 4.
Liver biopsy specimens.
Liver biopsies were performed percutaneously under local anesthesia. A 2- to 3-mm portion of the needle biopsy liver cylinder was immediately preserved in RNAlater reagent (Ambion), kept at 4°C for 24 hours, and then cryopreserved in liquid nitrogen after removal of the RNAlater reagent. The remaining cylinder was formalin fixed and paraffin embedded (FFPE). In non-transplanted control patients, surgical liver biopsy samples of non-tumoral livers were obtained and processed as previously described.
Histological assessment of liver biopsies.
For histological assessment, 3-μm-thick slides were stained using hematoxylin and eosin, Masson’s trichrome for connective tissue analysis, and Perls staining for iron content. For the current study, all histological examinations were performed by the same pathologist, who was blinded to all clinical and biological data. Iron content was assessed employing the total iron scoring system described by Deugnier et al. (48). This scoring system (range, 0–60) takes into consideration iron deposition into three compartments: hepatocytic, sinusoidal, and portal. Although it is a semiquantitative score, this method has been previously validated and shows a good correlation to the biochemical quantification of hepatic iron content.
Liver tissue RNA extraction and processing.
For total RNA extraction, cryopreserved liver tissue samples were homogenized in TRIzol reagent (Invitrogen) using a pestle and nuclease-free 1.5-ml reaction tubes (Ambion). Total RNA was then extracted following the manufacturers guidelines, and quality was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies). The same procedure was employed to extract RNA from Ficoll-isolated PBMCs.
Liver tissue microarray experiments.
One hundred and six liver tissue RNA samples (from 20 operationally tolerant and 26 non-tolerant recipients, 18 non-tolerant recipients at the time of rejection, 3 operationally tolerant recipients at the end of the study, 12 recipients with recurrent HCV infection (HEPC), 9 recipients with acute cellular rejection (Rej), 8 recipients under maintenance immunosuppression with normal liver histology (Ctrl-Tx), and 10 non-transplanted patients undergoing liver resection (Ctrl); all of them from Hospital Clinic Barcelona) were processed into cRNA and hybridized onto Illumina HumanHT-12 Expression BeadChips containing 48,771 probes corresponding to 25,000 annotated genes. In a selected group of 9 operationally tolerant and 10 non-tolerant recipients from the training set, microarray experiments were replicated onto Affymetrix Human Genome U133 Plus 2.0 arrays covering 47,000 annotated genes by 54,675 probes. In 5 of the 9 operationally tolerant recipients, additional Affymetrix experiments were conducted employing RNA extracted from liver biopsy samples obtained 12 months after drug withdrawal.
Liver tissue microarray gene expression data analysis.
For Illumina microarrays, expression data were computed using BeadStudio data analysis software (Illumina) and subsequently processed employing quantile normalization using the Lumi bioconductor package (49). Next, we conducted a conservative probe-filtering step excluding those probes with a coefficient of variation of 5%, which resulted in the selection of a total of 33,062 probes from the original set of 48,771. Affymetrix gene expression data were normalized using the guanine-cytosine content–adjusted robust multiarray algorithm (50). Thereupon, we employed a conservative probe-filtering step excluding probes not reaching a log2 expression value of 5 in at least one sample, which resulted in the selection of a total of 18,768 probes from the original set of 54,675. To identify genes differentially expressed between the different microarray study groups, we employed significant analysis of microarray (SAM) (13). To graphically represent global gene expression differences between the different study groups, we used the entire filtered probe list to perform a correspondence analysis employing the between-group-analysis (BGA) function included in the MADE4 package (51). To visualize the magnitude of the differential gene expression between tolerant and non-tolerant samples, we computed Q-Q plots for the Illumina and Affymetrix microarray expression datasets. Q-Q plots provide a visual comparison of two populations by displaying the sampled t statistic versus a theoretical t statistic. In these plots, points that deviate from the linear relationship to the theoretical t statistic correspond to genes exhibiting differential expression. All liver tissue microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE26627). Functional analysis of gene expression data was conducted using the R/Bioconductor package GOstats and the GO database ( http://www.geneontology.org). Only probes that could be associated with a unique EntrezGene ID were used. In cases where genes were targeted by multiple probes, genes were selected if at least one probe was significantly differentially expressed at an FDR of less than 25%. The hypergeometric distribution was used to evaluate the probability of randomly observing the enrichment for each GO term (52).
Liver tissue qPCR experiments.
The expression of 104 target genes and 3 housekeeping genes (Supplemental Table 3) was measured by qPCR on 46 (18 operationally tolerant and 28 non-tolerant) of the 48 recipients constituting the Barcelona cohort, and in an independent group of 21 (10 operationally tolerant and 11 non-tolerant) of the 23 recipients from Rome and Leuven. Four samples (2 from Barcelona and 2 from Rome) were excluded from the analysis due to quality control issues. Experiments were conducted employing the ABI 7900 Sequence Detection System and TaqMan LDA microfluidic plates (Applied Biosystems). DNA was removed from total RNA preparations using Turbo DNA-free DNAse treatment (Ambion), and RNA was then reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). To quantify transcript levels, target gene Ct values were normalized to the housekeeping genes to generate ΔCt values. The results were then computed as relative expression between cDNA of the target samples and a calibrated sample according to the ΔΔCt method. The following three samples were employed as calibrators: (a) pooled RNA from the 8 Ctrl-Tx samples; (b) pooled RNA from the 10 Ctrl samples; and (c) commercially available liver RNA (Human Liver Total RNA, Ambion). Target genes were selected based on: (a) Illumina and Affymetrix microarray experiment results; (b) blood transcriptional biomarkers previously described by our group as being associated with liver operational tolerance (19); and (c) prominent immunoregulatory genes described in the literature.
Identification and validation of gene classifiers on liver tissue samples.
To develop biopsy-based qPCR gene expression predictive classifiers, we employed the linear discriminant analysis and logistic regression algorithms implemented in misclassification penalized posterior (MiPP) software (53). MiPP is based on a stepwise incremental classification modeling for discovery of the most parsimonious models and employs both a 10-fold cross-validation and a random splitting cross-validation strategy to predict model performance and misclassification error rates. This was conducted on the whole data set of 67 samples (46 from Barcelona and 21 from Rome and Leuven) by repeatedly partitioning it into training set (two-thirds) and an independent test set (one-third) for external model validation. The prediction performance and mean misclassification error rates were obtained for each of the candidate classifiers. Finally, for each candidate model identified, the optimal probability cutoff of operational tolerance was computed through a ROC analysis. To demonstrate that the performance of the models was not center dependent, we then computed SN, SP, NPV, PPV, and overall error rates for the samples collected from Barcelona and those obtained from Rome and Leuven. To demonstrate that the random splitting strategy did not yield unrealistically high prediction accuracies, we conducted a new search for genetic classifiers on the training set of 18 operationally tolerant and 28 non-tolerant recipients employing Fisher’s linear discriminant analysis (LDA), and validated them on the 21-sample independent test set. This resulted in the identification of 5 additional candidate models that classified samples with less than 15% error rates in both the training and the test set (data not shown).
Serum ferritin and HFE genotyping.
Serum ferritin was measured at the Hospital Clinic Barcelona central laboratory using a commercially available automated ELISA assay (ADVIA Centaur, Siemens Healthcare Diagnostics). DNA samples from peripheral blood samples were genotyped for the nucleotide changes that correspond to the amino acid substitutions C282Y and H63D in the HFE protein using TaqMan (Applied Biosystems) qPCR probes as described previously (54).
Serum hepcidin measurements.
Baseline pre-weaning serum samples were available from 57 liver recipients (24 operationally tolerant and 33 non-tolerant) before the initiation of drug minimization, from 14 operationally tolerant recipients 12 months after complete drug withdrawal, and from 28 non-tolerant recipients 12 months after the rejection episode. Quantitative serum hepcidin measurements were conducted by a combination of weak cation exchange chromatography and time-of-flight mass spectrometry (TOF-MS) (55). For quantification, a hepcidin analog (synthetic hepcidin-24; Peptide International Inc.) was employed as internal standard (56). Peptide spectra were generated on a Microflex LT matrix-enhanced laser desorption/ionization TOF-MS platform (Bruker Daltonics). Serum hepcidin-25 concentrations were expressed as nmol/l. The lower limit of detection of this method was 0.5 nM; average coefficients of variation were 2.7% (intra-run) and 6.5% (inter-run) (57). Hepcidin serum levels were also obtained from a group of 80 non-transplanted individuals with normal ALT levels who were randomly selected from the 2,998 participants in the population-based Nijmegen Biomedical Study (58) after matching with operationally tolerant and non-tolerant recipients for age and sex.
Immunofluorescence quantification of intra-graft lymphocyte subsets.
Up to 3 FFPE sequential sections were analyzed per patient (15 operationally tolerant and 13 non-tolerant). Following deparaffinization and antigen retrieval steps, the following antibodies and reagents were employed: mouse anti–human CD4 (clone BC/1F6), rabbit anti–human CD8 (clone SP16) (both from Abcam), goat CY5-conjugated anti-mouse (115-176-071, Jackson ImmunoResearch Laboratories Inc.), goat AF488-conjugated anti-rabbit (A11034, Invitrogen), rat biotin-conjugated anti–human FoxP3 (PCH101, eBioscience), Cy3-conjugated streptavidin (016-160-084, Jackson ImunoResearch Laboratories Inc.), DAPI. Blocking, staining, and washing steps were performed with Tris-buffered saline with 0.05% Tween 20 at room temperature. To avoid unspecific binding, we blocked first with goat serum (5%) and, prior to the biotinylated antibody and streptavidin, separately with avidin (0.01%), biotin (0.005%) and mouse serum (5%). Fluorescence microscopy was carried out with an Axio Imager M1 at a magnification of ×200 (Zeiss). Evaluation of portal infiltration and quantification of CD4-, CD8-, FoxP3-positive cells was performed with AxioVision 4.6 software (Zeiss) in a blinded fashion.
Flow cytometric experiments on peripheral blood samples.
Whole blood samples were collected from all enrolled recipients at baseline to conduct flow cytometry immunophenotyping experiments. Titrated amounts of fluorochrome-conjugated monoclonal antibodies were employed to identify naive and memory T cells (CD3, CD8, CD4, CD45RB, CD62L, CCR7, γδ TCR, αβ TCR, Vδ1 TCR, Vδ2 TCR), B cells (CD19), NK cells (CD3, CD56, CD16), dendritic cells (CD14, CD3, CD19, CD20, CD123, CD11c, HLA-DR), and regulatory T cells (CD4, CD127, Foxp3). Cells were fixed in 1% paraformaldehyde/PBS, and data were acquired on a BD FACSCanto flow cytometer (BD) and analyzed employing FlowJo software (Tree Star).
Blood gene expression profiling experiments.
To define a blood transcriptional fingerprint of operational tolerance, we first conducted an exploratory analysis employing Affymetrix Human Genome U133 Plus 2.0 arrays on PBMCs collected before the initiation of drug minimization from 20 operationally tolerant and 25 non-tolerant recipients (all from Hospital Clinic Barcelona). In 12 operationally tolerant and 14 non-tolerant recipients from the same group of 45 patients, microarray experiments were also conducted on RNA samples collected 12 months after drug withdrawal or rejection. Due to the inter-laboratory variability inherent in the PBMC isolation procedure, these experiments were restricted to samples collected from patients enrolled at Hospital Clinic Barcelona. Total RNA was extracted from cryopreserved PBMCs using the TRIzol method, and Affymetrix microarray experiments were conducted as previously described (19). To identify genes differentially expressed between the different study groups, we employed RankProd (59) with a FDR less than 5% statistical threshold. Microarray data were deposited in NCBI’s Gene Expression Omnibus (GEO GSE28842). To identify PBMC-related biological pathways significantly associated with the operationally tolerant state, we employed Gene Set Enrichment Analysis (GSEA) (60, 61). For the current analysis, we used gene sets extracted from: (a) GO (62); (b) Molecular Signatures Database subset of canonical pathways (MSigDB version 3-0) (63); and (c) HaemAtlas blood cell lineage–specific gene set collection (64). Only gene sets with more than 15 genes were included in the analysis. Analyses were based on a t test and a weighted scoring scheme with 1,000 permutations.
The expression levels of a set of 94 target genes and 4 housekeeping genes (Supplemental Table 11) were measured in the same set of 45 baseline RNA samples employing dynamic arrays and the Fluidigm BioMark qPCR platform with a pre-amplification step. The set of target genes included the 34 operational tolerance–related genes previously described by Martínez-Llordella et al. (19) together with some of the most informative targets derived from the Affymetrix microarray experiments conducted at baseline on PBMC samples. To quantify transcript levels, we normalized the expression of the target genes to the mean of the housekeeping genes, and presented data as relative expression between cDNA of the target samples and a calibrated sample according to the ΔΔCT method. Differences in gene expression levels between tolerant and non-tolerant recipients were explored employing Student’s t test. Specific predictive models incorporating Fluidigm qPCR expression markers were defined employing the resulting posterior probabilities from leave-one-out cross-validation included into the linear discrimination analysis function from the Modern Applied Statistics with S (MASS) package (65). DiagnosisMed ( http://r-forge.r-project.org/projects/diagnosismed) and pROC libraries were employed to compute and plot ROC curves.
Statistics.
Statistical significance was analyzed using GraphPad software, and a P value less than 0.05 was considered statistically significant. Normal distribution of samples was determined employing Kolmogorov-Smirnov, D’Agostino-Pearson omnibus, and Shapiro-Wilk normality tests. Upon normal distribution, an unpaired Student’s t test was used; in the other cases, a nonparametric Mann-Whitney U test was employed. If not otherwise indicated, data are presented as mean ± SEM. The influence of different variables on the probability of tolerance was adjusted in a multivariable logistic regression analysis using SPSS statistical software (IBM SPSS Inc.). To determine whether the group of genes of interest was significantly associated with clinical parameters, we employed global test software (66), which is based on an empirical Bayesian generalized linear model where the regression coefficients between expression data and clinical outcome are the random variables estimated using a goodness-of-fit test. A syntax within this software was also employed to correct the association found between gene expression and clinical outcome for the possible confounding effects of nuisance clinical covariates. To assess whether the relationship between ferritin and hepcidin was different in operationally tolerant recipients, non-tolerant recipients, and control individuals, we assessed the homogeneity of slopes in an ANCOVA analysis by testing the significance of the interaction in a model with hepcidin as a dependent variable, operational tolerance status as a fixed factor, and ferritin as a covariable.
Study approval.
The study was approved by the institutional review boards of the three participating institutions, and written informed consent was obtained from all study patients.
Supplementary Material
Acknowledgments
We would like to acknowledge the scientific and technical assistance provided by Marta Martínez and Anna Rodríguez of the Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) Liver Transplant Immunology Laboratory; Miquel Bruguera of the Hospital Clinic Liver Unit; Pedro Jares of IDIBAPS Genomics Unit; and Erwin Wiegerinck, Coby Laarakkers, Siem Klaver, and others of Hepcidinanalysis.com. The study was conducted as part of the European Commission–supported RISET (Reprogramming the Immune System for the Establishment of Tolerance) consortium. Furthermore, the work was supported by TcLand Expression SA and by grants from the Ministry of Education and Science, Spain (SAF2008-04092), and the Roche Organ Transplantation Research Foundation (ROTRF), all of them to A. Sánchez-Fueyo. F. Bohne was supported by a research fellowship of the Deutsche Forschungsgemeinschaft (DFG reference BO 3370/1-1). Centro de Investigación Biomédica en Red: Enfermedades Hepáticas y Digestivas (CIBEREHD) is funded by the Instituto de Salud Carlos III Spain.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):368–382. doi:10.1172/JCI59411.
References
- 1.Sanchez-Fueyo A, Strom TB. Immunologic basis of graft rejection and tolerance following transplantation of liver or other solid organs. Gastroenterology. 2011;140(1):51–64. doi: 10.1053/j.gastro.2010.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turka LA, Lechler RI. Towards the identification of biomarkers of transplantation tolerance. Nat Rev Immunol. 2009;9(7):521–526. doi: 10.1038/nri2568. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, et al. HLA-mismatched renal transplantation without maintenance immunosuppression. . N Engl J Med. 2008;358(4):353–361. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devlin J, et al. Defining the outcome of immunosuppression withdrawal after liver transplantation. Hepatology. 1998;27(4):926–933. doi: 10.1002/hep.510270406. [DOI] [PubMed] [Google Scholar]
- 5.Lerut J, Sanchez-Fueyo A. An appraisal of tolerance in liver transplantation. Am J Transplant. 2006;6(8):1774–1780. doi: 10.1111/j.1600-6143.2006.01396.x. [DOI] [PubMed] [Google Scholar]
- 6.Brouard S, et al. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci U S A. 2007;104(39):15448–15453. doi: 10.1073/pnas.0705834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazariegos GV, et al. Weaning of immunosuppression in liver transplant recipients. Transplantation. 1997;63(2):243–249. doi: 10.1097/00007890-199701270-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pons JA, et al. Endothelial cell chimerism does not influence allograft tolerance in liver transplant patients after withdrawal of immunosuppression. Transplantation. 2003;75(7):1045–1047. doi: 10.1097/01.TP.0000058472.71775.7D. [DOI] [PubMed] [Google Scholar]
- 9.Tisone G, et al. Complete weaning off immunosuppression in HCV liver transplant recipients is feasible and favourably impacts on the progression of disease recurrence. J Hepatol. 2006;44(4):702–709. doi: 10.1016/j.jhep.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Llordella M, et al. Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant. 2007;7(2):309–319. doi: 10.1111/j.1600-6143.2006.01621.x. [DOI] [PubMed] [Google Scholar]
- 11.Newell KA, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120(6):1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagoo P, et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest. 2010;120(6):1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Annu Rev Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 15.Milman N, Kirchhoff M. Iron stores in 1433, 30- to 60-year-old Danish males. Evaluation by serum ferritin and haemoglobin. Scand J Clin Lab Invest. 1991;51(7):635–641. doi: 10.3109/00365519109104574. [DOI] [PubMed] [Google Scholar]
- 16.Milman N, Byg KE, Ovesen L. Iron status in Danes 1994. II: Prevalence of iron deficiency and iron overload in 1319 Danish women aged 40-70 years. Influence of blood donation, alcohol intake and iron supplementation. Ann Hematol. 2000;79(11):612–621. doi: 10.1007/s002770000209. [DOI] [PubMed] [Google Scholar]
- 17.Cable RG, et al. Iron deficiency in blood donors: analysis of enrollment data from the REDS-II Donor Iron Status Evaluation (RISE) study. Transfusion. 2011;51(3):511–522. doi: 10.1111/j.1537-2995.2010.02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minervini MI, et al. Liver biopsy findings from healthy potential living liver donors: reasons for disqualification, silent diseases and correlation with liver injury tests. J Hepatol. 2009;50(3):501–510. doi: 10.1016/j.jhep.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Llordella M, et al. Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients. J Clin Invest. 2008;118(8):2845–2857. doi: 10.1172/JCI35342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 21.Thomson AW, Knolle PA. Antigen-presenting cell function in the tolerogenic liver environment. Nat Rev Immunol. 2010;10(11):753–766. doi: 10.1038/nri2858. [DOI] [PubMed] [Google Scholar]
- 22.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142(1):24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 23.Weiss G. Iron and immunity: a double-edged sword. Eur J Clin Invest. 2002;32(suppl 1):70–78. doi: 10.1046/j.1365-2362.2002.0320s1070.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Cherayil BJ. Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity. J Innate Immun. 2009;1(5):455–464. doi: 10.1159/000210016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinder D, Ayonrinde OT, Olynyk JK. HCV, iron, and oxidative stress: the new choreography of hepcidin. Gastroenterology. 2008;134(1):348–351. doi: 10.1053/j.gastro.2007.11.048. [DOI] [PubMed] [Google Scholar]
- 26.Girelli D, et al. Reduced serum hepcidin levels in patients with chronic hepatitis C. J Hepatol. 2009;51(5):845–852. doi: 10.1016/j.jhep.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portugal S, et al. Host–mediated regulation of superinfection in malaria. Nat Med. 2011;17(6):732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6(7):541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 29.Goma J, Renia L, Miltgen F, Mazier D. Iron overload increases hepatic development of Plasmodium yoelii in mice. Parasitology. 1996;112(pt 2):165–168. doi: 10.1017/S0031182000084729. [DOI] [PubMed] [Google Scholar]
- 30.Chhabra D, Grafals M, Skaro AI, Parker M, Gallon L. Impact of anemia after renal transplantation on patient and graft survival and on rate of acute rejection. Clin J Am Soc Nephrol. 2008;3(4):1168–1174. doi: 10.2215/CJN.04641007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molnar MZ, et al. Anemia is associated with mortality in kidney-transplanted patients—a prospective cohort study. Am J Transplant. 2007;7(4):818–824. doi: 10.1111/j.1600-6143.2006.01727.x. [DOI] [PubMed] [Google Scholar]
- 32.Pagani A, et al. Low hepcidin accounts for the proinflammatory status associated with iron deficiency. Blood. 2011;118(3):736–746. doi: 10.1182/blood-2011-02-337212. [DOI] [PubMed] [Google Scholar]
- 33.De Domenico, I, et al. Hepcidin mediates transcriptional changes that modulate acute cytokine-induced inflammatory responses in mice. J Clin Invest. 2010;120(7):2395–2405. doi: 10.1172/JCI42011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunz JG 3rd, Specht SM, Murase N, Isse K, Demetris AJ. Gut-derived commensal bacterial products inhibit liver dendritic cell maturation by stimulating hepatic interleukin-6/signal transducer and activator of transcription 3 activity. Hepatology. 2007;46(6):1946–1959. doi: 10.1002/hep.21906. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Feng D, Wang Y, Luo Q, Xu L. STAT3 mediates protection from liver inflammation after partial hepatectomy. Cell Physiol Biochem. 2009;23(4–6):379–386. doi: 10.1159/000218184. [DOI] [PubMed] [Google Scholar]
- 36.Lafdil F, et al. Myeloid STAT3 inhibits T cell–mediated hepatitis by regulating T helper 1 cytokine and interleukin–17 production. Gastroenterology. 2009;137(6):2125–2135. doi: 10.1053/j.gastro.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plum W, et al. Lack of glycoprotein 130/signal transducer and activator of transcription 3-mediated signaling in hepatocytes enhances chronic liver injury and fibrosis progression in a model of sclerosing cholangitis. Am J Pathol. 2009;176(5):2236–2246. doi: 10.2353/ajpath.2010.090469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gri G, et al. CD4+CD25+ regulatory T cells suppress mast cell degranulation and allergic responses through OX40-OX40L interaction. Immunity. 2008;29(5):771–781. doi: 10.1016/j.immuni.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimoto M, Naka T. SOCS1, a negative regulator of cytokine signals and TLR responses, in human liver diseases. Gastroenterol Res Pract. 2010;2010(pii): 470468. doi: 10.1155/2010/470468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott MJ, Liu S, Shapiro RA, Vodovotz Y, Billiar TR. Endotoxin uptake in mouse liver is blocked by endotoxin pretreatment through a suppressor of cytokine signaling-1-dependent mechanism. Hepatology. 2009;49(5):1695–1708. doi: 10.1002/hep.22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latiano A, et al. Evaluating the role of the genetic variations of PTPN22, NFKB1, and FcGRIIIA genes in inflammatory bowel disease: a meta-analysis. Inflamm Bowel Dis. 2007;13(10):1212–1219. doi: 10.1002/ibd.20185. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Fueyo A, Weber M, Domenig C, Strom TB, Zheng XX. Tracking the immunoregulatory mechanisms active during allograft tolerance. J Immunol. 2002;168(5):2274–2281. doi: 10.4049/jimmunol.168.5.2274. [DOI] [PubMed] [Google Scholar]
- 43.Sho M, et al. Physiological mechanisms of regulating alloimmunity: cytokines, CTLA-4, CD25+ cells, and the alloreactive T cell clone size. J Immunol. 2002;169(7):3744–3751. doi: 10.4049/jimmunol.169.7.3744. [DOI] [PubMed] [Google Scholar]
- 44.Sandner SE, et al. Role of the programmed death-1 pathway in regulation of alloimmune responses in vivo. J Immunol. 2005;174(6):3408–3415. doi: 10.4049/jimmunol.174.6.3408. [DOI] [PubMed] [Google Scholar]
- 45.Fan Z, et al. In vivo tracking of ‘color-coded’ effector, natural and induced regulatory T cells in the allograft response. Nat Med. 2010;16(6):718–722. doi: 10.1038/nm.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood KJ, Sawitzki B. Interferon gamma: a crucial role in the function of induced regulatory T cells in vivo. Trends Immunol. 2006;27(4):183–187. doi: 10.1016/j.it.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Ormonde DG, et al. Banff schema for grading liver allograft rejection: utility in clinical practice. Liver Transpl Surg. 1999;5(4):261–268. doi: 10.1002/lt.500050418. [DOI] [PubMed] [Google Scholar]
- 48.Deugnier YM, et al. Preneoplastic significance of hepatic iron-free foci in genetic hemochromatosis: a study of 185 patients. Hepatology. 1993;18(6):1363–1369. [PubMed] [Google Scholar]
- 49.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z, Irizarri R, Gentleman R, Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;2004:909–917. [Google Scholar]
- 51.Culhane AC, Thioulouse J, Perriere G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21(11):2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- 52.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 53.Soukup M, Cho H, Lee JK. Robust classification modeling on microarray data using misclassification penalized posterior. Bioinformatics. 2005;21(suppl 1):i423–i430. doi: 10.1093/bioinformatics/bti1020. [DOI] [PubMed] [Google Scholar]
- 54.Allen KJ, et al. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358(3):221–230. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 55.Kroot JJ, et al. Immunochemical and mass-spectrometry-based serum hepcidin assays for iron metabolism disorders. Clin Chem. 2010;56(10):1570–1579. doi: 10.1373/clinchem.2010.149187. [DOI] [PubMed] [Google Scholar]
- 56.Swinkels DW, et al. Advances in quantitative hepcidin measurements by time-of-flight mass spectrometry. PLoS One. 2008;3(7):e2706. doi: 10.1371/journal.pone.0002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroot JJ, et al. (Pre)analytical imprecision, between-subject variability, and daily variations in serum and urine hepcidin: implications for clinical studies. Anal Biochem. 2009;389(2):124–129. doi: 10.1016/j.ab.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 58.Galesloot TE, et al. Serum hepcidin: reference ranges and biochemical correlates in the general population. Blood. 2011;117(25):e218–e225. doi: 10.1182/blood-2011-02-337907. [DOI] [PubMed] [Google Scholar]
- 59.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 60.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watkins NA, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113(19):e1–e9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Venables WN, Ripley BD.Modern Applied Statistics with S . 4th ed. Springer: New York, New York, USA; 2002. [Google Scholar]
- 66.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20(1):93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.