Abstract
Cells have evolved complex regulatory networks that reorganize gene expression patterns in response to changing environmental conditions. These changes often involve redundant mechanisms that affect various levels of gene expression. Here, we examine the consequences of enhanced mRNA degradation in the galactose utilization network of Saccharomyces cerevisiae. We observe that glucose-induced degradation of GAL1 transcripts provides a transient growth advantage to cells upon addition of glucose. We show that the advantage arises from relief of translational competition between GAL1 transcripts and those of cyclin CLN3, a translationally regulated initiator of cell division. This competition creates a translational bottleneck that balances the production of Gal1p and Cln3p and represents a posttranscriptional control mechanism that enhances the cell's ability to adapt to changes in carbon source. We present evidence that the spatial regulation of GAL1 and CLN3 transcripts is what allows growth to be maintained during fluctuations of glucose availability. Our results provide unique insights into how cells optimize energy use during growth in a dynamic environment.
Keywords: protein synthesis, mRNA stability, mRNA localization, biodynamics, yeast
Environmental change invariably elicits alterations in gene expression as a cell tunes its enzymatic repertoire to a new set of challenges (1–3). The time required to alter patterns of gene expression and create a new array of functional proteins limits the speed at which a cell can respond to changes in its growth environment. The concentrations of mRNA transcripts have a strong influence on the dynamics of gene networks. In dynamic environments, the rate of transcript turnover is one factor that controls the rapidity and duration of gene expression responses to stimuli (4, 5). In yeast, the transcripts of many glucose-repressed genes become unstable when cells are returned to growth on glucose. Sequences in the 5′-UTRs of several transcripts are known to convey glucose sensitivity (6–9).
The galactose network is a well characterized metabolic pathway that converts galactose to glucose 6-phosphate. In the presence of galactose, the genes of this pathway are highly expressed; however, like other secondary metabolic pathways, GAL genes are suppressed by glucose at multiple levels (10). In addition to being silenced at the transcriptional level, the half-life of GAL1 mRNA, which encodes the first enzyme in the pathway, is reduced fourfold when glucose is introduced into the medium (6). This observation is in contrast to that for the Gal1 protein, which is highly stable in both galactose and glucose environments (11). The mechanism behind the regulated degradation of GAL1 mRNA has not been reported. As part of this study, we found that, consistent with other glucose-sensitive genes, the 5′-UTR of GAL1 destabilizes the transcript in the presence of glucose.
The primary response to glucose availability is that yeast cells rapidly increase their growth rate. The decision to divide or not is contemplated during G1 and yeast cells increase their division rate by shortening the length of this cell cycle phase (12). Once past a certain point in G1, called START, yeast cells are committed to completing the division cycle. START was originally described as the point at which a threshold capacity for protein synthesis is reached (13, 14). This point is sensed by the translationally regulated transcript of CLN3, which encodes the earliest cyclin in the cascade that drives the cycle forward (15).
In this study, we analyzed the biological consequence of the glucose-mediated degradation of GAL1 mRNA. We first altered the 5′-UTR sequence of GAL1 to create a strain that expresses a glucose-resistant GAL1 transcript. We then used microfluidic technology (16) to measure the dynamics of the galactose network in single cells expressing this stabilized variant of GAL1 mRNA. Our results indicate that GAL1 mRNA is rapidly degraded in response to glucose to allow the cell to quickly increase its growth rate by shortening the length of G1. In subsequent experiments, we observed a reciprocally antagonistic relationship between the synthesis of Gal1p and Cln3p. When GAL1 translation was increased, CLN3 translation was reduced and vice versa, suggesting that these transcripts share a limited supply of translation factors. Finally, we show that the temporal coordination of Gal1p and Cln3p synthesis may arise from spatial regulation, a common mechanism in biological signaling pathways and an emerging theme in translational regulation.
Results
5′-UTR of GAL1 Conveys Glucose Sensitivity.
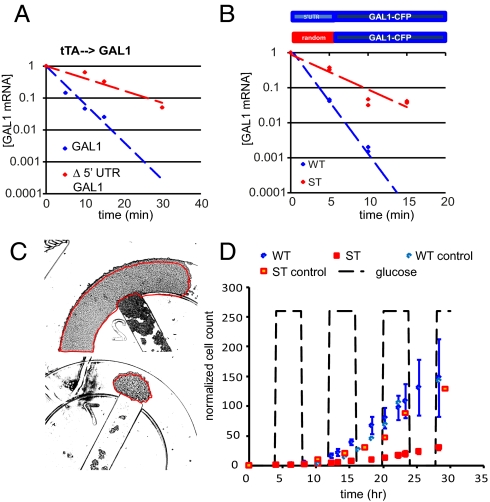
We used the tet-transactivator (tTA) expression system (17) to achieve regulated galactose-independent expression of GAL1 and then measured the half-lives of variants of GAL1 mRNA in cells grown in either glucose or galactose by quantitative RT-PCR (6). We found that deletion of the 300 bp upstream of the first ATG of GAL1 (Δ5′-GAL1) was sufficient to stabilize GAL1 mRNA by threefold in cells grown on glucose (half-life increased from ∼3 min to ∼9 min) (Fig. 1A). Likewise, fusion of the same 300-bp sequence to the YFP gene (5′-UTRGAL1-YFP) resulted in an mRNA that was threefold more stable in cells grown in galactose than in those grown in glucose (Fig. S1). These results suggest that the region immediately upstream of GAL1 is both necessary and sufficient for conferring glucose sensitivity, as is the case for other glucose-sensitive transcripts. We next used a PCR-based method to determine that the endogenous GAL1 transcript contains a 5′-UTR of ∼100 nt (Fig. S1). We then replaced the endogenous GAL1 gene with an allele harboring either the wild type or a randomized 100-bp sequence immediately upstream of the first ATG and a CFP tag at the 3′ end [strains WT and ST (stable), respectively]. We induced the expression of each allele from the native GAL1 promoter (PGAL1) by growing the two strains in galactose and then measured the half-life of GAL1 mRNA in WT and ST immediately after glucose addition. In the presence of glucose the randomized 5′-UTR stabilized the PGAL1-driven GAL1-CFP transcript in the ST strain by threefold compared with WT (Fig. 1B). We used these two strains to analyze the biological effects of glucose-mediated mRNA degradation on the dynamics of the galactose network.
Fig. 1.
Cells expressing stable GAL1 transcripts do not grow well in a dynamic environment. (A) Decay of tTA-driven GAL1transcripts, with (blue) and without (red) the 5′-UTR, in cells growing in glucose. Deletion of the 5′-UTR increased the half-life of the GAL1 transcript in glucose. (B) A schematic of the GAL1 transcript produced in WT and ST strains. The GAL1 mRNA levels measured by qRT-PCR in WT (blue) and ST (red) strains are plotted as normalized mRNA concentration vs. time after addition of glucose to the growth medium. GAL1-CFP mRNA decayed with a half-life of ∼3 min in WT and ∼9 min in ST. (C) Individual WT and ST colonies were grown in eight separate traps of a single microfluidic chip. There were three traps for each strain fed with galactose plus 4-h pulses of glucose every 4 h and one trap each for the galactose-only controls. Each colony was started from 25 to 75 cells. The growth of each colony was monitored over time by measuring the total area of the cells in each trap. The micrographs shown are final frames for one WT (Upper) and one ST (Lower) trap, both started from 31 cells, and were grown for 30 h under the dynamic conditions. (D) Results of the dynamic growth experiment. The number of cells in each trap was estimated from the total area of the colony and normalized to the number of cells in frame 1. The average sizes of the WT and ST colonies were plotted vs. time; error bars = 1 SD.
ST Cells Have a Growth Disadvantage in a Dynamic Environment.
Because glucose-sensitive transcripts are a part of multiple glucose-repressed pathways, it seems clear that the degradation of certain transcripts in response to glucose provides a selective advantage for cells. However, we found that the ST strain grew at least as well as the WT strain in conditions of constant glucose or constant galactose (Fig. S1). We hypothesized that the WT growth advantage would be realized only when cells were presented with a dynamic environment where glucose is only transiently available. To test this, we grew individual WT and ST colonies in separate chambers of a single microfluidic chemostat device that allows precise control over the growth medium (Fig. S2) (18), which was alternated between galactose only and galactose plus glucose medium every 4 h. We monitored the size of each colony by time-lapse microscopy as they grew in this dynamic environment. At regular time intervals, the area of each colony was measured (Fig. 1C) and the approximate number of cells in each frame was extrapolated on the basis of the average area of a single cell (the sizes of WT and ST cells were not found to be different in either static or dynamic growth conditions). WT and ST colonies grew at the same rate when fed only galactose (control colonies) (Fig. 1D). We found that WT cells grew as well as the controls under conditions of dynamic glucose availability. However, the periodic influx of glucose impaired the growth of ST cells. These results show that the selective advantage conferred by having a glucose-sensitive GAL1 transcript is realized only in conditions of transient glucose availability.
Stable GAL1 mRNA Causes a Prolonged G1 Phase After a Shift to Glucose.
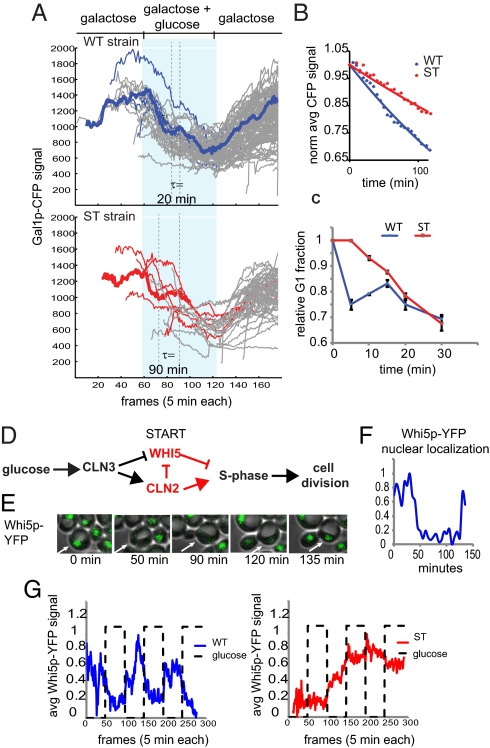
Cells growing in galactose medium respond to glucose by inhibiting the expression of GAL genes and by increasing their growth rate. We began by studying the effect of glucose-mediated degradation of GAL1 mRNA on the inhibition of the galactose network in cells growing in a dynamic environment. We grew the WT and ST strains in a microfluidic chemostat and recorded the level of Gal1p-CFP in single cells using time-lapse fluorescence microscopy. Consistent with Gal1p being a highly stable protein in both glucose and galactose, in both strains Gal1p-CFP was depleted primarily through dilution via cell division. This process produced a step-like decrease of fluorescence in the single-cell trajectories (Fig. 2A). On average the Gal1p-CFP signal decayed ∼2.5 times more slowly in ST cells than in WT cells (Fig. 2B). As highlighted in Fig. 2A (thick trajectories), the plateaus in the CFP trajectories show that the WT cells spent less time between cell divisions than ST cells during the glucose phase of the experiment (∼20 min vs. ∼90 min). Collectively, the results of the microfluidics experiments suggest that the primary difference between the strains is that WT cells divide more often in glucose than ST cells, causing them to deplete the Gal1p-CFP at a faster rate. On the basis of the observation that both WT and ST cells accumulated the same amount of Gal1p during growth in galactose and that Gal1p had not depleted in either cell type until the first cell division after glucose addition, we concluded that the ST phenotype was due to excess GAL1 mRNA, not protein.
Fig. 2.
Cells expressing stable GAL1 transcripts are impaired in the cell cycle response to glucose. (A) The cells were grown in a microfluidic chemostat under conditions of constant 0.5% galactose, supplemented with a 5-h square wave impulse of 0.25% glucose every 5 h. Single-cell Gal1p-CFP trajectories were recorded in individual cells by time-lapse fluorescence microscopy. The period of glucose addition is represented by the light blue shaded region. For each strain, the trajectories of five cells are highlighted in blue (WT) or red (ST). Cell divisions can be detected in the individual trajectories as sharp dips in fluorescence. The time elapsed between divisions (τ) is indicated for a representative trajectory for each strain, shown as a thick line. (B) The average Gal1p-CFP level for a population of cells (n = 25) after glucose addition is plotted vs. time (min). (C) xy plot of the change in the relative size of the G1 population over time after glucose addition, as measured by flow cytometry. (D) Increased global levels of protein synthesis in response to glucose lead to the production of Cln3p. Cln3p enters the nucleus and causes Whi5p, an inhibitor of S phase, to be expelled from the nucleus and induces the expression of a number of cell cycle genes, such as CLN2, that promote S phase and reinforce the cytoplasmic retention of Whi5p. (E) Cells expressing Whi5p-YFP were grown in a microfluidic chemostat and imaged every 5 min, using time-lapse fluorescence microscopy. Whi5p-YFP is easily detected in the nuclei of G1 cells, but the signal is weak when Whi5p is dispersed in the cytoplasm upon cell cycle entry. The fluorescent micrographs show changes in Whi5p-YFP localization as a WT cell divides in galactose medium. (F) The Whi5p-YFP trajectory of the cell shown in E, scaled from 0 to 1. (G) The average Whi5p-YFP signal plotted against time for single WT (n = 224) and ST (n = 493) cells growing in constant galactose with 4-h pulses of glucose every 4 h. See Fig. S3 for individual trajectories.
The results of the microfluidics experiments indicated that ST cells had a growth defect following a galactose-to-glucose switch; however, it was difficult to extract cell cycle data from the trajectories presented in Fig. 2A. To analyze this phenomenon more precisely, we conducted flow cytometry experiments to determine the cell cycle profiles of both strains grown under conditions of sudden glucose availability. As expected, both ST and WT cultures had larger G1 fractions when grown in galactose (37% and 36%) than when grown in glucose (25%) (Fig. S3). We next measured the rate at which the G1 fraction of each galactose culture was reduced after glucose addition. Here, we observed a considerable difference in the growth characteristics of the two strains: Whereas the G1 cells of the WT culture responded to glucose immediately by entering the division cycle, the ST culture was slower to respond (Fig. 2C). We then confirmed this result in synchronized cultures (Fig. S3). These observations suggest that the ST cells have lost the ability to tightly regulate cell cycle entry in response to glucose when growing in these dynamic conditions. To identify the relevant aspects of GAL1 repression, we assayed the cell cycle response in cells expressing a variety of tTA-driven GAL1 alleles. We found that both transcriptional repression and enhanced mRNA degradation were required for the normal response to glucose; however, most of the phenotype could be attributed to the decay of mRNA transcripts. The cell cycle dynamics were sensitive to overexpression of GAL1 transcript, as well as to the length of the GAL1 ORF, but did not require that the transcript encode a functional Gal1 protein (Fig. S3). These results are consistent with the hypothesis that the GAL1 transcript, not its protein product, interferes with cell cycle entry when glucose becomes available.
The events leading to cell cycle entry in yeast have been well characterized and involve the activation of a pathway that leads to the nuclear export of a cell cycle inhibitor, Whi5p (19–21) (Fig. 2D). We decided to monitor cell division in our WT and ST cells by following changes in the cellular localization of Whi5p-YFP. Whi5p-YFP was readily detected in the nucleus of unbudded cells, and as expected, just before bud formation the signal was dispersed throughout the cytoplasm. We made WT and ST versions of this strain and grew them in our microfluidic device while monitoring the Whi5p-YFP signal by time-lapse fluorescence microscopy (Fig. 2E). The WT and ST strains behaved similarly under conditions of constant galactose (Fig. S4). In the dynamic glucose experiments, the length of G1 in WT cells was strongly influenced by the presence of glucose. Under these conditions, G1 phases were long during the galactose phase (∼70 min), but rapidly shortened when glucose was added (to ∼10–20 min). In contrast, in ST cells the length of G1 did not respond to changes in carbon source. In both the galactose and the glucose phases, G1 was long and highly variable in duration. The scaled average Whi5p-YFP signal for each population is plotted over time in Fig. 2G. The results indicate that ST cells are defective in the growth response to glucose, upstream of Whi5p in the pathway that promotes cell cycle entry. (See Fig. S4 and SI Text for complete dataset and further discussion.)
Stable GAL1 Interferes with CLN3 Translation.
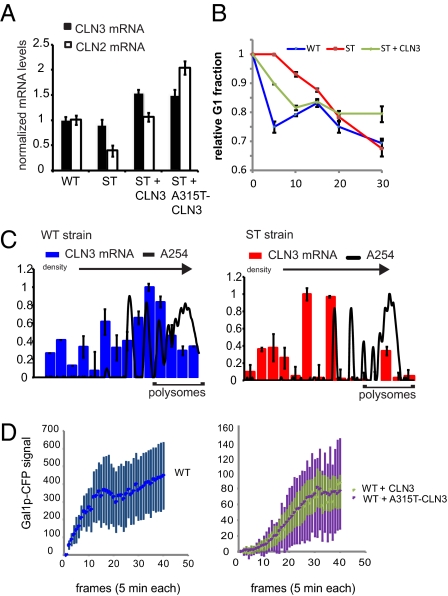
We next assayed the G1 pathway upstream of Whi5p (Fig. 2D). Cln3p activation leads to the transcription of the CLN2 gene, the product of which reinforces Whi5p nuclear export and S-phase entry. We found that 10 min after glucose was added to galactose cultures, WT cells expressed twofold more CLN2 mRNA than ST cells, as determined by qRT-PCR (Fig. 3A). When we raised the level of CLN3 expression in ST cells by deleting the endogenous gene and replacing it with a plasmid-borne copy, we found that the level of CLN2 mRNA detected in this assay was restored to that of WT cells. Increasing the expression of Cln3p even further by mutating the regulatory upstream ORF of the CLN3 plasmid (A315T-CLN3), which raises the translation efficiency of CLN3 without affecting the amount of mRNA, increased the level of CLN2 expression to twofold above the level measured in WT cells. These results show that the pathway connecting CLN3 to CLN2 expression is intact in ST cells. Because WT and ST cells had the same amount of CLN3 mRNA, and the pathway downstream of Cln3p appeared to be functional in ST, we concluded that the defect was in the up-regulation of CLN3 translation that occurs in response to glucose. We corroborated these findings by measuring the cell cycle response of ST + CLN3 cells in a flow cytometry assay (Fig. 3B and Fig. S5). The added CLN3 allowed the ST cells to respond to glucose as quickly as the WT cells. This result was then confirmed in synchronous cultures (Fig. S3A).
Fig. 3.
GAL1 and CLN3 transcripts are mutually antagonistic. (A) Summary of CLN3 and CLN2 mRNA levels, as measured by qRT-PCR in WT, ST, ST + CLN3, and ST + A315T-CLN3 strains 10 min after glucose addition. (B) xy plot of the change in the relative size of the G1 population over time after glucose addition, as measured by flow cytometry. (C) Polysome distribution of CLN3 mRNA in WT (Left) and ST (Right) cells just after glucose addition; error bars indicate ±1 SD. The sucrose gradient increases from left to right. A254 signal identifies the fractions containing translation complexes with increasing numbers of ribosomes. These graphs are representative of results obtained from at least three independent experiments. See SI Text for polysome distributions of cells growing in galactose. (D) Average Gal1p-CFP accumulation in WT (Left) and WT + CLN3[A315T] (Right) cells growing in a microfluidic chemostat; error bars are ±1 SD.
We next assayed the translation of CLN3 mRNA in WT and ST cells immediately after the addition of glucose, following the experimental design diagrammed in Fig. S5. First WT and ST cultures were grown for 2.5 h in galactose medium and then glucose was added. Immediately following the addition of glucose, cycloheximide and ice were added to the cultures to arrest translation. Cell lysates were prepared and fractionated over sucrose gradients by ultracentrifugation to separate translation complexes by density. The column graphs in Fig. 3C show the distribution of CLN3 mRNA in the fractionated WT and ST lysates, increasing in density from left to right, after glucose was added. The A254 traces show the polysome profiles for each strain and the polysome peaks, representing transcripts that were being translated by multiple ribosomes, are indicated (see Fig. S5D for direct comparison of WT and ST absorbance spectra). The results show that under these conditions, CLN3 transcripts exist in polysome complexes in WT cells, but in ST cells CLN3 messages are primarily associated with only single ribosomes. In contrast, the distribution of GAL1 transcripts was shifted toward higher ribosome occupancy in response to glucose in both strains, although the ST strain maintained high levels of this mRNA, whereas in WT cells GAL1 was quickly depleted (see Fig. S5 for GAL1 polysome distributions). These results confirm that the induction of CLN3 translation that normally occurs in response to glucose is impaired in the ST strain. We conclude that this is the source of the cell cycle defect exhibited by these cells when they are grown under dynamic conditions.
CLN3 Translation Interferes with Gal1p Synthesis.
Although overexpression of CLN3 was able to hasten the response of ST cells to glucose, it had the unexpected consequence of slowing the accumulation of Gal1p-CFP in cells during growth in galactose (Fig. S6). qRT-PCR results showed that the level of GAL1 mRNA was not decreased in this strain (Fig. S6), suggesting that the ability to efficiently produce Cln3p came at the expense of GAL1 translation, not transcription. It appeared that the relationship of GAL1 and CLN3 was reciprocal and that excess CLN3 transcripts could interfere with the translation of GAL1 mRNA when cells were switched from glucose to galactose. To explore this, we created a set of WT strains that harbor one of the CLN3 plasmids in addition to their endogenous CLN3 gene. We studied the accumulation of Gal1p-CFP in these strains by time-lapse microscopy. Fig. 3D presents the average trajectories for WT and WT + CLN3 cells growing in galactose under dynamic conditions. Upon being presented with galactose medium, WT cells immediately began to accumulate Gal1p-CFP. Despite expressing WT levels of GAL1 mRNA (determined by qRT-PCR), WT + CLN3 cells were delayed in Gal1p-CFP expression by almost 50 min. In addition, the steady-state levels of Gal1p-CFP were fivefold higher in WT cells relative to WT + CLN3 cells. Furthermore, WT cells reached steady-state levels in half the amount of time (75 min vs. 150 min). Eliminating the translational regulation of the plasmid-derived CLN3 (WT + A315T) had the same effect on the average Gal1p-CFP accumulation, but dramatically increased the variation between individual cells, illustrated by the error bars in Fig. 3D. In fact, >10% of these cells never induced expression of Gal1p-CFP over the course of the experiment (300 min).
We considered the possibility that GAL1 translation may be subject to cell cycle position and that, by decreasing the G1 fraction of the cell population, increased CLN3 expression may have indirectly prevented Gal1p-CFP accumulation. However, the single-cell trajectories do not support this hypothesis. In both WT and WT + CLN3, the cell cycle was responsive to Gal1p levels rather than the other way around (Fig. S6 and SI Text). To confirm that CLN3 expression was not inhibiting Gal1p accumulation through its effect on cell cycle position, we replaced the CLN3 plasmid with one encoding a nonfunctional protein, Cln3p-ΔNLS, which is incapable of regulating cell cycle entry (22). The dynamics and steady-state levels of Galp1-CFP in these cells were similar to what we observed in WT + CLN3 cells (Fig. S6).
GAL1 and CLN3 Transcripts Are Spatially Coregulated in Dynamic Environments.
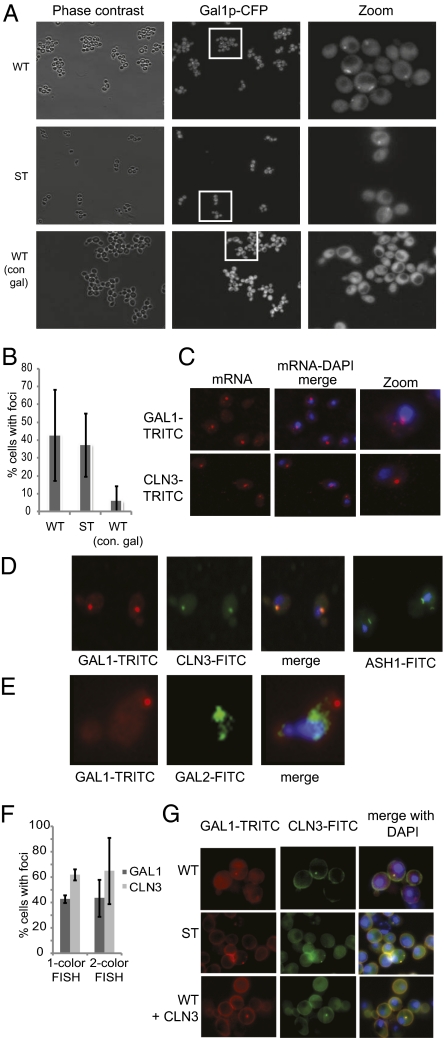
The GAL1 and CLN3 transcripts were each exquisitely sensitive to the expression level of the other in cells that were growing in dynamic glucose conditions. Considering that the genetic changes that revealed this relationship would not significantly impact the overall level of mRNA in the cell, it is unlikely that this sensitivity was due to a general phenomenon of protein synthesis. We began to contemplate possible scenarios that might cause these transcripts to be specifically influenced by each other. The first clue came from live cell images of WT and ST cells growing in galactose medium. We frequently observed one or two bright spots in the CFP channel in these cells as they induced the expression of Gal1p-CFP (WT images in Fig. S5 and Fig. 4A). This observation suggested that Gal1p synthesis may be spatially regulated. The number of cells with Gal1p-CFP foci was sensitive to the history of growth conditions before galactose induction. In cultures that had received only galactose in the past 24 h, cells with Gal1p-CFP foci were rare (6.5% vs. 42%) (Fig. 4B).
Fig. 4.
GAL1 and CLN3 mRNA are spatially coregulated. (A) Live cell imaging. Gal1p-CFP was detected in one to two foci in WT and ST cells growing under dynamic conditions. In contrast, Gal1p-CFP foci were rarely seen in WT cells that had been growing in galactose for >24 h. (B) Quantitation of Gal1p-CFP foci in WT and ST cells. (C) FISH of GAL1 and CLN3 transcripts in K699 cells (parent strain), 2.5 h after being switched from glucose to galactose. GAL1 mRNA was detected as one to two large brightly stained foci, with several smaller foci distributed throughout the cytoplasm. CLN3 mRNA was mostly detected in one to two large foci. (D) Two-color FISH of GAL1(TRITC) and CLN3(FITC) in K699 cells, 2.5 h after being switched from glucose to galactose; 99% of the CLN3 foci overlapped with a GAL1 focus, in cells for which both signals were detected. As a control, we observed the expected localization pattern for ASH1 mRNA (bud neck) (23). (E) Two-color FISH of GAL1(TRITC) and GAL2(FITC) in K699 cells, 2.5 h after being switched from glucose to galactose. No overlap between the two transcripts was observed. (F) Quantitation of foci detected in one-color and two-color FISH experiments. These results are averaged from at least three independent hybridizations. (G) Two-color FISH results from experiments in which the detergent wash was omitted to preserve the three-dimensional shape of the cells. GAL1(TRITC) and CLN3(FITC) foci overlapped in WT and ST strains, 2.5 h after being switched from glucose to galactose.
We used fluorescence in situ hybridization (FISH) to determine whether the observed Gal1p-CFP spots could result from the spatial regulation of GAL1 transcripts. We found that GAL1 transcripts were often clustered into one or two large foci in the cytoplasm (43% of cells that showed any TRITC hybridization). We also detected smaller signals dispersed throughout the cytoplasm, indicating that not all GAL1 transcripts were contained in these foci (Fig. 4C). Similarly, when we probed cells for CLN3 transcripts, we detected one or two large foci in many cells (62% of cells that showed any TRITC hybridization); however, in this case there was little signal outside of foci. In two-color FISH experiments where we simultaneously probed for GAL1 (TRITC) and CLN3 (FITC), we found near perfect overlap in 99% of the cells that displayed both GAL1 and CLN3 foci (Fig. 4D). On the other hand, the transcript of another galactose-induced gene, GAL2, did not colocalize with GAL1 spots (Fig. 4E). Consistently, the expression of Gal2p was not inhibited by CLN3 overexpression (Fig. S6E).
We repeated the two-color FISH experiment in the WT and ST strains, but modified the protocol so that the three-dimensional shape of the cells was better preserved. We found that 95% of WT cells in which both GAL1 and CLN3 foci were detected had near perfect fluorescence overlap (Fig. 4G), whereas 77% of the CLN3 foci overlapped with GAL1 in ST cells. WT + CLN3 cells also displayed colocalizing GAL1 and CLN3 foci; however, we did not observe Gal1p-CFP spots in these cells during galactose induction in the microfluidics experiments (Fig. S6).
Consistent with the live cell imaging of Gal1p-CFP foci, we found that GAL1 mRNA foci were rare when WT cells were grown in a static galactose environment (Fig. S7A). However, the few GAL1 mRNA foci we did detect in these cells were colocalized with CLN3 mRNA. In all cases where GAL1 mRNA was found outside of foci, it was not associated with CLN3 mRNA (Fig. S7A, Lower). The apparent lack of spatial regulation of GAL1 and CLN3 under static conditions suggests that these foci form in response to dynamic conditions to allow cells to rapidly respond to glucose. In support of this, we have found that WT cells do not respond to glucose, either by rapidly degrading GAL1 transcripts or by shortening the length of G1 phase, without prior exposure (Fig. S7 D and E). Although our results are not definitive, there is a trend consistent with the dynamic response to glucose arising from the spatial colocalization of GAL1 and CLN3 transcripts.
Discussion
GAL1 Transcripts Are Degraded in Response to Glucose to Optimize Growth in Dynamic Environments.
We began this study with the goal of understanding why glucose-induced GAL1 mRNA degradation leads to a selective advantage in cells growing in environments where glucose is only transiently available. The results of polysome fractionation experiments revealed that persistent GAL1 mRNA can inhibit CLN3 translation, which is normally induced by glucose. The observation of Whi5p-YFP strains showed that the rapid cell cycle response to glucose allowed WT cells to divide more often during bursts of glucose availability. In the absence of this regulation, ST cells missed the opportunity to increase their growth rate in glucose. We also discovered that when glucose was suddenly removed from the galactose-rich environment, cell division was stalled in both strains to allow Gal1p to accumulate again. For WT cells, this decrease in growth rate was balanced by the extra cell divisions in the glucose phase, and these cells wound up breaking even; i.e., their net growth in dynamic conditions was equivalent to their growth in a static galactose environment. Because ST cells did not gain a growth advantage in glucose, the occasional burst of glucose into the galactose-rich environment ultimately inhibited the growth of this strain. We conclude that the glucose-induced decay of metabolic transcripts is what allows cells to maintain optimal growth rates when carbon sources are fluctuating.
CLN3 Could Scavenge Ribosomes from GAL1 When Glucose Becomes Available.
In this study, we present evidence that the synthesis of Gal1p is spatially regulated under certain conditions. First, concentrated Gal1p-CFP signals were often detected in cells that were inducing GAL1 expression. The percentage of cells in the population that exhibited CFP foci was sensitive to prior growth conditions. We observed the largest number of foci when the culture had recent exposure to both galactose and glucose media. Likewise, we observed few foci in cultures that had been exposed only to galactose. These results suggest that the spatial regulation of Gal1p synthesis is something that occurs in response to dynamic shifts in glucose availability. Consistent with the apparent spatial regulation of Gal1p, GAL1 mRNA was detected in large foci in cells that were expressing high levels of Gal1p. This result, along with the early appearance of foci during Gal1p-CFP induction (Fig. S5), implies that these are sites of protein synthesis rather than aggregates of mature protein, although this hypothesis remains to be formally tested. The results of FISH experiments show that GAL1 and CLN3 transcripts are spatially coregulated when the environment is alternated between galactose and glucose. We propose that these transcripts compete for a local pool of translation components. Our hypothesis is supported by the mutual sensitivity of GAL1 and CLN3 translation to reciprocal changes in mRNA levels. When glucose becomes available, GAL1 transcripts are rapidly degraded, freeing up ribosomes (and other translation factors) in the region, which CLN3 transcripts can take advantage of. However, if GAL1 degradation is blocked, CLN3 translation continues to lag and the cell cycle response is delayed, as we saw in the ST strain. In this scenario, the region of concentrated GAL1 transcripts acts like a ribosome bank for CLN3 in the event that glucose is suddenly added to the environment (Fig. S8).
Materials and Methods
Plasmids and Strain Construction.
The plasmids used in this study are described in Table S1; strains are listed in Table S2. All cloned genes were amplified by PCR, using primers to incorporate the necessary flanking restriction sites for subsequent ligations. See SI Text for details.
Measurement of mRNA Decay Rates.
Total RNA preparations and experiments to determine the half-lives of specific mRNAs were performed as described in ref. 6. Primers used for qRT-PCR are listed in Table S3.
Microfluidics.
Microfluidics devices were prepared as described in ref. 6 and Fig. S2. Medium containing glucose was labeled by trace amounts of a fluorescent dye, sulforhodamine 101 (0.01 mg/mL). Image acquisition was performed on a Nikon Eclipse TI epifluorescent inverted microscope outfitted with fluorescence filter cubes optimized for CFP, YFP, and rhodamine imaging and a phase-contrast–based autofocus algorithm. Images were acquired using a Photometrics CoolSNAP HQ2 cooled CCD camera, controlled by Nikon Elements software. The cell chamber was photographed every 5 min in three channels to record cell morphology (40× phase), Gal1p-CFP (434 nm) or Whi5-YFP (520 nm), and glucose (560 nm). Single cell trajectories were generated using custom cell-tracking software developed in our laboratory (18), and CFP or YFP vs. time plots was created using Matlab or Microsoft Excel.
Flow Cytometry.
Cells were processed for flow cytometry, as follows: Cells were briefly pelleted and resuspended in 2 mL 70% ethanol (EtOH) and stored at −20 °C overnight. The next day, the EtOH-fixed cells were rehydrated in 5 mL dH2O, sonicated for 10 s at 40% power, and washed twice in 50 mM Tris, pH 8.0. Cell pellets were then resuspended in RNaseA (2 mg/mL) and incubated overnight at 37 °C. Cells were pelleted again, treated with pepsin (5 mg/mL) for 30 min at 37 °C, and then stained with propidium iodide (50 μg/mL) and run through a FACScan flow cytometer (Becton Dickinson). Flow cytometry data were analyzed using Gatelogic software. The size of the G1 fraction in each sample was estimated by measuring the percentage of cells in the 1N peak; the position of the G1 gate was kept constant for all samples. The reported results are averages of three independent experiments run in parallel. See SI Text for detailed protocol.
Gene Expression Measurements.
GAL1, CLN2, and CLN3 mRNA levels were measured by quantitative RT-PCR, as described for Fig. 1. Each experiment was performed in triplicate. Gene expression is displayed relative to results from WT. Measurements of Gal1p levels were derived from Gal1p-CFP signals in single cells at t = 150 min in the microfluidics experiments and are represented in the graph relative to the WT sample. See SI Text for detailed protocol.
Polysome Fractionation.
Yeast polysomes were prepared and fractionated according to the protocol at http://openwetware.org/images/4/47/PlyriRNA.pdf. Polysomes were fractionated on 36-mL 10–47% sucrose gradients by ultracentrifugation (27 K, 4 h, 4 °C) in an SW28 Beckman rotor. Fractions (1 mL) were collected from the flow-through into an equal volume of cold 100% EtOH in 2-mL microfuge tubes, stored overnight at −20 °C, and pelleted by centrifugation (14 K, 30 min, 4 °C), and the RNA was purified using the RNeasy kit (QIAGEN) following the manufacturer's protocol for RNA cleanup. Equal volumes of purified RNA (usually 10 μL) were used for qRT-PCR, as described above, to measure the relative amounts of GAL1 and CLN3 in the fractions of that region of the gradient containing ribosomes (usually fractions 17–32). See SI Text for detailed protocol; the primer sequences are listed in Table S3.
FISH.
For experiments shown in Fig. 4 C and D, cells were fixed and hybridized according to the protocol at (http://www.singerlab.org/protocols). Fluorescent probes were labeled using ULYSIS Nucleic Acid labeling kits (Alexa Fluor-546 and Alexa Fluor-488), according to the manufacturer's instructions (Invitrogen). Images were processed with ImageJ. For experiments shown in Fig. 4F, a modified version of the FISH protocol for hybridization of yeast in solution found at http://web.mit.edu/biophysics/data.html was used, except we used the same fluorescent probes for GAL1 and CLN3 described above instead of the oligo probes recommended in the published protocol and included an extra high-stringency wash step in 2× SSC/40% formamide for 30 min at 37 °C. Quantitation represents results from 100 to 300 individual cells per hybridization. See SI Text for detailed protocol.
Supplementary Material
Acknowledgments
We thank Dr. Simpson Joseph (University of California at San Diego) and his group for technical advice and for allowing us to perform the polysome fractionation experiments in his laboratory. We also thank Dr. Michael Polymenis (Texas A&M University) for the A315T-CLN3 plasmid. This work was supported by the National Institutes of Health and General Medicine (R01GM079333), the San Diego Center for Systems Biology (P50GM085764), and Welch Foundation Grant C-1729 (to M.R.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111408109/-/DCSupplemental.
References
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Youk H, van Oudenaarden A. Growth landscape formed by perception and import of glucose in yeast. Nature. 2009;462:875–879. doi: 10.1038/nature08653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronen M, Botstein D. Transcriptional response of steady-state yeast cultures to transient perturbations in carbon source. Proc Natl Acad Sci USA. 2006;103:389–394. doi: 10.1073/pnas.0509978103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalem O, et al. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Ortín JE, Alepuz PM, Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet. 2007;23:250–257. doi: 10.1016/j.tig.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Bennett MR, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffler IE, de la Cruz BJ, Prieto S. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Int J Biochem Cell Biol. 1998;30:1175–1193. doi: 10.1016/s1357-2725(98)00086-7. [DOI] [PubMed] [Google Scholar]
- 8.Andrade RP, Kötter P, Entian KD, Casal M. Multiple transcripts regulate glucose-triggered mRNA decay of the lactate transporter JEN1 from Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2005;332:254–262. doi: 10.1016/j.bbrc.2005.04.119. [DOI] [PubMed] [Google Scholar]
- 9.de la Cruz BJ, Prieto S, Scheffler IE. The role of the 5′ untranslated region (UTR) in glucose-dependent mRNA decay. Yeast. 2002;19:887–902. doi: 10.1002/yea.884. [DOI] [PubMed] [Google Scholar]
- 10.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast gal gene family: A complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 11.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 12.Johnston G, Singer R, Sharrow S, Slater M. Cell division in the yeast Saccharomyces cerevisiae growing at different rates. Microbiology. 1980;118:479–484. [Google Scholar]
- 13.Popolo L, Vanoni M, Alberghina L. Control of the yeast cell cycle by protein synthesis. Exp Cell Res. 1982;142:69–78. doi: 10.1016/0014-4827(82)90410-4. [DOI] [PubMed] [Google Scholar]
- 14.Moore SA. Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J Biol Chem. 1988;263:9674–9681. [PubMed] [Google Scholar]
- 15.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MR, Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet. 2009;10:628–638. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 18.Ferry MS, Razinkov IA, Hasty J. Microfluidics for synthetic biology: From design to execution. Methods Enzymol. 2011;497:295–372. doi: 10.1016/B978-0-12-385075-1.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 20.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 21.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 23.Beach DL, Bloom K. ASH1 mRNA localization in three acts. Mol Biol Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.