Abstract
Diabetes mellitus is the most common metabolic disorder worldwide and a major risk factor for cardiovascular disease. MicroRNAs are negative regulators of gene expression that have been implicated in many biological processes, including metabolism. Here we show that the Let-7 family of microRNAs regulates glucose metabolism in multiple organs. Global and pancreas-specific overexpression of Let-7 in mice resulted in impaired glucose tolerance and reduced glucose-induced pancreatic insulin secretion. Mice overexpressing Let-7 also had decreased fat mass and body weight, as well as reduced body size. Global knockdown of the Let-7 family with an antimiR was sufficient to prevent and treat impaired glucose tolerance in mice with diet-induced obesity, at least in part by improving insulin sensitivity in liver and muscle. AntimiR treatment of mice on a high-fat diet also resulted in increased lean and muscle mass, but not increased fat mass, and prevented ectopic fat deposition in the liver. These findings demonstrate that Let-7 regulates multiple aspects of glucose metabolism and suggest antimiR-induced Let-7 knockdown as a potential treatment for type 2 diabetes mellitus. Furthermore, our Cre-inducible Let-7-transgenic mice provide a unique model for studying tissue-specific aspects of body growth and type 2 diabetes.
Keywords: metabolic syndrome, insulin receptor, insulin receptor substrate 2, β-cell
According to a World Health Organization estimate, diabetes mellitus affects 346 million people worldwide (1), and its prevalence continues to increase due to advancing age, inactivity, and increasing obesity rates (2). In industrialized countries, diabetes mellitus is the leading cause of blindness, renal failure, and lower limb amputations and is a major risk factor for cardiovascular disease. Diabetes mellitus encompasses a group of etiologically different metabolic diseases characterized by high blood glucose levels. Type 1 diabetes results in elevated blood glucose levels due to a lack of insulin-producing pancreatic β-cells. Type 2 diabetes (T2D), the predominant form of diabetes mellitus, is often part of the so-called “metabolic syndrome,” which describes the co-occurrence of elevated blood pressure, dyslipoproteinemia, and obesity (2).
Patients with T2D have elevated blood glucose levels due to decreased insulin activity (i.e., insulin resistance) in peripheral organs, such as liver and muscle, with a consequent reduction in glucose uptake in these organs. Concomitantly, pancreatic insulin production/secretion is often reduced and is insufficient to meet the higher demand in T2D patients. Indeed, a number of lines of evidence, including genome-wide association studies, suggest dysfunction of pancreatic β-cells as a major pathogenic mechanism in T2D (3). T2D is often preceded by impaired glucose tolerance, which is characterized by abnormally increased postprandial blood glucose levels. Although fasting blood glucose levels often are only mildly elevated, impaired glucose tolerance is associated with increased cardiovascular mortality (4).
T2D can be initially treated with drugs that increase pancreatic insulin secretion and/or insulin sensitivity of peripheral tissues. Eventually, however, many patients require treatment with insulin to sufficiently control blood glucose levels. Novel therapies are needed to normalize pancreatic β-cell function and insulin sensitivity of peripheral tissues in patients with T2D.
MicroRNAs (miRNAs) are small, non–protein-coding RNAs that negatively regulate gene expression by promoting degradation and/or inhibiting translation of target mRNAs. MiRNAs have been shown to play important roles during development and disease, including T2D (5–7); for example, miR-375 regulates pancreatic insulin secretion (8), and miR-103 and miR-107 influence insulin sensitivity in peripheral tissues (9).
Let-7, one of the first miRNAs discovered, was initially shown to control developmental timing in Caenorhabditis elegans (10), but recently has been implicated in cancer and pluripotency as well (11). In mice, 12 genes encode members of the Let-7 family (Fig. S1), which includes nine slightly different miRNAs [Let-7a, Let-c, Let-7f (all encoded by two genes), and Let-7b, Let-7d, Let-7e, Let-7g, Let-7i, and miR-98 (all encoded by one gene)]. All Let-7 family members are believed to exert similar functions because they share a common seed region (nucleotides 2–8), which mediates miRNA interaction with target mRNAs. Processing of Let-7 can be inhibited by LIN28, an RNA-binding protein that is highly expressed in embryonic stem cells (12). Interestingly, ectopic global LIN28a overexpression in mice was found to result in increased body size and crown–rump length, as well as increased glucose metabolism and insulin sensitivity (13). However, Lin28a transgenic mice showed down-regulation (up to 50%) of Let-7 expression only in some adult organs, and whether the observed effects were mediated by Let-7 is unclear. While the present paper was in preparation, Zhu et al. (14) reported that global overexpression of Let-7g in mice resulted in growth retardation and impaired glucose tolerance. Extrapolating from data in mice with muscle-specific gain and loss of function of Lin28, the authors concluded that Let-7 regulates blood glucose levels at least in part via regulation of insulin signaling in muscle (14).
To study the function of Let-7 in mammals, we generated transgenic mice with Cre-inducible activation of Let-7a, Let-7d, and Let-7f expression, which enabled us to overexpress Let-7 in a tissue-specific manner. Mice with global overexpression of Let-7 were viable, but had reduced body size and weight. Furthermore, Let-7 transgenic mice exhibited impaired glucose tolerance due to diminished glucose-induced insulin secretion. To test whether global knockdown of the Let-7 family was sufficient to prevent or treat impaired glucose tolerance in mice, we developed a locked nucleic acid (LNA)-modified antimiR that could inhibit Let-7 function. We found that antimiR-induced knockdown of Let-7 improved blood glucose levels and insulin resistance in obese mice. We conclude that Let-7 regulates glucose metabolism in multiple organs, and that antimiR-induced knockdown of Let-7 could provide an approach to treating T2D.
Results and Discussion
Let-7 Regulates Adult Body Weight and Size.
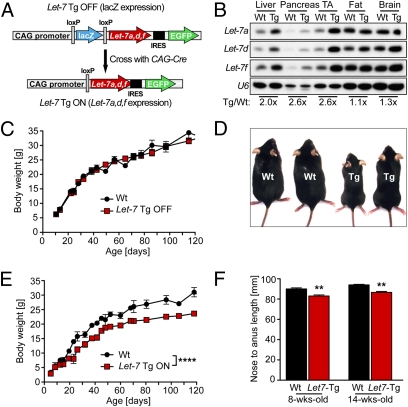
Five of 16 (31%) loci that have been associated with body length in humans have been identified as predicted Let-7 targets (15). Among these are confirmed Let-7 target genes such as high-mobility group AT-hook2 (Hmga2) (16), cyclin-dependent kinase 6 (Cdk6) (17), DOT1-like histone H3 methyltransferase (Dot1l) (18), and Lin28 (19). To study whether Let-7 is sufficient to regulate body size, we sought to globally overexpress Let-7 in mice. Because Let-7a, Let-7d, and Let-f (all of which are encoded by a single genomic cluster on chromosome 13) are among the most highly expressed Let-7 family members in some organs, we used this genomic cluster to generate transgenic mice with Cre-inducible expression of Let-7 (Fig. 1A). To achieve global overexpression of Let-7, we crossed transgenic mice carrying the inactive transgene construct (Let-7 Tg OFF) with transgenic mice expressing CAG-Cre. Subsequent removal of the CAG-Cre transgene by backcrossing to C57BL/6 mice produced Let-7 Tg ON mice. We performed Northern blot analysis to detect Let-7 expression and observed overexpression of the members of the transgenic Let-7 cluster in all tissues analyzed (Fig. 1B). The level of overexpression varied among tissues, with relatively stronger overexpression in organs with weaker endogenous Let-7 expression, such as liver and muscle.
Fig. 1.
Global Let-7 overexpression results in reduced body weight and body size. (A) Let-7 transgene. The “inactive” construct (Let-7 Tg OFF) expresses lacZ globally under the control of the CAG promoter. After Cre mediated excision of the lacZ gene, the transgene expresses the chromosome 13 Let-7 cluster comprising Let-7a, Let-7d, and Let-7f. Global overexpression of Let-7 (Let-7 Tg ON) was achieved by crossing the Let-7 Tg OFF mice to CAG-Cre transgenic mice. Let-7 Tg ON mice were then back-crossed to C57BL/6 mice to obtain Let-7 Tg ON mice without the CAG-Cre transgene. Because of an internal ribosome entry site (IRES), Let-7 Tg ON mice should also express GFP; however, the expression level was too low to enable detection by fluorescence microscopy. (B) Total RNA was isolated from multiple organs of 4-wk-old WT (Wt) and Let-7 Tg ON mice and used for Northern blot analysis to detect Let-7a, Let-7d, and Let-7f expression. Detection of U6 was used as a loading control. The ratios of Let-7 expression in Tg vs. WT organs are shown beneath each pair of lanes. (C) Body weight of WT mice vs. Let-7 Tg OFF littermates at different ages. Values are mean ± SEM; n = 3–9 males per time point. (D) Two WT and two Let-7 Tg ON male littermates at age 6 wk. (E) Body weight of WT mice vs. Let-7 Tg ON littermates. Values are mean ± SEM; n = 3–14 males for each time point. ****P < 0.0001 for WT vs. Let-7 Tg ON by ANOVA with Bonferroni posttest. (F) Reduced body length in Let-7 Tg ON mice; n = 3 males per time point. **P < 0.01 for WT vs. Tg by ANOVA with Bonferroni posttest.
Let-7 Tg OFF mice did not demonstrate an overt phenotype and had comparable body weight to WT littermates (Fig. 1C). Transgenic mice globally expressing Let-7 (Let-7 Tg ON) were viable and exhibited only a mildly reduced fertility rate with smaller litter sizes; however, Let-7 Tg ON mice were significantly smaller than their WT littermates (Fig. 1D). We observed this phenotype with 100% penetrance despite the mixed genetic background of the transgenic mice. The body weight of Let-7 Tg ON mice was similar to that of WT mice in the first 2 wk after birth, but then increased slower than the rate seen in WT mice and was further diminished in adult mice older than age 10 wk (Fig. 1E). Nose-to-anus lengths of adult Let-7 Tg ON mice overexpressing Let-7a, Let-7d, and Let-f were ∼8% less than WT littermates at age 8 wk and 14 wk (Fig. 1F). Given that Zhu et al. (14) also found a growth-inhibiting effect of global overexpression of Let-7g, we assume that most, if not all, Let-7 family members have a similar effect on adult body size.
Let-7 Transgenic Mice Have Reduced Fat Mass.
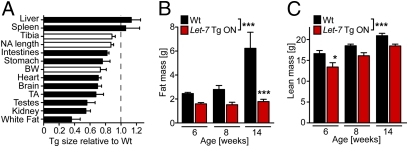
Analysis of isolated individual organs from 6-wk-old Let-7 Tg ON mice revealed that most organs were reduced in weight (or size) relative to WT littermates (Fig. 2A). Only liver and spleen were not reduced in weight, suggesting that Let-7 does not control the size of these organs. One reason for the reduced organ size could be that transgenic overexpression of Let-7 reduces proliferation and enhances differentiation of progenitor cell populations, which might express only low amounts of endogenous Let-7. Interestingly, body weight reductions seemed to be more pronounced than tibia or nose-to-anus length reductions in transgenic mice. This was most likely due to a disproportionate reduction in fat mass in the Let-7 Tg ON mice, as measured by the weight of epididymal fat (Fig. 2A). Therefore, we performed body composition measurements using EchoMRI (Echo Medical Systems) and found that fat mass did not increase with age in the Let-7 Tg ON mice as it did in their WT littermates (Fig. 2B). When fat mass was normalized to body weight, we found a 15% ± 4% lower fat mass in Let-7 Tg ON animals vs. WT littermates at age 6 wk, a 32% ± 6% lower fat mass at age 8 wk, and a 60% ± 5% lower fat mass at age 14 wk. Lean mass was also lower in Let-7 Tg ON mice, but it was proportionate to the decreased body size and exhibited normal age-dependent increases (Fig. 2C).
Fig. 2.
Reduced fat mass in Let-7 transgenic mice. (A) Relative organ weight or length [for tibia, nose-to-anus (NA) length, and intestine] of globally Let-7–expressing Tg mice vs. WT mice; n = 3. BW, body weight; TA, tibialis anterior; white fat, epididymal fat. (B and C) Body composition was measured in WT mice and Let-7 -Tg ON littermates at age 6, 8, and 14 wk by EchoMRI; n = 3 or 4 for each time point. *P < 0.05; ***P < 0.001 for WT vs. Let-7 Tg ON by ANOVA with Bonferroni posttest.
Given the important role of the hypothalamus in regulating body size and metabolism, we sought to generate transgenic mice with CNS-enriched Let-7 overexpression. Toward this end, we crossed Let-7 Tg OFF mice with transgenic mice expressing Cre under control of the Nestin promoter. Let-7 Tg/Nestin-Cre double-transgenic mice showed a similar reduction in body length as that resulting from global Let-7 overexpression at age 6 wk (Fig. S2). This might suggest that Let-7 controls body size predominantly via regulation of neurohormones. Interestingly, Let-7 Tg/Nestin-Cre double-transgenic mice exhibited only an attenuated reduction in body weight (Fig. S2), which suggests that fat mass was not affected, and that reductions in body size and fat mass by Let-7 are independent events.
Let-7 Transgenic Mice Exhibit Impaired Glucose Tolerance.
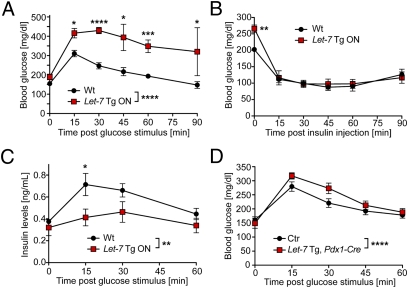
Given that changes in body composition can be caused by alterations in glucose metabolism, we investigated whether Let-7 expression is regulated in high-fat diet (HFD)-induced obesity, a well-characterized model of obesity-induced glucose intolerance. We assessed Let-7 expression by Northern blot analysis in multiple organs from mice on an HFD vs. mice on standard chow and found no major regulation of Let-7 expression in the investigated organs (Fig. S3). However, we cannot exclude the possibility that cellular subpopulations such as pancreatic β-cells display altered Let-7 expression on HFD. To study potential effects of global Let-7 overexpression on glucose metabolism, we performed glucose tolerance tests (GTTs) in 8-wk-old Let-7 Tg ON mice and WT littermates. Glucose stimulation produced abnormally elevated blood glucose levels in the Let-7 Tg ON mice (Fig. 3A). Glucose levels in these mice were highest 30 min after glucose stimulation and decreased slowly thereafter, suggesting impaired glucose-induced pancreatic insulin secretion and/or insulin resistance of peripheral tissues. We observed this effect with 100% penetrance despite the mixed genetic background of the transgenic mice.
Fig. 3.
Impaired glucose tolerance in Let-7 transgenic mice. (A) A GTT was performed in 7-wk-old WT mice and Let-7 Tg ON littermates after a 7-h fast. Glucose levels were determined at baseline and at the indicated times after an i.p. glucose stimulus (1.5 mg/g body weight); n = 7 (3 for 45 min and 90 min). *P < 0.05; ***P < 0.001; ****P < 0.0001 for WT vs. Let-7 Tg ON by ANOVA with Bonferroni posttest. (B) An ITT was performed in 8-wk-old fed Let-7 Tg ON mice and WT littermates. Blood glucose levels were measured at baseline and 15, 30, 45, 60, and 90 min after i.p. injections of insulin (1.0 U/kg body weight); n = 7. **P < 0.01 for WT vs. Let-7 Tg at baseline by ANOVA with Bonferroni posttest. (C) 7-wk-old Let-7 Tg ON mice and WT littermates were fasted for 7 h and then received a glucose stimulus (1.5 mg/g body weight i.p.). Blood was collected at baseline and after glucose stimulation and used for determination of insulin levels by ELISA; n = 4. *P < 0.05; **P < 0.01 by ANOVA with Bonferroni posttest. (D) Let-7 Tg OFF mice were crossed to Pdx1-Cre mice to induce pancreas-restricted Let-7 overexpression. A GTT was performed in 6-wk-old offspring after a 7 h fast to determine blood glucose levels after i.p. glucose stimulation (1.5 mg/g body weight). n = 5; ****P < 0.0001 for control (WT, Pdx1-Cre Tg, or Let-7 Tg OFF) vs. Let-7 Tg, Pdx1-Cre by ANOVA with Bonferroni posttest.
To assess insulin sensitivity of peripheral tissues, we performed insulin tolerance tests (ITTs) in 8-wk-old fed transgenic mice (Fig. 3B). The Let-7 Tg ON mice started with significantly higher blood glucose levels than their WT littermates (266 ± 11 mg/dL vs. 202 ± 7 mg/dL), but demonstrated reductions to similar levels as in WT littermates after insulin stimulation. This suggests that Let-7 Tg ON mice are not resistant to insulin under basal conditions, but rather have insufficient insulin secretion. To study glucose-induced insulin secretion, we performed a GTT and collected blood samples at baseline and at 15, 30, and 60 min after i.p. glucose injection. Measurement of insulin levels by ELISA found similar baseline insulin levels (after a 7-h fast) in WT and Let-7 Tg ON mice (Fig. 3C); however, the increase in insulin levels on glucose stimulation was severely attenuated and delayed in the Let-7 Tg ON mice, likely accounting for the elevated glucose levels. Surprisingly, insulin levels returned to baseline levels by 60 min after glucose stimulation in the Let-7 Tg ON mice, even though blood glucose levels were still severely elevated at this time (Fig. 3A). This suggests altered glucose sensing in pancreatic β-cells of Let-7 Tg ON mice. Immunofluorescent staining of pancreatic slices for insulin and glucagon showed similar gross morphology of islets in WT and Let-7 Tg ON mice (Fig. S4).
To study the specific effects of Let-7 overexpression in the pancreas, we crossed Let-7 Tg OFF mice with transgenic mice expressing Cre under control of the Pdx1 promoter. These mice also displayed impaired glucose tolerance on the GTT, confirming that Let-7 overexpression attenuates the pancreatic response to glucose (Fig. 3D). However, the observed effect was less pronounced in these mice than in Let-7 Tg ON mice. This suggests that Pdx1-Cre was not sufficient to turn on Let-7 in all pancreatic β-cells and/or that other cell types/organs also contributed to the impaired glucose tolerance. Thus, we activated Let-7 specifically in muscle (MCK-Cre), liver (Albumin-Cre), neurons (Nestin-Cre), and adipocytes (Fabp4-Cre), but did not observe impaired glucose tolerance after glucose stimulation (Fig. S5 A–D). This finding might indicate that Let-7 does not regulate glucose metabolism in these organs, or that other organs can counteract the effects of tissue-restricted overexpression of Let-7 in a healthy mouse.
Mice with muscle-specific gain and loss of function of the Let-7 inhibitor LIN28 display improved and diminished insulin sensitivity, respectively (14). Given that finding, the lack of insulin resistance of peripheral tissues in transgenic mice with global or muscle-specific Let-7 overexpression in the present study is surprising. One reason for this could be that we studied blood glucose levels in only young transgenic mice (age 6–8 wk), whereas insulin resistance typically worsens with age. Furthermore, we did not stress the transgenic mice with a HFD, which might have revealed differences in insulin resistance. Transgenic mice with global Let-7 overexpression also exhibited a significant reduction in fat mass, which might have reduced insulin resistance.
Insulin suppresses lipolysis. Consequently, patients with type 1 diabetes lacking insulin often have less adipose tissue, whereas those with T2D that receive insulin treatment often have more adipose tissue. Therefore, we wondered whether the decrease in adipose tissue in mice with global Let-7 overexpression is secondary to disturbances in glucose metabolism. To test this possibility, we determined body composition by EchoMRI in transgenic mice with pancreas-restricted Let-7 overexpression (Pdx1-Cre). These mice showed similar lean and fat mass as control mice (WT and single-transgenic mice) (Fig. S5E), suggesting that the loss in adipose tissue in mice with global Let-7 overexpression is not secondary to impaired glucose metabolism. An alternative explanation would be that the milder disturbances in glucose metabolism in mice with pancreas-specific Let-7 overexpression are insufficient to affect fat metabolism. The latter hypothesis is consistent with our finding that overexpression of Let-7 only in fat tissue (Fabp4-Cre) also did not result in changes in fat mass (Fig. S5F). This suggests that the loss of fat mass with global Let-7 overexpression is not due to a direct effect of Let-7 in (FABP4-expressing) adipocytes, even though Let-7 can regulate adipogenic differentiation in 3T3-L1 cells (20).
AntimiR-Induced Knockdown of Let-7 Is Sufficient to Prevent Obesity-Induced Glucose Intolerance.
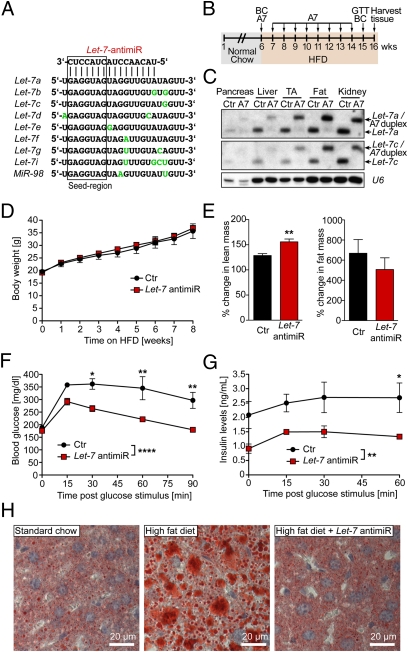
As noted earlier, the Let-7 family comprises a group of nine miRNAs encoded by 12 genes (Fig. S1), which poses a significant challenge to complete genetic deletion of this miRNA family. In an effort to inactivate the Let-7 family in vivo, we designed a DNA/LNA antimiR with sequence complementarity to nucleotides 2–17 of Let-7a/c (Fig. 4A). This region contains the conserved seed region of all Let-7 family members plus an additional eight nucleotides toward the 3′ end, which exhibit similar sequence in all Let-7 family members. To test whether antimiR knockdown of Let-7 was sufficient to prevent diet-induced glucose intolerance, we performed a prevention study in which C57BL/6 mice were placed on a HFD and injected once weekly for 8 consecutive weeks with Let-7 antimiR (Fig. 4B). At 2 wk after the last injection, we harvested multiple tissues from these mice and assessed Let-7 expression by Northern blot analysis. Mature Let-7 was strongly reduced in all tissues analyzed, suggesting that antimiR-induced knockdown of Let-7 is sustained for at least 14 d (Fig. 4C).
Fig. 4.
AntimiR-induced repression of Let-7 expression prevents impaired glucose tolerance. (A) Sequences of the Let-7 family. Nucleotide differences to Let-7a are labeled with green. The box indicates the seed region (nucleotides 2–8). We designed a 16mer LNA-DNA antimiR with complimentarity to nucleotides 2–17 of Let-7a and Let-7c. (B) Design of the diabetes prevention study. The 6-wk-old C57BL/6 mice were maintained on an HFD and injected s.c. once weekly with saline as a control or a specifically designed Let-7 antimiR (A7; 20 mg/kg body weight) for 8 wk. At 1 wk after the last A7 injection, a GTT was performed. Body composition (BC) was measured before the onset of HFD and at 1 wk after the last A7 injection. Tissues were harvested at 2 wk after the last A7 injection. (C) Tissues were harvested at the end of the prevention study, which was 2 wk after the last Let-7 antimiR injection. Pooled total RNA of 4 Ctr and, respectively, antimiR-treated animals was used for Northern blot analysis to determine expression of Let-7 family members. An upshift of mature Let-7 was detected in tissues of Let-7-antimiR–treated mice, likely representing a Let-7/Let-7 antimiR duplex. U6 was used as a loading control. (D) Body weight was determined weekly during the prevention study. (E) Body composition was assessed before the initiation of the HFD and at 1 wk after the last Let-7 antimiR injection by EchoMRI. The percent changes in lean and fat mass during the study period are depicted. n = 4. **P < 0.01, t test. (F and G) A GTT was performed at 1 wk after the last Let-7 antimiR injection after a 7-h fast to determine blood glucose and insulin levels upon i.p. glucose stimulation (2 mg/g lean mass). (F) Blood glucose levels were measured at baseline and at 15, 30, 60, and 90 min after glucose stimulation. (G) For assessment of insulin levels, plasma was collected at baseline and at 15, 30, and 60 min after glucose stimulation. Consecutive insulin levels were determined by ELISA. *P < 0.05; **P < 0.01; ****P < 0.0001 for control vs. Let-7 antimiR–treated mice by ANOVA followed by Bonferroni posttest. (H) At the end of the prevention study, frozen liver sections were stained by Oil Red O to detect ectopic lipid deposition. Representative sections are shown.
The Let-7 antimiR–injected mice and control mice (saline-injected) demonstrated similar increases in body weight during the treatment period (Fig. 4D), which is not surprising given that we started the Let-7 antimiR treatment only in adult mice (at age 6 wk). Furthermore, antimiRs do not cross the blood–brain barrier, whereas Let-7 overexpression with Nestin-Cre suggests that Let-7 regulates body size, at least in part, via gene regulation in neuronal cells (Fig. S2). As discussed earlier, the reduced body weight and fat mass in the Let-7 Tg ON mice might result secondarily from disturbances in glucose metabolism. We determined body composition before and after Let-7 antimiR treatment by EchoMRI and found a significantly stronger increase in lean mass during Let-7 antimiR treatment than during control treatment (Fig. 4E, Left). Consistently, individual muscles (gastrocnemius and tibialis anterior) also gained weight during Let-7 antimiR treatment, whereas muscles of obese mice on control treatment lost weight (Fig. S6). In contrast, fat mass tended to increase more slowly in Let-7 antimiR-treated mice (Fig. 4E, Right), accounting for the unchanged body weight. The smaller gain in fat mass might be secondary to higher energy consumption of the larger muscles of the Let-7 antimiR-treated mice.
At the end of the prevention study, we fasted the mice for 7 h and performed a GTT. Because the Let-7 antimiR-treated mice had greater lean mass compared with control mice, we adjusted the glucose injection to lean mass so that the Let-7 antimiR-treated mice received slightly more glucose per gram of body weight than the control mice. As expected, the control mice displayed impaired glucose tolerance, with blood glucose levels still significantly above baseline at 90 min after glucose injection (Fig. 4F). In contrast, the Let-7 antimiR-treated mice showed a normal response to glucose stimulation, with normalized blood glucose levels at 90 min after glucose injection. This finding suggests either that pancreatic insulin secretion is increased on glucose stimulation in Let-7 antimiR-treated mice or that peripheral tissues of Let-7 antimiR-treated mice are more sensitive to insulin. To distinguish between these possibilities, we measured blood insulin levels before and after glucose stimulation. We found significantly lower insulin levels in Let-7 antimiR-treated mice (Fig. 4G), which points toward improved insulin sensitivity of peripheral tissues but does not rule out the possibility of improved β-cell function in the Let-7 antimiR-treated mice.
Ectopic fat deposition in liver and other organs is one cause of reduced insulin sensitivity in obesity. Thus, we investigated lipid accumulation in the liver by Oil Red O staining. Although the saline-injected mice accumulated a significant amount of fat during the study period, the Let-7 antimiR-treated mice had a similar liver fat content as age-matched control animals on a standard diet (Fig. 4H).
AntimiR-Induced Knockdown of Let-7 Is Sufficient to Treat Obesity-Induced Glucose Intolerance.
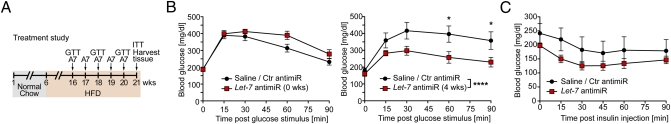
To test whether Let-7 antimiR treatment is sufficient to treat impaired glucose tolerance, we put C57BL/6 mice on an HFD for 10 wk and then initiated weekly treatment with Let-7 antimiR (20 mg/kg body weight) for 4 wk (Fig. 5A). Control mice received injections of either saline only or equal amounts of a control antimiR. GTTs were performed before antimiR treatment and at 2 and 4 wk after the onset of Let-7 antimiR treatment. There was no significant difference in blood glucose levels at baseline (Fig. 5B, Left); however, blood glucose levels were significantly reduced in Let-7 antimiR-treated mice compared with control mice after only 2 wk of treatment (Fig. S7). This effect was even more pronounced after 4 wk of Let-7 antimiR treatment (Fig. 4B, Right). These data demonstrate that treatment with Let-7 antimiR is sufficient not only to prevent, but also to treat impaired glucose tolerance. Analysis of insulin sensitivity of peripheral organs with ITTs showed that insulin injection reduced blood glucose to lower levels in Let-7 antimiR-treated mice than in control mice; however, the control mice started with higher glucose levels (Fig. 5C).
Fig. 5.
AntimiR-induced repression of Let-7 expression improves impaired glucose tolerance. (A) Design of the diabetes treatment study. Six-wk-old C57BL/6 mice were fed for 10 wk with HFD. Subsequently, the mice were maintained on an HFD for 5 more wk, but half of the mice (n = 5) were treated with Let-7 antimiR (once weekly 20 mg/kg body weight) whereas control animals (Ctr) received either saline (n = 3) or control antimiR (n = 2). (B) GTTs (1.5 mg glucose/g body weight) were performed before initiation of antimiR treatment (Left) and at 4 wk after the first antimiR injection (Right). *P < 0.05; ****P < 0.0001 for Ctr (saline + control antimiR) vs. Let-7 antimiR– treated mice by ANOVA with Bonferroni posttest. (C) An ITT was performed at 1 wk after the last Let-7 antimiR injection. Blood glucose levels were measured at baseline and at 15, 30, 45, 60, and 90 min after i.p. insulin injection (1.0 U/kg body weight).
Let-7 AntimiR Treatment Restores Insulin Signaling in Muscle and Liver.
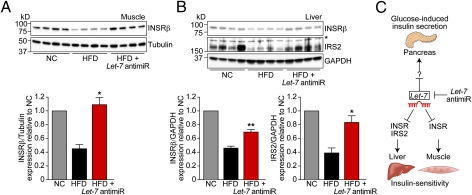
Individual miRNAs can target multiple genes within biological pathways, which enables the miRNAs to modulate complex disease processes. Target genes can be identified using prediction algorithms, which rank potential targets based on the complementarity between miRNA and mRNA 3′UTRs and the degree of conservation of the 3′UTR target sequence across species. Using one of these programs (targetscan; www.targetscan.org), we found that Let-7 is predicted to target the insulin receptor (Insr), which is responsible for insulin action in peripheral tissues. Let-7 is further predicted to target insulin receptor substrate 2 (Irs2), which is phosphorylated by INSR on insulin stimulation and mediates the insulin signal to downstream effectors. Up-regulation of INSR and/or IRS2 by antimiR knockdown of Let-7 could explain the enhanced insulin sensitivity that we observed during the Let-7 antimiR treatment. Furthermore, because Let-7 antimiR seems to work as a scaffold that binds Let-7 rather than inducing its degradation, up-regulation of Let-7 target genes serves as a necessary readout of Let-7 inhibition. Therefore, we performed Western blot analysis of INSR and IRS2 expression in mice from our prevention study. As an additional control group, we used age-matched mice that were fed normal chow. The HFD reduced expression of INSR compared with normal chow in muscle and liver, but Let-7 antimiR treatment restored INSR expression to levels found in healthy mice on normal chow (Fig. 6 A and B). In addition, IRS2 expression was down-regulated in mice on an HFD, and Let-7 antimiR treatment prevented down-regulation of IRS2 expression in the liver (Fig. 6B).
Fig. 6.
Let-7 regulates insulin signaling in liver and muscle. (A) Gastrocnemius muscle was harvested from mice at the end of the prevention study and from age-, sex-, and strain-matched mice fed with normal chow (NC). (Upper) Protein lysates from those mice were used to detect expression levels of the insulin receptor β (INSRβ) by Western blot analysis. Tubulin expression was used as a loading control. (Lower) Densitometric and statistical analysis. *P < 0.05 for HFD vs. HFD + Let-7 antimiR, t test. (B) Detection of INSRβ and insulin receptor substrate 2 (IRS2) by Western blot analysis of liver lysates from the same mice. GAPDH expression was used as loading control. The asterisk indicates an unspecific band detected by the IRS2 antibody. *P < 0.05; **P < 0.01 for HFD vs. HFD + Let-7 antimiR, t test. (C) Model of Let-7–induced modulation of glucose metabolism. Let-7 inhibits glucose-induced insulin secretion from pancreatic β-cells as demonstrated in Let-7 transgenic mice. In addition, antimiR knockdown of Let-7 improves insulin sensitivity in liver and muscle, at least in part by restoring expression levels of INSR and IRS2.
Potential Clinical Relevance.
Our results demonstrate that knockdown of Let-7 with an antimiR is sufficient to prevent and treat impaired glucose tolerance by improving insulin sensitivity in peripheral tissues. In addition, data from our transgenic mice indicate that Let-7 can block glucose-induced insulin secretion from the pancreas, suggesting that knockdown of Let-7 also might improve pancreatic β-cell function (Fig. 6C). Whether Let-7 is causatively involved in the pathogenesis of T2D is unclear, given that Let-7 expression was not altered in the mice on an HFD. Nevertheless, in principle Let-7 antimiR treatment could provide a unique approach to treating T2D, for several reasons. First, Let-7 likely regulates multiple components of glucose metabolism; second, antimiRs often result in long-lasting knockdown of miRNAs with sustained effects; and third, Let7 antimiR decreased fat mass in obese settings, in contrast to most drugs currently used to treat T2D. However, it also should be noted that Let-7 is associated with the occurrence of tumors, although we found no obvious signs of cancer during antimiR treatment periods of up to 10 wk. Furthermore, localized knockdown of Let-7 in skeletal muscle (a tissue rarely affected by tumors) might be sufficient to improve glucose metabolism. Our Cre-inducible Let-7-transgenic mice provide a unique model for studying tissue-specific aspects of body growth and T2D and for testing the effects of drugs on the pathogenesis of diabetes.
Methods
Mice.
For generation of the Let-7 transgenic mice, a genomic region on chromosome 13 comprising the genes for Let-7a, Let-7d, and Let-7f was amplified from C57BL/6 DNA and cloned into the CAG-Z-EGFP 1.0 vector (Invitrogen), which allows activation of transgene expression by Cre-mediated deletion of an inhibitory DNA element. Pdx1-Cre mice were kindly provided by Doug Melton (Harvard University, Cambridge, MA). All experimental animal procedures were approved by the University of Texas Southwestern Medical Center's Institutional Animal Care and Research Advisory Committee.
Let-7 Northern Blot Analysis.
Northern blots were performed as described previously (21).
Body Composition Analysis.
Body composition was measured using an EchoMRI-100 body composition analyzer (Echo Medical Systems) at the University of Texas Southwestern Medical Center's Metabolic Core Facility.
AntimiR.
The Let-7 antimiR was synthesized by miRagen Therapeutics. 20 mg antimiR per kg body weight dissolved in 100 μL of saline was injected s.c. once weekly. The control antimiR had an identical chemistry and comprised a sequence directed against a C. elegans-specific miRNA (5′-TccTAgAAaGAgtAgA-3′, with uppercase indicating LNA and lowercase indicating DNA).
Glucose and Insulin Measurements.
For GTTs, mice were fasted for 7 h before i.p. glucose injection. Insulin levels in plasma were measured by ELISA (Chrystal Chem).
Western Blot Analysis.
IRS2 and INSRβ antibodies were obtained from Cell Signaling and used at 1:1,000 dilutions as recommended by the manufacturer.
Statistical Analysis.
All data are shown as mean ± SEM. Data were analyzed using the Student t test or ANOVA followed by a Bonferroni posttest as appropriate. Experiments are described in more detail in SI Methods.
Supplementary Material
Acknowledgments
We thank Chad Grueter, Doug Millay, and the other members of the E.N.O. laboratory for many helpful discussions; Jose Cabrera for help with graphics; Xunshan Ding for help with insulin measurement; Eva van Rooij and miRagen Therapeutics for the Let-7 antimiR; Doug Melton for the Pdx1-Cre mice; Ann-Christin Pecher for help with cloning the transgene construct; John McAnally for the transgene construct injections; John Shelton for help with Oil Red O staining; and Gianluigi Condorelli, Scott Hammond, and Joshua Mendell for critical comments on the manuscript. Work in the E.N.O. laboratory was supported by grants from the National Institutes of Health, the Foundation Leducq TransAtlantic Network of Excellence Program, the American Heart Association–Jon Holden DeHaan Foundation, and the Robert A. Welch Foundation. R.J.A.F. was supported by a Beginning Grant-in-Aid from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118922109/-/DCSupplemental.
References
- 1.World Health Organization Diabetes fact sheet no. 312. 2011. Available at http://www.who.int/mediacentre/factsheets/fs312/en/index.htmlAccessed August, 2011.
- 2.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 3.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr EL, et al. Risk of cardiovascular and all-cause mortality in individuals with diabetes mellitus, impaired fasting glucose, and impaired glucose tolerance: The Australian Diabetes, Obesity, and Lifestyle Study (AusDiab) Circulation. 2007;116:151–157. doi: 10.1161/CIRCULATIONAHA.106.685628. [DOI] [PubMed] [Google Scholar]
- 5.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 6.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60:1825–1831. doi: 10.2337/db11-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 9.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 10.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 11.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 12.Newman MA, Hammond SM. Lin-28: An early embryonic sentinel that blocks Let-7 biogenesis. Int J Biochem Cell Biol. 2010;42:1330–1333. doi: 10.1016/j.biocel.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, et al. DIAGRAM Consortium. MAGIC Investigators The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lettre G, et al. Diabetes Genetics Initiative. FUSION. KORA. Prostate, Lung Colorectal and Ovarian Cancer Screening Trial. Nurses’ Health Study. SardiNIA Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 18.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23:925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost RJ, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.