Abstract
Accumulation of cadmium (Cd) in rice (Oryza sativa L.) grains poses a potential health problem, especially in Asia. Most Cd in rice grains accumulates through phloem transport, but the molecular mechanism of this transport has not been revealed. In this study, we identified a rice Cd transporter, OsLCT1, involved in Cd transport to the grains. OsLCT1-GFP was localized at the plasma membrane in plant cells, and OsLCT1 showed Cd efflux activity in yeast. In rice plants, strong OsLCT1 expression was observed in leaf blades and nodes during the reproductive stage. In the uppermost node, OsLCT1 transcripts were detected around large vascular bundles and in diffuse vascular bundles. RNAi-mediated knockdown of OsLCT1 did not affect xylem-mediated Cd transport but reduced phloem-mediated Cd transport. The knockdown plants of OsLCT1 accumulated approximately half as much Cd in the grains as did the control plants. The content of other metals in rice grains and plant growth were not negatively affected by OsLCT1 suppression. These results suggest that OsLCT1 functions at the nodes in Cd transport into grains and that in a standard japonica cultivar, the regulation of OsLCT1 enables the generation of “low-Cd rice” without negative effects on agronomical traits. These findings identify a transporter gene for phloem Cd transport in plants.
Keywords: heavy metals, food safety
Cadmium (Cd) is a heavy metal harmful to human health. The biological half-life of Cd in the body is estimated to be nearly 30 y (1), which leads to chronic toxicity. The adverse effect of Cd has been a worldwide concern since the outbreak of “Itai-Itai disease” in the mid-20th century in Japan that was caused by the daily consumption of Cd-contaminated rice (2, 3). Recently, the average dietary intake of Cd in Japan was estimated to be 3.0 μg Cd/kg body weight per week. This value is nearly 50% of a provisional tolerable monthly intake established by the Joint Food and Agriculture Organization/World Health Organization (FAO/WHO) Expert Committee on Food Additives and Contaminants (JEFCA) and higher than the tolerable weekly intake (2.5 μg Cd/kg body weight) set by the European Food Safety Authority (EFSA). Reflecting this greater Cd intake, the internal Cd level among Japanese people is high among Asians (4). Recent surveys in Sweden (5), the United Kingdom (6), and the United States (7, 8) also showed measurable internal Cd levels within the general population, indicating the importance of reducing Cd exposure in the general population (9). In Asian countries, including Japan, up to 50% of the ingested Cd comes from rice and its products (4, 10). The Food and Drug Administration's (FDA) Total Diet Study reported that the dietary intake of Cd among Americans increased by 26% from 1990 through 2003 and that this rise was correlated with an increased consumption of rice and other grains (11).
The source of Cd in rice grains is soil. Cd is absorbed by rice roots and transported to the grains, resulting in considerable Cd accumulation even when grown on slightly or moderately Cd-polluted soil (12, 13). Transporters for mineral nutrients, such as Zn or Fe, are partly responsible for Cd transport in plants (14). To improve the health of people who depend on rice as a staple, the establishment of a low-Cd rice cultivar is desirable. A first step in this direction is to clarify the mechanism of Cd transport in rice, including identification of the responsible molecule(s).
Recent physiological studies have advanced our understanding of how Cd is transported and accumulated in rice. The main determinant of the Cd concentration in shoot tissues is the ability to translocate Cd from root to shoot through the xylem, rather than Cd uptake by the roots (12). Quantitative trait loci (QTL) analyses have indicated several chromosomal regions controlling Cd accumulation in rice shoots; one of these loci regulates the loading of Cd into the xylem (15–19). Recently, OsHMA3, a rice P-type ATPase, was identified as a regulator of the xylem loading of Cd in roots (20, 21).
Despite its importance, little is known about the molecular mechanism of phloem Cd transport in plants. Phloem-mediated Cd transport to grains following xylem-mediated root-to-shoot translocation is critical for the accumulation of Cd in rice grains. Nearly 100% of the Cd in rice grains is attributable to phloem transport (22, 23). Fujimaki et al. (22) used a noninvasive live imaging technique to follow the transport of 107Cd in intact rice plants. They demonstrated the importance of shoot nodes for the transfer of Cd from the xylem to the phloem. For some minerals, the contribution of transporters is crucial for this kind of redirection of solute transport at nodal regions (24, 25). The Cd translocation ability into grains varies among rice genotypes (12). Kato et al. (26) also reported variable Cd concentrations in phloem sap among cultivars exhibiting different grain Cd levels and showed the correlation between grain and phloem sap Cd concentrations. These studies indicated the existence of transporters mediating Cd translocation into grains and suggested that regulation of the transporters can alter the level of Cd deposition in grains.
In this study, we focused on LOC_Os06g38120 as a possible Cd transporter gene expressed in rice shoots during grain ripening. This gene, OsLCT1, is predicted to encode the only rice homolog of the low-affinity cation transporter 1 (TaLCT1) found in the wheat cDNA library. TaLCT1 enhanced the intake of various cations, including Cd2+ in yeast (27). Here, we present evidence that OsLCT1 is a plasma membrane-localized Cd exporter involved in phloem Cd transport. By down-regulating OsLCT1 expression, we generated rice plants with reduced grain Cd levels, demonstrating the importance of OsLCT1 in Cd transport into grains.
Results
OsLCT1 Is a Plasma Membrane-Localized Efflux Transporter of Cd.
OsLCT1 cDNA cloned from a japonica rice cultivar, Nipponbare, was deduced to encode 511 aa with 11 transmembrane domains (Fig. S1). The primary sequence of OsLCT1 is 23% identical to that of TaLCT1. A Basic Local Alignment Search Tool (BLAST) search indicated that OsLCT1 is a single-copy gene in the rice genome, and no homolog of OsLCT1 was found in the Arabidopsis thaliana genome.
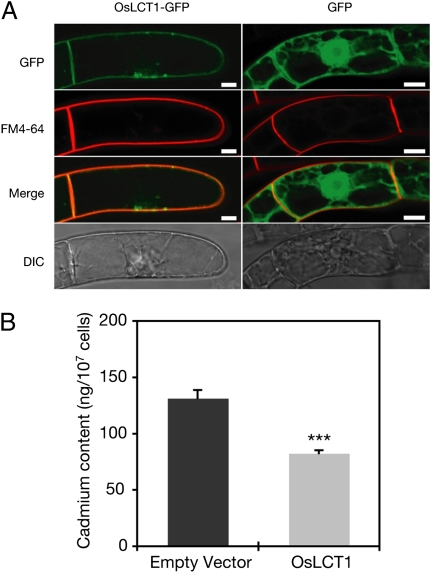
The subcellular localization of OsLCT1 was then investigated in tobacco BY-2 cells. OsLCT1 fused with GFP was primarily colocalized with the plasma membrane marker FM4-64, whereas in cells expressing GFP alone, green fluorescence was observed mainly in the cytosol and nucleus (Fig. 1A). This strongly suggests that OsLCT1 is a plasma membrane protein.
Fig. 1.
(A) Subcellular localization of OsLCT1-sGFP. 35S-OsLCT1-sGFP (OsLCT1-sGFP) and 35S-sGFP (sGFP) were introduced separately into cultured tobacco BY2 cells. FM4-64 (25 μM) was used as a plasma membrane marker. (Scale bars = 10 μm.) (B) Cd transport activity of OsLCT1. Yeast (strain WΔycf1) expressing OsLCT1 or harboring the empty vector were incubated for 75 min with arginine-phosphate medium containing 20 μM CdCl2. The concentrations of Cd in the harvested cells were determined by ICP-MS after nitric acid digestion. Asterisks represent a significant difference from the empty vector cells (P < 0.001, t test). The data are presented as means ± SD (n = 3).
TaLCT1 enhanced the transport of various cations, including Cd2+, in yeast (27). We tested whether OsLCT1 has Cd transport activity in a yeast heterologous expression system using WΔycf1. This yeast strain has a defect in the gene encoding a vacuolar transporter, Ycf1p, which mediates the transport of a Cd–glutathione conjugate into vacuoles (28). Yeast cells were transformed with a vector containing Myc-tagged OsLCT1 cDNA, and the expression of OsLCT1 was confirmed by Western blot analysis using anti-Myc antibodies (Fig. S2A). OsLCT1 expression in the yeast did not affect the Cd sensitivity of the cells (Fig. S2B). However, OsLCT1 expression significantly reduced Cd accumulation in the cells compared with vector control cells in 10 and 20 μM Cd treatments (Fig. 1B and Fig. S2C), suggesting that OsLCT1 is an efflux-type Cd transporter. We also analyzed the concentrations of several minerals in the cells. Yeast cells expressing OsLCT1 showed significantly reduced Mg, Ca, K, and Mn accumulation compared with the vector control cells, with no significant difference in the concentrations of Na, Fe, Zn, or Cu (Fig. S2D). Treatment with 1 mM CaCl2 or 10 mM MgSO4 with 10 μM CdCl2 reduced the Cd transport activity of OsLCT1-expressing cells (Fig. S2E) compared with that under the normal Ca (0.1 mM) and Mg (1 mM) conditions (Fig. 1B and Fig. S2C).
OsLCT1 Is Strongly Expressed in Nodes and Leaf Blades During Grain Ripening.
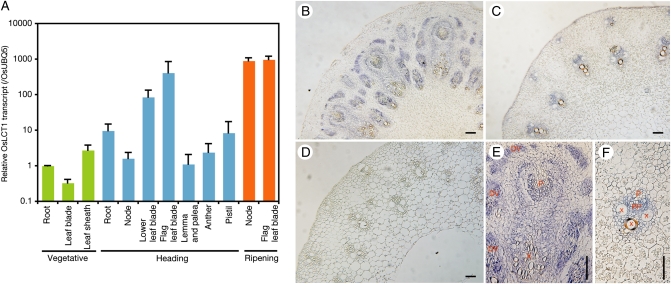
To determine the expression profile of OsLCT1 in rice plants, we first analyzed microarray data from the rice gene expression database RiceXPro (29). The analysis of a dataset obtained from various rice tissues at different growth stages demonstrated that OsLCT1 is strongly expressed in leaf blades during reproductive stages, rather than during vegetative stages (Fig. S3). Further analysis was conducted using real-time PCR analysis to examine the specificity of OsLCT1 expression in more detail (Fig. 2A). In this experiment, nodes were added as a sample because they were not included in the RiceXPro microarray data. The real-time PCR analysis showed strong OsLCT1 expression in both leaf blades and nodes during the grain-ripening stage, whereas its expression in shoots and roots in the vegetative growth stage was relatively lower (Fig. 2A).
Fig. 2.
Expression profiles of OsLCT1 in rice. (A) Real-time PCR analysis. cDNAs were synthesized from total RNA extracted from various tissues of rice grown in a greenhouse, and the mRNA levels were quantified by real-time PCR. The data were normalized to OsUBQ5 and are shown relative to the vegetative root sample. The data are presented as means ± SD (n = 3). (B–F) In situ hybridization of OsLCT1 in node I. (B–D) Cross-sections of the middle of node I (B), the border region of node I and internode II (C), and internode II (D). (E and F) Enlarged images of vascular bundles in the middle of node I (E) and in the border region of node I and internode II (F). (Scale bars = 10 μm.) DV, diffuse vascular bundles; P, phloem regions of large vascular bundles; PP; phloem parenchyma cells; X, xylem regions of large vascular bundles.
We then examined the cell type specificity of OsLCT1 expression by in situ hybridization in node I, the uppermost node connected to the flag leaf and the panicle (Fig. 2 B–F). OsLCT1 expression was detected specifically in vascular bundles in the middle of node I (Fig. 2B) and in the border region between node I and internode II (Fig. 2C), but expression was not observed in internode II (Fig. 2D). The accumulation pattern observed in the cross-sectioned samples was confirmed in the longitudinal sections of node I and internode II (Fig. S4B). Furthermore, high-magnification observation of the middle of node I identified the accumulation of OsLCT1 transcripts in the cells around enlarged large vascular bundles and more strongly in the cells of diffuse vascular bundles (Fig. 2E). OsLCT1 was also expressed in the phloem parenchyma cells surrounding the phloem of enlarged large vascular bundles (Fig. 2E). OsLCT1 transcript accumulation in the phloem parenchyma cells was evident in the border region of node I and internode II (Fig. 2F), where the vascular bundles were not enlarged.
Down-Regulation of OsLCT1 Decreases Grain Cd Levels in Rice Plants.
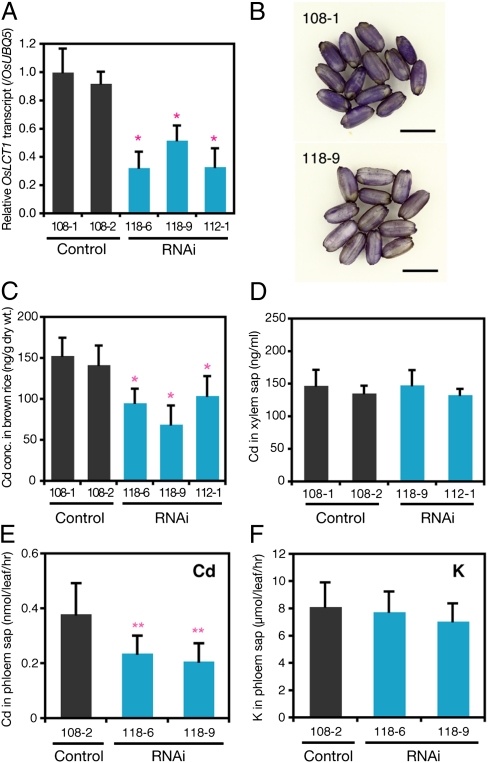
To examine the role of OsLCT1 in rice Cd accumulation, we generated RNAi-mediated OsLCT1 knockdown rice plants using a japonica cultivar, Nipponbare. OsLCT1 mRNA level in rice plants transformed with the RNAi vector was ∼30–50% of that in vector control plants (Fig. 3A). Plants were then grown until grain ripening in soil containing 0.198 mg of Cd/kg dry weight (Table S1), which is comparable to the average Cd concentration in Japanese soils (30). Intermittent flooding was introduced throughout cultivation to enhance Cd availability in the soil and promote Cd accumulation in the grains (12). Grains were harvested from ripened plants, and histochemical Cd staining with 2-(8-quinolylazo)-4,5-diphenylimidazole (QAI) was conducted on the brown rice. QAI can specifically detect Cd in the tissue, in combination with reagents used to mask other heavy metals such as Zn and Cu (31). Less staining was observed in brown rice from the knockdown plants than in those from the control plants (Fig. 3B), suggesting less grain Cd accumulation in the knockdown line. For confirmation, the Cd content of the grains was quantified by inductively coupled plasma mass spectrometry (ICP-MS). The grains of the control plants contained nearly 0.15 μg of Cd/g dry weight (Fig. 3C), which is greater than the average Cd concentration in brown rice produced in Japan (0.04–0.07 μg/g) (32), suggesting that our cultivation condition reflected those that produce moderately Cd-polluted rice. Notably, under this condition, the OsLCT1 knockdown plants showed up to 50% less Cd in brown rice than the control plants (Fig. 3C). These results suggest that OsLCT1 is critical for the level of grain Cd accumulation when grown in soil.
Fig. 3.
Accumulation and transport of Cd in the RNAi-mediated knockdown rice plants of OsLCT1. (A) Generation of OsLCT1 RNAi-mediated knockdown plants. The OsLCT1 transcript was quantified by real-time PCR. Total RNAs were prepared from flag leaf blades of the plants (T1). The data are presented as means ± SD (n = 3). Asterisks represent a significant difference from both control lines (P < 0.05; t test). (B and C) Cd accumulation in grains. OsLCT1 RNAi and control plants (T1) were grown in pots containing soil until grain ripening, and the harvested grains were used for histochemical Cd staining and elemental analysis. (B) Histochemical Cd staining of brown rice by QAI. Brown rice grains were stained with QAI after dehydration and masking. (Scale bars = 5 mm.) (C) Cd concentration in brown rice of control plants and OsLCT1 RNAi plants. The data are presented as means ± SD (n = 4–6). Asterisks represent a significant difference from both control lines (P < 0.05, t test). (D) Cd concentration in the xylem sap of vector control and RNAi plants. Plants (T1) were grown hydroponically for 3 wk and then exposed to 0.2 μM CdCl2 for 6 h; after which, xylem sap was collected. The data are presented as means ± SD (n = 4–7). No significant difference was observed between the control and RNAi plants (P > 0.05, t test). (E and F) Quantification of Cd (E) and K (F) in phloem exudates of vector control and RNAi plants. Plants (T1) were grown hydroponically for 2 mo with 0.2 μM CdCl2; after which, phloem exudate was collected from the seventh and eighth leaf blades. The data are presented as means ± SD (n = 8).
OsLCT1 Is Involved in Phloem Cd Transport.
In rice, xylem-mediated root-to-shoot Cd translocation activity (12) or phloem-mediated Cd transport (26) determine grain Cd levels. First, to examine the possibility that OsLCT1 regulates grain Cd level by affecting xylem-mediated Cd transport, we analyzed the Cd concentration in xylem sap of the vector control and RNAi plants. Compared with the control plants, the RNAi plants showed no significant difference in root-to-shoot Cd translocation mediated by xylem (Fig. 3D).
To examine the involvement of OsLCT1 in phloem Cd transport, phloem sap was collected by the EDTA method (33), and the level of Cd in the phloem sap was determined in addition to that of K (Fig. 3 E and F). K is a major cationic element in phloem sap (34). The Cd level in phloem exudates of RNAi plants was almost half that of the control plants (Fig. 3E), although the level of K in phloem exudates did not differ significantly among tested lines (Fig. 3F). These results show the involvement of OsLCT1 in phloem Cd transport rather than in xylem transport.
Suppression of OsLCT1 Expression Does Not Have Negative Effects on Growth and Grain Nutrient Accumulation in Rice Plants.
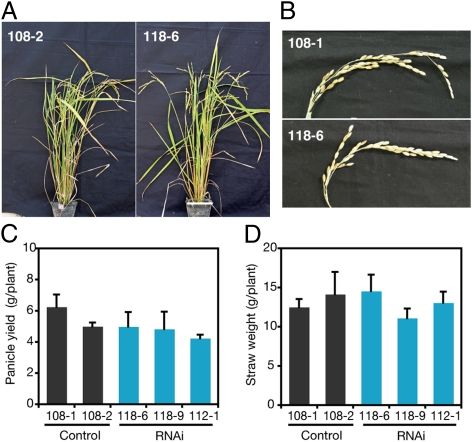
Three independent RNAi lines exhibited no apparent phenotypic difference from two vector control lines (Fig. 4 A and B). No significant or consistent difference in straw weight or grain yield was observed among the three RNAi lines and two control lines (Fig. 4 C and D). These results show that OsLCT1 suppression had a negligible effect on vegetative and reproductive growth, at least under our cultivation conditions.
Fig. 4.
Growth of OsLCT1 RNAi plants. (A and B) Phenotype of whole shoots (A) and panicles (B) of control (108-1 or 108–2) and RNAi plants (118-6). Representative images of the respective lines are shown. (C and D) Panicle yields and straw weight of the control and RNAi plants. The data are presented as means ± SD (n = 4–6). No significant difference was observed between the RNAi plants and two control lines (P > 0.05; t test).
The yeast assay demonstrated that OsLCT1 acts as an efflux transporter of Ca, Mg, K, and Mn, as well as of Cd (Fig. S2D), indicating that these nutrients may be substrates of this transporter in planta. Thus, suppression of OsLCT1 was expected to disturb nutrient accumulation and distribution. However, in contrast to the Cd results, no significant decrease occurred in the concentrations of minerals in the brown rice of these plants (Fig. S5A). Unexpectedly, the Fe content in the brown rice was significantly higher in the RNAi plants (Fig. S5A), and the Fe concentration in the leaf blades was significantly lower in the RNAi plants (Fig. S5B). Low Ca or Mg treatment, as well as exposure to high Na or Cd, did not affect OsLCT1 expression (Fig. S6). These results indicate little contribution of OsLCT1 in grain mineral accumulation except for Fe.
Discussion
In this study, we characterized OsLCT1 as a possible rice Cd transporter gene and demonstrated that OsLCT1 is localized to the plasma membrane (Fig. 1A) and can export Cd, as well as K, Ca, Mg, and Mn (Fig. 1B and Fig. S2D). Previous reports showed that wheat TaLCT1 also mediates the transport of various cations, such as Cd, Ca, Na, Rb, and K, but does not mediate that of Zn (27, 35). The Cd transport activity of OsLCT1 was inhibited by high Ca or Mg levels (Fig. S2E), as with TaLCT1 (27). These results suggest that, like TaLCT1, OsLCT1 possesses cation transport activity with broad substrate specificity and is not specific to Cd transport. It is well known that Cd2+ and divalent cations (e.g., Zn2+, Fe2+, and Ca2+) are chemically analogous and that Cd2+ is transported by transporters for such ions as Zn2+ or Ca2+ (36–42). The present and previous results (27, 35) suggest that both OsLCT1 and TaLCT1 have a higher transport activity for Ca and Cd than for Zn. However, whereas wheat TaLCT1 is indicated as an influx transporter (27, 35), rice OsLCT1 appears to have efflux activity, indicating a possibility that the differentiation of rice and wheat resulted in divergent functions for OsLCT1 and TaLCT1. Overall, the present results suggest that rice LCT1 is a plasma membrane-localized cation exporter with broad substrate specificity and that OsLCT1 possesses Cd transporter activity.
In rice, xylem-mediated root-to-shoot Cd translocation activity (12) and phloem-mediated Cd transport activity into grains (26) are both known to determine grain Cd levels. OsHMA3, a rice P-type ATPase, was demonstrated to regulate xylem-mediated translocation of Cd (20, 21). However, to date, a transporter involved in phloem Cd transport has not been identified in plants. We generated RNAi-mediated OsLCT1 knockdown plants (Fig. 3A) and showed that OsLCT1 is involved in phloem Cd transport rather than in xylem Cd transport (Fig. 3 D and E). These results suggest that OsLCT1 is a transporter regulating phloem Cd transport and that it, thereby, possibly regulates Cd deposition into grains. Significant reduction of grain Cd levels in RNAi plants (Fig. 3 B and C) strongly supports this hypothesis.
Previous studies on Cd behavior and the structure of nodal vascular bundles in rice have suggested a pathway for Cd transport from xylem to phloem. Cd is translocated from roots to shoots by the xylem (12) and after xylem-to-phloem transfer at nodes, Cd is preferentially transported to the upper nodes and finally into a panicle rather than into leaves (22). Large vascular bundles mediate major xylem transport from roots to leaf blades, whereas diffuse vascular bundles function in transporting nutrients into the upper nodes (25, 43). These suggest intervascular Cd transfer from enlarged large vascular bundles to diffuse vascular bundles in nodes. In the case of node I, the uppermost node, large vascular bundles connect to the flag leaf and diffuse vascular bundles connect to the panicle (25, 43). Therefore, intervascular Cd transfer at node I appears to be critical for grain Cd accumulation. The fact that our OsLCT1-RNAi plants accumulated less Cd in grains suggests that OsLCT1 plays a role in this intervascular Cd transport.
Cell type specificity of OsLCT1 transcript accumulation further supports this conclusion. The several layers of cells between the enlarged large vascular bundles and diffuse vascular bundles are termed the parenchyma cell bridge, which mediates intervascular solute transport. OsLCT1 is expressed in cells surrounding enlarged large vascular bundles (Fig. 2E). These cells appear to be part of the parenchyma cell bridge. A silicon influx transporter in rice, Lsi6, is also localized to the outer boundary region of enlarged large vascular bundles and mediates the first step in silicon transport across the parenchyma cell bridge (25). This transport process mediated by Lsi6 in node I is essential for silicon accumulation in panicles (25). OsLCT1 expressed around enlarged large vascular bundles may have a similar role in intervascular Cd transfer as Lsi6 has for silicon. Moreover, stronger expression of OsLCT1 was observed in the diffuse vascular bundles (Fig. 2E). Diffuse vascular bundles contain large numbers of phloem parenchyma cells surrounded by numerous phloem sieve elements, and these tissues have been suggested to mediate active phloem transport (44, 45). In particular, in node I, diffuse vascular bundles connect to the panicle, and, thus, Cd transfer into diffuse vascular bundles in node I is likely to be crucial for Cd deposition into grains. Strong expression of OsLCT1 in the diffuse vascular bundles of node I suggests that OsLCT1 expression in this region is important for Cd transport into grains.
Temporal patterns of OsLCT1 expression also support the function of OsLCT1 in Cd transport into grains. Expression of OsLCT1 in nodes during the grain-ripening stage is >100-fold higher than in nodes at the heading stage (Fig. 2A). Considering the fact that the grain ripening stage is a critical period for grain Cd accumulation (46), this further suggests the significance of OsLCT1 in Cd transport into grains.
In contrast to the clear involvement of OsLCT1 in Cd transport in rice plants, its involvement in transport of other metals is not clear, because RNAi plants did not show significant reduction in mineral nutrient contents in grains (Fig. S5A). However, the transport activity for such elements as Ca and Mg, as well as for Cd, was observed in yeast cells expressing OsLCT1 (Fig. S2D), leaving the possibility that OsLCT1 mediates transport of these minerals in planta.
Conclusions
In the present study, we demonstrated that OsLCT1 plays an important role in Cd loading to the phloem and grain Cd accumulation. A rice Cd transporter regulating xylem-mediated translocation of Cd has already been identified and explains the high Cd accumulation in some high Cd-accumulating cultivars (20, 21). This study demonstrates a transporter gene that is crucial for phloem Cd transport and grain Cd accumulation in a standard japonica cultivar. The manipulation of mineral transporters often causes adverse effects on plant growth and nutrient accumulation (39, 41, 47). However, RNAi-mediated OsLCT1 knockdown plants showed a substantial reduction in the Cd content in brown rice, without reduction in mineral nutrients or defects in growth or grain yield under the relatively lower soil Cd condition (Fig. 4 and Fig. S5A). These results indicate the significant potential of OsLCT1 alleles for the practical engineering or breeding of low-Cd rice plants. Considering that ∼40% of the Cd intake among Japanese people originates from rice (4), a 50% reduction of the Cd content in rice by down-regulating OsLCT1 expression will have a major impact on human Cd intake. The success of this approach with rice may be extended to other staple crops, such as wheat and maize.
Materials and Methods
Plant Materials.
Rice (Oryza sativa L., cv. Nipponbare) was used for the cloning of OsLCT1, expression analysis, and transformation. For in situ hybridization, the japonica rice cultivar Shiokari was used.
Subcellular Localization Analysis.
The transformation of cultured tobacco BY2 cells was conducted as reported previously (48) with modifications. The cells transformed with pSU40 (35S-LCT1-sGFP) or pTS100 (35S-sGFP) were stained with 25 μM FM4-64 just before imaging and were observed using an FV1000 laser-scanning confocal microscope (Olympus).
Yeast Cd Transport Activity Assay.
Saccharomyces cerevisiae strain WΔycf1 was used for the yeast assay. WΔycf1 was transformed with pESC-URA (Agilent Technologies) or pSU28 (pESC-URA carrying the OsLCT1 ORF with some modifications in the codon use). The obtained transformants were subjected to Cd transport-activity assay according to previous reports (49, 50). After three washes with ice-cold 2 mM CaCl2, the element concentrations in the acid-digested cells were determined by ICP-MS (model SPQ9700; SII NanoTechnology, Seiko).
Real-Time PCR.
Total RNAs were extracted from various tissues of rice grown in a pot in a greenhouse. cDNA synthesis and real-time PCR were conducted as described previously (24). The primers used for the quantification were 5′-GAGTTCTTCGTCAGAGCTAC-3′ and 5′-CAGTGCTGGATGACGAATTG-3′ for OsLCT1 and 5′-GAAGGAGGAGGAAATCGAAC-3′ and 5′-CTTCACAGAGGTGATGCTAAGG-3′ for OsUBQ5.
In situ Hybridization.
The ORF of OsLCT1 was amplified by PCR and cloned into pST19 (Roche Diagnostics). The obtained plasmid was used to prepare digoxygenin-labeled antisense probe after linearization. In situ hybridization and immunological detection of the signals were carried out according to a previous report (51). The sense probe was also prepared and used as a negative control. No visible signal was observed in the sense probe-treated samples (Fig. S4A).
Generation of OsLCT1 Knockdown Plants.
pIG121-RNAi-DEST (52) (vector control) and pSU19 (pIG121-RNAi-DEST carrying a 527-bp fragment of the N-terminal region of the OsLCT1 ORF) for RNAi were introduced separately into Agrobacterium tumefaciens strain EHA101 (Rifr) pEHA101 (Kmr). Rice plants were transformed according to a previous report (53). The transcription of OsLCT1 in flag leaf blades of each line (T0 and T1 generations) was evaluated by real-time PCR with OsUBQ5 as a control. The primers used for the quantification of OsLCT1 and OsUBQ5 are described above.
Soil Culture Conditions and Elemental Analysis.
A commercial soil for rice cultivation (Kumiai Chemical Industry Co.) was used in our soil culture experiment. The soil Cd, Fe, Mn, Cu, and Zn concentrations were determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES) (Varian) after extraction with 0.1 M HCl (12).
For the growth experiments, two independent vector control lines (108-1 and 108–2; T1 generation) and three independent RNAi lines (118-6, 118-9, and 112–1; T1 generation) were used. After confirming the T-DNA insertion by PCR, seedlings were transplanted into plastic pots filled with 0.4 kg of soil in a greenhouse under natural lighting conditions. Four grams of fertilizer (MagAmp K; Hyponex) were applied to each pot as a top dressing. Intermittent irrigation was conducted throughout the cultivation (12). Plants were harvested after grain ripening. The concentrations of heavy metals in the plant tissues were evaluated by ICP-MS (model SPQ9700; SII NanoTechnology, Seiko) after digestion with nitric acid and hydrogen peroxide (12).
Histochemical Cd Staining.
Histochemical staining of Cd was performed according to a previous method (31) with a slight modification using QAI. Grains of vector control (108-1) and RNAi (118-9) plants harvested from pot experiments were subjected to the staining.
Xylem and Phloem Sap Analyses.
Plant culture, Cd exposure, and xylem sap collection were carried out according to a previous report (12, 54). Phloem sap collection was conducted according to the EDTA method (33). Determination of element concentration in the sap was conducted by ICP-MS.
For further details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge S. Clemens for technical advice on the yeast assay, N. K. Nishizawa and H. Nakanishi for pIG121Hm-RNAi-DEST, T. Nakagawa for pGWB505, Y. Sumi for QAI, S. Arimura and Y. Hayashi for BY-2, and K. Aizawa for technical assistance. We also thank N. Suzui and S. Fujimaki for helpful suggestions and T. Hakoyama for constructive comments on the manuscript. This work was supported, in part, by grants from the Japanese Society for the Promotion of Science to S.U. (22-8989) and from the Ministry of Agriculture, Forestry, and Fisheries of Japan to T.F. (Genomics for Agricultural Innovation Grant IPG-0005) and Y.N. (Genomics for Agricultural Innovation Grant RTR-0002).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116531109/-/DCSupplemental.
References
- 1.Friberg L, Piscator M, Nordberg GF, Kjellstörm T. Cadmium in the Environment. 2nd Ed. Cleveland, OH: CRC Press Inc.; 1974. [Google Scholar]
- 2.Yamagata N, Shigematsu I. Cadmium pollution in perspective. Koshu Eisei In Kenkyu Hokoku. 1970;19:1–27. [Google Scholar]
- 3.Horiguchi H, et al. Hypoproduction of erythropoietin contributes to anemia in chronic cadmium intoxication: Clinical study on Itai-itai disease in Japan. Arch Toxicol. 1994;68:632–636. doi: 10.1007/BF03208342. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T, et al. Cadmium exposure of women in general populations in Japan during 1991-1997 compared with 1977-1981. Int Arch Occup Environ Health. 2000;73:26–34. doi: 10.1007/pl00007934. [DOI] [PubMed] [Google Scholar]
- 5.Järup L, et al. Low level exposure to cadmium and early kidney damage: The OSCAR study. Occup Environ Med. 2000;57:668–672. doi: 10.1136/oem.57.10.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas LDK, Hodgson S, Nieuwenhuijsen M, Jarup L. Early kidney damage in a population exposed to cadmium and other heavy metals. Environ Health Perspect. 2009;117:181–184. doi: 10.1289/ehp.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, Magnus JH, Hentz JG. Urinary cadmium, osteopenia, and osteoporosis in the US population. Osteoporos Int. 2010;21:1449–1454. doi: 10.1007/s00198-009-1111-y. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher CM, Kovach JS, Meliker JR. Urinary cadmium and osteoporosis in U.S. Women >or= 50 years of age: NHANES 1988-1994 and 1999-2004. Environ Health Perspect. 2008;116:1338–1343. doi: 10.1289/ehp.11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Järup L, Akesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol. 2009;238:201–208. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Tsukahara T, et al. Rice as the most influential source of cadmium intake among general Japanese population. Sci Total Environ. 2003;305:41–51. doi: 10.1016/S0048-9697(02)00475-8. [DOI] [PubMed] [Google Scholar]
- 11.Egan SK, Bolger PM, Carrington CD. Update of US FDA's Total Diet Study food list and diets. J Expo Sci Environ Epidemiol. 2007;17:573–582. doi: 10.1038/sj.jes.7500554. [DOI] [PubMed] [Google Scholar]
- 12.Uraguchi S, et al. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot. 2009;60:2677–2688. doi: 10.1093/jxb/erp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arao T, Ae N. Genotypic variations in cadmium levels of rice grain. Soil Sci Plant Nutr. 2003;49:473–479. [Google Scholar]
- 14.Clemens S, Palmgren MG, Krämer U. A long way ahead: Understanding and engineering plant metal accumulation. Trends Plant Sci. 2002;7:309–315. doi: 10.1016/s1360-1385(02)02295-1. [DOI] [PubMed] [Google Scholar]
- 15.Ueno D, et al. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa) New Phytol. 2009;182:644–653. doi: 10.1111/j.1469-8137.2009.02784.x. [DOI] [PubMed] [Google Scholar]
- 16.Ueno D, et al. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant Cell Physiol. 2009;50:2223–2233. doi: 10.1093/pcp/pcp160. [DOI] [PubMed] [Google Scholar]
- 17.Tezuka K, et al. A single recessive gene controls cadmium translocation in the cadmium hyperaccumulating rice cultivar Cho-Ko-Koku. Theor Appl Genet. 2010;120:1175–1182. doi: 10.1007/s00122-009-1244-6. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa S, Ae N, Yano M. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa) New Phytol. 2005;168:345–350. doi: 10.1111/j.1469-8137.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa S, et al. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7. J Exp Bot. 2010;61:923–934. doi: 10.1093/jxb/erp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno D, et al. Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA. 2010;107:16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyadate H, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189:190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 22.Fujimaki S, et al. Tracing cadmium from culture to spikelet: Noninvasive imaging and quantitative characterization of absorption, transport, and accumulation of cadmium in an intact rice plant. Plant Physiol. 2010;152:1796–1806. doi: 10.1104/pp.109.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka K, Fujimaki S, Fujiwara T, Yoneyama T, Hayashi H. Quantitative estimation of the contribution of the phloem in cadmium transport to grains in rice plants (Oryza sativa L.) Soil Sci Plant Nutr. 2007;53:72–77. [Google Scholar]
- 24.Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell. 2008;20:2860–2875. doi: 10.1105/tpc.108.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaji N, Ma JF. A transporter at the node responsible for intervascular transfer of silicon in rice. Plant Cell. 2009;21:2878–2883. doi: 10.1105/tpc.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M, et al. Possible chemical forms of cadmium and varietal differences in cadmium concentrations in the phloem sap of rice plants (Oryza sativa L.) Soil Sci Plant Nutr. 2010;56:839–847. [Google Scholar]
- 27.Clemens S, Antosiewicz DM, Ward JM, Schachtman DP, Schroeder JI. The plant cDNA LCT1 mediates the uptake of calcium and cadmium in yeast. Proc Natl Acad Sci USA. 1998;95:12043–12048. doi: 10.1073/pnas.95.20.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li ZS, et al. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato Y, et al. RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda A, Kimura K, Yamasaki S. Analysis of 57 elements in Japanese soils, with special reference to soil group and agricultural use. Geoderma. 2004;119:291–307. [Google Scholar]
- 31.Sumi Y, Itoh MT, Muraki T, Suzuki T. Histochemical staining of cadmium with 2-(8-quinolylazo)-4,5-diphenylimidazole. Histochem Cell Biol. 1996;106:223–227. doi: 10.1007/BF02484404. [DOI] [PubMed] [Google Scholar]
- 32.Shimbo S, et al. Cadmium and lead contents in rice and other cereal products in Japan in 1998-2000. Sci Total Environ. 2001;281:165–175. doi: 10.1016/s0048-9697(01)00844-0. [DOI] [PubMed] [Google Scholar]
- 33.King RW, Zeevaart JA. Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol. 1974;53:96–103. doi: 10.1104/pp.53.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giaquinta RT. Phloem loading of sucrose. Annu Rev Plant Physiol Plant Mol Biol. 1983;34:347–387. [Google Scholar]
- 35.Schachtman DP, Kumar R, Schroeder JI, Marsh EL. Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc Natl Acad Sci USA. 1997;94:11079–11084. doi: 10.1073/pnas.94.20.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vert G, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. Expression of arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 2000;124:125–133. doi: 10.1104/pp.124.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CKE, Cobbett CS. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009;181:71–78. doi: 10.1111/j.1469-8137.2008.02638.x. [DOI] [PubMed] [Google Scholar]
- 40.Verret F, et al. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Hussain D, et al. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanikenne M, et al. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- 43.Kawahara H, Chonan N, Matsuda T. Studies on morphogenesis in rice plants 7. The morphology of vascular bundles in the vegetative nodes of the culm. Jpn J Crop Sci. 1974;43:389–401. [Google Scholar]
- 44.Zee SY. Transfer cells and vascular tissue distribution in vegetative nodes of rice. Aust J Bot. 1972;20:41–48. [Google Scholar]
- 45.Chonan N, Kawahara H, Matsuda T. Ultrastructure of elliptical and diffuse bundles in the vegetative nodes of rice. Jpn J Crop Sci. 1985;54:393–402. [Google Scholar]
- 46.Arao T, Kawasaki A, Baba K, Mori S, Matsumoto S. Effects of water management on cadmium and arsenic accumulation and dimethylarsinic acid concentrations in Japanese rice. Environ Sci Technol. 2009;43:9361–9367. doi: 10.1021/es9022738. [DOI] [PubMed] [Google Scholar]
- 47.Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 48.Mayo KJ, Gonzales BJ, Mason HS. Genetic transformation of tobacco NT1 cells with Agrobacterium tumefaciens. Nat Protoc. 2006;1:1105–1111. doi: 10.1038/nprot.2006.176. [DOI] [PubMed] [Google Scholar]
- 49.Kiyono M, et al. Engineering expression of the heavy metal transporter MerC in Saccharomyces cerevisiae for increased cadmium accumulation. Appl Microbiol Biotechnol. 2010;86:753–759. doi: 10.1007/s00253-009-2402-0. [DOI] [PubMed] [Google Scholar]
- 50.Papoyan A, Kochian LV. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004;136:3814–3823. doi: 10.1104/pp.104.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kyozuka J, et al. LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci USA. 2003;94:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogo Y, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- 53.Toki S, et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 54.Uraguchi S, Fujiwara T. Significant contribution of boron stored in seeds to initial growth of rice seedlings. Plant Soil. 2011;340:435–442. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.