Abstract
Tectonic processes drive megacycles of atmospheric carbon dioxide (CO2) concentration, ca, that force large fluctuations in global climate. With a period of several hundred million years, these megacycles have been linked to the evolution of vascular plants, but adaptation at the subcellular scale has been difficult to determine because fossils typically do not preserve this information. Here we show, after accounting for evolutionary relatedness using phylogenetic comparative methods, that plant nuclear genome size (measured as the haploid DNA amount) and the size of stomatal guard cells are correlated across a broad taxonomic range of extant species. This phylogenetic regression was used to estimate the mean genome size of fossil plants from the size of fossil stomata. For the last 400 Myr, spanning almost the full evolutionary history of vascular plants, we found a significant correlation between fossil plant genome size and ca, modelled independently using geochemical data. The correlation is consistent with selection for stomatal size and genome size by ca as plants adapted towards optimal leaf gas exchange under a changing CO2 regime. Our findings point to the possibility that major episodes of change in ca throughout Earth history might have selected for changes in genome size, influencing plant diversification.
Keywords: C-value, 1C DNA amount, plant evolution, phenotypic plasticity, evolution of correlated characters, correlated traits
1. Introduction
Wilson cycles (‘megacycles’) describe continental break-up, dispersal and reassembly occurring with a period of several hundred million years [1,2]. Volcanic carbon dioxide (CO2) outgassing during continental fragmentation is proposed to create greenhouse climates by increasing atmospheric CO2 concentration, ca, while CO2 drawdown owing to the predominance of weathering processes switches the planet to an icehouse mode (figure 1) [3,4]. Over the Phanaerozoic period (past 540 Myr), continental movements and greenhouse–icehouse cycles have influenced the evolution and adaptation of marine [5] and terrestrial primary producers [6–10]. On land, they have been linked through morphological information in the fossil record to plant evolution and speciation [11], transforming the landscape and its biota [12,13]. If ca has influenced plant evolution, then adaptation and selection may have acted on subcellular characters also, but this information is not readily preserved in the fossil record. Consequently, there has been little evidence of a linkage between the large geological-scale changes in ca and changes in subcellular plant characters over geological time. Here, we show that tectonically driven megacycles of ca may have selected for changes in plant genome size (the haploid nuclear DNA amount or C-value)—a fundamental subcellular feature of land plant evolution.
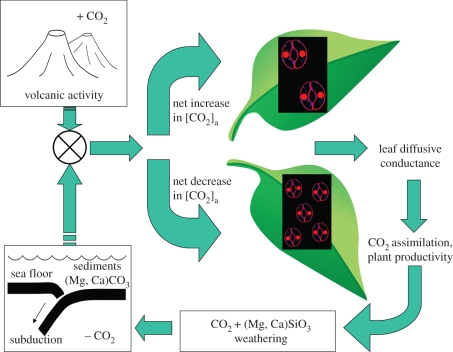
Figure 1.
Schematic diagram of coupling between leaf CO2 assimilation and the geochemical carbon cycle. Atmospheric CO2 concentration, ca, on geological timescales is determined by the balance between major sources (volcanism) and sinks (deep ocean sedimentation). The fossil record suggests that leaves have adapted to megacycles in atmospheric CO2 concentration by adjusting leaf diffusive conductance to CO2, achieved through changing the size and density (number) of stomatal guard cells on the leaf surface. Stomata on the leaf surface are represented as guard cell pairs in outline containing circular red nuclei.
(a). Atmospheric carbon dioxide and stomata
There is coupling between ca and the physiology of plant gas exchange, illustrated in figure 1a. Atmospheric CO2 is the substrate for photosynthesis, and its concentration on geological timescales is strongly influenced by major geological sources and sinks, including the action of plant photosynthetic productivity on weathering of Ca–Mg silicate rocks [12]. In geological intervals when ca was low (as inferred from geochemical data [4]), fossil leaves exhibit more numerous, smaller stomata, and the reverse pattern is seen in periods of high ca [14]. These changes in stomatal size and number (density) have unambiguous functional consequences for leaves owing to the basic physics of diffusion: higher average maximum leaf diffusive conductance to CO2 under low ca, and lower maximum diffusive conductance under higher ca [14]. Although played out at the global scale and over geological time, the pattern mimics the well-characterized feedback interaction between ca and leaf diffusive conductance observed routinely in the laboratory [15–18], albeit through more subtle adjustments of stomatal aperture rather than stomatal size and density. At the geological and experimental scale, both phenomena exhibit characteristics of an adaptive response that tends towards (though not necessarily achieves) optimization of leaf gas exchange [16,19]. We cannot be certain that the apparent feedback response of stomata to ca at the geological scale is indeed an adaptive feedback response, or a fortuitous coevolution, because we are unable to physically duplicate the past 400 Myr of the Earth system under controlled conditions. But the pattern, suggesting adaptation of stomatal size and density to ca, can be simulated mathematically with a feedback model built on established physiological theory that minimizes the perturbation incurred on CO2 assimilation rate by a change in ca [20]. Furthermore, the geological-scale pattern is mirrored in growth chamber experiments, with plants grown under different ca treatments showing in many cases a qualitatively similar change in stomatal size and density [21–23]. For the purpose of our investigation, we therefore began with the premise that there is a strong functional connection between ca and stomatal dimensions, i.e. plants exhibit a predictable pattern of phenotypic plasticity in stomatal size and density in response to a change in ca.
(b). Genome size and stomata
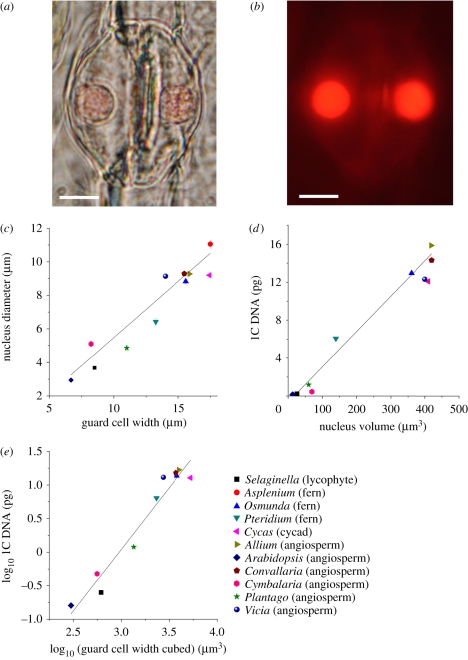
The ratio of guard cell to nucleus width is relatively constant for taxa from across the phylogenetic spectrum (figure 2a–c). Assuming that nuclear DNA amount is proportional to the volume of the nucleus [24,25] and that stomatal guard cells are typically diploid [26], the size of fossil guard cells is potentially a proxy for estimating nuclear DNA amounts of fossil species. This principle has been applied in relation to the study of ploidy in fossil angiosperms [27], but here we develop the methodology further by accounting for phylogenetic effects to investigate the relationship between plant nuclear genome size and ca. Across a diverse group of 11 extant plant genera, we observed a strong positive correlation between nuclear genome size (1C DNA amount), nucleus volume and a simple volumetric measure of guard cell size, guard cell width cubed (figure 2d,e). This strong geometric scaling between genome size and guard cell size in land plants, similar to that observed between genome size and guard cell length in a large group of angiosperms [28], indicates that guard cell size can act as a proxy for genome size in fossil plants.
Figure 2.
Guard cell size, nucleus size and genome size are strongly correlated across species. (a) Two guard cells forming a single stoma in the epidermis of Allium cepa. (b) Fluorescing guard cell nuclei from (a) after preparation with a DNA fluorophore. Scale bar in (a) and (b): 10 µm. (c–e) Linear relationship between (c) guard cell nucleus diameter versus guard cell width, (d) genome size, as 1C DNA amount, versus guard cell nucleus volume and (e) log10 1C DNA versus log10 guard cell width cubed across phylogenetically diverse species. (d,e) Without Asplenium because its 1C DNA amount is unknown. Linear regressions (solid lines): (c) y = 0.67x − 1.24, r2 = 0.93, p < 0.001; (d) y = 0.037 − 0.65, r2 = 0.96, p < 0.001; (e) y = 1.83x − 5.46, r2 = 0.94, p < 0.001.
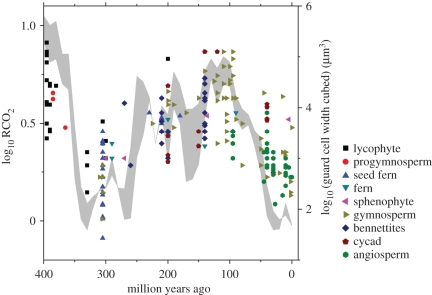
Guard cell size in 211 fossil plant species spanning the last 400 Myr shows a clear pattern of covariance with large, long-term changes in ca calculated from a coupled carbon–sulphur cycle geochemical model [4] (figure 3). Assuming that guard cell size and genome size are correlated (figure 2e), the relationship in figure 3 suggests that 1C DNA and ca are correlated through the fossil record. This hypothesis was evaluated by reconstructing changes in ancestral C-values for long-extinct plant lineages and testing for correlation between fossil plant C-values and ca over the last 400 Myr. Our approach uses evolutionary comparative analyses of the relationship between guard cell size and plant nuclear genome size in a way that parallels the reconstruction of dinosaur nuclear genome sizes from fossil bone cell sizes [29]. The method enables a regression model to be fitted to comparative data that are inherently non-independent owing to evolutionary relatedness [30]. Our goal is not to identify, nor does it require, a mechanism for change in or evolution of plant genome size. However, an understanding of the selective forces involved requires a model that tells us what to expect from a given set of assumptions [31], so to this end, we describe a simple model to accompany our findings.
Figure 3.
Covariation of relative atmospheric CO2 concentration (RCO2) and fossil guard cell size (as guard cell width cubed) over the last 400 Myr. The shaded envelope represents uncertainties owing to the weathering rates of basalt rocks. Each point is an individual fossil species, listed in the electronic supplementary material, table A1. RCO2 data are from Berner [4], where RCO2 is ca relative to the pre-industrial low of approximately 280 ppm.
2. Materials and methods
(a). Observation of guard cell nuclei
Epidermal peels were bathed for 10 min in LB01 buffer containing the DNA fluorophore propidium iodide, prepared as described for suspensions of isolated nuclei for flow cytometry [32]. Peels were mounted in water on glass slides and guard cell nuclei were observed using standard fluorescence microscopy (excited with 488 nm wavelength light, and detected with a 562–588 nm band pass filter). Mean ± s.e.m. of 3–15 stomata were measured for each species. Allium cepa, Arabidopsis thaliana, Osmunda regalis, Plantago lanceolata, Selaginella uncinata and Vicia faba were grown in controlled environment cabinets under typical natural conditions: 1000 µmol m−2 s−1 photosynthetically active radiation, ambient CO2 concentration, 10 h photoperiod, 25°C, well-watered, commercial compost soil. Asplenium sp., Convallaria majalis, Cycas revoluta, Cymbalaria muralis and Pteridium aquilinum were collected from urban gardens in Sheffield. The 1C DNA amount for C. muralis and S. uncinata was measured as described earlier [32] using LB01 buffer and propidium iodide stain; Solanum lycopersicum ‘Gardner's Delight’ was the standard for C. muralis [33], and Oryza sativa cv. IR36 was the standard for S. uncinata [34]. All other 1C values are from the plant DNA C-values database (http://data.kew.org/cvalues/).
(b). Phylogenetic comparative method
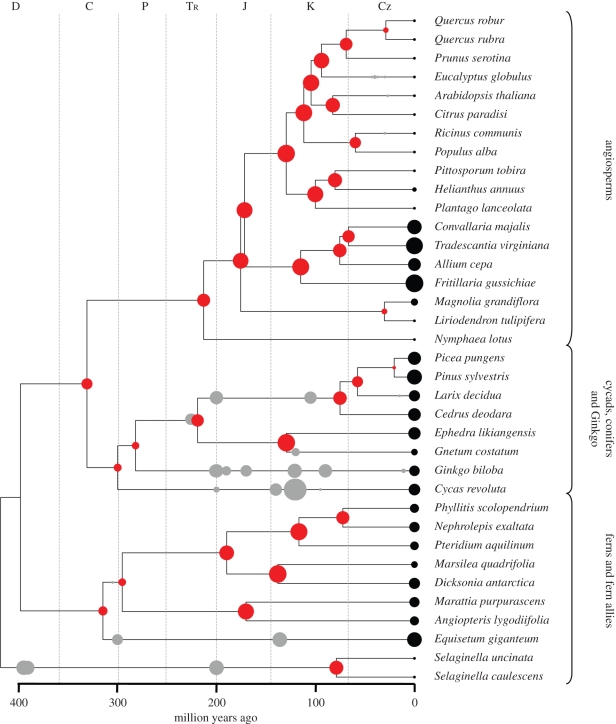
We first constructed a chronogram (figure 4) for 36 extant species of broad phylogenetic diversity (electronic supplementary material, table A2) using sequences of atpB, rbcL and 18S rDNA deposited in GenBank, which are known to produce a phylogenetic tree that adequately represents the current hypothesis of relationships among the major land plant groups [35,36]. We carried out a simultaneous divergence-time and phylogenetic analysis using Markov chain Monte Carlo methods (MCMC) implemented in BEAST (v. 1.5.4; [37,38]). BEAST employs an uncorrelated relaxed clock model that draws substitution rates from a lognormal distribution and estimates divergence times from fossil calibrations that are treated as probabilistic priors. We attached a lognormal prior probability to several minimum-age estimates representing the first fossil occurrences of several of the major land plant groups (i.e. Tracheophyta, Euphyllophyta, Acrogymnospermae, Eudicotyledonae etc.; see Smith et al. [39] for details regarding the fossils used in this step). We also imposed several topological constraints to ensure that the topology matched the current hypothesis for relationships among land plants. The most notable constraint forced the monophyly of the Monocotyledonae and Eudicotyledonae to reflect recent genome-scale chloroplast analyses that have indicated some support for this relationship [40,41]. We ran four independent MCMC runs of 10 million generations each, sampling every 1000 generations and using an unpartitioned GTR + Γ substitution model. The first 2 500 000 generations were discarded as burn-in. To improve the certainty of dated nodes in the phylogenetic tree, we used the age of 30 fossil plant clades, spanning 400 Myr, which are linked to known stem lineages of the 36 extant species (see electronic supplementary material, table A3 for a list of the 30 fossil clades used in this step). The 30 fossil clades were assumed to lie on or close to the ancestral branch (the grey points in figure 4). We checked this assumption by moving the points backwards in time (i.e. simulating earlier branching), however the results were not materially affected. The 30 fossil clades (electronic supplementary material, table A3; grey points in figure 4) were assumed to be direct ancestors on the lineages leading to extant species (black dots on the phylogenetic tree; figure 4). The alternative was to assume an earlier splitting of the fossils from these lineages. However, there was no obvious algorithm for doing this and, given the broad and coarse resolution of the phylogeny, our approach is pragmatic.
Figure 4.
Timing of key changes in ancestral plant genome size. The dated phylogenetic tree, spanning major land plant clades, shows trends in genome size evolution reconstructed using information from fossil and extant stomatal guard cells. Relative C-value is indicated by the size of the dots (red, reconstructed ancestral node; black, extant, listed in the electronic supplementary material, table A2; grey, dated fossil clade, listed in the electronic supplementary material, table A3).
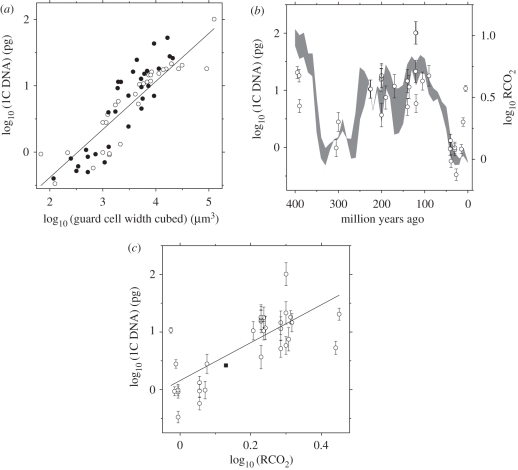
We then fitted a regression model, incorporating phylogeny, to 1C DNA amount versus guard cell width cubed for the 36 extant species in the chronogram (black symbols in figure 5a; data listed in electronic supplementary material, table A2) using standard methods, with the λ-statistic [43] being used to account for phylogenetic dependence. The phylogenetic regression (line in figure 5a) predicts C-value from guard cell size and explains 67 per cent of the variation in extant species. This regression was used to estimate the mean 1C-values of the 30 fossil plant clades spanning 400 Myr from their mean guard cell width cubed (figure 5a, open symbols; fossil plant clades are listed in electronic supplementary material, table A3). To account for phylogenetic non-independence, the inferred 1C DNA amount included phylogenetic covariance using the methods described earlier [30,43,44]. The λ-statistic was not significantly different from 1, indicating a strong phylogenetic signal in the residuals of the regression.
Figure 5.
Nuclear genome size of fossil plants correlates with atmospheric CO2 concentration over 400 Myr. (a) The positive relationship between genome size (as 1C DNA amount) and stomatal size (as guard cell width cubed). Black symbols are data from direct measurements on the 36 extant species in figure 4 (species listed in electronic supplementary material, table A2). The solid line is the phylogenetic regression through the extant data used to predict fossil genome size (y = 0.70x − 1.8; r2 = 0.67, p = 8.1 × 10−10). Open symbols (n = 30) are the genome size (1C DNA amount) of extinct fossil clades (electronic supplementary material, table A3) predicted from fossil guard cell size using the phylogenetic regression and phylogenetically based statistical methods (s.e.m. omitted for clarity, but see (b,c)). (b) Covariation of fossil genome size (open symbols in (a)) and RCO2 through geological time (RCO2 is ca relative to the pre-industrial low, taken as approx. 280 ppm [4,42]). Each point is the mean ± s.e.m. for a clade of extinct fossil plants. (c) Correlation between fossil genome size (open symbols in (b)) and RCO2 over 400 Myr (solid line, y = 3.3x + 0.16; r2 = 0.52, p = 2.1 × 10−6). Included for illustration (black symbol) is the mean ± s.e.m. 1C DNA amount for approximately 6700 extant species in the plant DNA C-values database (http://data.kew.org/cvalues/).
3. Results and discussion
Our reconstructions of genome size for key clades of vascular land plants reveal a significant correlation between the inferred 1C DNA amount and ca over 400 Myr (figure 5b,c). In order to confirm that phylogenetic uncertainty was not an influence on the outcome of our analyses, we repeated the entire comparative analysis for 1000 phylogenetic trees randomly selected from the posterior distribution. The 95% confidence interval for the slope of the regression in figure 5a was 0.63–0.91, and for the slope of the relationship in figure 5c it was 1.56–2.20. The results are thus highly robust to phylogenetic uncertainty.
We interpret the pattern in figures 3–5 as the result of natural selection on correlated characters [45]. To clarify our approach, we distinguish between natural selection and evolutionary response [45]: phenotypic selection occurs regardless of genetic basis, but evolutionary response to selection comprises genetic change. Though not without its criticisms [46] (see also the historical discussion by Mayr [47]), we apply the assumption that adaptation can be explained by natural selection. In this sense, we are viewing plant nuclear genome size as the phenotype [48] and therefore assume that it exhibits variation and adaptation upon which natural selection can act [31,45,49,50]. Our goal in this discussion is only to consider how ca, as a global environmental factor, might select for stomatal size and genome size as characters, and not to propose how this may ultimately have contributed to speciation through geological time.
We hypothesize that guard cell size and genome size are correlated traits that may be subject to selection by ca. To explain how ca selects for stomatal size, we propose a simple feedback model, based on the short-term stomatal feedback response to ca, whereby maximum leaf diffusive conductance adapts towards a new optimum following a sustained shift in ca. The shift in maximum leaf diffusive conductance, a function of the size and density of stomata [14], comprises a change in stomatal guard cell size. The individuals that adapt maximum leaf diffusive conductance are more competitive in the new global ca, leading to selection (differential reproductive success). The central tenet here is developmental plasticity: the capacity to reorganize the phenotype in response to environmental stimuli, producing the variants upon which natural selection can act [51].
Selection for stomatal size is accompanied by a correlated change in genome size (as guard cell nucleus size), but unlike the physiological foundations for predicting the response of stomatal size to ca, we have no a priori physiological model that explains the change in genome size. The correlation between the size of guard cells and their nuclear genomes (figures 2c,e and 5a) is likely to be related to a general phenomenon of the same form, i.e. the correlation between cell size (or more precisely, cell cytoplasmic volume) and genome size. A considerable body of work has focused on this fundamental general relationship (see review and analysis in Cavalier-Smith [52]). However, as with evolutionary theory [47], the field of genomics and genome evolution, within which the explanation to this puzzle lies, has been deeply divided along theoretical grounds [52]. We will therefore not attempt to offer a subcellular mechanism for the correlation between guard cell size and genome size, except to say that it is consistent with at least one theory, the skeletal DNA theory [52]. This states that a change in cell cytoplasmic volume is accompanied by a change in nucleus volume in order to balance the rate of protein synthesis (cytoplasm-based) with the rate of RNA synthesis and processing (nucleus-based) [52].
We recognize that no single environmental factor like atmospheric CO2 can entirely explain variations in genome size, and that other interactions between plants and their environment are likely to have contributed, including ecological and life-history traits [53]. Nevertheless, over the 400 Myr encompassed by the fossils, our results suggest that plant C-value has covaried with changes in ca (figure 5c). This could be linked to selection for altered guard cell size and leaf diffusive conductance by large, long-term changes in ca [14] (figure 1). Our fossil data and the ca model of Berner [4] limit the analysis to a temporal resolution of no less than 10 Myr and encompass changes in ca of several hundred parts per million in amplitude, but the same process may operate over ca cycles of smaller periodicity and amplitude. The molecular and genetic processes involved in the evolution of smaller or larger genomes are complex [54,55] and beyond the scope of this study.
4. Conclusion
We conclude that the nuclear genome sizes of fossil plant clades, derived using phylogenetic comparative methods, correlate with tectonically induced megacycles in ca, modelled independently using geochemical data [4]. The underlying mechanism is unclear. The correlation is consistent with selection towards optimal leaf gas exchange, via guard cell size, under a changing ca regime driven by continental fragmentation and reassembly over the last 400 Myr. This is not to say that selection is favouring only a change in genome size in order to enhance stomatal size. Genome size and guard cell size may be under selection by several factors [28,56–59]. It also does not suggest that at any time plant nuclear genome size and maximum leaf diffusive conductance were optimal. The assumption of optimality is a common and misdirected criticism of the optimization interpretation [31]. Our findings and hypothesis are compatible with the observation of a broad range of genome sizes in extant flora [53]. A selective disadvantage under a new ca regime is unlikely to be lethal, and there is little evidence that ca has ever fallen below the compensation concentration, where respiration exceeds photosynthesis. Selection by ca could however affect the frequency distribution of genome size and is perhaps a contributing factor in the predominance of smaller plant genome sizes under the low-ca conditions of recent times.
Given that natural selection acts on many characters simultaneously, and phenotypic correlations between traits are ubiquitous [45], we do not presuppose that our findings explain the variation of plant genome size. However, selection acts with varying strength on different traits [60,61], and on the basis of our results, we suggest that megacycles in ca are a major selective force on the adaptation of stomata and plant genome size. Our findings, and accompanying model, point to the additional possibility that major episodes of change in ca throughout Earth history [4], linked for example to abrupt volcanism, such as the Triassic–Jurassic transition 210 Myr ago [62], or meteorite impacts at the Cretaceous–Tertiary transition, 65 Myr ago [63], might have selected for changes in genome size that promoted subsequent bursts of plant diversification. Such genomic consequences of cell size evolution may be analogous to the evolution of small bone-cells and genomes in non-avian dinosaurs that led to the origin of flight in birds [29]. Finally, we recognize that our derivation of plant genome size and explanation of the observed correlation in terms of selective forces are, in the words of Parker & Maynard Smith [31], the consequence of a given set of assumptions, some well-founded, others emergent. Herein lies the challenge.
Acknowledgements
We thank R. Berner, M. W. Chase, T. Cavalier-Smith, M. J. Donoghue and J. E. Gray for helpful comments and discussion on this work, supported by funding from the Australian Research Council and the University of Sheffield (P. J. F. and D.J.B.). D. J. B. gratefully acknowledges a Royal Society-Wolfson Research Merit Award.
References
- 1.Wilson J. T. 1966. Did the Atlantic close and then reopen? Nature 211, 676–681 10.1038/211676a0 (doi:10.1038/211676a0) [DOI] [Google Scholar]
- 2.Worsley T. R., Nance R. D., Moody J. B. 1986. Tectonic cycles and the history of the Earth's biogeochemical and paleoceanographic record. Paleoceanography 1, 233–263 10.1029/PA001i003p00233 (doi:10.1029/PA001i003p00233) [DOI] [Google Scholar]
- 3.Fischer A. G. 1986. Climatic rhythms recorded in strata. Annu. Rev. Earth Planet. Sci. 14, 351–376 10.1146/annurev.ea.14.050186.002031 (doi:10.1146/annurev.ea.14.050186.002031) [DOI] [Google Scholar]
- 4.Berner R. A. 2006. GEOCARBSULF: a combined model for Phanerozoic atmospheric O2 and CO2. Geochim. Cosmochim. Acta 70, 5653–5664 10.1016/j.gca.2005.11.032 (doi:10.1016/j.gca.2005.11.032) [DOI] [Google Scholar]
- 5.Katz M. E., Wright J. D., Miller K. G., Cramer B. S., Fennel K., Falkowski P. G. 2005. Biological overprint of the geological carbon cycle. Mar. Geol. 217, 323–338 10.1016/j.margeo.2004.08.005 (doi:10.1016/j.margeo.2004.08.005) [DOI] [Google Scholar]
- 6.Raven P. H., Axelrod D. I. 1974. Angiosperm biogeography and past continental movements. Ann. Missouri Botanic. Garden 61, 539–673 10.2307/2395021 (doi:10.2307/2395021) [DOI] [Google Scholar]
- 7.McElwain J. C., Chaloner W. G. 1995. Stomatal density and index of fossil plants track atmospheric carbon dioxide in the Palaeozoic. Ann. Bot. 76, 389–395 10.1006/anbo.1995.1112 (doi:10.1006/anbo.1995.1112) [DOI] [Google Scholar]
- 8.Beerling D. J., Osborne C. P., Chaloner W. G. 2001. Evolution of leaf-form in land plants linked to atmospheric CO2 decline in the Late Palaeozoic era. Nature 410, 352–354 10.1038/35066546 (doi:10.1038/35066546) [DOI] [PubMed] [Google Scholar]
- 9.Osborne C. P., Beerling D. J., Lomax B. H., Chaloner W. G. 2004. Biophysical constraints on the origin of leaves inferred from the fossil record. Proc. Natl Acad. Sci. USA 101, 10 360–10 362 10.1073/pnas.0402787101 (doi:10.1073/pnas.0402787101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beerling D. J. 2007. The emerald planet: how plants changed Earth's history. New York, NY: Oxford University Press [Google Scholar]
- 11.Knoll A. H., Niklas K. J., Tiffney B. H. 1979. Phanerozoic land plant diversity in North America. Science 206, 1400–1402 10.1126/science.206.4425.1400 (doi:10.1126/science.206.4425.1400) [DOI] [PubMed] [Google Scholar]
- 12.Berner R. A. 1997. The rise of plants and their effect on weathering and atmospheric CO2. Science 276, 544–546 10.1126/science.276.5312.544 (doi:10.1126/science.276.5312.544) [DOI] [Google Scholar]
- 13.Willis K. J., McElwain J. C. 2002. The evolution of plants. Oxford, UK: Oxford University Press [Google Scholar]
- 14.Franks P. J., Beerling D. J. 2009. Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc. Natl Acad. Sci. USA 106, 10 343–10 347 10.1073/pnas.0904209106 (doi:10.1073/pnas.0904209106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raschke K. 1975. Stomatal action. Ann. Rev. Plant Physiol. Plant Mol. Biol. 26, 309–340 [Google Scholar]
- 16.Farquhar G. D., Dubbe D. R., Raschke K. 1978. Gain of the feedback loop involving carbon dioxide and stomata. Theory and measurement. Plant Physiol. 62, 406–412 10.1104/pp.62.3.406 (doi:10.1104/pp.62.3.406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morison J. I. L., Gifford R. M. 1983. Stomatal sensitivity to carbon dioxide and humidity: a comparison of two C3 and two C4 grass species. Plant Physiol. 71, 789–796 10.1104/pp.71.4.789 (doi:10.1104/pp.71.4.789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morison J. I. L. 1998. Stomatal response to increased CO2 concentration. J. Exp. Bot. 49, 443–452 10.1093/jexbot/49.suppl_1.443 (doi:10.1093/jexbot/49.suppl_1.443) [DOI] [Google Scholar]
- 19.Cowan I. R. 1977. Stomatal behaviour and environment. Adv. Bot. Res. 4, 117–228 10.1016/S0065-2296(08)60370-5 (doi:10.1016/S0065-2296(08)60370-5) [DOI] [Google Scholar]
- 20.Franks P. J., Beerling D. J. 2009. CO2-forced evolution of plant gas exchange capacity and water-use efficiency over the Phanerozoic. Geobiology 7, 227–236 10.1111/j.1472-4669.2009.00193.x (doi:10.1111/j.1472-4669.2009.00193.x) [DOI] [PubMed] [Google Scholar]
- 21.Woodward F. I. 1987. Stomatal numbers are sensitive to increases in CO2 from preindustrial levels. Nature 327, 617–618 10.1038/327617a0 (doi:10.1038/327617a0) [DOI] [Google Scholar]
- 22.Farnsworth E. J., Ellison A. M., Gong W. K. 1996. Elevated CO2 alters anatomy, physiology, growth, and reproduction of red mangrove (Rhizophora mangle L.). Oecologia 108, 599–609 10.1007/BF00329032 (doi:10.1007/BF00329032) [DOI] [PubMed] [Google Scholar]
- 23.Kürschner W. A., Stulen I., Wagner F., Kuiper P. J. C. 1998. Comparison of palaeobotanical observations with experimental data on the leaf anatomy of durmast oak [Quercus petraea (Fagaceae)] in response to environmental change. Ann. Bot. 81, 657–664 10.1006/anbo.1998.0605 (doi:10.1006/anbo.1998.0605) [DOI] [Google Scholar]
- 24.Cavalier-Smith T. 1985. Cell volume and the evolution of genome size. In The evolution of genome size (ed. Cavalier-Smith T.), pp. 105–184 Chichester, UK: Wiley [Google Scholar]
- 25.Jovtchev G., Schubert V., Meister A., Barow M., Schubert I. 2006. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114, 77–82 10.1159/000091932 (doi:10.1159/000091932) [DOI] [PubMed] [Google Scholar]
- 26.Melarango J. E., Mehrotra B., Coleman A. W. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. The Plant Cell 5, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masterson J. 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424 10.1126/science.264.5157.421 (doi:10.1126/science.264.5157.421) [DOI] [PubMed] [Google Scholar]
- 28.Beaulieu J. M., Leitch I. J., Patel S., Pendharkar A., Knight C. A. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986 10.1111/j.1469-8137.2008.02528.x (doi:10.1111/j.1469-8137.2008.02528.x) [DOI] [PubMed] [Google Scholar]
- 29.Organ C. L., Shedlock A. M., Meade A., Pagel M., Edwards S. V. 2007. Origin of avian genome size and structure in non-avian dinosaurs. Nature 446, 180–184 10.1038/nature05621 (doi:10.1038/nature05621) [DOI] [PubMed] [Google Scholar]
- 30.Martins E. P., Hansen T. F. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of inter-specific data. Am. Nat. 149, 646–667 10.1086/286013 (doi:10.1086/286013) [DOI] [Google Scholar]
- 31.Parker G. A., Maynard Smith J. 1990. Optimality theory in evolutionary biology. Nature 348, 27–33 10.1038/348027a0 (doi:10.1038/348027a0) [DOI] [Google Scholar]
- 32.Dolezel J., Greilhuber J., Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protocols 2, 2233–2244 10.1038/nprot.2007.310 (doi:10.1038/nprot.2007.310) [DOI] [PubMed] [Google Scholar]
- 33.Dolezel J., Sgorbati S., Lucretti S. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 85, 625–631 10.1111/j.1399-3054.1992.tb04764.x (doi:10.1111/j.1399-3054.1992.tb04764.x) [DOI] [Google Scholar]
- 34.Bennett M. D., Leitch I. J. 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann. Bot. 95, 45–90 10.1093/aob/mci003 (doi:10.1093/aob/mci003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pryer K. M., Schneider H., Smith A. R., Cranfill R., Wolf P. G., Hunt J. S., Sipes S. D. 2001. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409, 618–622 10.1038/35054555 (doi:10.1038/35054555) [DOI] [PubMed] [Google Scholar]
- 36.Soltis P. S., Soltis D. E., Chase M. W. 1999. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404 10.1038/46528 (doi:10.1038/46528) [DOI] [PubMed] [Google Scholar]
- 37.Drummond A. J. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith S. A., Beaulieu J. M., Donoghue M. J. 2010. An uncorrelated relaxed clock analysis suggests an earlier origin for flowering plants. Proc. Natl Acad. Sci. USA 107, 5897–5902 10.1073/pnas.1001225107 (doi:10.1073/pnas.1001225107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen R. K., et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl Acad. Sci. USA 104, 19 369–19 374 10.1073/pnas.0709121104 (doi:10.1073/pnas.0709121104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore M. J., Bell C. D., Soltis P. S., Soltis D. E. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl Acad. Sci. USA 104, 19 369–19 374 10.1073/pnas.0709121104 (doi:10.1073/pnas.0709121104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etheridge D. M., Steele L. P., Langenfelds R. L., Francey R. J., Barnola J.-M., Morgan V. I. 1996. Natural and anthropogenic changes in atmospheric CO2 over the last 1000 years from air in Antarctic ice and firn. J. Geophys. Res. 101, 4115–4128 10.1029/95JD03410 (doi:10.1029/95JD03410) [DOI] [Google Scholar]
- 43.Freckleton R. P., Harvey P. H., Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of the evidence. Am. Nat. 160, 716–726 [DOI] [PubMed] [Google Scholar]
- 44.Garland T., Midford P. E., Ives A. R. 1999. An introduction to phylogenetically-based statistical methods with a new method for confidence intervals on ancestral values. Am. Zool. 39, 374–388 [Google Scholar]
- 45.Lande R., Arnold S. J. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226 10.2307/2408842 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 46.Lewontin R. C. 1978. Adaptation. Sci. Am. 239, 212–230 10.1038/scientificamerican0978-212 (doi:10.1038/scientificamerican0978-212) [DOI] [PubMed] [Google Scholar]
- 47.Mayr E. 2004. 80 years of watching the evolutionary scenery. Science 305, 46–47 10.1126/science.1100561 (doi:10.1126/science.1100561) [DOI] [PubMed] [Google Scholar]
- 48.Gregory T. R. 2004. Macroevolution, hierarchy theory, and the C-value enigma. Paleobiology 30, 179–202 (doi:10.1666/0094-8373(2004)030<0179:MHTATC>2.0.CO;2) [DOI] [Google Scholar]
- 49.Darwin C. 1859. On the origin of species by means of natural selection, 1st edn. London, UK: John Murray [Google Scholar]
- 50.Mayr E. 1942. Systematics and the origin of species. New York, NY: Columbia University Press [Google Scholar]
- 51.West-Eberhard M. J. 2005. Developmental plasticity and the origin of species differences. Proc. Natl Acad. Sci. USA 102, 6543–6549 10.1073/pnas.0501844102 (doi:10.1073/pnas.0501844102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 95, 147–175 10.1093/aob/mci010 (doi:10.1093/aob/mci010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leitch I. J., Soltis D. E., Soltis P. S., Bennett M. D. 2005. Evolution of DNA amounts across land plants. Ann. Bot. 95, 207–217 10.1093/aob/mci014 (doi:10.1093/aob/mci014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrov D. A. 2001. Evolution of genome size: a new approach to an old problem. Trends Genet. 17, 23–28 10.1016/S0168-9525(00)02157-0 (doi:10.1016/S0168-9525(00)02157-0) [DOI] [PubMed] [Google Scholar]
- 55.Kazazian J. H. H. 2004. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 10.1126/science.1089670 (doi:10.1126/science.1089670) [DOI] [PubMed] [Google Scholar]
- 56.Beaulieu J. M., Leitch I. J., Knight C. A. 2007. Genome size evolution in relation to leaf strategy and metabolic rates revisited. Ann. Bot. 99, 495–505 10.1093/aob/mcl271 (doi:10.1093/aob/mcl271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knight C. A., Beaulieu J. M. 2008. Genome size scaling through phenotype space. Ann. Bot. 101, 759–766 10.1093/aob/mcm321 (doi:10.1093/aob/mcm321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lomax B., Woodward F. I., Leitch I. J., Knight C. A., Lake J. A. 2009. Genome size as a predictor of guard cell length in Arabidopsis thaliana is independent of environmental conditions. New Phytol. 181, 311–314 10.1111/j.1469-8137.2008.02700.x (doi:10.1111/j.1469-8137.2008.02700.x) [DOI] [PubMed] [Google Scholar]
- 59.Hodgson J. G. 2010. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Ann. Bot. 105, 573–584 10.1093/aob/mcq011 (doi:10.1093/aob/mcq011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kingsolver J. G., Hoekstra H. E., Hoekstra J. M., Berrigan D., Vignieri S. N., Hill C. H., Hoang A., Gilbert P., Beerli P. 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261 10.1086/319193 (doi:10.1086/319193) [DOI] [PubMed] [Google Scholar]
- 61.Rieseberg L. H., Widmer A., Arntz A. M., Burke J. M. 2002. Directional selection is the primary cause of phenotypic diversification. Proc. Natl Acad. Sci. USA 99, 12 242–12 245 10.1073/pnas.192360899 (doi:10.1073/pnas.192360899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McElwain J. C., Beerling D. J., Woodward F. I. 1999. Fossil plants and global warming at the Triassic-Jurassic boundary. Science 285, 1386–1390 10.1126/science.285.5432.1386 (doi:10.1126/science.285.5432.1386) [DOI] [PubMed] [Google Scholar]
- 63.O'Keefe J. D., Ahrens T. J. 1989. Impact production of CO2 by the Cretaceous/Tertiary extinction bolide and the resultant heating of the Earth. Nature 338, 247–249 10.1038/338247a0 (doi:10.1038/338247a0) [DOI] [Google Scholar]