SUMMARY
To fulfill the bioenergetic and biosynthetic demand of proliferation, T cells reprogram their metabolic pathways from fatty acid β-oxidation and pyruvate oxidation via the TCA cycle to the glycolytic, pentose-phosphate, and glutaminolytic pathways. Two of the top-ranked candidate transcription factors potentially responsible for the activation-induced T cell metabolic transcriptome, HIF1α and Myc, were induced upon T cell activation, but only the acute deletion of Myc markedly inhibited activation-induced glycolysis and glutaminolysis in T cells. Glutamine deprivation compromised activation-induced T cell growth and proliferation, and this was partially replaced by nucleotides and polyamines, implicating glutamine as an important source for biosynthetic precursors in active T cells. Metabolic tracer analysis revealed a Myc-dependent metabolic pathway linking glutaminolysis to the biosynthesis of polyamines. Therefore, a Myc-dependent global metabolic transcriptome drives metabolic reprogramming in activated, primary T lymphocytes. This may represent a general mechanism for metabolic reprogramming under patho-physiological conditions.
INTRODUCTION
Proliferating animal cells consume considerable amounts of energy and require de novo synthesis of macromolecules to support their growth and proliferation (DeBerardinis et al., 2008; Newsholme et al., 1985b). Consequently, one of the fundamental problems in animal cells is coordination of the metabolic program with cell growth- and proliferation-associated bioenergetic demand (Jorgensen and Tyers, 2004). While prokaryotes and unicellular eukaryotes rely primarily on homeostatic regulation of cell metabolism, metabolic programs in multicellular eukaryotes have evolved to be actively controlled by extracellular signals (Deberardinis et al., 2008).
Upon stimulation of antigen receptors, quiescent naïve T cells undergo a growth phase of approx. 24 hr, followed by massive clonal expansion and differentiation phases that are essential for appropriate immune defense and regulation. During proliferation, T cells can undergo a cell division every 4–6 hr (van Stipdonk et al., 2003). Coinciding with the clearance of antigen, effector T cells undergo an abrupt contraction phase in which the majority of T cells are eliminated by apoptosis (Green, 2003; Schumacher et al., 2010). Accumulation of cell biomass during the initial growth and rapid proliferation during the expansion phase is associated with dramatically increased bioenergetic and biosynthetic demand. Thus, T cell activation provides a unique physiological cellular model to help us understand how animal cells coordinate their metabolic program with cell growth and proliferation in response to extracellular fate-decisive signals.
Early studies showed that polyclonal mitogens such as concanavalin A and phytohaemagglutinin drive the up-regulation of glycolysis and glutamine oxidation in thymocyte and mature lymphocyte (Brand et al., 1984; Newsholme et al., 1985b). Consistent with these observations, the uptake of glutamine and glucose, and the consumption of the latter, mainly through glycolysis, are substantially up-regulated upon stimulation with anti-CD3 and anti-CD28 (Carr et al., 2011; Frauwirth et al., 2002). Signaling via ERK and AKT pathways has been implicated in the uptake of glutamine and glucose, respectively (Carr et al., 2011; Frauwirth et al., 2002; Wofford et al., 2008), and the mammalian target of rapamycin (mTOR), an essential component of PI3K-Akt pathway, has recently been implicated in regulating fatty acid metabolism in memory T cells (Pearce et al., 2009); (Araki et al., 2009). However, the robustness of metabolic changes following T cell activation suggests the presence of additional regulatory steps in the T cell metabolic program, and the molecular mechanisms behind these unrevealed regulatory steps remain to be explored. The metabolic changes associated with T cell activation are reminiscent of the characteristic metabolic activities of tumor cells and may represent a general metabolic reprogramming during cell growth and proliferation (Kim and Dang, 2006; Vander Heiden et al., 2009; Warburg, 1956).
Results
T cell activation induces metabolic reprogramming
To understand how T cells reprogram metabolic pathways to fulfill the bioenergetic and biosynthetic demand for the burst of proliferation upon activation, we used a well established ex vivo model to recapitulate the process of antigen stimulation of T cells without a requirement for additional cells. Freshly isolated murine primary T cells were either maintained in interleukin 7 (IL-7) (hereafter “resting T cells”) (Vella et al., 1997) or were stimulated with anti-CD3 plus anti-CD28 (hereafter “active T cells”). Following activation, both CD4+ and CD8+ T cells increased their cell size over a 24 hr period (Fig. S1A), followed by rapid cell division over the ensuing 24–72 hr (Fig. S1B). This provided a 24 hr “window” in which to interrogate changes in metabolism upon activation, without the potentially confounding effects of proliferation.
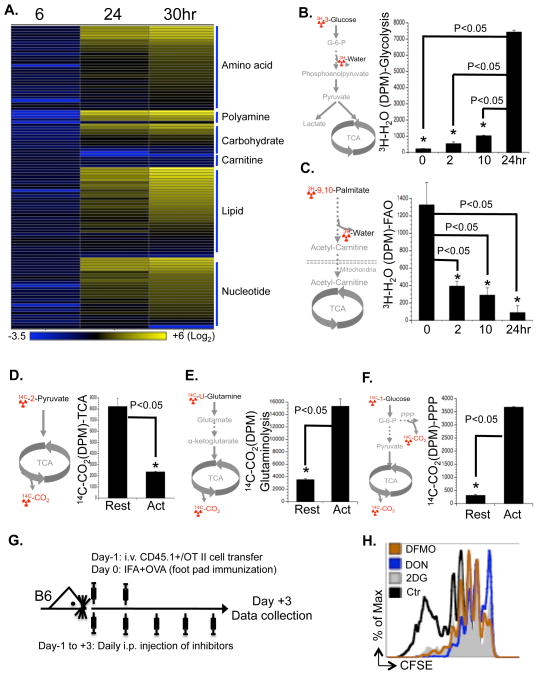
We applied a mass-spectrum based metabolomic approach to determine the relative intracellular concentration of metabolites in T cells following activation. While at an early time point (6 hr after activation), activated T cells contained lower amounts of metabolites than resting T cells, T cells that had been activated for longer times (24 hr and 30 hr) accumulated markedly higher amounts of metabolites involved in the anabolic pathways of lipid, amino acid and nucleotide (Fig. 1A and Table S1). This may reflect cell growth before entering into the cell cycle (Fig. S1A). In contrast, carnitines, which are required for fatty acid β-oxidation, were down-regulated after activation (Fig. 1A).
Figure 1. Stimulation of TCR and CD28 drives T cell metabolic reprogramming.
(A) Intracellular metabolites in T cells collected at the indicated times after activation were profiled by mass spectroscopy. The value for each metabolite represents the average of triplicates and the amount of each metabolite in resting T cells was set to 1. The heat map represents the log2 value of the relative amount of each metabolite, which was grouped in the indicated metabolic pathways (see color scale). The complete metabolomic profile is provided in Table S1.
(B–F) Metabolic assays, with the isotopically-labeled tracer used highlighted in red (left panel). Resting T cells (Rest) and activated T cells (collected at 24h after activation in D, E and F or at the indicated times after activation in B and C) were used for measuring the generation of 3H2O from [3-3H]-glucose (glycolysis, right panel in B) or from [9,10-3H]-palmitic acid (FAO, right panel in C); the generation of 14CO2 from [2-14C]-pyruvate (TCA, right panel in D), from [U-14C]-glutamine (glutaminolysis, right panel in E), or from [1-14C]-glucose (PPP, right panel in F). Error bars represent standard deviation from the mean of triplicate samples. Data are representative of three independent experiments.
(G) Overview of the experimental procedure used for assessing OT-II T cell proliferation in vivo (Fig. 1H).
(H) Naïve OT-II T cells (CD45.1+) cells were CFSE labeled and transferred into C57BL/6 mice, which were immunized with OVA and treated daily with the indicated inhibitors or vehicle controls. Splenocytes were analyzed 3 days after immunization and OT-II cell proliferation was determined by flow cytometry. Data represent two independent experiments.
Consistent with earlier studies (Frauwirth et al., 2002), activation of T cells dramatically up-regulated glycolysis (Fig. 1B). In contrast, both mitochondria-dependent fatty acid β-oxidation (FAO) and pyruvate oxidation through the tricarboxylic acid cycle (TCA cycle) were markedly down-regulated upon activation (Fig. 1C and D). Similarly, glutamine consumption through oxidative catabolism was significantly up-regulated upon activation (Fig. 1E), as suggested by earlier observations (Newsholme et al., 1985a). Glucose consumption via the pentose phosphate pathway (PPP) (Fig. 1F), and oxygen consumption (Fig. S1C) were also significantly elevated in activated T cells. Therefore, activation rapidly switches T cell metabolism from FAO and pyruvate oxidation via the TCA cycle to aerobic glycolysis, PPP and glutamine oxidation (hereafter, “activation-induced metabolic reprogramming”).
Glucose and glutamine catabolism are required for activation-induced T cell proliferation in vitro and in vivo
To explore the impact of metabolic reprogramming on T cell growth and proliferation, we stimulated T cells in either complete or nutrient-free media. While deprivation of glutamine but not glucose led to an impairment of activation-induced cell growth (Fig. S1D) associated with a reduction of lipid and protein biosynthesis (Fig. S1E and F), neither condition affected viability or appearance of the activation markers CD25 and CD69 (Fig. S1G and H). However, both conditions resulted in impaired activation-induced cell proliferation (Fig. S1I). Consistent with previous work (Rowell and Wells, 2006), T cell activation was associated with down-regulation of the cell cycle inhibitor p27, the up-regulation of cyclins and cyclin-dependent kinases (CDKs) and the appearance of markers of S-phase, such as the phosphorylation of RB and Cdc6 (Fig. S1J). The stabilization of p27 and the absence of S-phase markers indicated a G1 cell cycle arrest under glutamine starvation. However, the similar expression and phosphorylation pattern of cell cycle regulators in control and glucose-starved cells suggested that glucose deprivation might inhibit cell proliferation through a mechanism distinct from that of glutamine starvation.
We next applied a panel of potent chemical inhibitors targeting different metabolic pathways. Inhibition of FAO by the carnitine palmitoyltransferase-I (CPT-I) inhibitor Etomoxir (Eto) did not affect T lymphocyte proliferation (Fig. S1K), consistent with the low FAO flux in active T cells. In contrast, the hexokinse (HK) inhibitor 2-deoxy-D-glucose (2DG), the glucose-6-phosphate dehydrogenase (G6PD) inhibitor dehydroepiandrosterone (DHEA), the glutaminase (Gls) inhibitor 6-diazo-5-oxo-l-norleucine (DON) or the transaminase inhibitor aminooxyacetate (AOA), each of which inhibits activation-induced metabolic pathways, blocked T cell proliferation (Fig. S1K).
To further test the dependency of T cell proliferation on glucose and glutamine catabolism in vivo, we applied 2DG and DON in an adoptive transfer model using ovalbumin (OVA)-specific, TCR-transgenic OT-II T cells (Fig. 1G) (Shi et al.). In agreement with our in vitro results, both 2DG and DON partially inhibited the proliferation of antigen-specific active T cells after OVA immunization in vivo (Fig 1H). Therefore, both glucose and glutamine catabolic pathways are required for activation-induced T cell proliferation in vitro and in vivo.
T cell metabolic reprogramming is associated with global changes in the metabolic gene transcriptome
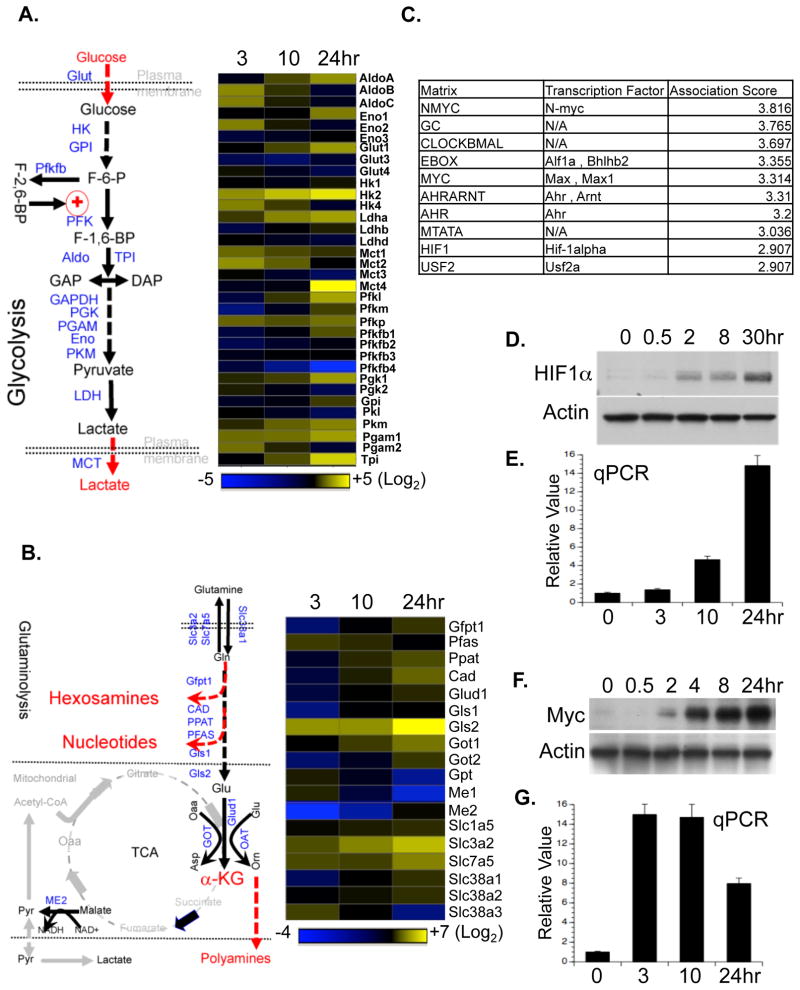
Antigen activation of T lymphocytes drives distinct transcriptional programs including up-regulation of cell cycle and cytokine genes (Lu et al., 2004; Teague et al., 1999). Real time quantitative PCR (qPCR) analysis revealed a time-dependent up-regulation of mRNAs encoding metabolic enzymes and transporters involved in glycolysis (Fig. 2A) and in both arms of the PPP (Fig. S2A). Notably, only one member in each metabolic enzyme gene family was selectively up-regulated (Fig. 2A). Immunoblot analysis further confirmed the up-regulation of selected metabolic genes at the protein level (Fig. S2B).
Figure 2. T cell activation drives the transcription of a distinct set of metabolic genes.
(A–B) The glycolytic pathway (left panel in A) and the glutaminolytic pathway (left panel in B), with the metabolic genes measured highlighted in blue. RNA was isolated from T cells collected at the indicated times after activation, and used for qPCR analyses of metabolic genes in the glycolytic pathway (right panel in A) and the glutaminolytic pathway (right panel in B). mRNA levels in resting T cells were set to 1. The heat map represents the log2 value of the relative mRNA expression level (see color scale). Values and standard deviations are provided in Table S2. Data are representative of two independent experiments.
(C) The top ten predicted corresponding transcription factors and motifs were ranked in descending order of association score, which indicates the likelihood of the binding of a particular transcription factor to the input gene promoters correlating with the expression levels. The input gene list and expression profile is provided in Table S3.
(D–G) Protein and mRNA expression of HIF1α (D and E, respectively) and of Myc (F and G, respectively) in T cells collected at the indicated times after activation were determined by Immunoblot and qPCR. mRNA levels in resting T cells were set to 1. Error bars represent standard deviation from the mean of triplicate samples. Data are representative of two independent experiments.
Glutamine catabolism is not only tightly coupled to different biosynthetic pathways, but also generates the anaplerotic substrate, α-ketoglutarate (α-KG), which can be metabolized through the TCA cycle to generate either citrate or pyruvate (“glutaminolysis”) (Newsholme et al., 1985a). While the conversion of glutamine (Gln) to glutamate (Glu) is controlled by glutamine-fructose-6-phosphate transaminase 1(Gfpt1), carbamoyl-phosphate synthetase (CAD), phosphoribosyl pyrophosphate amidotransferase (Ppat) and glutaminase 2 (Gls2), the conversion of glutamate (Glu) to α-KG is controlled by glutamate dehydrogenase 1 (Glud1), glutamate oxaloacetate transaminase (Got) and ornithine aminotransferase (Oat). Consistent with the idea that α-KG is an anaplerotic substrate of the TCA cycle, metabolic enzymes in the TCA cycle including isocitrate dehydrogenase-3 (IDH3), malate dehydrogenase (MDH), succinate dehydrogenase complex subunit C (SDHc) and malic enzyme 2 (ME2), were also up-regulated (Fig. 2B, S2A and S2B).
We compared our metabolic gene expression data of active mouse T lymphocytes with metabolic gene expression data from a recently published microarray study of active human T lymphocytes (Wang et al., 2008). A marked correlation between these two data sets, indicated by the Spearman rank-order correlation coefficient, suggests that metabolic genes in activated human T lymphocytes are similarly regulated (Fig. S2C). Taken together, these results are consistent with our metabolic flux data and indicate that global regulation of metabolic gene transcription is involved in activation-induced T cell metabolic reprogramming.
Myc, but not HIF1α is required for activation-induced T cell metabolic reprogramming
Using an in silico approach (Roider et al. 2009), transcription factors possibly involved in activation-induced T cell metabolic reprogramming were ranked by the likelihood of a particular transcription factor binding to the input gene promoters (Fig. 2C and Table S3). Among the top ten candidates (Fig. 2C), hypoxia inducible factor 1, alpha subunit (HIF1α) and myelocytomatosis oncogene (Myc) were chosen for further investigation. Both HIF1α and Myc were recently suggested to regulate cell metabolism in transformed cells (Dang and Semenza, 1999; Gordan et al., 2007), and were highly induced at both the transcription and protein levels upon T cell activation (Fig. 2D–G). The induction of these two transcription factors preceded the induction of most metabolic genes examined (Fig. 2A and B) and the up-regulation of glycolysis and glutaminolysis (Fig. 1B and E). In contrast, the unperturbed expression of the endogenous Myc antagonist, Mad, suggests that this may not be instrumental in the observed changes (Fig S2D).
Early studies have implicated the involvement of ERK-, AKT- and mTOR-mediated signaling pathways in regulating T cell metabolism (Carr et al., 2011; Frauwirth et al., 2002; Pearce et al., 2009). We next applied a panel of potent chemical inhibitors targeting different activation-induced signaling pathways and followed the expression of Myc and HIF1α. Inhibition of any of these kinases significantly reduced the induction of Myc and HIF1α after T cell activation (Fig. S2F), which was associated with the reduction of glycolytic activitiy (Fig. S2E). Therefore, ERK-, AKT- and mTOR-mediated signaling pathways may regulate T cell metabolism at least partially through regulating the expression of Myc and HIF1α.
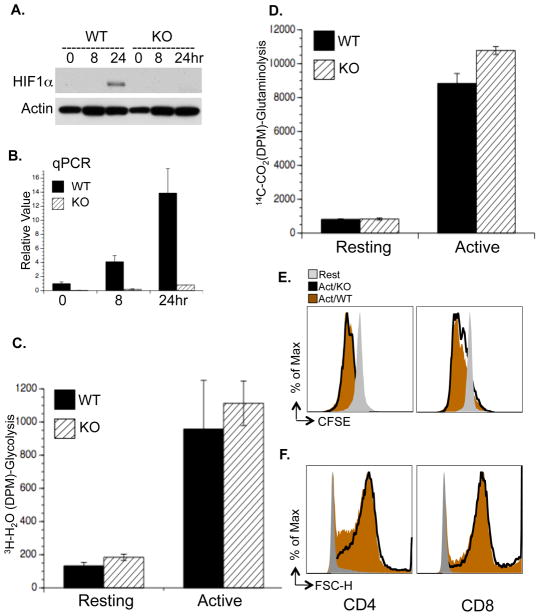
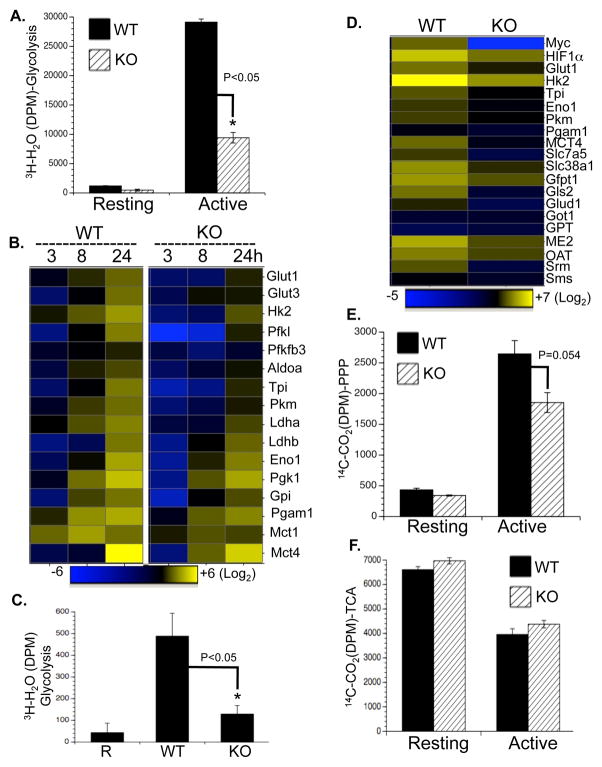
We generated a mouse model carrying a conditional HIF1α allele (Hif1a flox/flox) and a tamoxifen-inducible Cre recombinase (CreERT2) transgene (de Luca et al., 2005; Ryan et al., 2000) to delete HIF1α flox alleles in an acute manner. Freshly isolated T cells were cultured with IL-7 for two days with or without 4-Hydroxytamoxifen (4OHT), following which cells were either maintained in IL-7 (resting) or activated with anti-CD3 and anti-CD28 (active) (Fig. S3A). As controls, comparable cell proliferation and glycolytic rate were observed in T cells containing the CreER transgene only, and treated with vehicle or 4OHT (Fig. S3B and C). Efficient deletion of HIF1α was observed at the protein (Fig. 3A) and mRNA (Fig. 3B) levels after 4OHT treatment. However, glycolysis, glutaminolysis and FAO were unaffected by acute HIF1α deletion 24 hr after T cell activation (Fig. 3C, D and S3E). While the induction of two known HIF1α target genes, LDHa and HK2, was moderately impaired by the deletion of HIF1α, the expression of other activation-induced metabolic genes was comparable in WT and HIF1α deficient T cells (Fig. S3F). Moreover, activation-associated cell proliferation (Fig. 3E), cell growth (Fig. 3F) and cell surface appearance of the activation marker CD25 (Fig. S3D) were unaffected in HIF1α-deficient cells, and mild impairment of glycolysis was observed only at 72 hr post-activation (Fig. S3G). Therefore, HIF1α is not required for activation-induced metabolic reprogramming prior to T cell proliferation, but may be involved in optimally maintaining T cell glycolytic activities following entry into the cell cycle.
Figure 3. HIF1α is not required for activation-induced metabolic reprogramming.
(A–B) T cells were isolated from RosaCreERT2/HIF1αflox/flox mice and pretreated with vehicle (WT) or with 500 nM 4OHT (KO). HIF1α protein and mRNA levels in T cells collected at the indicated times after activation were determined by Immunoblot (A) and qPCR (B). mRNA levels in resting WT T cells were set to 1.
(C–D) Glycolysis and glutaminolysis as determined by the generation of 3H2O from [3-3H]-glucose (C) and the generation of 14CO2 from [U-14C]-glutamine (D), respectively. Error bars in B–D represent standard deviation from the mean of triplicate samples. Data are representative of two independent experiments.
(E) Cell proliferation of resting (Rest), active WT and KO T cells (48hr) was determined as CFSE dilution.
(F) Cell size of resting (Rest), active WT and KO T cells (24hr) was determined as forward light scatter.
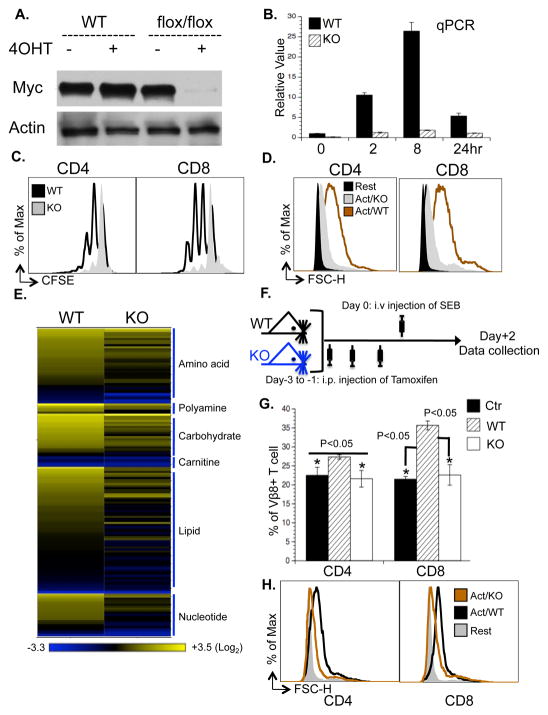
We then generated mice carrying a conditional Myc allele (Myc flox/flox) and a tamoxifen-inducible Cre recombinase (CreERTam) (Badea et al., 2003; de Alboran et al., 2001). Following addition of 4OHT, efficient deletion of the Myc gene was observed at the protein (Fig. 4A) and mRNA (Fig. 4B) levels. While Myc was suggested to be required for T cell proliferation and growth under most experimental conditions (Dose et al., 2006; Iritani et al., 2002), one study indicated that it is essential for cell proliferation but not cell growth (Trumpp et al., 2001). We found that acute deletion of Myc blocked activation-associated cell proliferation and growth in vitro (Fig. 4C and D). Metabolomic profiling further revealed that Myc-deficient T cells accumulated significantly lower levels of lipids, amino acids, and nucleotides (Fig. 4E). We then examined the role of Myc in regulating T cell proliferation and growth in vivo in response to Staphylococcal Enterotoxin B (SEB) (Fig. 4F), which as a superantigen, engaged sufficiently large numbers of naïve Vβ8+ T cells to perform metabolic studies (see below). Consistent with our in vitro results, the in vivo deletion of Myc blocked SEB-induced Vβ8+ T cell proliferation and growth (Fig. 4G and H). We observed that acute depletion of Myc, in vivo, did not compromise the health of the animals over a period of one week, consistent with a recent report (Soucek et al., 2008).
Figure 4. Myc is required for T cell growth and proliferation.
(A) T cells isolated from either RosaCreERTam-MycWT/WT mice (WT) or RosaCreERTam-Mycflox/flox mice (flox/flox) were pretreated with vehicle (−4OHT) or with 500 nM 4OHT (+4OHT). Myc protein levels were determined at the indicated times after activation by immuoblot.
(B) Myc mRNA levels in T cells collected at the indicated times after activation as determined by qPCR. mRNA levels in resting WT T cells were set to 1. Error bars represent standard deviation from the mean of triplicate samples. Data are representative of two independent experiments.
(C) Cell proliferation of active T cells (48hr) was determined as CFSE dilution.
(D) Cell size of resting (Rest) and active T cells (24hr) was determined as forward light scatter.
(E) The level of intracellular metabolites in the indicated samples; the value of each metabolite was obtained from the average of triplicates and the level of each metabolite in resting WT T cells was set to 1. The heat map represents the log2 value of the relative level of each metabolite, which was grouped in the indicated metabolic pathway (see color scale). The complete metabolomic profile is provided in Table S6.
(F–H) The experimental procedure (F) used for the SEB superantigen challenge model. Myc flox/flox mice (WT) or RosaCreERTam-Mycflox/flox mice (KO) were treated with tamoxifen (i.p. 1mg/mouse for three days), and then were injected with SEB (i.v. 100μg/mouse). The percentage (G) and the cell size (H) of Vβ8+ T cells were determined two days after injection of SEB.
Myc regulates cell cycle progression either directly or indirectly through controlling the expression of several essential cell cycle regulators including p27, cyclins and CDKs (Dang et al., 2006; Eilers and Eisenman, 2008). While the down-regulation of p27 and up-regulation of Cyclin D3 were largely undisturbed, induction of Cyclin A, cdk2, cdk4 and cdc25a were impaired to varying degrees in Myc-deficient T cells (Fig. S4A). Phosphorylation of RB and Cdc6, two S-phase markers, was delayed and abolished, respectively (Fig. S4A). In contrast, the up-regulation of the activation markers CD69 and CD25 (Fig. S4B), was comparable between Myc-deficient and WT cells, as was the phosphorylation of AKT and ERK (Fig. S4C). The expression of several cytokines was unaffected or elevated in Myc-deficient cells (Fig. S4D and E). Thus, the general signaling pathways mediated by TCR and cytokines are largely intact in T cells lacking Myc, and the acute deletion of Myc abrogates activation-induced T cell growth and proliferation but not other activation-induced events in T cells.
Myc-dependent induction of glucose metabolic genes drives glucose catabolism
Deletion of Myc significantly impaired activation-induced glycolytic flux at both early and late time points (Fig. 5A and S5A). Consistent with this, intracellular lactate in Myc-deficient T cells was lower than in WT control cells (Fig. S5B). Moreover, the partial reduction of Myc expression in T cells containing heterozygous Myc flox/wt alleles led to a moderate impairment of activation-induced glycolysis (Fig S5C and D). This suggests that the maximum expression of Myc is required for the maximum increase in glycolysis following activation of T cells. The up-regulation of several glycolytic enzymes and transporters was impaired to varying degrees in Myc-deficient T cells upon activation (Fig. 5B). Further, Myc deficiency led to a reduction of HK2 and pyruvate kinase muscle isoform 2 (PKM2) at the protein level (Fig. S5F). Finally, we tested the relevance of Myc in regulating T cell metabolism in response to SEB, as described above (Fig. 4F). Consistent with our in vitro results, Myc is required for the induction of enhanced glycolytic activity and metabolic gene expression in SEB-responsive T cells in vivo (Fig. 5C and D).
Figure 5. Myc drives a transcription program that regulates glucose catabolism upon T cell activation.
(A) Glycolytic flux as determined by the generation of 3H2O from [3-3H]-glucose.
(B) qPCR analyses of metabolic genes. mRNA levels in resting WT T cells were set to 1. The heat map represents the log2 value of the relative mRNA expression level (see color scale). Values and standard deviations are provided in Table S5. Data are representative of two independent experiments, performed in triplicate.
(C–D) Naïve CD4+Vβ8+ T cells (R) were sorted from mice without SEB immunization; WT and acutely Myc-deleted CD4+Vβ8+ T cells (KO) were isolated from mice two days after SEB immunization (Fig. 4F). Glycolysis (C) and mRNA expression (D and Table S5) in the indicated groups were determined by the generation of 3H2O from [3-3H]-glucose and qPCR.
(E–F) PPP flux (E) and pyruvate oxidation through the TCA cycle (F) were determined by the generation of 14CO2 from [1-14C]-glucose and from [2-14C]-pyruvate, respectively.
Since glycolysis interconnects with other metabolic pathways, such as the PPP, the TCA cycle and FAO, we investigated the metabolic flux through these pathways. Acute deletion of Myc moderately inhibited activation-induced up-regulation of metabolic activity through the PPP (Fig. 5E), and the induction of two key metabolic enzymes in the PPP, Tkt and G6PDx, was compromised in Myc-deficient T cells upon activation (Fig. S5E). In contrast, pyruvate metabolic flux, FAO flux, and oxygen consumption rates were unaffected by Myc deletion (Fig. 5F, S5G and S5H).
Myc-dependent glutaminolysis cross-talks with the mTOR pathway
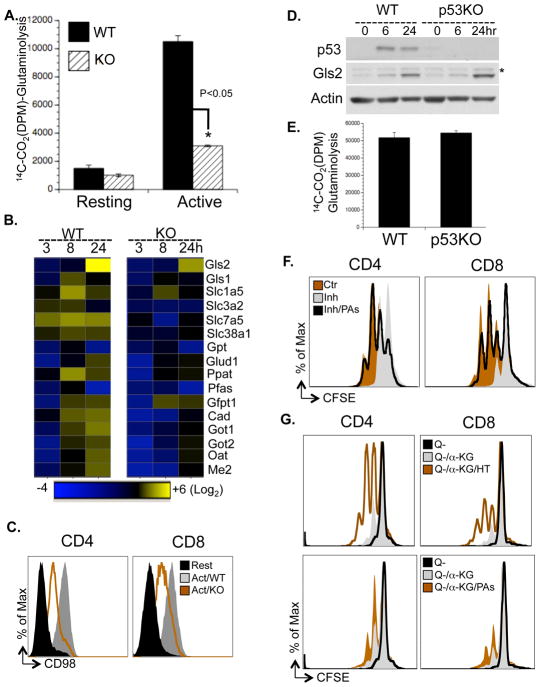
Deletion of Myc profoundly inhibited the up-regulation of glutamine oxidation in active T cells (Fig. 6A). Recent studies in human cell lines have suggested that enforced expression of Myc regulates the rate-limiting glutaminolyic enzyme glutaminase 1 (Gls1) through both transcription and post-transcriptional mechanisms (Gao et al., 2009; Wise et al., 2008), and p53 regulates the transcription of glutaminase 2 (Gls2) (Hu et al., 2010; Suzuki et al., 2010). We observed that neither the mRNA nor protein levels of Gls1 were elevated upon T cell activation (Fig. 2B and S2B), while the transcription and translation of glutaminase 2 (Gls2) along with other metabolic genes in glutamine catabolic pathways were induced in a Myc-dependent manner in vitro and in vivo (Fig. 5D, 6B and S5F). However, we found that p53 was dispensable for the induction of Gls2 and glutaminolysis upon T cell activation (Fig. 6D and 6E).
Figure 6. Myc drives a transcriptional program coupling glutaminolysis to biosynthetic pathways.
(A) Rates of glutaminolysis, determined by the generation of 14CO2 from [U-14C]-glutamine. Data are representative of two independent experiments.
(B) qPCR analyses of metabolic genes. mRNA levels in resting WT T cells were set to 1. The heat map represents the log2 value of the relative mRNA expression level (see color scale). Values and standard deviations are provided in Table S5. Data are representative of two independent experiments.
(C) Cell surface expression of CD98 was determined by flow cytometry.
(D) Protein levels of Gls2 and p53 were determined by immunoblot. The asterisk indicates a non-specific band.
(E) Glutaminolytic flux was determined by the generation of 14CO2 from [U-14C]-glutamine.
(F) Cell proliferation of active T cells (48hr), treated with vehicle (Ctr), with 1mM DFMO (Inh) or with 1mM DFMO plus polyamine mixture (Inh/PAs), determined by CFSE dilution. The composition and concentration of polyamine mixture are provided in Table S7.
(G) T cells were collected at 72hr after activation in glutamine free media (Q−), in glutamine free media supplemented with 2μM α-KG (Q−/αKG), in glutamine free media supplemented with 2μM α-KG and 100μM hypoxanthine and 16μM thymidine (Q−/αKG/HT) or in glutamine free media supplemented with 2μM α-KG and polyamine mixture (Q−/αKG/PAs). The composition and concentration of the polyamine mixture is provided in Table S7. Cell proliferation was determined as CFSE dilution by flow cytometry.
Either glutamine starvation or Myc deletion was sufficient to abrogate activation-induced T cell growth (Fig. S1D, 4D and H). As glutamine flux might regulate cell growth through the mTOR pathway (Nicklin et al., 2009), glutamine-dependent modulation of mTOR activation might partially account for the regulation of T cell growth by Myc. While the phosphorylation kinetics of upstream components (mTOR and AMPK) were unaffected, acute deletion of Myc impaired the activation of downstream effectors in the mTOR pathway, as indicated by the reduction of phosphorylation of P70K and the reduction of phosphorylation and protein levels of 4E-BP and S6 in Myc deficient cells (Fig. S6A). Consistent with an indispensable role for the glutamine antiporter CD98, a heterodimer of Slc3a2/Slc7a5, in modulating the mTOR pathway (Nicklin et al., 2009), both the transcription of Slc3a2/Slc7a5 and the cell surface appearance of CD98 were impaired in Myc-deficient T cells upon activation in vitro and in vivo (Fig. 6B, 6C, S6C and 5D). Moreover, glutamine starvation largely recapitulated the impairment of the activation of downstream effectors in the mTOR pathway as observed in Myc deficient cells (Fig. S6B). These results demonstrate an essential role for Myc in the up-regulation of glutaminolysis upon activation and imply that cross-talk with the mTOR pathway via amino-acid uptake may be involved in the Myc-dependent regulation of T cell growth.
Myc driven glutamine catabolism couples with multiple biosynthetic pathways
Myc drives the expression of Gfpt1, Cad, Ppat and Oat (Fig. 2B and 6B), which are not only required for glutaminolysis but also control critical steps in the hexosamine, pyrimidine, purine, and polyamine biosynthetic pathways (Fig. 2B, left panel). While we failed to detect any intermediate metabolites in the hexosamine biosynthetic pathway due to technical limitations, the levels of intermediate metabolites in the polyamine and nucleotide synthetic pathways were lower in Myc-deficient cells (Fig. S6D, S6E and Table S6). Consistent with a previous study (Bowlin et al., 1987), difluoromethylornithine (DFMO), a potent inhibitor of ornithine decarboxylase (ODC) and polyamine biosynthesis, inhibited activation-induced T cell proliferation in vitro and in vivo (Fig. 6F and 1H), and the addition of exogenous polyamines completely restored proliferation in the presence of DFMO in vitro (Fig. 6F).
Addition of either hypoxanthine and thymidine (HT) or polyamines (PAs) alone was insufficient to permit cell proliferation in the absence of glutamine (Fig. S7A). As α-KG is required to maintain cell viability and minimal cell proliferation in the absence of glutamine (Wise et al., 2008), addition of α-KG alone was sufficient to drive only one cell division in the absence of glutamine. However, the combination of α-KG with either HT or with PAs promoted additional cell divisions (Fig. 6G, Fig. S7B). Consistent with a direct role of Myc in regulating cell proliferation (Dang et al., 2006; Eilers and Eisenman, 2008), addition of α-KG with HT and PAs failed to promote cell proliferation in Myc deficient T cells (Fig S7C). Taken together, our results indicate that Myc-driven glutaminolysis coordinates multiple biosynthetic pathways to support activation-induced cell proliferation and growth.
A Myc-dependent metabolic pathway linking glutaminolysis to ornithine and polyamine biosynthesis
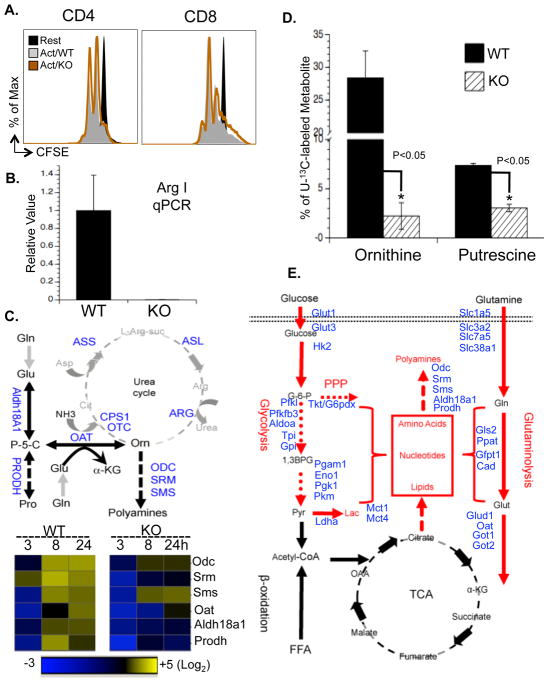
Although ornithine is produced largely from arginine via the action of arginase as part of the urea cycle (Morris, 2004), urea cycle enzymes including arginase were not induced upon activation (Fig. S7D) nor were any defects in proliferation observed in T cells lacking arginase I (Fig. 7A and B), the dominant isoform in the thymus (Yu et al., 2003). These results indicate that arginine is not the only precursor for ornthine in T cells. Previous studies in bovine endothelial and intestinal epithelial cells suggested the possible presence of a noncanonical metabolic pathway, which generates ornithine from glutamine (Wu et al., 2000; Wu and Morris, 1998). This putative pathway is composed of two key steps: aldehyde dehydrogenase 18 family member A1 (Aldh18a1) converts glutamate to (S)-1-pyrroline-5-carboxylate (P-5-C), following which OAT converts P-5-C to ornithine (Fig. 7C). Alternatively, proline has also been suggested as a source for ornithine through a two-step reaction involving proline dehydrogenase (Prodh) and OAT (Phang et al., 2008). Intriguingly, the metabolic enzymes Aldh18A1, Prodh and OAT that link glutamine and proline to ornithine synthesis were induced in a Myc-dependent manner upon T cell activation (Fig. 7C and S5F), as were the metabolic enzymes, ODC, spermine synthase (SRM) and spermidine synthase (SMS), that convert ornithine to polyamines (Fig. 7C). Therefore, we followed U-13C-isotope labeled glutamine (U-13C-glutamine) flux and observed the incorporation of 13C into all five carbons of ornithine (U-13C-ornithine), which accounted for up to 30% of total ornithine in active T cells but only 1% in resting T cells (Fig. S7E). Finally, deletion of Myc partially abolished the incorporation of glutamine-derived carbons into both ornithine and putrescine (Fig. 7D). Taken together, our results indicate a Myc-dependent non-canonical ornithine biosynthetic pathway coupling glutaminolysis to ornithine and polyamine biosynthesis in activated T cells (Fig. 7E).
Figure 7. Myc-driven glutaminolysis fuels polyamine biosynthesis upon T cell activation.
(A) Arginase I WT and KO T cells were collected 48hr after activation (Act/WT and Act/KO, respectively). Cell proliferation was determined by CFSE dilution.
(B) WT and arginase I KO active T cells were analyzed by qPCR for arginase I mRNA expression.
(C) (Upper panel) Potential metabolic steps linked to the polyamine biosynthetic pathway, with metabolic genes measured highlighted in blue. (Lower panel) WT and Myc KO T cells were collected at the indicated times after activation and qPCR analyses of metabolic genes was performed. mRNA levels in resting T cells were set to 1. The heat map represents the log2 value of the relative mRNA expression level (see color scale). Values and standard deviations are provided in Table S5. Data are representative of two independent experiments, performed in triplicate.
(D) T cells were cultured in media containing U-13C-glutamine and collected at the indicated times after activation. The intracellular levels of ornithine and putrescine including U-13C- and 12C-labeled forms were determined by mass spectroscopy.
(E) Myc-dependent metabolic reprogramming upon T cell activation, with upregulated metabolic pathways highlighted in red, downregulated metabolic pathways highlighted in black and Myc-dependent metabolic genes highlighted in blue.
DISCUSSION
Extracellular signals play an essential role in dictating eukaryotic cell growth, proliferation and metabolism. TCR and costimulator-mediated signals drive T cell growth and clonal expansion, and coordinately reprogram metabolic cascades to fulfill the ensuing bioenergetic and biosynthetic demands (Fox et al., 2005; Krauss et al., 2001). While ectopic overexpression of Myc in transformed cell lines coordinately drives cell proliferation and proliferation-associated metabolic activities, including the catabolism of glucose and glutamine (Dang and Semenza, 1999; Gordan et al., 2007), whether the induction of endogenous Myc upon physiological stimulation plays a similar role has not been explored. We show that the activation of T cells rapidly switches metabolic programs from fatty acid β-oxidation and pyruvate oxidation via the TCA cycle to aerobic glycolysis, PPP and glutaminolysis. This metabolic reprogramming is associated with a global change in the metabolic transcriptome, which follows the induction of endogenous Myc and HIF1α upon activation. Based on the effects of acute genetic ablation of these transcription factors, we identified Myc as a critical metabolic reprogramming factor in T cells. Our studies shed light on the complex utilization of metabolic resources, pathways and intermediates that rationalize the strict dependence of T cell growth and proliferation on glutaminolysis, and indicate a novel glutaminolysis- and Myc-dependent de novo polyamine biosynthetic pathway essential for T cell proliferation. While previous studies clearly demonstrate that Myc directly regulates T cell proliferation mainly through transcriptional control of cell cycle regulators (Dose et al., 2006; Iritani et al., 2002), our studies implicate Myc as an essential coordinator of T cell activation-induced cell growth, proliferation and several aspects of metabolic reprogramming.
T cell activation-induced glycolysis and glutaminolysis is reminiscent of the metabolic changes in tumor cells, where both aerobic glycolysis (the Warburg effect) and glutaminolysis are driven by aberrant oncogenic signals (Dang and Semenza, 1999; Vander Heiden et al., 2009). Notably, ectopic overexpression of Myc not only alters the transcription of several glycolytic genes but also preferentially switches the expression of PKM1 to PKM2 by modulating the alternative splicing of the PKM transcript (Dang and Semenza, 1999; David et al., 2010). While the transcription of metabolic enzymes and transporters involved in almost every step of glycolysis including PKM2 are induced in a Myc-dependent manner upon T cell activation, whether active T cells preferentially express PKM2 through alternative splicing remains to be determined. In contrast, the metabolic program associated with glutaminolysis in tumor cell lines and activated T cells is slightly different. In tumor cells, Myc controls glutamine uptake through transcription of SLC3a2, SLC5A1 and SLC7A1 and regulates the conversion of glutamine to glutamate through both transcriptional and posttranslational regulation of GLS1 (Gao et al., 2009; Wise et al., 2008). Corresponding steps in active T cells, however, are controlled by Myc-mediated transcription of SLC32a1, SLC32a2 and GLS2. Although recent studies indicate that the tumor suppressor p53 is required for the induction of GLS2 in a human tumor cell line (Hu et al., 2010; Suzuki et al., 2010), we have found that the up-regulation of GLS2 in murine T cells is p53-independent. Although the regulatory mechanisms involved are slightly different, the resemblance of tumor-associated and T cell activation-associated metabolic programs indicate that a general metabolic switch is required to support cell growth and proliferation in both pathological and physiological situations.
Glutamine has long been known to be an indispensible nutrient for proliferating cells (Kovacevic and McGivan, 1983). Enhanced glutamine utilization in rat lymphocytes upon polyclonal mitogen stimulation is responsible for providing precursors for nucleotide biosynthesis (Newsholme et al., 1985a). Metabolic flux analysis in a glioblastoma cell line further revealed that a major portion of glutamine-derived nitrogen and carbon are incorporated into amino acids and lactate, a fraction of which are secreted as metabolic by-products (DeBerardinis et al., 2007). These findings indicate that glutamine is an important nitrogen- and carbon-donor for a variety of biosynthetic precursors. Supporting this notion, we found that activation of T cells triggers a Myc-dependent induction of glutaminolysis, which not only leads to the production of α-KG in the TCA cycle but is also coupled to nucleotide biosynthesis. Furthermore, the depletion of intracellular ornithine and polyamines in Myc-deficient activated T cells implicates a noncanonical, Myc-dependent polyamine biosynthetic pathway upon activation.
Early studies showed that overexpression of Myc in transformed cell lines induces the expression of ODC (Bello-Fernandez, et al., 1993). While the protein level of ODC was not elevated, the mRNAs of all three metabolic enzymes downstream of ornithine were induced in a Myc-dependent manner upon T cell activation, leading to a massive increase in polyamine synthesis. The Myc-dependent induction of Aldh18a1, Prodh and OAT further indicates a novel Myc-dependent metabolic route upstream of ornithine that links glutamine and possibly proline to ornithine. Consistent with this, the Myc-dependent incorporation of 13C-labeled glutamine carbon into ornithine after T cell activation indicates that activated T cells may acquire an alternative Myc-dependent pathway coupling glutaminolysis to polyamine biosynthesis to meet the markedly increased polyamine demands required for proliferation. Our results suggest that Myc-dependent glutamine catabolism not only replenishes the intermediate metabolites of the TCA cycle by producing the anaplerotic substrate α-KG, but also coordinates with Myc-dependent glucose catabolism to support amino acid, nucleotide and lipid biosynthesis. In addition, glutamine catabolism is tightly coupled to the biosynthesis of polyamines that are essential for T cell proliferation.
EXPERIMENTAL PROCEDURES
Animals
Hif1aflox/flox and p53 null mice on the C57BL/6 background were purchased from the Jackson Laboratory (Jacks et al., 1994; Ryan et al., 2000) and Hif1aflox/flox were crossed with ROSA26CreERT2 (de Luca et al., 2005). Mycflox/flox mice were obtained from Dr. Fred Alt (Harvard) (de Alboran et al., 2001), and were backcrossed five generations onto the C56BL/6 background before breeding to ROSA26CreER(Tam) (Badea et al., 2003). Resulting offspring were interbred to obtain experimental animals. Arg1flox/flox; Tie2Cre mice that were backcrossed to the C57BL/6 background were obtained from Dr. Peter Murray (SJCRH, Memphis) (El Kasmi et al., 2008). Mice at 7–12 weeks of age were used. All mice were kept in specific pathogen–free conditions within the Animal Resource Center at St. Jude Children’s Research Hospital. Animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
RNA isolation, reverse transcription and qPCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and was reverse transcribed using random hexamer primers and M-MLV Reverse Transcriptase (Invitrogen). Real-time qPCR for cytokines was performed using a TaqMan Low Density Array (4367786, ABI). The comparison of cytokine gene expression in resting and active T cells was visualized as heat maps using Cluster (Eisen and Brown, 1999) and TreeView (Eisen et al., 1998). SYBR Green-based quantitative RT-PCR for other genes was performed using the Applied Biosystems 7900 Real Time PCR System. Samples for each experimental condition were run in triplicate and were normalized to β-2-microglobulin to determine relative expression levels. Primer sequences were obtained from PrimerBank (Spandidos et al., 2010) except for Gls1 and Gls2, which were manually designed. Primer sequences are listed in Table S7. Comparison of the expression of selected metabolic genes in resting and active T cell was visualized as heat maps using Spotfire.
Promoter analyses
Transcription factor motifs enriched in the promoters were identified by using the motif analysis algorithm in the PASTAA webserver (http://trap.molgen.mpg.de/cgi-bin/pastaa.cgi). This method considers the input order of promoters and uses a biophysical model of promoter-transcription factor interaction to detect motifs (Roider et al., 2009). The input order was that of decreasing fold-change in expression and default settings of the server were used.
Metabolic assays
Glycolytic flux was determined by measuring the detritiation of [3-3H]-glucose. Fatty acid β-oxidation flux was determined by measuring the detritiation of [9,10-3H]-palmitic acid. Glutamine oxidation flux was determined by the rate of 14CO2 released from [U-14C]-glutamine. Pyruvate oxidation flux was determined by the rate of 14CO2 released from [2-14C]-pyruvate. Glucose oxidation flux through the pentose phosphate pathway (PPP) was determined as previously described with some modifications described in the supplemental information. Respiration was measured in intact T cells using the Seahorse XF24 analyzer. Metabolomic profiling was performed by Metabolon, Inc (Durham, NC). Metabolic flux analysis of 13C-glutamine in active T cells was determined by LC-MS.
Statistical analysis
P-values were calculated with Student’s t test. P-values <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Zhaohui Feng for providing the anti-Gls2 antibody and Dr. Peter Murray for providing Arginase-I knockout mice. We thank Haopeng Wang, Diana Spierings, Tudor Moldoveanu, Helen Beere, Lisa Bouchier-Hayes, David Smithson, Joseph Qualls, Chitra Subramanian and Charles Rock for valuable discussions and technical assistance. We thank the members of the St. Jude Immunology FACS core facility, as well as the St. Jude Hartwell Center. This work was supported by the George J. Mitchell fellowship from St Jude Children Hospital (R.W.), a Damon Runyon-Rachleff Innovator Award (J.M.), and NIH grants AI081773 (J.M.), S064599 and AR053573 (H.C.), AI40646 and GM52735 (D.R.G.) and the American Lebanese and Syrian Associated Charities. We declare no competing financial interests.
Footnotes
Supplemental information includes Extended Experimental Procedures, seven figures, and seven tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlin TL, McKown BJ, Babcock GF, Sunkara PS. Intracellular polyamine biosynthesis is required for interleukin 2 responsiveness during lymphocyte mitogenesis. Cell Immunol. 1987;106:420–427. doi: 10.1016/0008-8749(87)90184-5. [DOI] [PubMed] [Google Scholar]
- Brand K, Williams JF, Weidemann MJ. Glucose and glutamine metabolism in rat thymocytes. Biochem J. 1984;221:471–475. doi: 10.1042/bj2210471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2011;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- de Luca C, Kowalski TJ, Zhang Y, Elmquist JK, Lee C, Kilimann MW, Ludwig T, Liu SM, Chua SC., Jr Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest. 2005;115:3484–3493. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M, Khan I, Guo Z, Kovalovsky D, Krueger A, von Boehmer H, Khazaie K, Gounari F. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108:2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. Introduction: apoptosis in the development and function of the immune system. Semin Immunol. 2003;15:121–123. doi: 10.1016/s1044-5323(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani BM, Delrow J, Grandori C, Gomez I, Klacking M, Carlos LS, Eisenman RN. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 2002;21:4820–4830. doi: 10.1093/emboj/cdf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- Krauss S, Brand MD, Buttgereit F. Signaling takes a breath--new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- Lu B, Zagouras P, Fischer JE, Lu J, Li B, Flavell RA. Kinetic analysis of genomewide gene expression reveals molecule circuitries that control T cell activation and Th1/2 differentiation. Proc Natl Acad Sci U S A. 2004;101:3023–3028. doi: 10.1073/pnas.0307743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM., Jr Recent advances in arginine metabolism. Curr Opin Clin Nutr Metab Care. 2004;7:45–51. doi: 10.1097/00075197-200401000-00009. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q J Exp Physiol. 1985a;70:473–489. doi: 10.1113/expphysiol.1985.sp002935. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985b;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang JM, Pandhare J, Liu Y. The metabolism of proline as microenvironmental stress substrate. J Nutr. 2008;138:2008S–2015S. doi: 10.1093/jn/138.10.2008S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roider HG, Manke T, O’Keeffe S, Vingron M, Haas SA. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell EA, Wells AD. The role of cyclin-dependent kinases in T-cell development, proliferation, and function. Crit Rev Immunol. 2006;26:189–212. doi: 10.1615/critrevimmunol.v26.i3.10. [DOI] [PubMed] [Google Scholar]
- Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Schumacher TN, Gerlach C, van Heijst JW. Mapping the life histories of T cells. Nat Rev Immunol. 2010;10:621–631. doi: 10.1038/nri2822. [DOI] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1{alpha}-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague TK, Hildeman D, Kedl RM, Mitchell T, Rees W, Schaefer BC, Bender J, Kappler J, Marrack P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc Natl Acad Sci U S A. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- van Stipdonk MJ, Hardenberg G, Bijker MS, Lemmens EE, Droin NM, Green DR, Schoenberger SP. Dynamic programming of CD8+ T lymphocyte responses. Nat Immunol. 2003;4:361–365. doi: 10.1038/ni912. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella A, Teague TK, Ihle J, Kappler J, Marrack P. Interleukin 4 (IL-4) or IL-7 prevents the death of resting T cells: stat6 is probably not required for the effect of IL-4. J Exp Med. 1997;186:325–330. doi: 10.1084/jem.186.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics. 2008;9:225. doi: 10.1186/1471-2164-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–2111. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Haynes TE, Li H, Meininger CJ. Glutamine metabolism in endothelial cells: ornithine synthesis from glutamine via pyrroline-5-carboxylate synthase. Comp Biochem Physiol A Mol Integr Physiol. 2000;126:115–123. doi: 10.1016/s1095-6433(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Wu G, Morris SM., Jr Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Yoo PK, Aguirre CC, Tsoa RW, Kern RM, Grody WW, Cederbaum SD, Iyer RK. Widespread expression of arginase I in mouse tissues. Biochemical and physiological implications. J Histochem Cytochem. 2003;51:1151–1160. doi: 10.1177/002215540305100905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.