Background: We monitored photosystem II (PSII) protein lifetimes with or without small Cab-like proteins (SCPs) by 15N labeling and mass spectrometry.
Results: Without SCPs, nascent PSII complexes were destabilized, and early chlorophyll precursors were reduced.
Conclusion: SCPs play a major role in chlorophyll biosynthesis and nascent PSII stabilization.
Significance: Proteins temporarily associated with a protein complex may affect biosynthesis of the complex and cofactors.
Keywords: Cyanobacteria, Photosynthesis, Photosynthetic Pigments, Photosystem II, Protein Turnover, Thylakoid Membranes
Abstract
To gain insight in the lifetimes of photosystem II (PSII) chlorophyll and proteins, a combined stable isotope labeling (15N)/mass spectrometry method was used to follow both old and new pigments and proteins. Photosystem I-less Synechocystis cells were grown to exponential or post-exponential phase and then diluted in BG-11 medium with [15N]ammonium and [15N]nitrate. PSII was isolated, and the masses of PSII protein fragments and chlorophyll were determined. Lifetimes of PSII components ranged from 1.5 to 40 h, implying that at least some of the proteins and chlorophyll turned over independently from each other. Also, a significant amount of nascent PSII components accumulated in thylakoids when cells were in post-exponential growth phase. In a mutant lacking small Cab-like proteins (SCPs), most PSII protein lifetimes were unaffected, but the lifetime of chlorophyll and the amount of nascent PSII components that accumulated were decreased. In the absence of SCPs, one of the PSII biosynthesis intermediates, the monomeric PSII complex without CP43, was missing. Therefore, SCPs may stabilize nascent PSII protein complexes. Moreover, upon SCP deletion, the rate of chlorophyll synthesis and the accumulation of early tetrapyrrole precursors were drastically reduced. When [14N]aminolevulinic acid (ALA) was supplemented to 15N-BG-11 cultures, the mutant lacking SCPs incorporated much more exogenous ALA into chlorophyll than the control demonstrating that ALA biosynthesis was impaired in the absence of SCPs. This illustrates the major effects that nonstoichiometric PSII components such as SCPs have on intermediates and assembly but not on the lifetime of PSII proteins.
Introduction
Cyanobacteria, algae, and plants can use sunlight and water to carry out oxygenic photosynthesis. In these organisms, linear photosynthetic electron transfer is catalyzed by the thylakoid-embedded protein complexes photosystem II (PSII),3 cytochrome b6f, and photosystem I (PSI). Linear electron transfer provides electrons to NADP producing NADPH and transfers protons across the thylakoid membrane leading to a proton gradient that is used for ATP synthesis. NADPH and ATP can be used for carbon fixation producing organic compounds. These organic compounds, along with oxygen produced in water splitting in PSII, enable heterotrophic, aerobic life on Earth.
The photosystems are multiprotein subunits that noncovalently bind different cofactors, including chlorophyll a, carotenoids, quinones, lipids, and several inorganic ions. During photosynthesis, components of PSII complexes turn over rapidly, at least in comparison with PSI complexes (1, 2). Of the proteins in the PSII complex, the PsbA (D1) protein turns over most rapidly in the light (3, 4). This rapid turnover presumably is due to redox chemistry at the water-splitting complex and/or to reactive oxygen species that are generated from oxygen reacting with the triplet state chlorophyll formed upon charge recombination between the primary donor P680+ and the primary acceptor pheophytin (Phe)− (5–7). According to pulse-chase experiments, the D1 protein has a half-time of 30 min to 1 h under intense illumination (8). However, the other PSII components appear to have a much lower turnover rate. For example, the half-time of the PsbB (CP47) protein was estimated to be about 12 h (9, 10), and the lifetime of total chlorophyll in Synechocystis cells was over a week (11). If this vast disparity in the lifetime of PSII components indeed is true, then careful orchestration of the synthesis, assembly, and repair of photosynthetic complexes required as free chlorophyll in the cell would be harmful in the light and in the presence of oxygen and as PSII polypeptides that are not incorporated in a complex may not be stable in the membrane (12–14).
In the cyanobacterium Synechocystis sp. PCC 6803, there are five small Cab-like proteins (ScpA–E), which are single helix membrane proteins that are located in the thylakoid membrane (15). The presence of the CAB (chlorophyll a/b-binding) motif in SCPs suggests that SCPs bind chlorophyll molecules at motifs similar to those of LHCII in plants (16–19). SCPs appear to play an important role in early stages of tetrapyrrole biosynthesis and may regulate chlorophyll availability (20). However, unlike CAB proteins that are associated with functional PSII in plants and are involved in light harvesting and nonphotochemical quenching, at least two of the SCPs (ScpC and ScpD) have been found to be associated with damaged and/or nascent PSII complexes (21). One SCP (ScpA) is fused with ferrochelatase, suggesting a regulatory role in tetrapyrrole biosynthesis (22). Furthermore, SCPs may prevent the formation of reactive oxygen species by serving as transient carriers of chlorophyll (23), and SCPs appear to be involved in PSII re-assembly and/or repair processes by temporarily binding chlorophyll, whereas PSII protein components are being replaced (18).
Here, we expand on the role of SCPs and show that they stabilize nascent PSII complexes and increase the presence of early chlorophyll biosynthesis precursors in the cell. Stable isotope labeling and mass spectroscopy allow for a detailed analysis of lifetimes of components of the PSII complex and illustrate that although different components degrade at different rates, degradation of only chlorophyll and to some degree D1 is significantly affected by SCPs.
EXPERIMENTAL PROCEDURES
Growth Conditions
Synechocystis sp. PCC 6803 strains, which included the PSI-less (ΔpsaAB) (24), PSI-less/SCP-less (scpABCDE−) (20, 23), CP47-His PSI-less (carrying a His tag at the C terminus of the CP47 (PsbB) protein) (see mutant construction), and CP47-His PSI-less/SCP-less strains, were cultivated at 30 °C in BG-11 medium (25) with 5 mm glucose and buffered with 10 mm TES-NaOH (pH 8.0). Because of the light sensitivity of PSI-less strains, cells were cultured at a light intensity of 4 μmol photons m−2 s−1. Cell growth was monitored by measuring the optical density at 730 nm in a 1-cm cuvette using a Shimadzu UV-160 spectrophotometer.
Mutant Construction
To generate strains with His-tagged PsbB (CP47), a pUC19-psbB-His6-gentamycin (Gm)R plasmid was constructed. A DNA region upstream of the His6 tag, including part of the psbB gene from the HT-3 strain (26) and a DNA region of wild-type Synechocystis downstream of the psbB gene stop codon, were amplified by PCR with artificially generated restriction sites for EcoRI right after the stop codon of psbB. These two PCR fragments were digested with EcoRI and ligated. The ligated DNA fragment containing natural KpnI sites at both ends was cloned into the KpnI site of the pUC19 plasmid. The GmR gene from the pHP45-GmR plasmid was introduced into this plasmid by using the EcoRI restriction site. The plasmid was introduced into the PSI-less and PSI-less/SCP-less strains to create the CP47-His PSI-less and CP47-His PSI-less/SCP-less strains. Insertion of the psbB-His gene at the desired location, replacing the native psbB gene, was confirmed by DNA sequencing, and segregation of the mutant genome in Synechocystis was confirmed by PCR.
Isotope Labeling and Isolation of His-tagged Complexes
CP47-His PSI-less cultures were grown to OD730 ∼0.65 (exponential phase) or 0.9 (post-exponential phase) and were diluted 4-fold in BG-11 medium containing 4.5 mm Na15NO3 and 2 mm 15NH4Cl. Cell samples were collected at 1, 3, 9, 24, and 48 h after the dilution. Cell pellets were resuspended in Buffer A (50 mm MES-NaOH (pH 6.0), 10 mm MgCl2, and 25% glycerol), and broken by Bead Beater (BioSpec Products, Bartlesville, OK). Cell homogenates were prepared as described (26). The cell homogenate (at 0.2 mg/ml chlorophyll) was brought to 1% β-dodecyl maltoside and incubated for 35 min at 4 °C and centrifuged. The supernatant was then loaded on an affinity column with 3 ml of nickel-nitrilotriacetic acid-agarose (Qiagen). The column was washed with 10 bed volumes of Buffer A containing 0.04% β-dodecyl maltoside and 10 mm imidazole. CP47-His and its associated proteins were eluted with 0.04% β-dodecyl maltoside and 100 mm imidazole in Buffer A. The eluted samples were precipitated with an equal volume of 50 mm MES-NaOH and 25% PEG 6000 (pH 6.0) and centrifuged. Isolated CP47-His complex samples were resuspended in Buffer A with 0.04% β-dodecyl maltoside. The oxygen evolution yield of the preparations was about 1250 μmol of O2 (mg of chlorophyll)−1 h−1.
Pigment and Protein Analysis
Isolated CP47-His complexes corresponding to 2 μg of chlorophyll were resuspended in 10 volumes of ice-cold 100% acetone with 0.1% NH4Cl and then incubated at −80 °C for 2 h and centrifuged. Chlorophyll from the supernatant was purified by HPLC using a Waters Spherisorb S10ODS2 semi-prep column (250 × 10 mm) eluted with a water/methanol-acetone gradient, and the mass distribution was determined by MALDI-TOF (11). The pellet, which contained proteins, was dissolved in SDS sample buffer (86 mm Tris-HCl (pH 8.0), 2.5% SDS, 20 mm dithiothreitol, and 0.25 m sucrose) and loaded on an SDS-12–20% polyacrylamide gradient gel containing 7 m urea (27). The gel was stained with 0.15% Coomassie Brilliant Blue R-250 in a solution of 50% methanol and 10% acetic acid. In-gel digestion to produce peptides for analysis by mass spectrometry (LC-MS/MS) was carried out essentially as described (28) using sequencing-grade modified trypsin (Promega/SDS Bioscience). For Blue Native (BN) two-dimensional gels, isolated CP47-His complex samples corresponding to 2 μg of chlorophyll were loaded. BN-PAGE was performed on a 5–14% polyacrylamide gradient gel as described (29). For separation of proteins in the second dimension, the lanes of the BN gel were excised and incubated with 25 mm Tris-HCl (pH 7.5) containing 1% SDS (v/v) for 30 min at room temperature. The lanes were then layered onto 1.5-mm-thick SDS-polyacrylamide gels. SDS-PAGE and gel staining were performed as described above.
Peptides in trypsin digests were separated using a Dionex Ultimate 3000 liquid chromatography system equipped with both a HPG 3400 M high pressure gradient pump and an LPG 3400 MB low pressure gradient pump together with a WPS3000TB autosampler and an FLM 3100B column compartment. Solvents used for peptide chromatography were X, water with 0.1% formic acid, and Y, acetonitrile with 0.1% formic acid. The LPG 3400 MB pump supplied 5% Y and 95% X at a constant flow rate of 5 μl/min and was used to load 3 μl of trypsin-digested samples onto a Dionex Acclaim PepMap 100 C18, 5 μm, precolumn cartridge (300 μm inner diameter × 5 mm length). Sample loading proceeded for 6 min, after which a valve in the column compartment placed the precolumn cartridge in line with a Dionex Acclaim PepMap 100 C18, 3-μm capillary column (75 μm inner diameter × 15 cm length) operated at a flow rate of 300 nl/min. Tryptic peptides were then separated using the following linear gradient: 0–4 min, 5% Y; 4–11.5 min, 5–20% Y; 11.5–51.5 min, 20–50% Y; 51.5–59 min, 50–65% Y; 59–69 min, 65–95% Y; 69–72 min, 95% Y; 72–74 min, 95 to 5% Y, with the remainder at each time being solvent X.
Peptides eluting from the column were analyzed using a Bruker MicroTOF-Q mass spectrometer equipped with an on-line nanospray source. Calibration was performed prior to running the first sample using sodium iodide clusters sprayed from a 200 μm solution in acetone. The inlet capillary of the mass spectrometer was set at −1500 V relative to the spray needle, and nitrogen drying gas was supplied at 180 °C and a flow rate of 3 liters/min. Spectra were acquired over an m/z range of 50–1800. Automatic MS/MS analysis with argon as the collision gas occurred at peak intensities greater than 2000 counts, with doubly charged precursors preferred. Collision energy settings for doubly charged ions were 16 eV at m/z = 350, 28 eV at m/z = 800, and 44 eV at m/z = 1200 and beyond. Collision energy settings for triply charged ions were 14 eV at m/z = 350, 24 eV at m/z = 800, and 45 eV at m/z = 1200 or greater. Data were acquired with a digitizer rate of 2 GHz with a spectra summation rate of 2 Hz. After summation of two spectra, acquisition of MS/MS spectra on each precursor was excluded for 1 min.
Data analysis, including deconvolution, was performed using Bruker Data Analysis 3.4 or 4.0 software and compound mass lists exported to Biotools 3.1 as Mascot Generic (mgf) files. Peak lists were then submitted on line to the Matrix Science website to search databases for peptide identification using the Mascot search engine. The Mascot search engine was only able to recognize unlabeled peptides for correct protein identification. For the samples with very little remaining unlabeled peptides that were not picked up by Bruker Data Analysis, we used the LC elution time to track the peptides.
The percentage of unlabeled protein remaining as a function of time was corrected for growth of the culture (OD730) during the time of labeling to be able to compare all data to those at time 0. For example, if cells doubled every 24 h and unlabeled protein at 24 h was 30% of the total intensity for that protein fragment (the remaining 70% of the protein being labeled), the % of unlabeled protein (relative to the amount at time 0) was entered as 2 × 30% = 60% in Figs. 3 and 4. The percentage of unlabeled proteins came from the average of several peptides of the whole proteins that were identified by the Mascot search engine except for the PsbF protein, for which only one peptide was detected.
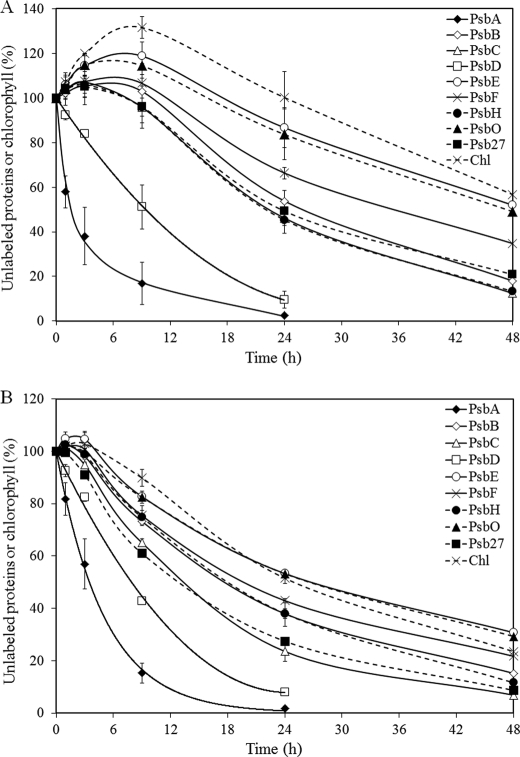
FIGURE 3.
Turnover of PSII components from PSI-less (A) and PSI-less/SCP-less (B) cells that were harvested in post-exponential growth phase (OD730∼ 0.9). The amount of unlabeled proteins and chlorophyll in these strains was followed during a 48-h period after the start of 15N labeling. 100% indicates the amount present at the start of labeling. PsbA, ♦ with solid line; PsbB, ♢ with solid line; PsbC, △ with solid line; PsbD, □ with solid line; PsbE, ○ with solid line; PsbF, × with solid line; PsbH, ● with dashed line; PsbO, ▴ with dashed line; Psb27, ■ with dashed line; and chlorophyll (Chl),: × with dashed line. Numbers on the y axis represent the percentage of unlabeled proteins/chlorophyll relative to time 0. Shown are the average results of two independent experiments ± S.D.
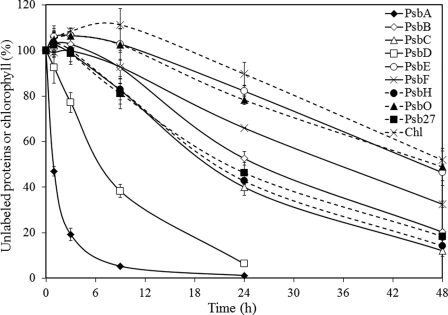
FIGURE 4.
Turnover of PSII components from PSI-less cells that were harvested in the exponential growth phase (OD730∼ 0.65). The amount of unlabeled proteins and chlorophyll in the strain is followed during a 48-h period after the start of 15N labeling. 100% indicates the amount present at the start of labeling. PsbA, ♦ with solid line; PsbB, ♢ with solid line; PsbC, △ with solid line; PsbD, □ with solid line; PsbE, ○ with solid line; PsbF, × with solid line; PsbH, ● with dashed line; PsbO, ▴ with dashed line; Psb27, ■ with dashed line; and chlorophyll (Chl), × with dashed line. Numbers on the y axis represent the percentage of unlabeled proteins/chlorophyll relative to time 0. Shown are the average results of three independent experiments ± S.D.
Chlorophyll Synthesis upon Illumination
PSI-less/chlL− and PSI-less/SCP-less/chlL− strains were grown in regular BG-11 medium with 5 mm glucose and 10 mm TES/NaOH (pH 8.0) in darkness for a week (30). Subsequently, cells were diluted 4-fold in BG-11 medium with 5 mm glucose and 10 mm TES/NaOH (pH 8.0) and also containing 4.5 mm Na15NO3 and 2 mm 15NH4Cl. Cells were then exposed to continuous illumination at an intensity of 4 μmol photons m−2 s−1. Culture samples were collected after 1, 3, 6, 9, 12, and 24 h. Pigments were extracted from the cells with 100% methanol. Chlorophyll was purified by HPLC and analyzed by MALDI-TOF.
Oxygen Evolution
Oxygen evolution measurements were performed on intact cells at 30 °C using a Clark-type electrode (Hansatech, Cambridge, UK). Electron acceptors were 2.0 mm K3Fe(CN)6 and 0.4 mm 2,5-dimethyl-p-benzoquinone. The light intensity (after filtering through a water filter and a filter transmitting >550 nm light) was saturating (2500 μmol photons m−2 s−1).
Fluorescence Spectroscopy
Fluorescence emission spectra of intact cells were measured at 77 K using a SPEX Fluorolog 2 instrument (SPEX Industries, Edison, NJ). Measurements were carried out with excitation and emission slit widths of 1 and 0.25 mm, respectively, which correspond to bandwidths of 4 and 1 nm. The excitation wavelength was 435 nm.
Aminolevulinic Acid (ALA) Supplementation
PSI-less and PSI-less/SCP-less strains were propagated for 2 weeks in 15N-BG-11 medium lacking unlabeled nitrate but containing 9 mm Na15NO3, 10 mm TES-NaOH (pH 8.0), and 5 mm glucose. As needed, cell cultures were diluted from OD730 = 0.9 to an OD730 of about 0.3 with fresh 15N-BG-11 medium. After 2 weeks, 4 mm ALA (14N) was added to the culture. After cells were grown for an additional 24 h, pigments were extracted from the cells with 100% methanol. Chlorophyll was purified by HPLC and analyzed by MALDI-TOF.
RESULTS
Identification of PSII Components
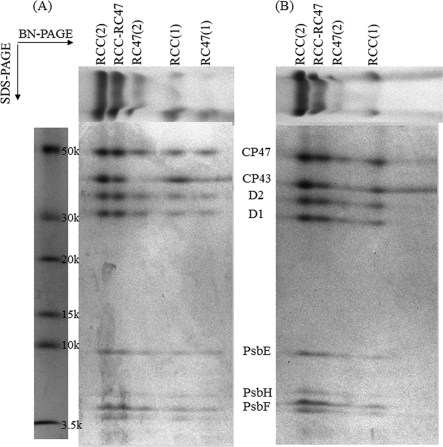
To determine the lifetime of PSII components, CP47-His PSI-less Synechocystis cells were grown in the presence of 15NO3− and 15NH4+ for a specific time period. After harvesting and breaking the cells, the total membrane fraction was solubilized using β-dodecyl maltoside, and PSII complexes were isolated via nickel column chromatography. Subsequently, PSII proteins were separated by SDS-PAGE and analyzed by LC-MS/MS mass spectrometry. The PsbA (D1), PsbB (CP47), PsbC (CP43), PsbD (D2), PsbE and PsbF (cytochrome b559), PsbH, PsbO, and Psb27 proteins were identified in the gel (Fig. 1), and their identity was confirmed by mass spectrometry analysis. Mascot scores for PSII proteins are indicated in Table 1. More detail on MS/MS analysis of individual tryptic fragments of PSII proteins is provided in supplemental Table 1. All proteins were identified reliably, but the score for PsbF was rather low as only a single peptide was identified for this component. In addition, Slr0909, a protein of unknown function whose gene is located 3.5 kbp downstream of the psbB gene in the Synechocystis genome, and the translation elongation factor Tu (Sll1099) were identified in the fraction purified on the nickel column. The pattern of proteins co-isolating with the His-tagged CP47 was identical in the PSI-less strain and the PSI-less/SCP-less strain (Fig. 1).
FIGURE 1.
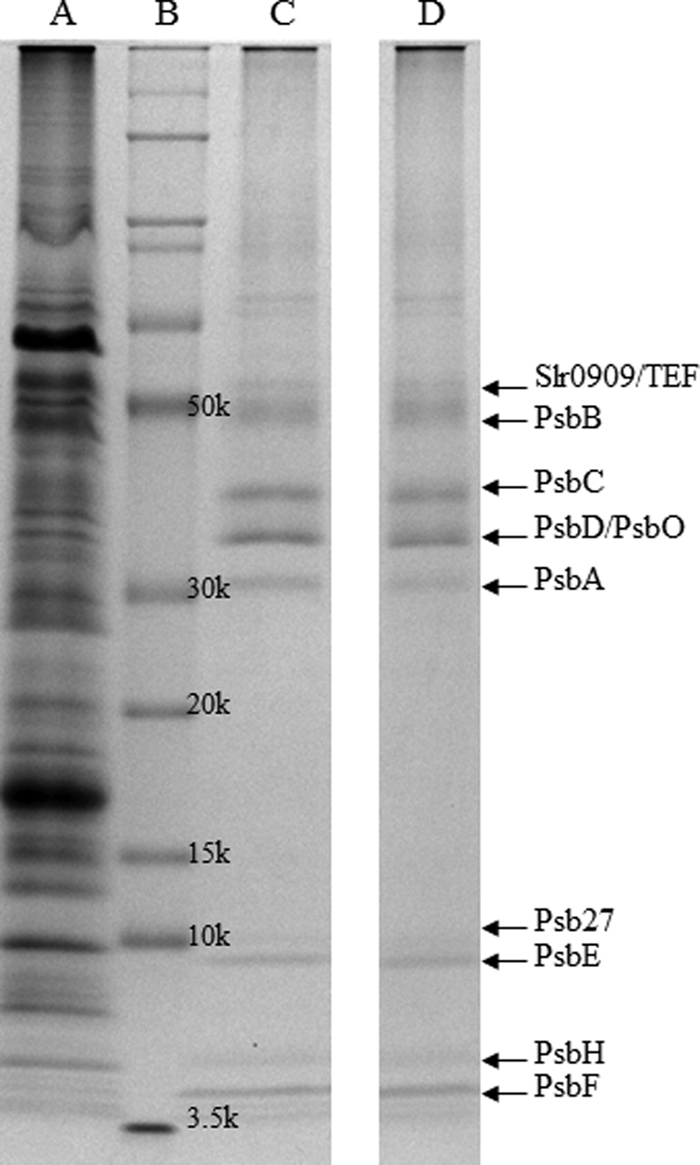
Coomassie Brilliant Blue-stained SDS-polyacrylamide gel of components co-isolating with CP47-His-purified via nickel affinity chromatography. A, fraction from PSI-less cells obtained during the washing step (0.04% β-dodecyl maltoside and 10 mm imidazole in Buffer A). B, protein ladder. C and D, fractions from PSI-less cells (C) and PSI-less/SCP-less cells (D) obtained in the elution step (0.04% β-dodecyl maltoside and 100 mm imidazole in Buffer A). Proteins were identified by LC-MS/MS. TEF (translation elongation factor-Tu, Sll1099) and Slr0909 co-purified with the PSII proteins.
TABLE 1.
Average Mascot scores of mass spectrometric identification of tryptic peptides of PSII proteins
The average Mascot score for each protein was calculated from the individual Mascot scores of each sample collected within 9 h of the start of 15N labeling for all proteins except the PsbA protein; for PsbA, only samples collected within 3 h of 15N labeling were used. Longer labeling times had insufficient unlabeled peptides that are used for determining the Mascot score.
| Protein | PsbA | PsbB | PsbC | PsbD | PsbE | PsbF | PsbH | PsbO | Psb27 |
|---|---|---|---|---|---|---|---|---|---|
| Average Mascot score | 245 ± 93 | 689 ± 222 | 540 ± 152 | 382 ± 131 | 191 ± 45 | 40 ± 14 | 72 ± 28 | 330 ± 114 | 169 ± 87 |
PSII Dynamics
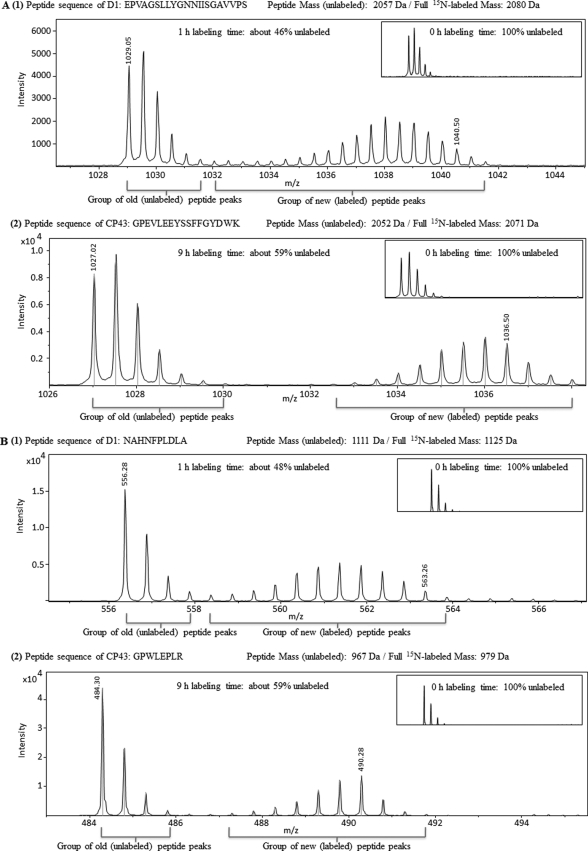
When using a stable isotope (15N) rather than traditional pulse-chase labeling with a radiolabeled tracer, both old (unlabeled) and new (labeled) peptides can be monitored at the same time using mass spectrometry. In Fig. 2, an example of the analysis is shown for D1 and CP43 peptides. MS/MS spectra are presented in supplemental Fig. 1. In Fig. 2A, the peptides are from close to the N terminus (residues 65–85 of D1, unlabeled base mass of 2057; and 123–139 of CP43, unlabeled base mass of 2052). These masses increase to 2080 and 2071, respectively, if these peptides are fully 15N-labeled. Fig. 2A shows LC-MS mass spectra of the D1 peptide after 1 h of 15N labeling and of the CP43 peptide after 9 h of labeling. The group of peaks on the left side of the spectra represents old peptides (unlabeled); slightly heavier molecules are due to natural abundance of isotopes. The group of peaks to the right side of the spectra consists of newly synthesized peptides (fully and partially 15N-labeled). The m/z values are half the theoretical mass due to the double charge of the peptide: (M + 2H)2+. The spectra of these peptides at the 0-h labeling time (Fig. 2, insets) show a distribution of peaks contributed mainly by the natural abundance of 13C. The distinction between peaks with 15N label and isotope peaks due to natural isotope abundance is clear (Fig. 2). By comparing the summed amplitudes of the peaks of old versus new peptides, the ratio between the two can be determined. The N-terminal peptide of the D1 protein had only 46% unlabeled peptides left after 1 h of 15N labeling, indicating rapid turnover of this protein even at low light intensity (4 μmol photons m−2 s−1) (Fig. 2A). In contrast, CP43 peptides still were 59% unlabeled after 9 h of 15N labeling (Fig. 2A).
FIGURE 2.
LC-MS/MS spectra of peptides from near the N terminus (A) and C terminus (B) of PsbA (D1) that was 15N-labeled for 1 h (panel 1) and of PsbC (CP43) that was 15N-labeled for 9 h (panel 2) in PSI-less cells. The insets show controls that were labeled for 0 h. All peptides were detected as doubly charged (z = 2) molecules.
Ribosome pausing has been postulated to occur upon translation of psbA message (31). If so, then the labeling rate of a C-terminal region of the D1 protein is expected to have a delay relative to that of the N-terminal region. To test whether significant ribosome pausing occurs that would delay the labeling of C-terminal regions of the D1 protein, the labeling of a D1 peptide near the C terminus (Fig. 2B) was compared with that shown in Fig. 2A. After 1 h of labeling, the amount of labeled D1 near the C terminus was 52%, whereas it was 54% near the N terminus. This suggests that if ribosome pausing occurs, it is no more than a couple of minutes. A similar observation was made for labeling of a CP43 peptide near the C terminus; after 9 h, the amount of labeling (41%) was identical for a peptide near the C terminus versus near the N terminus (Fig. 2).
Labeling of the PSII proteins as well as chlorophyll was followed over time (Fig. 3). The cell number increased during the 15N-labeling period, and therefore, the total amount of PSII proteins increased as well. What is indicated in Fig. 3A is the amount of remaining unlabeled protein in the cells over the labeling period relative to the amount of unlabeled protein at time 0. Interestingly, the amount of unlabeled material initially increased in the first 6–9 h for many of the PSII proteins (except PsbA and PsbD) as well as for chlorophyll. This increase in the amount of unlabeled protein largely was absent in the PSI-less/SCP-less strain (Fig. 3B). The half-lives of PSII components were determined by monitoring the disappearance of old (unlabeled) peptides in the time period between 9 and 48 h after the start of labeling, whereas for PsbA and PsbD the time range starting at time 0 h was used (Table 2). Table 2 shows that the half-life time of most PSII proteins is independent of the presence of SCPs and that the various PSII proteins have greatly different lifetimes. The half-life of the D1 protein was 1.5 h in the PSI-less strain, and the D2 protein was 5-fold more stable with a half-time of 7.5 h. The CP47 and CP43 proteins and the PsbH protein that participates in the binding of chlorophyll together with CP47 (32) are about 2-fold more stable than D2, with half-times of 13–15 h. The cytochrome b559 proteins (PsbE and PsbF proteins), which are the anchor proteins for PSII (33), are the most stable intrinsic PSII proteins with half-times of about a day. The two luminal proteins, PsbO and Psb27, have lifetimes similar to that of some of the slower degrading integral membrane proteins. PsbO, the manganese-stabilizing protein in the oxygen-evolving complex, has a particularly long half-life (24–33 h), presumably because it can be dissociated from damaged PSII and reused for repaired/new PSII. The Psb27 protein, which is an assembly factor mainly associated with CP47 and CP43 of monomeric PSII and non-oxygen-evolving PSII complexes (34, 35), had a 13–15-h halftime. Interestingly, in the PSI-less background strain, chlorophyll had a half-life time of about 40 h, longer than any of the PSII proteins. This illustrates that chlorophyll is largely reutilized when chlorophyll-binding proteins turn over.
TABLE 2.
Comparison of half-life times of PSII components in PSI-less and PSI-less/SCP-less strains
The half-life times (in hours) were calculated from the decrease in the percentage of unlabeled protein correcting for the increase in unlabeled protein that occurred for the longer lived polypeptides and the chlorophyll, particularly in PSI-less cells in post-exponential phase (OD730 ∼0.9). Listed are the average results of two to five independent experiments ± S.D.
| Strains | Half-life time (h) |
|
|---|---|---|
| PSI-less | PSI-less/SCP-less | |
| PsbA (D1) | 1.5 ± 0.5a | 2.5 ± 0.5 |
| PsbB (CP47) | 15 ± 1 | 15 ± 2 |
| PsbC (CP43) | 13 ± 1 | 11 ± 2 |
| PsbD (D2) | 7.5 ± 1 | 7 ± 0.5 |
| PsbE | 28 ± 2 | 25 ± 2 |
| PsbF | 23 ± 3 | 20 ± 2 |
| PsbH | 14 ± 1 | 15 ± 1 |
| PsbO | 33 ± 4 | 24 ± 2 |
| Psb27 | 15 ± 1 | 13 ± 1 |
| Chlorophyll | 40 ± 2 | 20 ± 1 |
a In exponential growth phase (OD730 ∼0.65), the half-life time of the D1 protein is 1 h. This difference is due to a small contribution of unlabeled D1 that is incorporated into PSII complexes in cells at higher density (OD730 ∼0.9).
Absence of SCP has been shown to reduce the lifetime of chlorophylls (18). Removal of SCPs does not greatly alter the half-life time of most PSII proteins. In the PSI-less/SCP-less mutant, the lifetimes of the CP43, PsbE, PsbF, and Psb27 proteins were slightly shorter compared with those in the PSI-less strain (Table 2), and the PsbO lifetime seemed to have been affected a little more. However, the biggest effect of the SCP deletion was on the lifetime of chlorophyll, which was about 50% shorter relative to that in the PSI-less strain. The fact that absence of SCPs affects the lifetime of PSII-associated chlorophyll more than that of PSII proteins confirms the notion that SCPs aid in chlorophyll stabilization and recycling upon PSII degradation and reassembly (18). However, as the chlorophyll lifetime in the PSI-less/SCP-less strain exceeds that of the main chlorophyll-binding PSII proteins, CP47 and CP43, some chlorophyll recycling occurs even in the absence of SCPs.
Pool of Nascent PSII Components
As indicated in Fig. 3, in the PSI-less strain there was a 3–9-h period after the start of labeling during which the amount of unlabeled PSII proteins (except D1 and D2) as well as chlorophyll continued to increase. This cannot be due to a slow incorporation of label into amino acids as otherwise we should also have observed a significant lag for the D1 and D2 polypeptides. Our interpretation of the increase in unlabeled complexes is that there is a significant amount of PSII proteins and chlorophyll in thylakoid membranes that is not incorporated into mature PSII complexes and that provides parts for PSII assembly and repair.
To test our interpretation of the increase in unlabeled polypeptide being due to a reservoir of unfinished PSII complexes in the cell, the labeling experiment with PSI-less cells was repeated with cells from exponential phase (OD730 ∼0.65) instead of the post-exponential phase (OD730 ∼0.9) as in exponential phase the reservoir may be expected to be smaller due to a faster de novo PSII biosynthesis (Fig. 4). The lifetimes of the PSII components were similar to those measured in cells in post-exponential phase. However, the increase in the amount of unlabeled protein is much less, and the time over which an increase is observed is much shorter. These results support our interpretation and demonstrate that cells in the post-exponential growth phase tend to accumulate PSII proteins and chlorophyll to be ready for later use.
Role of SCPs
As indicated in Fig. 3B, the increase in the amount of unlabeled PSII protein in cells in the post-exponential phase is much smaller in the PSI-less/SCP-less strain relative to in the PSI-less control, whereas the half-life time of the PSII proteins is not much affected. This is very much comparable with what was shown above for cells in exponential phase. These data suggest that pools of nascent PSII proteins and protein-bound chlorophyll may be stabilized by SCPs.
To test the interpretation that SCPs are involved with stabilization of nascent PSII complexes and/or intermediates, isolated PSII samples from the PSI-less strain and the PSI-less/SCP-less strain were run on BN-SDS-PAGE. As indicated in Fig. 5, the main difference between PSII complexes from the PSI-less and PSI-less/SCP-less strains was that the RC47(1) complex, corresponding to the PSII monomer form without the CP43 protein, was missing in the PSII preparation from the PSI-less/SCP-less strain. However, the RC47(2) band, corresponding to the RC47 dimer, was present in preparations from both strains. In PSII biogenesis, RC47 complexes are assembled from RC complexes (D1, D2, cytochrome b559, and PsbI proteins) and sub-CP47 complexes (CP47, PsbH, and SCP proteins) (36, 37). SCP association could aid the stability of the RC47 complex before dimerization occurs. These observations suggest that our interpretation of SCPs stabilizing intermediates in formation of the mature PSII complex is reasonable. It should be noted that the horizontal smear that vertically comigrates with CP43 and that is to the right of CP43 incorporated in PSII complexes actually is not due to CP43. As indicated in supplemental Fig. 2, this band does not cross-react with CP43 antibodies, although immunoreaction is observed with CP43 itself. The identity of the band to the right of CP43 (Fig. 5) has not been determined as it is not relevant to this study.
FIGURE 5.
BN-PAGE followed by SDS-polyacrylamide gel for PSII complexes co-isolating with CP47-His from the PSI-less strain (A) and the PSI-less/SCP-less strain (B). The bands on BN-PAGE (top) are unstained and are colored by the native chlorophyll and Coomassie Brilliant Blue. The SDS-polyacrylamide gel was stained with Coomassie Brilliant Blue. RCC(2), mature PSII dimer; RCC-RC47, PSII dimer lacking CP43 in one of the monomers; RC47(2), PSII dimer lacking CP43 in both monomers; RCC(1), PSII monomer; and RC47(1), PSII monomer lacking CP43.
Another role of the SCPs involves chlorophyll binding and stabilization, and earlier work has shown that deletion of SCPs in the PSI-less background strain also affects the chlorophyll content per cell and the accumulation of chlorophyll precursors (20, 23). However, the location of this SCP effect has not yet been pinpointed. The stability of PSII is not affected by SCPs as shown in Table 2. In line with earlier observations (18), we do not see evidence of SCPs stably binding a large amount of chlorophyll as the oxygen evolution rate in the PSI-less strain was 2480 ± 80 μmol of O2 (mg of Chl)−1 h−1, whereas in the PSI-less/SCP-less strain, this rate was 2730 ± 180 μmol of O2 (mg of Chl)−1 h−1. Thus, the number of PSII reaction centers on a per chlorophyll basis is similar regardless of the presence of SCPs.
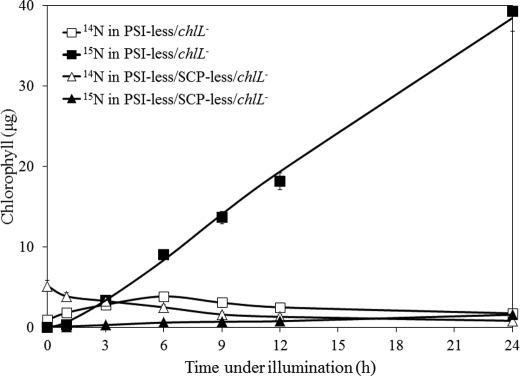
Earlier work had indicated the lack of accumulation of chlorophyll precursors in SCP-less strains (23) suggesting an early block in chlorophyll biosynthesis in the absence of SCPs. The 15N-labeling approach provides an opportunity to monitor the effects of the absence of SCPs in more detail. After growth of the PSI-less/chlL− and PSI-less/SCP-less/chlL− strains in darkness for 5 days, the strains had little chlorophyll left as ChlL is required for light-independent protochlorophyllide reduction and chlorophyll synthesis (Fig. 6). After incubation in darkness, the culture was diluted 4-fold with medium containing 4.5 mm Na15NO3 and 2 mm 15NH4Cl, and the culture was transferred to continuous illumination at 4 μmol photons m−2 s−1. As indicated in Fig. 6, during the first 6 h of illumination, the PSI-less/chlL− strain synthesized primarily labeled (15N) chlorophyll along with some unlabeled (14N) chlorophyll. Unlabeled chlorophyll may have been synthesized in part from accumulated protochlorophyllide or other available precursors. However, in the PSI-less/SCP-less/chlL− strain, the amount of unlabeled chlorophyll decreased, and labeled (15N) chlorophyll was synthesized very slowly (about 20-fold slower than in the PSI-less/chlL− strain). The decrease in unlabeled chlorophyll and the slow increase in labeled chlorophyll suggest that in the SCP-less mutant the amount of chlorophyll precursors is greatly diminished, and tetrapyrrole biosynthesis is impaired.
FIGURE 6.
Unlabeled (14N) and labeled (15N) chlorophyll from the PSI-less/chlL− and PSI-less/SCP-less/chlL− strains upon illumination. Cells were grown in the dark for 5 days. The cultures were illuminated after they had been diluted with 3 volumes of BG-11 medium containing Na15NO3 and 15NH4Cl. Chlorophyll was extracted from the cells for analysis at the times indicated. Open and closed symbols represent unlabeled (14N) and labeled (15N) chlorophyll, respectively, in the PSI-less/chlL− (squares) and PSI-less/SCP-less/chlL− (triangles) strains. Shown are the average results of two independent experiments ± S.D.
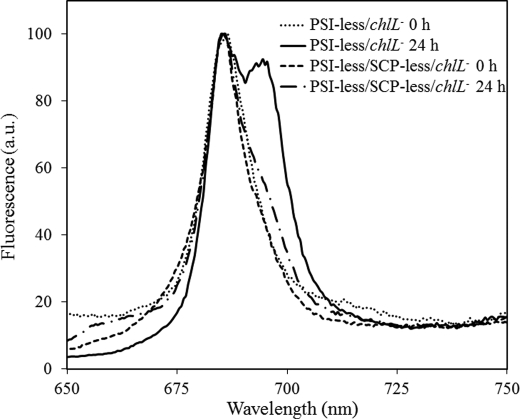
A very slow synthesis of PSII in the PSI-less/SCP-less/chlL− strain was confirmed by 77 K fluorescence emission spectra after 24 h of illumination (Fig. 7). A peak at 695 nm corresponds to CP47-associated chlorophyll and reflects intact PSII complexes. Although in the PSI-less/chlL− strain a significant amount of PSII complexes was present after 24 h of illumination, in the PSI-less/SCP-less/chlL− strain only very little 695 nm emission was observed.
FIGURE 7.
77 K fluorescence emission spectra of Synechocystis sp. PCC 6803 cells lacking PSI and ChlL. Cells had been grown in darkness for a week and were then transferred to continuous light (4 μmol photons m−2 s−1) for 0 h (PSI-less/chlL−, dotted line; PSI-less/SCP-less/chlL−, dashed line) or 24 h (PSI-less/chlL−, solid line; PSI-less/SCP-less/chlL−, dashed/dotted line). The spectra were normalized to 100 at 683 nm, where phycobilisomes and some chlorophylls emit maximally. The excitation wavelength was 435 nm. a.u., arbitrary units.
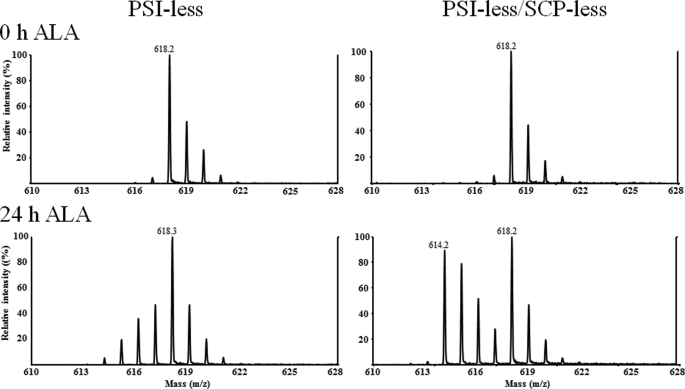
The very slow chlorophyll biosynthesis and the lack of a large amount of unlabeled chlorophyll synthesis upon illumination of the PSI-less/SCP-less/chlL− strain suggest that early intermediates in chlorophyll biosynthesis may have been depleted. Therefore, we wished to determine whether ALA, an early intermediate in tetrapyrrole biosynthesis, was depleted in strains lacking SCPs. As labeled ALA was not readily available, PSI-less and PSI-less/SCP-less strains, carrying normal ChlL, were grown in 15N medium for 2 weeks (the culture was diluted with fresh medium as needed) so that chlorophyll in the cells was fully labeled. The cultures were then supplemented with 4 mm 14N-ALA, and cells were grown for 24 h with added ALA. If cells can readily make their own ALA, then ALA and thereby chlorophyll will be 15N-labeled, and if cells are limited in their ALA supply and need to utilize exogenous ALA, then newly synthesized chlorophyll will be mostly unlabeled. Pigments were extracted from both strains, and chlorophyll was purified by HPLC, and chlorophyll was analyzed by MALDI-TOF. As shown in Fig. 8, chlorophyll from both strains was fully 15N-labeled at 0 h before the ALA supplementation, as expected. After 24 h of ALA supplementation, during which time the chlorophyll amount in the culture virtually doubled, the mass spectrum of chlorophyll from the PSI-less strain showed that the ALA used for chlorophyll synthesis primarily was [15N]ALA, which cells had synthesized themselves. In contrast, newly synthesized chlorophyll from the PSI-less/SCP-less strain was primarily unlabeled as the amounts of labeled (original) and unlabeled (newly synthesized) chlorophyll were about the same. These results suggest that the ALA amount is limiting as a consequence of the SCP deletion and therefore that SCPs appear to play an important role in the very early steps of tetrapyrrole biosynthesis (ALA formation).
FIGURE 8.
MALDI-TOF mass spectra of chlorophyll isolated from 15N-grown PSI-less cells with (left) or without (right) SCPs that were supplemented with 14N-ALA for 0 (top) or 24 (bottom) h. Prior to the experiment, cells were grown in 15N medium, containing Na15NO3, for 10 days, and the culture was diluted with 15N BG-11 to OD730 = 0.35 at time 0. Unlabeled (14N) aminolevulinic acid was added at time 0 to a final concentration of 4 mm.
DISCUSSION
PSII Protein and Chlorophyll Turnover
Previous work with radioisotopes had indicated that different PSII polypeptides differed in their lifetimes (9, 10), and work from our group had indicated that the chlorophyll lifetime in the cell was very long (11, 18). It is clear that careful orchestration of the synthesis, assembly, and repair of photosynthetic complexes is required, but until now very few data were available on the lifetimes of the longer lived protein components, and there is little known about how this orchestration may occur and how it is regulated. Using stable isotope labeling combined with mass spectrometry, both labeled (new) and unlabeled (old) peptides can be detected. The rate of disappearance of unlabeled peptides over time determines the turnover rate of the proteins, and this approach allowed an accurate and comprehensive determination of lifetimes of PSII components.
PSII complexes were isolated from PSI-less Synechocystis cultures grown at a light intensity of 4 μmol photons m−2 s−1, making use of a His tag attached to the PsbB (CP47) protein. Therefore, only polypeptides and complexes associated with PsbB with an exposed C-terminal His tag will be detected. The fraction obtained after His tag affinity purification contained at least nine PSII proteins (seven intrinsic proteins, including the reaction center proteins PsbA and PsbD, the chlorophyll-binding proteins PsbB and PsbC, the cytochrome b559 proteins PsbE and PsbF, PsbH, and two lumenal proteins (PsbO and Psb27)). Other PSII proteins may not have stained sufficiently well to be detected. Moreover, a translation elongation factor, Tu (Sll1099), and Slr0909 co-isolated with the PSII complex. In a previous study, Sll1099 was also seen from co-isolating with PSII-associated His-ScpB (38). Slr0909 is a protein of unknown function, but it is encoded about 3 kbp downstream from psbB, and slr0909 could possibly be co-transcribed with psbB and an intervening gene of unknown function.
As indicated in Table 2, PSII proteins have very different half-life times ranging from 1.5 to 33 h. Therefore, PSII proteins (or groups of such polypeptides) turn over and are replaced independently from each other, and the remaining PSII proteins appear to be reused for assembly into a functional PSII complex. Our results confirm the radiolabeling-based interpretations of other groups (3, 4) that the rapidly synthesized D1 protein, which is the PSII polypeptide with the shortest lifetime, is actually incorporated into PSII complexes. The fast turnover of D1 in the PSII complex (5–20 times faster than turnover rates of the other PSII proteins) poses a challenge for the PSII complex as D1 is a central part rather than a peripheral part of the complex. However, the lipids around the reaction center may facilitate the replacement of damaged D1 proteins (39). The other PSII proteins from the damaged complex may stay together and be re-assembled around a new D1 polypeptide, or subcomplexes may form a pool from which new complexes are formed (40–42). In the latter case, PSII complexes may be assembled from old polypeptides originating from different PSII complexes.
The light intensity used for this study is relatively low (4 μmol photons m−2 s−1) as the PSI-less strain used for these studies does not survive at light intensities above 10 μmol photons m−2 s−1. The turnover rate of the D1 protein increases with increasing light intensity, and D1 has a half-life time of less than an hour (about 30 min) in wild type grown at 75 μmol photons m−2 s−1 (data not shown). Therefore, the large change in light intensity does not affect the D1 turnover rate by more than about a factor of 2 (log phase cells) or 3 (older cultures) (Table 2).
The lifetime of chlorophyll in various mutant strains has been studied earlier by means of stable isotope labeling, and because chlorophyll is much more stable than chlorophyll-binding proteins according to the lifetimes of chlorophyll-binding proteins estimated from the pulse-chase method, we have suggested that chlorophyll is recycled upon degradation of chlorophyll-binding PSII components (18). In this study, we experimentally support the earlier interpretations by comparing the half-life times of the main chlorophyll-binding proteins in PSII (D1, D2, CP47, and CP43) (t½ = 1.5–15 h) with the half-life time of chlorophyll extracted from isolated PSII (t½ = 40 h) (Table 2). Therefore, chlorophyll in damaged chlorophyll-binding proteins can be re-utilized.
SCPs and Chlorophyll Reutilization
In cyanobacteria the SCP proteins bind chlorophyll, may associate with damaged PSII centers, and serve as a temporary pigment reservoir, whereas PSII components are being replaced (19, 21, 23, 36). We have now analyzed how SCPs affect the lifetimes of PSII polypeptides and chlorophyll. In line with earlier observations (18), the lifetime of PSII chlorophyll was reduced by half in the PSI-less/SCP-less strain compared with the PSI-less strain. However, the lifetimes of the chlorophyll-binding proteins in PSII (D1, D2, CP47, and CP43) and of PsbH that participates in the binding of chlorophyll remained essentially unchanged (Table 2). This indicates that SCPs function primarily in reutilization of chlorophyll and do not affect the stability of chlorophyll-binding proteins.
Interestingly, although the lifetimes of PSII proteins were not changed or were decreased slightly in the SCP-less strain, the lifetime of D1 was increased (Table 2 and Fig. 3). The longer lifetime of D1 may have been caused by slower assembly of PSII due to the lack of chlorophyll availability, which affects D1 translation and processing in Synechocystis (43). Chlorophyll availability is thought to be lower in the absence of SCPs as the rate of chlorophyll synthesis was decreased 4-fold in the PSI-less/SCP-less strain, and the chlorophyll content per cell was almost 4-fold less than in the PSI-less strain (Fig. 6) (18, 23). Therefore, during the replacement of the D1 protein in the PSII repair cycle, the proteins may be waiting for available chlorophylls to assemble PSII, which appears to take longer in the absence of SCPs.
SCPs Stabilize Nascent PSII Protein Complexes
As shown in Fig. 3, unlabeled PSII proteins (except D1 and D2) increased for the first 3–9 h of 15N labeling if late log cultures are used and SCPs are present. The D1 and D2 proteins did not show this increase in unlabeled protein, excluding the possibility that there is an extensive pool of unlabeled amino acids that remains available for many hours after the start of labeling. Instead, the most plausible explanation is that the increase in unlabeled protein originates from PSII proteins that were present at time 0 but that were not associated with PsbB with an exposed C-terminal His tag at that time. Fig. 5 shows that the His tag-isolated samples consist of mature PSII complexes in monomer and dimer forms as well as RC47 (PSII without CP43) dimers and RC47/mature PSII dimers. These complexes are in line with what was observed previously (44, 45). However, RC47 monomers were found to occur only in the presence of SCPs, suggesting that SCPs can stabilize biosynthetic intermediates of PSII complexes. Indeed, even when cells are in late log stage, in the PSI-less/SCP-less mutant very little synthesis of unlabeled PSII components is observed after the start of labeling (Fig. 3B), suggesting that no significant accumulation of such PSII biosynthesis intermediates occurs if SCPs are absent.
SCPs have been found to be associated with PSII (21, 38). Analysis of isolated His-tagged ScpD complexes suggests that ScpD binds to CP47 proteins in the vicinity of PsbH, and SCPs have been found to be associated with PSII throughout its biogenesis, from just CP47 proteins to monomeric PSII complexes (21, 36). Therefore, even though SCPs do not stabilize PSII components in mature PSII complexes (Table 2), there is significant SCP-induced stabilization of nascent PSII complexes.
SCPs and ALA Biosynthesis
In addition, SCPs also appear to be involved, directly or indirectly, in ALA biosynthesis. This effect results in slow chlorophyll biosynthesis in the absence of SCPs (Fig. 6) and in slow generation of PSII complexes (Fig. 7). In contrast to the PSI-less strain that does not use much exogenous ALA and that therefore can generate and utilize sufficient internal ALA, the PSI-less/SCP-less mutant readily uses exogenous ALA for chlorophyll biosynthesis (Fig. 8). The most straightforward interpretation of these results is that deletion of SCPs in the PSI-less background strain severely impairs the ALA biosynthesis pathway. However, ALA synthesis is thought to occur in the cytoplasm (46), and SCPs are transmembrane proteins located in thylakoid membranes. This suggests that the SCP-mediated regulation of ALA synthesis is not a direct effect but rather may be influenced by intermediate events.
ALA synthesis is a tightly regulated step that is negatively regulated by, for example, protochlorophyllide that accumulates in darkness in plants (47). However, based on results of earlier studies, there is no accumulation of protochlorophyllide or earlier intermediates such as Mg-protoporphyrin IX in PSI-less and PSI-less/SCP-less strains if chlorophyll biosynthesis has not been impaired (23). However, chlorophyllide, which is chlorophyll without the phytyl tail, accumulates as the chlorophyll biosynthesis rate and chlorophyll content per cell decrease in the PSI-less/SCP-less strain (23). It is possible that chlorophyllide, which is more hydrophilic than chlorophyll, may serve as a signal for ALA biosynthesis enzymes to negatively regulate the output of ALA, thus reducing overall chlorophyll biosynthesis in the PSI-less/SCP-less strain (Fig. 8). Indeed, in plants the reduced expression and activity of chlorophyll synthase also has been shown to cause a feedback-controlled inactivation of ALA synthesis (48).
In conclusion, stable isotope labeling (15N), mass spectrometry, and well defined mutants are a powerful combination to provide insights in the assembly and synthesis of PSII polypeptides and associated cofactors; chlorophyll already is associated with subcomplexes that are intermediates in PSII synthesis. SCPs aid the stability of nascent PSII complexes in PSII biogenesis and also affect the flux from ALA through the tetrapyrrole biosynthesis pathway to chlorophyll. This work illustrates the multiple levels of control and regulation that pigments and SCPs have in PSII synthesis and assembly.
Supplementary Material
This work was supported by Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy Grant DE-FG02-08ER15543.

This article contains supplemental Table 1 and Figs. 1 and 2.
- PSII
- photosystem II
- PSI
- photosystem I
- SCP
- small Cab-like protein
- ALA
- aminolevulinic acid
- TES
- N-tris(hydroxymethyl)-2-aminoethanesulfonic acid
- Chl
- chlorophyll
- BN
- Blue Native.
REFERENCES
- 1. Powles S. B. (1984) Annu. Rev. Plant Physiol. 35, 15–44 [Google Scholar]
- 2. Sonoike K. (2006) in Advances in Photosynthesis and Respiration (Golbeck J., ed) Vol. 24, pp. 657–668, Springer, Dordrecht, The Netherlands [Google Scholar]
- 3. Mattoo A. K., Hoffman-Falk H., Marder J. B., Edelman M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohad I., Kyle D. J., Arntzen C. J. (1984) J. Cell Biol. 99, 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vass I., Styring S., Hundal T., Koivuniemi A., Aro E., Andersson B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. MacPherson A. N., Telfer A., Barber J., Truscott T. G. (1993) Biochim. Biophys. Acta 1143, 301–309 [Google Scholar]
- 7. Krieger-Liszkay A., Fufezan C., Trebst A. (2008) Photosynth. Res. 98, 551–564 [DOI] [PubMed] [Google Scholar]
- 8. Prasil O., Adir N., Ohad I. (1992) in Topics in Photosynthesis (Barber J., ed) Vol. 2, pp. 293–348, Elsevier, Amsterdam [Google Scholar]
- 9. Schuster G., Timberg R., Ohad I. (1988) Eur. J. Biochem. 177, 403–410 [DOI] [PubMed] [Google Scholar]
- 10. Mattoo A. K., Giardi M. T., Raskind A., Edelman M. (1999) Physiol. Plant. 107, 454–461 [Google Scholar]
- 11. Vavilin D., Brune D. C., Vermaas W. (2005) Biochim. Biophys. Acta 1708, 91–101 [DOI] [PubMed] [Google Scholar]
- 12. Mullet J. E., Klein P. G., Klein R. R. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4038–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krieger-Liszkay A. (2005) J. Exp. Bot. 56, 337–346 [DOI] [PubMed] [Google Scholar]
- 14. Triantaphylidès C., Havaux M. (2009) Trends Plant Sci. 14, 219–228 [DOI] [PubMed] [Google Scholar]
- 15. Funk C., Vermaas W. (1999) Biochemistry 38, 9397–9404 [DOI] [PubMed] [Google Scholar]
- 16. Jansson S. (1994) Biochim. Biophys. Acta 1184, 1–19 [DOI] [PubMed] [Google Scholar]
- 17. Jansson S., Andersson J., Kim S. J., Jackowski G. (2000) Plant Mol. Biol. 42, 345–351 [DOI] [PubMed] [Google Scholar]
- 18. Vavilin D., Yao D., Vermaas W. (2007) J. Biol. Chem. 282, 37660–37668 [DOI] [PubMed] [Google Scholar]
- 19. Storm P., Hernandez-Prieto M. A., Eggink L. L., Hoober J. K., Funk C. (2008) Photosynth. Res. 98, 479–488 [DOI] [PubMed] [Google Scholar]
- 20. Xu H., Vavilin D., Funk C., Vermaas W. (2002) Plant Mol. Biol. 49, 149–160 [DOI] [PubMed] [Google Scholar]
- 21. Yao D., Kieselbach T., Komenda J., Promnares K., Prieto M. A., Tichy M., Vermaas W., Funk C. (2007) J. Biol. Chem. 282, 267–276 [DOI] [PubMed] [Google Scholar]
- 22. Sobotka R., McLean S., Zuberova M., Hunter C. N., Tichy M. (2008) J. Bacteriol. 190, 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu H., Vavilin D., Funk C., Vermaas W. (2004) J. Biol. Chem. 279, 27971–27979 [DOI] [PubMed] [Google Scholar]
- 24. Shen G., Boussiba S., Vermaas W. F. J. (1993) Plant Cell 5, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 26. Bricker T. M., Morvant J., Masri N., Sutton H. M., Frankel L. K. (1998) Biochim. Biophys. Acta 1409, 50–57 [DOI] [PubMed] [Google Scholar]
- 27. Komenda J., Lupínková L., Kopecký J. (2002) Eur. J. Biochem. 269, 610–619 [DOI] [PubMed] [Google Scholar]
- 28. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 29. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 30. Wu Q., Vermaas W. F. (1995) Plant Mol. Biol. 29, 933–945 [DOI] [PubMed] [Google Scholar]
- 31. Zhang L., Aro E. M. (2002) FEBS Lett. 512, 13–18 [DOI] [PubMed] [Google Scholar]
- 32. Müh F., Renger T., Zouni A. (2008) Plant Physiol. Biochem. 46, 238–264 [DOI] [PubMed] [Google Scholar]
- 33. Stewart D. H., Brudvig G. W. (1998) Biochim. Biophys. Acta 1367, 63–87 [DOI] [PubMed] [Google Scholar]
- 34. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 35. Cormann K. U., Bangert J. A., Ikeuchi M., Rögner M., Stoll R., Nowaczyk M. M. (2009) Biochemistry 48, 8768–8770 [DOI] [PubMed] [Google Scholar]
- 36. Promnares K., Komenda J., Bumba L., Nebesarova J., Vacha F., Tichy M. (2006) J. Biol. Chem. 281, 32705–32713 [DOI] [PubMed] [Google Scholar]
- 37. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kufryk G., Hernandez-Prieto M. A., Kieselbach T., Miranda H., Vermaas W., Funk C. (2008) Photosynth. Res. 95, 135–145 [DOI] [PubMed] [Google Scholar]
- 39. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2007) Biochim. Biophys. Acta 1767, 509–519 [DOI] [PubMed] [Google Scholar]
- 40. Smith D., Howe C. J. (1993) FEMS Microbiol. Lett. 110, 341–347 [Google Scholar]
- 41. Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nixon P. J., Barker M., Boehm M., de Vries R., Komenda J. (2005) J. Exp. Bot. 56, 357–363 [DOI] [PubMed] [Google Scholar]
- 43. He Q., Vermaas W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E. M. (2004) Plant Physiol. 134, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C. W., Tichy M., Nixon P. J. (2006) J. Biol. Chem. 281, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 46. Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009) Mol. Plant 2, 1154–1180 [DOI] [PubMed] [Google Scholar]
- 47. Richter A., Peter E., Pörs Y., Lorenzen S., Grimm B., Czarnecki O. (2010) Plant Cell Physiol. 51, 670–681 [DOI] [PubMed] [Google Scholar]
- 48. Shalygo N., Czarnecki O., Peter E., Grimm B. (2009) Plant Mol. Biol. 71, 425–436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.