Background: The endogenous l-α-lysophosphatidylinositol activates GPR55.
Results: Structural analogues of SR141716A act both as agonists alone and as inhibitors of l-α-lysophosphatidylinositol. Certain CB2 receptor agonists also modulate GPR55 activity.
Conclusion: Certain cannabinoids can both activate GPR55 and attenuate l-α-lysophosphatidylinositol-mediated phosphorylated ERK1/2 activation. This has mechanistic implications for the antinociceptive effects of certain CB2 agonists.
Significance: Cannabinoid ligands have complex interactions with the l-α-lysophosphatidylinositol/GPR55 signaling system.
Keywords: Allosteric Regulation, Cannabinoid Receptors, Cannabinoids, G Protein-coupled Receptors (GPCR), Pharmacology, Acomplia, AlphaScreen, Bitopic Ligand, GPR55, PLX-4720 BRAF Inhibitor
Abstract
GPR55 is activated by l-α-lysophosphatidylinositol (LPI) but also by certain cannabinoids. In this study, we investigated the GPR55 pharmacology of various cannabinoids, including analogues of the CB1 receptor antagonist Rimonabant®, CB2 receptor agonists, and Cannabis sativa constituents. To test ERK1/2 phosphorylation, a primary downstream signaling pathway that conveys LPI-induced activation of GPR55, a high throughput system, was established using the AlphaScreen® SureFire® assay. Here, we show that CB1 receptor antagonists can act both as agonists alone and as inhibitors of LPI signaling under the same assay conditions. This study clarifies the controversy surrounding the GPR55-mediated actions of SR141716A; some reports indicate the compound to be an agonist and some report antagonism. In contrast, we report that the CB2 ligand GW405833 behaves as a partial agonist of GPR55 alone and enhances LPI signaling. GPR55 has been implicated in pain transmission, and thus our results suggest that this receptor may be responsible for some of the antinociceptive actions of certain CB2 receptor ligands. The phytocannabinoids Δ9-tetrahydrocannabivarin, cannabidivarin, and cannabigerovarin are also potent inhibitors of LPI. These Cannabis sativa constituents may represent novel therapeutics targeting GPR55.
Introduction
The physiological roles of GPR55 and its possible involvement in the pathophysiology of medical conditions are emerging. Studies in mice lacking GPR55 have reported a reduction in inflammatory and neuropathic pain (1), and these mice have increased bone mass (2). Subsequently, studies have established that the endogenous lysophospholipid LPI2 activates GPR55 (3–10). In certain cancer cell lines, GPR55 is highly expressed, and LPI mediates increased cell migration, invasion, and proliferation (3, 8, 11). Moreover, increased circulating levels of LPI have been found in cancer patients and are associated with a poor prognosis (12). These observations suggest that modulation of GPR55 may have therapeutic implications for the treatment of pain, bone diseases, and cancer.
To date, certain arylpyrazole CB1 receptor antagonists such as Rimonabant® (also known as SR141716A or Acomplia®) and AM251 have been reported to be GPR55 agonists (5, 13, 14). However, other groups have suggested that Rimonabant® is a GPR55 antagonist (6, 8). Furthermore, reports on the behavior of Δ9-THC at GPR55 are also inconsistent. Lauckner et al. (6) have shown that Δ9-THC is a GPR55 agonist capable of stimulating calcium release, and Kapur et al. (5) did not detect β-arrestin-mediated activation of GPR55 with this phytocannabinoid. Another cannabis constituent, cannabidiol (CBD), is reported to be an antagonist of GPR55 (15). The GPR55 pharmacology of many other Cannabis sativa (C. sativa) constituents has still to be investigated. Furthermore, the pharmacology of various CB2 receptor-selective ligands at GPR55 has not been investigated. This is important because CB2 ligands are antinociceptive, a characteristic that may be shared by GPR55 ligands.
Activation of GPR55 by LPI, but also by certain cannabinoid ligands, initiates multiple signaling pathways distinct from those initiated by cannabinoid CB1 and CB2 receptors. GPR55 is predominantly coupled to Gα12/13 leading to activation of small G proteins (15). It has also been suggested to couple to Gαq to promote the activation of phospholipase C and the increase of intracellular calcium release from inositol triphosphate receptor-gated stores (6, 9). LPI-induced activation of GPR55 has been shown to lead to the recruitment of multiple nuclear transcription factors. Among these factors, the most investigated is ERK1/2 (5–9, 11, 15–17). Other nuclear transcription factors recruited by GPR55 activation are nuclear factor of activated T cells (NFAT) (9, 13), cAMP-response element-binding protein (CREB), nuclear factor κ-light chain enhancer of activated T cells (NF-κB) (4, 9, 13), p38 MAPK (18), and Akt serine/threonine protein kinase (8, 11).
Here, we report on the GPR55 pharmacology of arylpyrazole analogues, CB2 agonists, and a number of C. sativa constituents (for structures see Table 3). We show that arylpyrazole analogues act both as agonists alone and as inhibitors of LPI signaling. The compounds significantly decrease the Emax value for the GPR55 endogenous agonist, LPI, which is characteristic of a noncompetitive mode of action; this may suggest allostery. These results provide a possible explanation for the controversy surrounding the pharmacology of certain ligands at GPR55, which have been reported, by different groups, to behave as both agonists and antagonists. Here, we demonstrate for the first time that a single ligand can display both behaviors in the same assay. Furthermore, we show that certain CB2 receptor-selective agonists also act as antagonists of GPR55; this may have implications for the mechanism of action underlying the reported antinociceptive actions of these compounds (19).
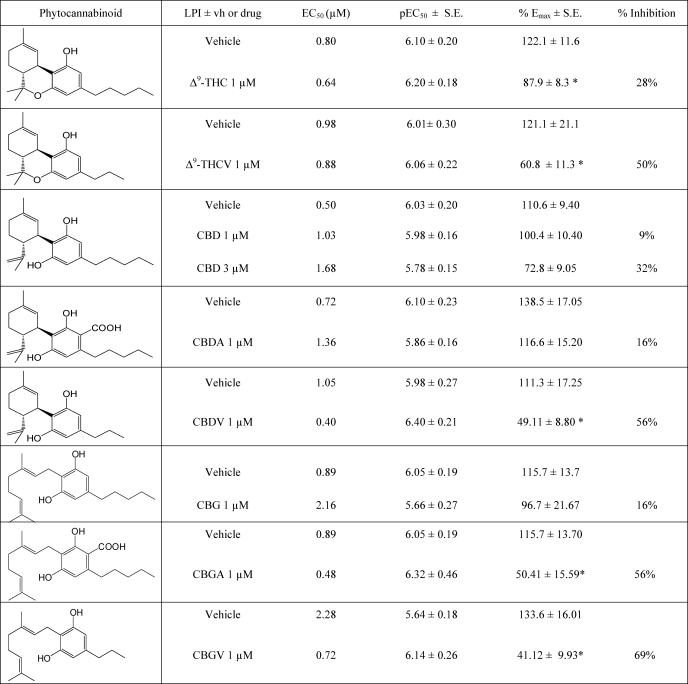
TABLE 3.
Effect of phytocannabinoids and CB2 receptor compounds on LPI-mediated stimulation of ERK1/2 phosphorylation in hGPR55-HEK293 cells
Cells were co-treated with LPI in the presence or absence of a given drug at 1 μm for 20 min at 37 °C. Final concentration of DMSO was 0.2%.
*, p < 0.05 versus LPI in each experiment. % inhibition is percentage inhibition of Emax relative to LPI (100%). **, p < 0.01 versus LPI in each experiment.
EXPERIMENTAL PROCEDURES
Materials
Cannabis constituents Δ9-THC, Δ9-THCV, (−)CBD, CBDA, CBDV, CBG, CBGA, and CBGV were supplied by GW Pharmaceuticals, and SR141716A was from Sanofi-Aventis (Montpellier, France). (−)CBD, AM251, AM281, CP55940, WIN55212-2, HU-308, GW405833, and JWH-133 were from Tocris Cookson (Avonmouth, UK). PLX-4720 was from Selleck, Houston, TX. ABD824 was synthesized using similar methodology to that described previously for SR141716A (Iain R. Greig, University of Aberdeen). (E)-β-Caryophyllene was gifted by Prof. Gertsch (Institute of Biochemistry and Molecular Medicine, University of Bern, Switzerland). PD98059 and AM1241 were from Cayman. G-418 was from PAA (United Kingdom), DMEM/F-12, DMEM, newborn calf serum, and penicillin/streptomycin solution were obtained from Fisher. l-Glutamine, LPI, LY294002, and Y27632 and all the other chemicals were obtained from Sigma-Aldrich (Dorset, U.K.). AlphaScreen® SureFire phospho-ERK kit (catalogue TGRES10K) was from PerkinElmer Life Sciences.
Cell Culture
Untransfected HEK293 cells were maintained in DMEM containing 2 mm l-glutamine medium supplemented with 10% fetal bovine serum. The preparation of HEK293 stably expressing the tagged human GPR55 receptor (hGPR55-HEK293) has been published previously by Henstridge et al. (4). Briefly, the GPR55 receptor was tagged with a triple hemagglutinin (HA epitope) at the N terminus (3×HA-GPR55), preceded by the signal sequence from the human growth hormone (residues 1–33), and subcloned into pcDNA 3.1 vector. The cells were maintained in Dulbecco's modified Eagle's medium DMEM/F-12 supplemented with 10% newborn calf serum, 0.5 mg/ml G-418, 60 units of penicillin, 60 μg of streptomycin, and 2 mm l-glutamine at 37 °C and 5% CO2. Transfected cells and untransfected cells were plated on the same plates for comparison.
ERK1/2 MAPK Phosphorylation Assay
For experimental studies of ERK1/2 MAPK phosphorylation, cells (40,000 cells/well) were plated onto 96-well plates and serum-starved for 48 h. hGPR55-HEK293 cells were serum-starved in DMEM/F-12 medium supplemented with G-418 and 2 mm l-glutamine. HEK293 cells were serum-starved in DMEM/F-12 medium supplemented with 2 mm l-glutamine. Cells were assayed in DMEM/F-12 medium containing l-glutamine and incubated for 20 or 60 min at 37 °C in a humidified atmosphere. Drugs were dissolved in DMSO, and stocks at a concentration of 10 mm were kept at −20 °C. LPI was stored at −80 °C for up to 3 months. Drugs were tested in the absence of LPI at a final concentration of 0.1% DMSO or in the presence of LPI at a final concentration of 0.2% DMSO, unless stated otherwise. At the end of the assay, the medium was removed, and cells were lysed with lysis buffer supplied in the AlphaScreen® SureFire® ERK kit.
AlphaScreen® SureFire® ERK Assay
The assay was performed in 384-well white Proxiplates according to the manufacturer's instructions. Briefly, 4 μl samples were incubated with 7 μl of mixture containing the following: 1 part donor beads, 1 part acceptor beads, 10 parts activation buffer, 60 parts reaction buffer. Plates were incubated at room temperature and read with the Envision system (PerkinElmer Life Sciences) using AlphaScreen® settings.
Analysis
Raw data were presented as “Envision units.” Basal level was defined as zero. Results were presented as means and variability as S.E. or 95% confidence limits of the percent stimulation of phosphorylated ERK1/2 above the basal level (in the presence of vehicle). Data were analyzed using nonlinear analysis of log agonist versus response curve using GraphPad Prism 5.0 (GraphPad, San Diego). The results of this analysis were presented as Emax ± S.E. and pEC50 ± S.E. (logEC50) or EC50 ± 95% confidence limits (CL; where appropriate). In Fig. 6A, for each kinase inhibitor, the values for percent stimulation of phosphorylated ERK1/2 were normalized to the mean value produced by 10 μm LPI (in the presence of vehicle) in matched experiments. Curves of LPI-induced response were not different between experiments; therefore, the data were pooled. Data were presented as “pERK” stimulation as percent of LPI. The statistical significance of Emax ± S.E. or logEC50 ± pEC50 was determined with an unpaired Student's t test (95% confidence interval). When curves could not be fitted on a nonlinear analysis of log agonist versus response, the statistical significance of the stimulation was determined with an unpaired Student's t test at each specific concentration. Results were considered significant only when the F-test comparing the variance was not significantly different.
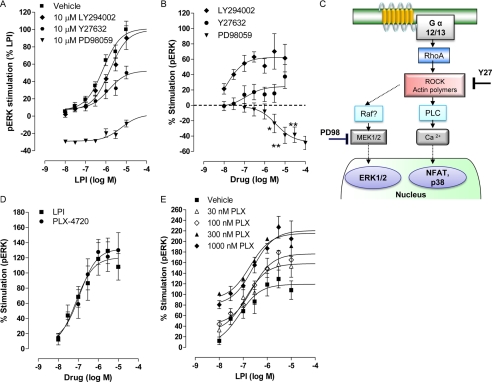
FIGURE 6.
Effect of kinase inhibitors. A, LPI-induced stimulation of ERK1/2 phosphorylation was attenuated by 10 μm PD98059, a MEK1 inhibitor, and inhibited by 10 μm Y27632, a Rho-associated protein kinase inhibitor (n = 5 each in duplicate), but not by 10 μm LY294002, a phosphatidylinositol 3-kinase inhibitor (n = 3 each in duplicate). B, effect of each inhibitor alone on phosphorylated ERK1/2 level (n = 4 for PD98059; n = 7 for Y27632; and n = 3 for LY294002, each in duplicate). PD98059 significantly reduced basal pERK levels; *, p < 0.05; **, p < 0.01, one-sample t test. C, diagram representing the putative GPR55/LPI signaling cascade. D, selective B-Raf inhibitor, PLX-4720, stimulated hGPR55-HEK293-expressing cells in a similar manner to LPI (n = 3 each in duplicate). E, increasing concentrations of PLX-4720 significantly increased the efficacy of LPI-induced response (100 nm, p < 0.05; 300 nm, p < 0.01; 1000 nm, p < 0.01) and the bottom of the curve (300 nm, p < 0.05), (n = 3 each in duplicate).
RESULTS
Studying the Pharmacology of GPR55 Using AlphaScreen® Surefire® pERK1/2 Assay
The phosphorylation of ERK1/2 protein has been reported as one of the main signaling pathways initiated upon stimulation of the GPR55 receptor; therefore, we focused our research on phosphorylated ERK1/2 protein and established a high throughput system using the AlphaScreen® SureFire® phospho-ERK assay. We validated the assay by time (data not shown) and vehicle responses and using untransfected HEK293 cells (Fig. 1 and Table 1).
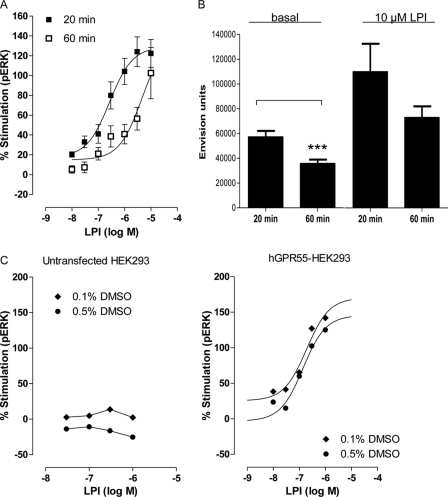
FIGURE 1.
Detection of LPI-induced ERK1/2 phosphorylation in hGPR55-HEK293 cells using the AlphaScreen® SureFire® ERK assay. A, mean log concentration-response curves for the effect of LPI on ERK1/2 phosphorylation in hGPR55-HEK293 cells after 20 min (n = 4 each in triplicate) or 60 min (n = 3 each 2–3 repeats) of incubation at 37 °C. Increasing the incubation time significantly (p < 0.05) reduced the potency of LPI-induced ERK1/2 phosphorylation but did not affect LPI-induced maximal stimulation (Emax) of the GPR55 receptor. B, compared with a 20-min incubation time at 37 °C, a 60-min incubation significantly (p < 0.05) reduced the basal level of ERK1/2 phosphorylation (n = 4, each in 12 repeats). Raw data of AlphaScreen® SureFire® ERK assay are presented as Envision units, Student's t test, ***, p < 0.05. C, no significant differences were observed in basal levels of phosphorylated ERK1/2 in untransfected HEK293 and hGPR55-HEK293 incubated for 20 min in 0.1 or 0.5% DMSO. C, effect of DMSO on LPI-induced ERK1/2 phosphorylation was assessed at 0.1 or 0.5% DMSO in either hGPR55-HEK293 cells or untransfected HEK293 cells. In untransfected HEK293 cells, no stimulation of ERK1/2 phosphorylation was detected at any concentration of LPI at any concentration of DMSO (untransfected and hGPR55-HEK293 cells were seeded on the same plate). Each symbol represents the mean percentage change in bound phosphorylated ERK1/2 protein.
TABLE 1.
Cannabinoid-mediated stimulation of ERK1/2 phosphorylation in hGPR55-HEK293 cells
Cells were treated with each drug for 20 min or for the indicated time at 37 °C. CL means confidence limits, and NA means not applicable.
| Compound | EC50 (95% CL) | % Emax (S.E.) |
|---|---|---|
| μm | ||
| LPI (0.1% DMSO) 20 min | 0.27 (0.10–0.76) | 129.0 ± 9.65 |
| LPI (0.2% DMSO) 20 min | 1.12 (0.59–2.16) | 126.0 ± 9.44 |
| LPI (0.1% DMSO) 60 min | 4.61 (0.75–28.26)a | 139.8 ± 45.70 |
| AM251 | 2.34 (0.99–5.56)b | 153.2 ± 22.10 |
| ABD824 | 0.7 (0.11–4.44) | (−)34.16 ± 22.4 (up to 1 μm) |
| AM281 | NA | ∼106 (10 μm) |
| SR141716A | NA | ∼160 (10 μm) |
| CP55940 | NA | ∼50 (10 μm) |
| JWH-133 | 0.16 ((−)30.98 to (−)4.41) | (−)29.8 ± 6.40 (up to 3 μm) |
| GW405833 | 1.87 (0.33 to 10.7) | 54.0 ± 13.04 |
| HU-308 | NA | ∼20 (10 μm) |
| AM1241 | NA | ∼15 (10 μm) |
| BCP | NA | ∼22 (10 μm) |
a p < 0.05 versus LPI in 0.1% DMSO for 20 min (first raw).
b p < 0.01 versus LPI in 0.1% DMSO for 20 min (first raw).
LPI (0.1% DMSO) produced a maximal stimulation of 129.0 ± 9.65% (Emax) (Fig. 1A), which was not significantly different from the maximal stimulation with 0.2% DMSO (n = 15 each in triplicate; Table 1; e.g. Fig. 2A). The potency of LPI was not different either (Table 1). In untransfected HEK293 cells, no stimulation of phosphorylated ERK1/2 was detected at any concentration of LPI nor in any given concentration of DMSO (Fig. 1C). These results are similar to the results published by other groups who reported the potency of LPI in ERK1/2 phosphorylation assays (5, 13) or other readouts (6).
FIGURE 2.
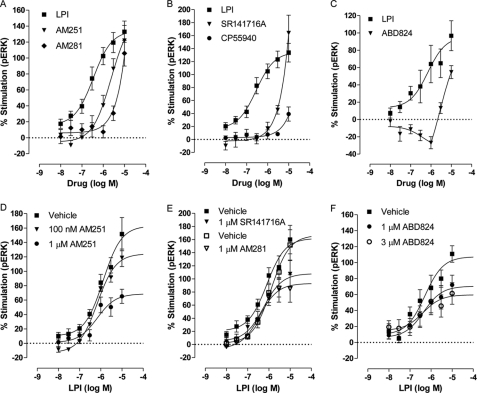
Structural analogues of SR141716A-induced ERK1/2 phosphorylation in hGPR55-HEK293 cells. A, mean log concentration-response curves for percent stimulation of ERK1/2 phosphorylation by LPI (n = 4), AM251 (n = 3), or AM281 (n = 3) after a 20 min stimulation at 37 °C. B, mean log concentration-response curves of LPI (n = 4), SR141716A (n = 3), or CP55940 (n = 3), a nonselective cannabinoid CB1/CB2 agonist. C, mean log concentration-response curves of LPI or ABD824, an AM251 analog in which iodine was substituted with a bromine (n = 3, each in duplicate). The effect of SR141716A analogues on LPI induced ERK1/2 phosphorylation in hGPR55-HEK293 cells. D, LPI (n = 4) in the presence or absence of 100 nm AM251 (n = 3) or 1 μm AM251 (n = 4). E, effect of LPI in the presence or absence of 1 μm SR141716A (n = 4) or 1 μm AM281 (n = 3). F, effect of LPI in the presence or absence of 1 μm and 3 μm ABD824 (n = 4, each in duplicate). Each symbol represents the mean percentage change in bound phosphorylated ERK1/2 ± S.E. over the basal level, and each independent experiment was performed in triplicate unless stated otherwise.
LPI Induces Sustained Activation of ERK1/2 Phosphorylation in hGPR55-HEK293 Cells
Sustained activation of ERK1/2 phosphorylation has been implicated as a measure for cancer progression, increase in cell metastasis, and invasiveness of tumor cells (20, 21). Importantly, GPR55-induced ERK1/2 phosphorylation regulates human cancer cell migration in vitro and proliferation in vivo (3, 8, 11). It also appears to govern the maintenance of persistent inflammatory pain (22, 23). Therefore, we investigated the ability of LPI to maintain ERK1/2 phosphorylation after 20 and 60 min of incubation (Fig. 1A). The potency of LPI after 20 min of incubation, with an EC50 of 0.27 μm (0.10–0.76), was significantly (p < 0.05) reduced to 4.61 μm (0.75–28.3) after 60 min. Prolonged incubation reduced both basal and LPI-induced stimulation (Fig. 1B and Table 1) but did not affect the net effect of LPI-induced maximal stimulation of ERK1/2 phosphorylation (Fig. 1A), showing that percent stimulation was sustained. This is in contrast, for example, to CBD (10 μm) that significantly increased the percent stimulation after 60 min (supplemental Fig. 2).
Structural Analogues of SR141716A Inhibit LPI-induced Activation of ERK1/2 Phosphorylation, Implications for Allosteric Inhibition
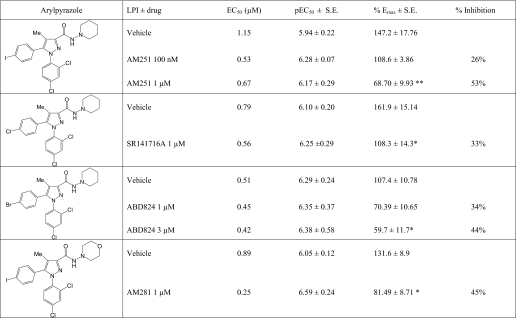
There is still a major controversy surrounding the profile of the arylpyrazoles at GPR55. In some studies, SR141716A is reported to be an inhibitor (6), and in other studies, the compound is reported to be an agonist of GPR55 (5, 13, 14). This is of major importance as SR141716A (Rimonabant®) was widely prescribed to patients as an anti-obesity agent and was withdrawn because of mental health issues. We compared the ability of AM251, AM281, SR141716A, and ABD824, an AM251 analog in which the iodine was substituted with bromine (Fig. 2, Table 1, and see Table 2 for structures), to induce ERK1/2 phosphorylation in hGPR55-HEK293 cells with that of LPI. The efficacy of AM251 was not significantly different from that of LPI, but the compound was significantly less potent (Fig. 2A and Table 1). It was not possible to obtain an accurate measurement of the Emax for SR141716A and AM281; however, the compounds had efficacy that was not lower than that of LPI (Fig. 2, A and B, and Table 1). Interestingly, ABD824 had a bi-phasic response in hGPR55-HEK293 cells (Fig. 2C).
TABLE 2.
Effect of arylpyrazoles on LPI-mediated stimulation of ERK1/2 phosphorylation in hGPR55-HEK293 cells
Cells were co-treated with LPI in the presence or absence of a given drug for 20 min at 37 °C. Final concentration of DMSO was 0.2%.
*, p < 0.05; **, p < 0.01 versus LPI in each experiment. % inhibition is percentage inhibition of Emax relative to LPI (100%).
Arylpyrazole-induced stimulation appears to be GPR55-mediated as these ligands do not stimulate ERK1/2 phosphorylation in non-expressing HEK293 cells (4), and CP55940 alone did not induce a significant stimulation of ERK1/2 phosphorylation at any concentration (Fig. 2B).
In some studies, SR141716A was reported to inhibit GPR55 activity (6), and we therefore assessed if certain arylpyrazoles could inhibit LPI-induced GPR55 stimulation, in the same experimental system. The arylpyrazoles AM251, AM281, SR141716A, and ABD824 reduced the maximal stimulation (Emax) by LPI of ERK1/2 phosphorylation but did not significantly alter the potency of this bioactive lipid (results are summarized in Fig. 2, E and F, and in Table 2).
Importantly, these data provide the first demonstration of a dual action (both agonist and inhibitor) of these compounds in the same assay. The inhibition appears noncompetitive and may indicate allostery. Of the tested analogues, AM251 was the most effective as an inhibitor of LPI (Fig. 2D). The Emax of LPI of 147.2 ± 17.8% was reduced to 108.6 ± 3.86% by 100 nm and to 68.7 ± 9.93% (p < 0.01) by 1 μm AM251.
We continued to explore the effects of high concentrations of AM251 and SR141716A. The maximal stimulation by LPI in the presence of high concentrations of AM251 or SR141716A was not significantly different from that of LPI only. This would be expected because the compounds are agonists alone at these concentrations (supplemental Fig. 1, A and B).
Previous studies reported that CP55940, a nonselective cannabinoid receptor agonist, is an inactive ligand at GPR55 that could not increase intracellular calcium level (6) but behaves as a competitive antagonist in the presence of LPI (4); another group showed it antagonized LPI, AM251, and SR141716A-induced β-arrestin trafficking and LPI-induced ERK1/2 phosphorylation (5). For comparison, in this study, LPI-induced stimulation was 144.0 ± 16.7% and CP55940 at 1 μm reduced it by only 13% (124.9 ± 16.0%), which was not significantly different from LPI alone (supplemental Fig. 1C).
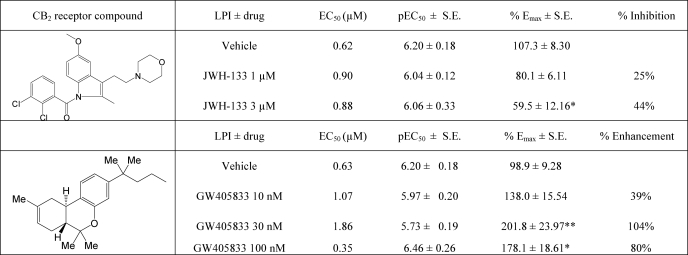
Effect of CB2 Agonists on LPI-induced Activation of ERK1/2 Phosphorylation, Implications for Neuropathic Pain and Positive Allosteric Modulation
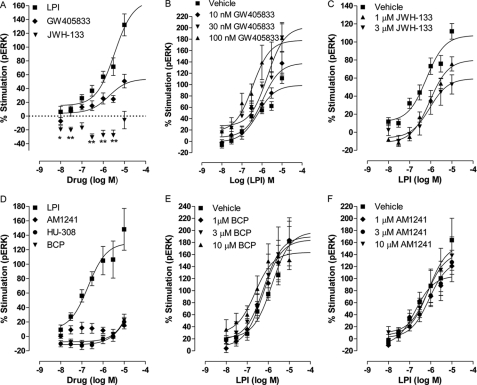
The antinociceptive effect of CB2 receptor agonists has been extensively investigated (reviewed in Ref. 24). Subsequently, CB2 receptor agonists have been developed by scientists and pharmaceutical companies as alternative treatments aimed at alleviation of neuropathic pain. Interestingly, a comparison of the behavioral responses of wild type mice with GPR55 knock-out mice in pain models revealed the involvement of GPR55 in the conduction of neuropathic pain (1). These results suggest that the analgesic effects of certain CB2 receptor-selective ligands may be mediated by GPR55. We compared several CB2 agonists that have been intensively investigated in the past and thus form a core of active structures for GPR55-mediated ERK1/2 phosphorylation. JWH-133 (up to 3 μm) significantly reduced basal pERK levels. GW405833 behaved as a partial agonist of GPR55 (Emax 54 ± 13.0% versus LPI 106.0 ± 10.5%; p < 0.05), with a similar potency to that of LPI (1.9 μm (0.3–10)) (Fig. 3A). In contrast to the arylpyrazoles, GW405833 enhanced LPI-induced ERK1/2 phosphorylation at a concentration that alone had no effect on pERK (Fig. 3B). Conversely, JWH-133 at 1 and 3 μm (Fig. 3C), but not at 10 μm (results not shown), inhibited the LPI-induced response. These results further support the data obtained with the arylpyrazoles, indicating that cannabinoids can act as noncompetitive inhibitors of LPI signaling. Of the tested compounds, (E)-β-caryophyllene (25), HU-308 (data not shown), and AM1241 had little/no effect up to a concentration of 10 μm (Fig. 3D) and did not significantly alter the LPI-induced effect on GPR55 (Fig. 3, E and F).
FIGURE 3.
Effect of CB2 receptor agonists on ERK1/2 phosphorylation in hGPR55-HEK293 cells. A, mean log concentration-response curves for percent stimulation of ERK1/2 phosphorylation by LPI (n = 7), GW405833 (n = 4), or JWH-133 (n = 3) after 20 min of stimulation at 37 °C. JWH-133 significantly reduced basal pERK levels, *, p < 0.05; **, p < 0.01, one-sample t test. B, GW405833 at 10, 30, and 100 nm enhanced the LPI-induced ERK1/2 phosphorylation (n = 4). C, JWH-133 at 1 and 3 μm inhibited the LPI-induced ERK1/2 phosphorylation (n = 4). D, percent stimulation of ERK1/2 phosphorylation by LPI (n = 3), AM1241 (n = 4), or HU-308 (n = 4) and (E)-β-caryophyllene (BCP) (n = 3) after 20 min of stimulation at 37 °C. E, (E)-β-caryophyllene at 1, 3, and 10 μm (n = 3) did not alter the LPI-induced ERK1/2 phosphorylation nor did AM1241 (F, n = 3). Each symbol represents the mean percentage change in bound phosphorylated ERK1/2 ± S.E. over the basal level, and each independent experiment was performed in duplicate.
Effect of Phytocannabinoids on LPI-induced Activation of ERK1/2 Phosphorylation, Comparison of Δ9-THC and Δ9-THCV
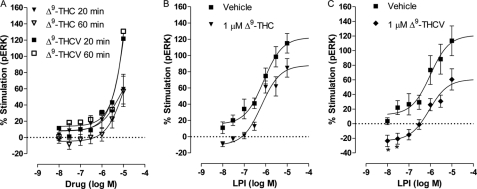
Similarly to arylpyrazoles, the effect of Δ9-THC, the main psychoactive constituent of C. sativa, on GPR55 is also controversial. It has been shown to increase calcium release (6) but not to mobilize β-arrestin (5) in GPR55-expressing cells. In this study, we compared the structurally related cannabis constituents, Δ9-THC and Δ9-THCV (Fig. 4A and Table 3). Both compounds stimulated ERK1/2 phosphorylation. However, for neither compound could the stimulation be fitted to a concentration-response curve. Compared with the maximal stimulation of LPI (129.0 ± 9.65%; Fig. 1A), these results suggest that Δ9-THC is a low affinity partial agonist, whereas Δ9-THCV is a low affinity, high efficacy, agonist of GPR55. The levels of ERK1/2 phosphorylation after prolonged incubation (60 min) with Δ9-THC or Δ9-THCV were not significantly different from those obtained after 20 min.
FIGURE 4.
Effect of Δ9-THC and Δ9-THCV on ERK1/2 phosphorylation in hGPR55-HEK293 cells. Mean log concentration-response curves for ERK1/2 phosphorylation after 20 or 60 min of stimulation at 37 °C with Δ9-THC (n = 3) or Δ9-THCV (n = 4) (A). B shows the effect of LPI in the presence or absence of 1 μm Δ9-THC after 20 min of stimulation at 37 °C (n = 4). C shows the effect of LPI in the presence or absence of 1 μm Δ9-THCV after 20 min of stimulation at 37 °C (n = 3). Each symbol represents the mean percentage change in bound phosphorylated ERK1/2 ± S.E. over the basal level, and each independent experiment was performed in triplicate.
We then assessed the modulation of LPI-induced stimulation with 1 μm Δ9-THC or 1 μm Δ9-THCV (produced stimulation of 22.2 ± 11.1% (not significant from zero) and 31.4 ± 10.0% (p < 0.01 versus 0), respectively). Incubation of LPI with 1 μm of Δ9-THC or Δ9-THCV significantly inhibited LPI-induced stimulation (Fig. 4, B and C, respectively, and Table 3). Neither ligand affected the potency of LPI (Table 3). These results show for the first time that the phytocannabinoids Δ9-THC and Δ9-THCV are inhibitors of LPI. Moreover, Δ9-THCV elicited a downward shift in the log concentration-response curve of LPI, such that LPI-induced pERK level was lower in the presence of Δ9-THCV, and this resembles negative cooperativity.
Structural Analogues of Cannabidiol
We have recently shown that CBD antagonizes LPI-induced stimulation of [35S]GTPγS binding in breast cancer MDA-MB-231 cells, which highly express GPR55 (3). CBD also inhibits the metastasis and aggressiveness of brain and breast cancer cells (26, 27) and LPI-induced calcium mobilization in prostate cancer cells (8). Therefore, we have evaluated two cannabis constituents that are structurally related to CBD, alone or in combination with LPI. Cannabidiolic acid (CBDA) has an acid group (COOH) on the benzene ring, whereas cannabidivarin (CBDV) has a shorter side chain compared with that of CBD (see Table 3 for structures).
We compared the effects of CBD from two sources (Tocris and GW Pharmaceuticals). The effect of CBD on ERK1/2 phosphorylation after 20 min was not significantly different between the two sources (supplemental Fig. 2A); However, after a 60 min incubation, CBD significantly increased ERK1/2 phosphorylation (100.5 ± 11.9% and 220.9 ± 33.2% each at 10 μm, Tocris or GW Pharmaceuticals, respectively). Neither CBD nor CBDA affected basal pERK after a 20 min incubation, whereas CBDV produced a maximal stimulation of 43.0 ± 23.0% and an EC50 of 1.9 μm (confidence intervals 0.13–27.9 μm) (supplemental Fig. 2, B and C, respectively).
The effects of CBD, CBDA, and CBDV on LPI-induced ERK1/2 phosphorylation are summarized in Table 3 and Fig. 5, A–C. CBD did not significantly alter LPI efficacy or potency at 1 or 3 μm (Table 3). However, the inhibition of 32% at 3 μm showed a trend toward antagonism. In fact, we have found similar results using the [35S]GTPγS binding assay with MDA-MB-231 breast cancer cells (3). In these endogenously expressing GPR55 receptor cells, LPI stimulates [35S]GTPγS binding, whereas 1 μm CBD produces a significant downward shift (6.6 ± 5.35% versus (−)17.5 ± 3.69 with CBD) but did not affect the maximal stimulation (Emax) of LPI-induced [35S]GTPγS binding (LPI, 50.1 ± 7.5%; 48.3 ± 21.2%). However, these cells express a spectrum of receptors making them not ideal for further characterization of the pharmacology of GPR55. Taken together, these results support the hypothesis that CBD is an inhibitor of GPR55.
FIGURE 5.
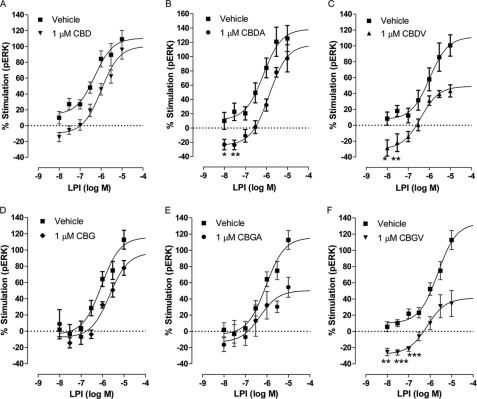
Effect of CBD and CBDA and CBDV on ERK1/2 phosphorylation in hGPR55-HEK293 cells. Mean log concentration-response curves for ERK1/2 phosphorylation after 20 min of stimulation at 37 °C. The effect of LPI in the presence or absence is shown as follows: 1 μm CBD (n = 5) (A); 1 μm CBDA (n = 3) (B); 1 μm CBDV (n = 3) (C); 1 μm CBG (n = 4) (D); 1 μm CBGA (n = 4) (E); 1 μm CBGV (n = 3) (F). CBDV and CGV pronouncedly inhibited LPI compared with their analogues. Each symbol represents the mean percentage change in bound phosphorylated ERK1/2 ± S.E. over the basal level (n = 3). CBDA, CBDV, and CBGV significantly reduced basal pERK levels, *, p < 0.05; **, p < 0.01; ***, p < 0.001 one-sample t test.
Of the tested analogues, CBDV (9.99% stimulation at 1 μm; not significant versus zero) significantly reduced the maximal stimulation of LPI by 56% (p < 0.05) (Fig. 5C and Table 3). Increasing the CBDV concentration to 3 μm did not inhibit or increase LPI stimulation (data not shown). CBDA did not significantly affect the efficacy or potency of LPI but produced a significant downward shift of its response (11.15 ± 10.1% versus (−)24.1 ± 5.81% with CBDA; p < 0.05) (Fig. 5B and Table 3); this may be indicative of negative cooperativity. In summary, the three compounds structurally related to CBD significantly inhibit LPI-induced ERK1/2 phosphorylation with a rank order of CBDV (56%) > CBDA (16%) > CBD (9%). Furthermore, both CBDA and CBDV displayed negative cooperativity with LPI. Basal pERK was significantly lower when either of these compounds was co-incubated with LPI. These results underlie a new structure-activity relationship of GPR55 ligands and suggest that the previously reported CBD antagonism of LPI-induced stimulation of ERK1/2 phosphorylation can be further enhanced by shortening its alkyl side chain from pentyl to propyl, although the additional acid group mainly affects the inhibition of ERK1/2 phosphorylation at a low concentration of LPI.
Structural Analogues of Cannabigerol
Following these findings, we sought to evaluate CBG and two structurally related cannabis constituents that either contain an acid group on the benzene ring (CBGA) or a propyl side chain (CBGV). After a 20 min incubation, CBG did not produce a significant stimulation (supplemental Fig. 2D). CBGA decreased basal pERK with an Emax of (−)79.9 ± 27.2% (20 min) and after a 60 min incubation, it was (−)31.4 ± 7.34% with an EC50 of 5.26 μm (confidence intervals 0.99–27.9) and 0.83 μm (confidence intervals 0.01–4.79), respectively (supplemental Fig. 2E).
We then assessed the effect of each compound on LPI-induced stimulation of GPR55 (Fig. 5, D and E, and Table 3). The effect of each compound at 1 μm was not significantly different versus zero (stimulation of (−)7.7% for CBG; (−)7.4% for CBGA; (−)6.2% for CBGV)). At 1 μm, CBGA (Fig. 5E) and CBGV (Fig. 5F) but not CBG (Fig. 5D) significantly reduced the maximal stimulatory effect of LPI on ERK1/2 phosphorylation (p < 0.05), suggesting that these compounds are also inhibitors of LPI-induced activation of the GPR55 receptor. We observed a similar structure-activity relationship to that found for CBD analogues, and thus the rank order of LPI inhibition was CBGV (69%) > CBGA (56%) > CBG (16%). Furthermore, CBGV displayed negative cooperativity with LPI, whereby basal pERK was significantly lower in the presence of both compounds.
GPR55 Signaling Involves Cross-talk between MAPK and Rho GTPases Signaling Pathways
We then tested if LPI-induced stimulation of ERK1/2 phosphorylation can be inhibited by the MEK1/2 noncompetitive inhibitor PD98059. LPI (10 nm to 10 μm) did not stimulate the phosphorylation of ERK1/2 proteins in the presence of PD98059, which significantly inhibited basal pERK alone (Fig. 6, A and B). These findings were in line with the receptor-independent mechanism of action of PD98059 by binding the inactive form of MEK1 and inhibiting its activation by upstream activators such as Raf kinases. However, this could not explain previous observations showing that GPR55 is predominantly coupled to Gα12/13. To test our hypothesis for cross-talk between MAPK and Rho GTPases signaling pathways, we co-incubated LPI with Y27632, a p160ROCK (Rho-associated kinase) inhibitor. After 20 min, 10 μm Y27632 significantly inhibited LPI-induced stimulation of ERK1/2 phosphorylation (Fig. 6A; p < 0.01), without inducing a significant change in LPI potency. Although 10 μm Y27632 or 10 μm LY294002, an inhibitor of phosphoinositide 3-kinase (PI3K), could equally stimulate ERK1/2 phosphorylation (Fig. 6B), only Y27632 significantly inhibited the response to LPI under these conditions.
Enhancement of GPR55 Signaling by PLX-4720 Oncogene B-Raf Inhibitor
B-Raf is a member of Raf kinase family and plays a major role in regulating ERK1/2 phosphorylation. Importantly, inherited and acquired mutations, such as V600E, in B-Raf are associated with various diseases of which most are cancers (28). Therefore, B-Raf inhibitors have been developed as anti-cancer therapeutics. PLX-4720, a potent and selective B-Raf inhibitor (29), induced a concentration-dependent stimulation of ERK1/2 phosphorylation at a magnitude similar to that induced by LPI (Fig. 6C), opposing the inhibitory effects of the MEK signaling pathway in BRAF(V600E)-expressing cells. In the presence of LPI, increasing concentrations of PLX-4720 (100, 300, 1000 nm) significantly increased the Emax of LPI (119.2 ± 10.53%) reaching 220.6 ± 13.97% at 1000 nm (p < 0.01, one-way analysis of variance) but did not alter its potency; this suggests an additive relationship between intracellular inhibition of B-Raf and extracellular stimulation of LPI-induced of ERK1/2 phosphorylation, supporting a cross-talk between MAPK and Rho GTPases signaling pathways.
DISCUSSION
The lack of appropriate radiolabeled ligands for GPR55 precludes the characterization of the binding of novel small molecules to this receptor. In this study, we established a new rapid and sensitive AlphaScreen® SureFire® assay as a strategy for studying the pharmacology of GPR55. This method has been used to explore ligands of the cytokine receptors (30), LDL endothelial receptor (31), and potassium channel TREK-1 receptors (32) and to develop antagonists for other intracellular targets (33–35).
LPI and Certain Cannabinoids Induce Sustained GPR55-mediated ERK1/2 Phosphorylation
Studies have shown that GPR55 induces maximal ERK1/2 phosphorylation response after 10–20 min (this study and see Refs. 4, 5, 7); here we show that although both basal and LPI-induced stimulation levels are reduced at 60 min (Fig. 1 compared with 20 min) the percent stimulation is sustained, but the potency of LPI is significantly decreased after 60 min.
Sustained ERK1/2 activation in injured neurons has been suggested to reflect alterations in the intracellular feedback regulators that normally function to terminate signaling responses (36) and has been associated with brain ischemia (37). ERK1/2 phosphorylation is also associated with a variety of human pathologies (38). In addition, many mutations of components upstream to ERK1/2 alter this signaling pathway and have been associated with increased cancer metastasis and invasiveness (20, 38). The combination of strength and duration of ERK1/2 signaling determines the distinct outcomes, ranging from sustained high activation that can lead to apoptosis or differentiation to sustained lower levels of activation that are correlated with cell proliferation (39). In our case, it appears that LPI-induced activation parallels the latter scenario, supporting the role of GPR55 in cancers. Therefore, it will be interesting to determine whether this mechanism involves the endogenous mitogen-activated protein kinase phosphatase 1 (MKP-1) that controls the constitutive activation of ERK1/2 (21).
GPR55 Pharmacology of Arylpyrazoles
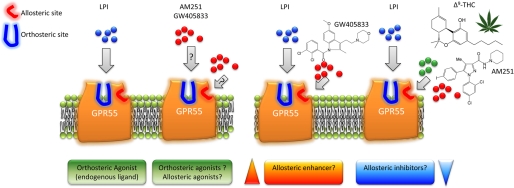
A key finding of this study is the demonstration that arylpyrazoles, e.g. SR141716A, can act both as agonists of GPR55 and inhibitors of LPI-induced activation of pERK. Thus, we demonstrate here for the first time that both types of behavior (agonism and inhibition) as reported by previous studies from different laboratories can exist in the same model system. Our data suggest that there may be two topographically distinct binding sites on GPR55 (Fig. 7). In the first scenario, certain ligands (e.g. AM251 and GW405833) could act as agonists alone (either via the orthosteric (LPI) binding site or a distinct (allosteric) binding site (40). In the second scenario, the same ligands could act as modulators of LPI-induced pERK activation. This suggests that the compounds may bind to an allosteric site to induce a conformational change in the orthosteric (LPI) binding site such that the efficacy of LPI is reduced (e.g. AM251) or enhanced (e.g. GW405833). These observations raise the possibility that certain arylpyrazole ligands may represent bitopic ligands of GPR55. Bitopic ligands have the capacity to interact with both the orthosteric site (as agonists) and the allosteric site (as modulators) through distinct chemical structures (41).
FIGURE 7.
Pharmacological mechanisms for the modulation of GPR55. LPI is suggested to primarily bind the GPR55 orthosteric binding site. GPR55 may also contain an allosteric binding site. These observations raise at least two possibilities as follows. Left, one possibility is that certain arylpyrazole ligands actually represent bitopic ligands of GPR55. These ligands may have the capacity to modulate both the orthosteric (agonists) and the allosteric site through different pharmacophores. The second possibility is that AM251 and certain arylpyrazole analogues are only allosteric ligands that are ago-allosteric alone. Right, in the presence of LPI, arylpyrazoles can also behave as allosteric inhibitors and GW405833 as an allosteric enhancer. In addition, a number of Cannabis sativa constituents appear to inhibit ERK1/2 phosphorylation in an allosteric manner.
Allosteric binding sites have been described for other lipid receptors, including allosteric modulators of anandamide at the cannabinoid CB1 receptor (42) and ago-allosteric ligands for short chain free fatty acids at FFA2 and FFA3 receptors (43). This study demonstrates the modulation of GPR55 by certain ligands, possibly by an allosteric mechanism. The first evidence is the reduction in Emax of LPI observed in the presence of various compounds. However, it is important to note that this study does not provide direct evidence of allostery (e.g. dissociation kinetics), and there is a possibility that the compounds may inhibit LPI signaling downstream of the receptor to disrupt the pERK signaling cascade. The second evidence is the apparent negative cooperativity between various ligands and LPI such that LPI-induced pERK level is significantly decreased in the presence of both ligands, an effect not observed with either ligand alone.
Clearly, this evidence is circumstantial, and dissociation kinetic analysis (which is not currently feasible because of the lack of appropriate radioligand) or extensive mutation studies, will be required to confirm the existence of topographically distinct binding sites at GPR55. Nevertheless, this study provides the first potential explanation for the considerable controversy in the pharmacology of cannabinoid ligands at GPR55 and has important therapeutic implications.
The arylpyrazoles belong to a subgroup of ligands that interact with both GPR55 and CB1 receptors. In this study, arylpyrazole analogues induced activation of pERK with a rank order of potency LPI ≫≫ AM251 > SR141716A > AM281. This rank order of potency is in agreement with the study by Henstridge et al. (4) using the same hGPR55-expressing HEK293 cells and is similar to the study by Kapur et al. (5) that used hGPR55-expressing U2OS cells and a β-arrestin mobilization assay. Although in the latter study arylpyrazoles could not induce the phosphorylation of ERK1/2, the difference between this and our study may be due to differences in method sensitivity (Western blot versus AlphaScreen®) or may reflect cell type differences.
Collectively, these results suggest that AM251, SR141716A, and AM281 are weak agonists of GPR55 as measured by their ability to induce ERK1/2 activation. Interestingly, ABD824, an AM251 analog, decreased basal pERK at concentrations below 3 μm. This suggests a role for the chemical group on the aryl ring in shifting the GPR55 receptor between inactive and active receptor conformations. As for the efficacy for the observed antagonism of LPI by arylpyrazoles, the rank order is AM251 > AM281 > ABD824 ≥ SR141716A, which does not correspond to the relative potencies of these ligands as GPR55 agonists (as detailed above). This suggests that the pharmacophore for inhibition of GPR55 by these pyrazoles may be different from that responsible for activation of GPR55.
SR141716A was developed as a cannabinoid CB1 receptor antagonist and progressed to clinical studies as a drug treatment for obesity-related disorders (44). Interestingly, SR141716A has been shown to reduce neuropathy associated with type-2 diabetes in patients (45). This has also been supported by in vivo studies in which SR141716A reduced neuropathic pain in a murine model for obesity (46) and in a rodent model for nerve injury (47). This is unexpected for an inverse agonist of CB1 because CB1 receptor agonists are effective in neuropathic pain. Collectively, these studies suggest that SR141716A may act on a target other than the CB1 receptor to inhibit pain. As GPR55 knock-out mice are less sensitive to the development of chronic pain (1), our findings that LPI-induced activation of GPR55 is inhibited by SR141716A suggests a novel mechanism of action for the regulation of pain and possibly for metabolic syndrome by this compound. This effect may be mediated by a putative allosteric binding site on GPR55.
GPR55 Pharmacology of CB2 Agonists
This study has identified GW405833 as a GPR55 partial agonist. JWH-133 significantly reduced basal pERK and as such may be an inverse agonist of GPR55. In line with our results, JWH-133 has been previously studied for its analgesic effects and has been shown to produce antinociception via nicotinic-dependent pathways (48). However, its effect could not always be reversed by SR144528, a CB2 receptor antagonist (49). Similarly, GW405833 was found to promote a pro-antinociceptive response in a rat model of osteoarthritis in contrast to its behavior as a CB2 receptor partial agonist (50). These studies suggest that JWH-133 and GW405833 have an additional target(s), and our results suggest that GPR55 signaling may be one of their targets.
Phytocannabinoids That Inhibit LPI-mediated Activation of GPR55
The pharmacological actions of several C. sativa constituents have been recently reviewed; both the psychoactive and nonpsychoactive compounds have multiple targets and convey a range of pharmacological actions (51). In this study, we have evaluated a range of C. sativa constituents for their ability to modulate GPR55. Although Δ9-THC activates CB1 and CB2 receptors and GPR55 and behaves as a partial agonist at these receptors, Δ9-THCV is a CB1 antagonist and a CB2 partial agonist. In this study, we found that Δ9-THCV is a weak agonist of GPR55 that can significantly inhibit LPI-induced stimulation of pERK in GPR55-expressing cells. Thus, the analgesic actions of Δ9-THCV in vivo (52) might, at least in part, be mediated by modulation of GPR55. Expression of GPR55 mRNA has been detected in adipose tissue (15), and a polymorphism in the GPR55 gene has been found in females with anorexia nervosa (53). Although the precise role of GPR55 in food intake and metabolism remains to be elucidated, it is possible that the inhibition of food intake reported for Δ9-THCV (54) and other cannabinoids may be mediated, at least in part, by modulation of GPR55.
Here, we report that the little investigated cannabis constituents CBDV, CBGA, and CBGV are potent inhibitors of LPI-induced GPR55 signaling. CBD is the most investigated nonpsychotropic constituent of cannabis and has a wide range of pharmacological targets (51); together with a safe profile in humans, this made it highly attractive for the development of drugs for neuropathic pain associated with multiple sclerosis (55) and cancer (56). Several studies have also shown that CBD is effective against proliferation, migration, and invasion of a variety of breast cancer, glioblastoma, prostate cancer, and human cervical cancer cell lines (3, 8, 26, 27, 57). In cancer cells, the target for CBD remains elusive, whereas the actions of Δ9-THC have so far been considered to be mediated by activation of cannabinoid receptors. Recently, GPR55 has been linked to inhibition of the migration and proliferation of cancer cells by CBD (3, 8). The results warrant further investigation into the potential therapeutic use of various phytocannabinoid. CBDA and CBG have been shown to activate TRPA1- and TRPV1-expressing cells and to antagonize TRPM8, implying a role in analgesia and prostate cancer for these compounds (58, 59). Our study suggests that GPR55 is an additional target for these compounds; inhibition of GPR55 would therefore further support their potential role in the treatment of carcinoma and pain (51).
GPR55-mediated MAPK Signaling
Importantly, our findings demonstrate that LPI-induced ERK1/2 phosphorylation is controlled, at least in part, by ROCK, supporting the role for Gα12/13 in LPI signaling and indicating cross-talk between MAPK and Rho GTPases signaling, which is in line with Andradas et al. (11). Such cross-talk may be communicated via the serine/threonine kinase Raf family, of which the expression level of the oncogene B-Raf in turn controls the stimulation of ROCK (60). In line with Hatzivassiliou et al. (16), the selective B-Raf inhibitor PLX-4720 unregulated ERK1/2 phosphorylation in hGPR55-HEK293 cells that do not carry the BRAF(V600E) mutation; interestingly, the magnitude of the response closely resembled that of LPI. Furthermore, ERK1/2 phosphorylation was unregulated following the co-incubation of PLX-4270 with LPI in an additive manner, supporting a cross-talk between B-Raf and ROCK signaling pathways in LPI-mediated GPR55 signaling. Further support for this interaction comes from B-Raf knock-out mice in which ROCKII expression is reduced (61).
Conclusions
We have established a rapid and sensitive method to study the pharmacology of the GPCR GPR55 using the AlphaScreen® SureFire® ERK assay. It is important to note that this study exclusively measures ERK1/2 phosphorylation as a readout of GPR55. This readout may not allow the distinction between various signaling pathways; thus, this work might be limited by the fact that the signaling cascade leading to ERK1/2 activation may vary with the compound used. That said, to our knowledge this study provides the first evidence that certain cannabinoids can display both activation of GPR55 and inhibition of LPI-mediated pERK stimulation in the same assay. Further studies should lead to a resolution of the controversy regarding the pharmacology of cannabinoids at GPR55. Furthermore, we show that certain ligands previously thought to be selective for the CB2 receptor also modulate GPR55. Our findings also suggest that GPR55 may be a new pharmacological target for the following C. sativa constituents: Δ9-THCV, CBDV, CBGA, and CBGV. Combined mutagenesis and pharmacological investigations will enable us to determine the pharmacophores responsible for cannabinoid binding to GPR55 to facilitate rational drug design (62). This study has implications for developing new therapeutics for the treatment of cancer, pain, and metabolic disorders.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DA-03672 and DA-09789 (to R. A. R. and R. G. P.). This work was also supported by a Knowledge Transfer Grant Award from the University of Aberdeen. R. G. P. and R. A. R. received funding from GW Pharmaceuticals.

This article contains supplemental Figs. 1 and 2.
- LPI
- l-α-lysophosphatidylinositol
- AM281
- 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-N-4-morpholinyl-1H-pyrazole-3-carboxamide
- AM251
- 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide
- CBD
- cannabidiol
- CBDA
- cannabidiolic acid
- CBDV
- cannabidivarin
- CBG
- cannabigerol
- CBGA
- cannabigerolic acid
- CBGV
- cannabigerovarin
- GPR55
- G protein-coupled receptor 55
- SR141716A
- 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide
- Δ9-THC
- Δ9-tetrahydrocannabinol
- Δ9-THCV
- Δ9-tetrahydrocannabivarin
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Staton P. C., Hatcher J. P., Walker D. J., Morrison A. D., Shapland E. M., Hughes J. P., Chong E., Mander P. K., Green P. J., Billinton A., Fulleylove M., Lancaster H. C., Smith J. C., Bailey L. T., Wise A., Brown A. J., Richardson J. C., Chessell I. P. (2008) Pain 139, 225–236 [DOI] [PubMed] [Google Scholar]
- 2. Whyte L. S., Ryberg E., Sims N. A., Ridge S. A., Mackie K., Greasley P. J., Ross R. A., Rogers M. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16511–16516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ford L. A., Roelofs A. J., Anavi-Goffer S., Mowat L., Simpson D. G., Irving A. J., Rogers M. J., Rajnicek A. M., Ross R. A. (2010) Br. J. Pharmacol. 160, 762–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henstridge C. M., Balenga N. A., Ford L. A., Ross R. A., Waldhoer M., Irving A. J. (2009) FASEB J. 23, 183–193 [DOI] [PubMed] [Google Scholar]
- 5. Kapur A., Zhao P., Sharir H., Bai Y., Caron M. G., Barak L. S., Abood M. E. (2009) J. Biol. Chem. 284, 29817–29827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lauckner J. E., Jensen J. B., Chen H. Y., Lu H. C., Hille B., Mackie K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oka S., Nakajima K., Yamashita A., Kishimoto S., Sugiura T. (2007) Biochem. Biophys. Res. Commun. 362, 928–934 [DOI] [PubMed] [Google Scholar]
- 8. Piñeiro R., Maffucci T., Falasca M. (2011) Oncogene 30, 142–152 [DOI] [PubMed] [Google Scholar]
- 9. Waldeck-Weiermair M., Zoratti C., Osibow K., Balenga N., Goessnitzer E., Waldhoer M., Malli R., Graier W. F. (2008) J. Cell Sci. 121, 1704–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bondarenko A., Waldeck-Weiermair M., Naghdi S., Poteser M., Malli R., Graier W. F. (2010) Br. J. Pharmacol. 161, 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andradas C., Caffarel M. M., Pérez-Gómez E., Salazar M., Lorente M., Velasco G., Guzmán M., Sánchez C. (2011) Oncogene 30, 245–252 [DOI] [PubMed] [Google Scholar]
- 12. Sutphen R., Xu Y., Wilbanks G. D., Fiorica J., Grendys E. C., Jr., LaPolla J. P., Arango H., Hoffman M. S., Martino M., Wakeley K., Griffin D., Blanco R. W., Cantor A. B., Xiao Y. J., Krischer J. P. (2004) Cancer Epidemiol. Biomarkers Prev. 13, 1185–1191 [PubMed] [Google Scholar]
- 13. Henstridge C. M., Balenga N. A., Schroder R., Kargl J. K., Platzer W., Martini L., Arthur S., Penman J., Whistler J. L., Kostenis E., Waldhoer M., Irving A. J. (2010) Br. J. Pharmacol. 160, 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin H., Chu A., Li W., Wang B., Shelton F., Otero F., Nguyen D. G., Caldwell J. S., Chen Y. A. (2009) J. Biol. Chem. 284, 12328–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ryberg E., Larsson N., Sjögren S., Hjorth S., Hermansson N. O., Leonova J., Elebring T., Nilsson K., Drmota T., Greasley P. J. (2007) Br. J. Pharmacol. 152, 1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hatzivassiliou G., Song K., Yen I., Brandhuber B. J., Anderson D. J., Alvarado R., Ludlam M. J., Stokoe D., Gloor S. L., Vigers G., Morales T., Aliagas I., Liu B., Sideris S., Hoeflich K. P., Jaiswal B. S., Seshagiri S., Koeppen H., Belvin M., Friedman L. S., Malek S. (2010) Nature 464, 431–435 [DOI] [PubMed] [Google Scholar]
- 17. Pietr M., Kozela E., Levy R., Rimmerman N., Lin Y. H., Stella N., Vogel Z., Juknat A. (2009) FEBS Lett. 583, 2071–2076 [DOI] [PubMed] [Google Scholar]
- 18. Oka S., Kimura S., Toshida T., Ota R., Yamashita A., Sugiura T. (2010) J. Biochem. 147, 671–678 [DOI] [PubMed] [Google Scholar]
- 19. Beltramo M. (2009) Mini. Rev. Med. Chem. 9, 11–25 [DOI] [PubMed] [Google Scholar]
- 20. Li H. F., Chen Y., Rao S. S., Chen X. M., Liu H. C., Qin J. H., Tang W. F., Yue-Wang Zhou X., Lu T. (2010) Curr. Med. Chem. 17, 1618–1634 [DOI] [PubMed] [Google Scholar]
- 21. Lin Y. W., Chuang S. M., Yang J. L. (2003) J. Biol. Chem. 278, 21534–21541 [DOI] [PubMed] [Google Scholar]
- 22. Adwanikar H., Karim F., Gereau R. W., 4th (2004) Pain 111, 125–135 [DOI] [PubMed] [Google Scholar]
- 23. Ji R. R., Befort K., Brenner G. J., Woolf C. J. (2002) J. Neurosci. 22, 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atwood B. K., Mackie K. (2010) Br. J. Pharmacol. 160, 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gertsch J., Leonti M., Raduner S., Racz I., Chen J. Z., Xie X. Q., Altmann K. H., Karsak M., Zimmer A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9099–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marcu J. P., Christian R. T., Lau D., Zielinski A. J., Horowitz M. P., Lee J., Pakdel A., Allison J., Limbad C., Moore D. H., Yount G. L., Desprez P. Y., McAllister S. D. (2010) Mol. Cancer Ther. 9, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McAllister S. D., Christian R. T., Horowitz M. P., Garcia A., Desprez P. Y. (2007) Mol. Cancer Ther. 6, 2921–2927 [DOI] [PubMed] [Google Scholar]
- 28. Shepherd C., Puzanov I., Sosman J. A. (2010) Curr. Oncol. Rep. 12, 146–152 [DOI] [PubMed] [Google Scholar]
- 29. Tsai J., Lee J. T., Wang W., Zhang J., Cho H., Mamo S., Bremer R., Gillette S., Kong J., Haass N. K., Sproesser K., Li L., Smalley K. S., Fong D., Zhu Y. L., Marimuthu A., Nguyen H., Lam B., Liu J., Cheung I., Rice J., Suzuki Y., Luu C., Settachatgul C., Shellooe R., Cantwell J., Kim S. H., Schlessinger J., Zhang K. Y., West B. L., Powell B., Habets G., Zhang C., Ibrahim P. N., Hirth P., Artis D. R., Herlyn M., Bollag G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osmond R. I., Das S., Crouch M. F. (2010) Anal. Biochem. 403, 94–101 [DOI] [PubMed] [Google Scholar]
- 31. Shih H. H., Zhang S., Cao W., Hahn A., Wang J., Paulsen J. E., Harnish D. C. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H1643–H1650 [DOI] [PubMed] [Google Scholar]
- 32. Mazella J., Pétrault O., Lucas G., Deval E., Béraud-Dufour S., Gandin C., El-Yacoubi M., Widmann C., Guyon A., Chevet E., Taouji S., Conductier G., Corinus A., Coppola T., Gobbi G., Nahon J. L., Heurteaux C., Borsotto M. (2010) PLoS Biol. 8, e1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uehara Y., Mochizuki M., Matsuno K., Haino T., Asai A. (2009) Biochem. Biophys. Res. Commun. 380, 627–631 [DOI] [PubMed] [Google Scholar]
- 34. Wigle T. J., Herold J. M., Senisterra G. A., Vedadi M., Kireev D. B., Arrowsmith C. H., Frye S. V., Janzen W. P. (2010) J. Biomol. Screen. 15, 62–71 [DOI] [PubMed] [Google Scholar]
- 35. Xu Z., Nagashima K., Sun D., Rush T., Northrup A., Andersen J. N., Kariv I., Bobkova E. V. (2009) J. Biomol. Screen. 14, 1257–1262 [DOI] [PubMed] [Google Scholar]
- 36. Chu C. T., Levinthal D. J., Kulich S. M., Chalovich E. M., DeFranco D. B. (2004) Eur. J. Biochem. 271, 2060–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szydlowska K., Gozdz A., Dabrowski M., Zawadzka M., Kaminska B. (2010) J. Neurochem. 113, 904–918 [DOI] [PubMed] [Google Scholar]
- 38. Kim E. K., Choi E. J. (2010) Biochim. Biophys. Acta 1802, 396–405 [DOI] [PubMed] [Google Scholar]
- 39. Boutros T., Chevet E., Metrakos P. (2008) Pharmacol. Rev. 60, 261–310 [DOI] [PubMed] [Google Scholar]
- 40. May L. T., Leach K., Sexton P. M., Christopoulos A. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 1–51 [DOI] [PubMed] [Google Scholar]
- 41. Valant C., Sexton P. M., Christopoulos A. (2009) Mol. Interv. 9, 125–135 [DOI] [PubMed] [Google Scholar]
- 42. Price M. R., Baillie G. L., Thomas A., Stevenson L. A., Easson M., Goodwin R., McLean A., McIntosh L., Goodwin G., Walker G., Westwood P., Marrs J., Thomson F., Cowley P., Christopoulos A., Pertwee R. G., Ross R. A. (2005) Mol. Pharmacol. 68, 1484–1495 [DOI] [PubMed] [Google Scholar]
- 43. Milligan G., Stoddart L. A., Smith N. J. (2009) Br. J. Pharmacol. 158, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Padwal R. S., Majumdar S. R. (2007) Lancet 369, 71–77 [DOI] [PubMed] [Google Scholar]
- 45. Scheen A. J., Finer N., Hollander P., Jensen M. D., Van Gaal L. F. (2006) Lancet 368, 1660–1672 [DOI] [PubMed] [Google Scholar]
- 46. Comelli F., Bettoni I., Colombo A., Fumagalli P., Giagnoni G., Costa B. (2010) Eur. J. Pharmacol. 637, 62–69 [DOI] [PubMed] [Google Scholar]
- 47. Costa B., Trovato A. E., Colleoni M., Giagnoni G., Zarini E., Croci T. (2005) Pain 116, 52–61 [DOI] [PubMed] [Google Scholar]
- 48. Jafari M. R., Golmohammadi S., Ghiasvand F., Zarrindast M. R., Djahanguiri B. (2007) Behav. Pharmacol. 18, 691–697 [DOI] [PubMed] [Google Scholar]
- 49. Elmes S. J., Jhaveri M. D., Smart D., Kendall D. A., Chapman V. (2004) Eur. J. Neurosci. 20, 2311–2320 [DOI] [PubMed] [Google Scholar]
- 50. Schuelert N., Zhang C., Mogg A. J., Broad L. M., Hepburn D. L., Nisenbaum E. S., Johnson M. P., McDougall J. J. (2010) Osteoarthritis Cartilage 18, 1536–1543 [DOI] [PubMed] [Google Scholar]
- 51. Izzo A. A., Borrelli F., Capasso R., Di Marzo V., Mechoulam R. (2009) Trends Pharmacol. Sci. 30, 515–527 [DOI] [PubMed] [Google Scholar]
- 52. Pertwee R. G. (2008) Br. J. Pharmacol. 153, 199–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ishiguro H., Onaivi E. S., Horiuchi Y., Imai K., Komaki G., Ishikawa T., Suzuki M., Watanabe Y., Ando T., Higuchi S., Arinami T. (2011) Synapse 65, 103–108 [DOI] [PubMed] [Google Scholar]
- 54. Riedel G., Fadda P., McKillop-Smith S., Pertwee R. G., Platt B., Robinson L. (2009) Br. J. Pharmacol. 156, 1154–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aragona M., Onesti E., Tomassini V., Conte A., Gupta S., Gilio F., Pantano P., Pozzilli C., Inghilleri M. (2009) Clin. Neuropharmacol. 32, 41–47 [DOI] [PubMed] [Google Scholar]
- 56. Johnson J. R., Burnell-Nugent M., Lossignol D., Ganae-Motan E. D., Potts R., Fallon M. T. (2010) J. Pain Symptom Manage 39, 167–179 [DOI] [PubMed] [Google Scholar]
- 57. Ramer R., Merkord J., Rohde H., Hinz B. (2010) Biochem. Pharmacol. 79, 955–966 [DOI] [PubMed] [Google Scholar]
- 58. De Petrocellis L., Vellani V., Schiano-Moriello A., Marini P., Magherini P. C., Orlando P., Di Marzo V. (2008) J. Pharmacol. Exp. Ther. 325, 1007–1015 [DOI] [PubMed] [Google Scholar]
- 59. Ligresti A., Moriello A. S., Starowicz K., Matias I., Pisanti S., De Petrocellis L., Laezza C., Portella G., Bifulco M., Di Marzo V. (2006) J. Pharmacol. Exp. Ther. 318, 1375–1387 [DOI] [PubMed] [Google Scholar]
- 60. Klein R. M., Spofford L. S., Abel E. V., Ortiz A., Aplin A. E. (2008) Mol. Biol. Cell 19, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pritchard C. A., Hayes L., Wojnowski L., Zimmer A., Marais R. M., Norman J. C. (2004) Mol. Cell. Biol. 24, 5937–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kotsikorou E., Madrigal K. E., Hurst D. P., Sharir H., Lynch D. L., Heynen-Genel S., Milan L. B., Chung T. D., Seltzman H. H., Bai Y., Caron M. G., Barak L., Abood M. E., Reggio P. H. (2011) Biochemistry 50, 5633–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.