Abstract
LRP5 and LRP6 are Wnt co-receptors essential for Wnt/β-catenin signaling. DKK1 inhibits Wnt signaling by interacting with the extracellular domain of LRP5/6, and is a drug target for multiple diseases. Here we present the crystal structures of the first and second halves of LRP6’s four propeller–EGF pairs (LRP6-E1E2 and LRP6-E3E4), and a LRP6-E3E4/DKK1 complex. Combined with EM analysis, these data demonstrate that LRP6-E1E2 and LRP6-E3E4 form two rigid structural blocks, with a short intervening hinge that restrains their relative orientation. DKK1c interacts with the top surface of the LRP6-E3 YWTD propeller, and likely also that of the LRP6-E1 propeller due to structural similarity, through conserved hydrophobic patches buttressed by a network of salt bridges and hydrogen bonds. Our work provides key insights for understanding LRP5/6 structure and the interaction of LRP5/6 with DKK, as well as for drug discovery.
Keywords: Wnt, receptor, LRP5, LRP6, LDL receptor-like protein, Dickkopf (DKK), crystal structure, YWTD β-propeller
Wnt proteins constitute a family of secreted glycoproteins that regulate the Wnt/β-catenin signaling pathway, as well as the planar cell polarity and Ca2+ signaling pathways. The Wnt/β-catenin signaling pathway controls cell proliferation and differentiation in diverse contexts, including embryonic development and adult homeostasis, and dysregulation is tightly associated with cancer, bone anomalies, and other diseases1–4. Signaling is initiated when Wnt proteins simultaneously interact with their receptor and co-receptor, Frizzled and LRP5/6. The formation of the Frizzled–Wnt–LRP5/6 ternary complex then leads to phosphorylation of the intracellular domain of LRP5/6 and to transduction of the Wnt signal inside the cell5,6.
Low-density-lipoprotein (LDL) receptor-related proteins 5 and 6 (LRP5 and LRP6) form a subfamily of the LDL receptor (LDLR) family, share 73% identity in their extracellular domains, and are essential for Wnt/β-catenin signaling7–13. The LRP5/6 extracellular domains consist of three types of subdomains: the YWTD-type β-propeller domain, the EGF-like domain, and the LDLR type A (LA) domain (Fig. 1a). Based on the crystal structures of the LDL receptor that contains a single propeller, the YWTD-type β-propeller domain has six YWTD repeats, which form a six-bladed β-propeller structure14–16. The four propeller domains in LRP5/6 share a relatively low identity among them (~19% for human LRP6; Supplementary Fig. 1), indicating the functional differences among these YWTD propellers. Each YWTD-propeller domain is followed by a ~40 residue EGF-like domain that may cover the bottom face of the YWTD-propeller, similar to LDLR14–16. The small LA domains in LRP5/6 appear to be dispensable for binding extracellular ligands Wnts and DKKs17. While understanding the structures of LRP5/6 is clearly important for understanding the mechanisms of signal transduction and for identifying candidate new therapeutics, there have been no previous reports of the structure of the extracellular domain of LRP5/6.
Figure 1.
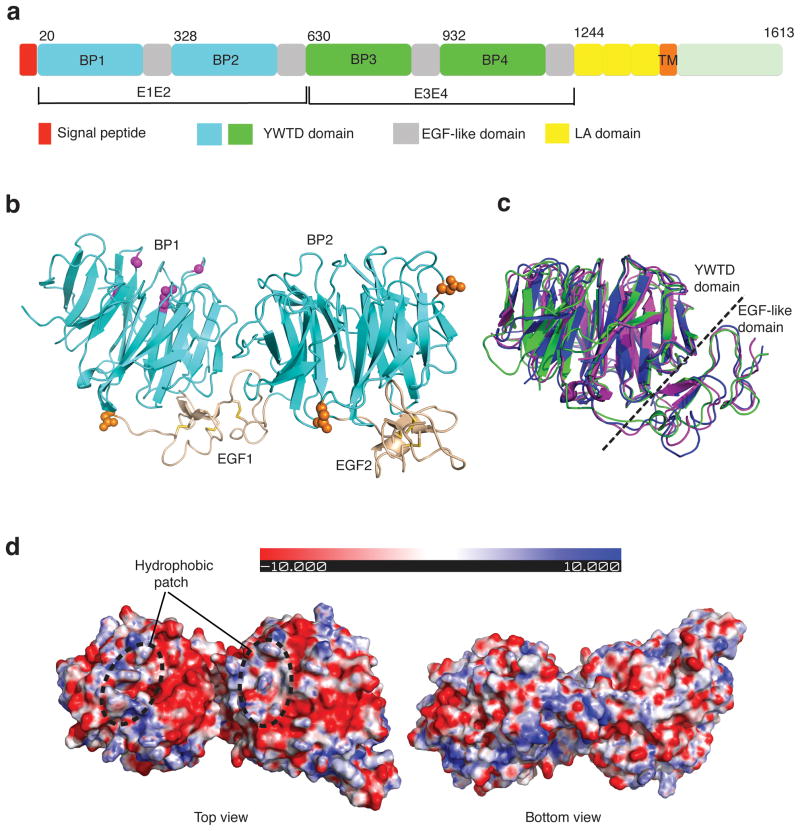
Crystal structure of LRP6-E1E2. (a) Schematic representation of the domain organization of human LRP6. “BP” stands for YWTD β-propeller domain. First and second YWTD domains are colored in cyan. Third and fourth YWTD domains are shown in green. EGF-like domains, LDLR type A repeats, and transmembrane region are shown in gray, yellow, and orange, respectively. (b) Overall structure of LRP6-E1E2 fragment. YWTD domains are shown in cyan, EGF-like domain in wheat. Observed N-glycosylation sites/moiety (in orange spheres), and four sites corresponding to LRP5 mutants associated with high bone density syndrome (in magenta balls) are shown. (c) Superposition of three YWTD–EGF pairs: LRP6-E1, LRP6-E2 and the LDLR YWTD–EGF3 pair. LRP6-E1 and LRP6-E2 are shown in magenta and green, respectively, while LDLR is in blue. (d) Electrostatic surfaces of LRP6-E1E2 (top and bottom views).
There is considerable interest in understanding how LRP5/6 interacts with its regulatory proteins. Dickkopf (DKK) proteins, exemplified by DKK1, are secreted glycoproteins that specifically inhibit Wnt/β-catenin signaling by antagonizing LRP5/6 (ref. 10–13). Structurally, DKKs contain two characteristic cysteine-rich domains, of which the C-terminal domain is highly conserved and belongs to the Colipase family18. Analysis of individual DKK domains and chimeric DKKs shows that the DKK C-terminal domains interact with LRP6 and are necessary and sufficient for Wnt inhibition19. DKKs mainly interact with the third and fourth propellers of LRP5/6 but can also bind to the first and second propellers17. Two mechanisms have been proposed for DKK1 antagonism of LRP5/6. The first supports DKK1 inhibition of Wnt/β-catenin signaling mainly by binding LRP5/6 and disrupting the Wnt-induced Fzd–Wnt–LRP6 complex10, while the second mechanism supports a DKK1 and Kremen-dependent induction of LRP5/6 endocytosis and degradation in specific tissues13,20,21.
Modulation of the interactions between LRP5/6 and DKK would be an attractive therapeutic goal for treating multiple diseases, including osteoporosis, multiple myeloma, osteosarcoma and melanoma22–24. LRP5 mutations are associated with osteoporosis pseudoglioma syndrome (OPPG) and autosomal dominant high bone mass (HBM)25–27, and LRP6 mutations have been linked to early coronary disease and metabolic risk factors28. Additionally, reduction of DKK1 results in increased bone mass in mice and has become an important drug target for the treatment of osteoporosis29,30. Finally, antibodies targeting DKK1 have been used to treat bone tissue loss in mouse models of multiple myeloma31 and osteoarthritis.
A detailed analysis of how LRP5/6 interacts with both activators and inhibitors of Wnt/β-catenin signaling, and elucidating whether these interactions induce conformational changes in LRP5/6, is critical for understanding how LRP5/6 functions as a Wnt co-receptor as well as validating it as a therapeutic target. To understand how LRP5/6 plays an essential role in transducing Wnt signal and how DKK1 inhibits Wnt signaling through its interaction with LRP5/6, we have determined crystal structures of the extracellular domain of human LRP6 and compare this with the structure of its complex with human DKK1. Combined with EM and mutation-function analysis, our work advances our understanding of the three-dimensional structure of the extracellular domain of LRP5/6, as well as providing novel insights into how DKK inhibits Wnt/β-catenin signaling by interacting with LRP5/6.
Results
Crystal structure of the first two YWTD–EGF pairs of LRP6
For simplicity we refer to the LRP6 fragment containing the first “YWTD-type β-propeller and EGF pair” as LRP6-E1, the second “YWTD-type β-propeller and EGF pair” as LRP6-E2, etc. (Fig. 1a). Human LRP6-E1E2 was overexpressed using a baculovirus secretion system and purified to homogeneity. The crystal structure of LRP6-E1E2 was then determined by molecular replacement using the LDLR YWTD β-propeller as the search model. The two EGF domains were manually built into the electron density and the structure was refined at 2.8 Å resolution (Table 1). There are two LRP6-E1E2 copies in the asymmetric unit, with almost identical structures (with a RMSD of 0.92 Å for 567 Cα’s in LRP6-E1E2). In each copy, the two YWTD β-propellers reside with the propeller axis roughly in parallel. Each of the two EGF domains pack tightly against the bottom surface of the preceding YWTD β-propellers in a manner almost identical to that of the YWTD–EGF pair in LDLR (Fig. 1b,c). In fact, all 10 YWTD–EGF pairs (2 copies for E1 and E2, and 3 copies for E3 and E4, respectively) in the three crystal structures reported here have the same YWTD–EGF inter-domain orientation, strongly indicating that this structural arrangement may be preserved in all LDLR family members. It should be noted that there are side-by-side interactions for E1 and E2, and the EGF domain in E1 interacts with both the YWTD propellers 1 and 2, which may be important for stabilizing the relative orientation of E1 and E2 (see below).
Table 1.
Data collection and refinement statistics
| Human LRP6-E1E2 | Human LRP6-E3E4 | Human LRP6- E3E4/Dkk1c | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 70.54, 144.51, 70.76 | 68.6, 83.75, 166.62 | 92.97, 104.76, 158.8 |
| α, β, γ (°) | 90, 96.0, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 30–2.8(2.95–2.8) | 30–2.8(2.95–2.8) | 30–3.1(3.21–3.1) |
| Rsym | 9.1(33.5) | 9.7(41.3) | 11.4(62) |
| I/σI | 7(2.3) | 4.8(1.8) | 11(1.7) |
| Completeness (%) | 100(100) | 99.9(100) | 98.4(92.8) |
| Redundancy | 3.9(4.0) | 6.8(7.1) | 3.3(2.9) |
| Refinement | |||
| Resolution (Å) | 30–2.8 | 30–2.8 | 30–3.1 |
| No. reflections | 32951 | 23097 | 27740 |
| Rwork/Rfree | 24.4/30.2 | 25.2/27.9 | 24.3/29.2 |
| No. atoms | |||
| Protein | 9336 | 4833 | 10178 |
| Ligand/ion | 70 | 70 | – |
| B-factors | |||
| Protein | 70.04 | 91.9 | 87.1 |
| Ligand/ion | 137.2 | 138.4 | – |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.0085 | 0.006 | 0.007 |
| Bond angles (°) | 1.21 | 1.1 | 1.08 |
One crystal was used for each dataset. Values in parentheses are for highest-resolution shell.
Consistent with the N-glycosylation site prediction, electron densities for sugar moieties are found for three glycosylation sites in LRP6-E1E2. These glycosylation sites are all on the propeller side surface or EGF repeats and are not expected to play a role in binding of LRP6 ligand for the top surface (Fig. 1b). The top surfaces of E1 and E2β-propellers contain a hydrophobic patch, while the rest of these top surfaces are largely negatively charged (Fig. 1d). Residues on the E1 and E2 top surfaces are conserved among corresponding LRP5/6 propellers. Interestingly, several missense mutations associated with high bone density syndrome, including D111Y, G171V, G171R, A214V, A214T and A242T, are mapped to the top surface of LRP6-E1 β-propeller (Fig. 1b). Since these mutants were shown to be defective in inhibition of Wnt signaling by DKK1 (ref. 32), the top surface of LRP6-E1 β-propeller is expected to be involved in DKK1 interaction.
Crystal structure of the third and fourth YWTD–EGF pairs of LRP6
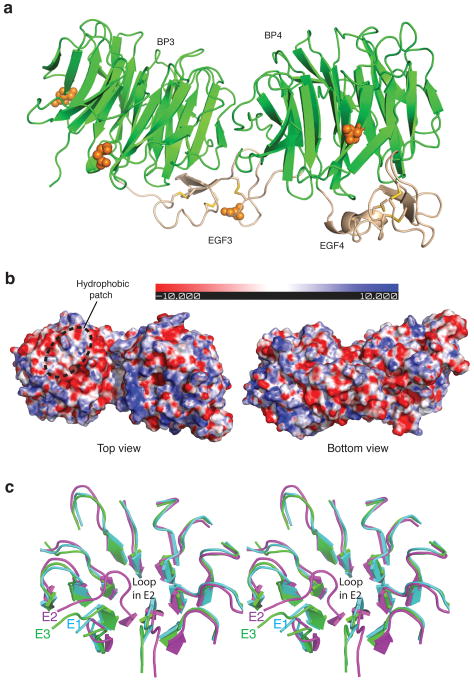
The distinct domain LRP6-E3E4 was then overexpressed using a baculovirus secretion system and purified to homogeneity. The crystal structure of LRP6-E3E4 was determined by molecular replacement and refined at 2.8 Å resolution (Table 1). There is one copy of E3E4 in the crystal asymmetric unit. Electron densities for sugar moieties are found for four glycosylation sites in LRP6-E3E4, with none of them on the propeller top surfaces. The overall structure of LRP6-E3E4 is very similar to that of LRP6-E1E2 (Fig. 2a). The top surface of LRP6-E3 is highly negatively charged. LRP6-E3, but not LRP6-E4, contains the hydrophobic patch on the top surface of propellers that is also observed in LRP6-E1 and E2 (Fig. 2b). Structural comparison demonstrates that the top surface of LRP6-E2 is significantly different from that of LRP6-E3 in our crystal structures. In contrast, LRP6-E1 has a very similar top surface with LRP6-E3 (Fig. 2c).
Figure 2.
Crystal structure of LRP6-E3E4. (a) Overall structure and observed N-glycosylation sites. YWTD domains are shown in green and EGF-like domain in wheat. N-glycosylation sites are shown in orange spheres. (b) Electrostatic surfaces of LRP6-E3E4 (top and bottom views). (c) Comparison of the top surfaces of LRP6-E3 vs E2, and E3 vs E1. Structures of LRP6-E3 and E1, E2 are superimposed. It is clear that LRP6-E1, but not LRP6-E2, has a very similar top surface with LRP6-E3. The difference in the E2-propeller top surface is mainly due to the “bulge out” of the loop between blades 5 and 6, which is observed in both copies of LRP6-E2.
LRP6-E1E4 contains two rigid structural blocks linked by a hinge
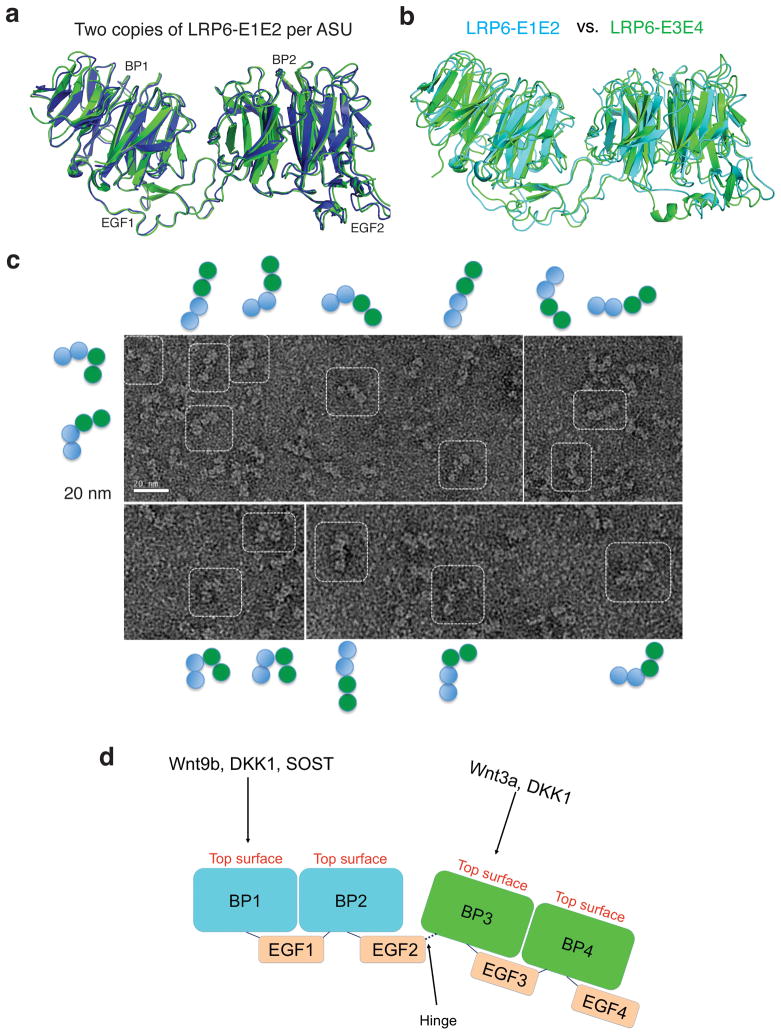
Including the two LRP6-E3E4 copies in the crystal structure of the LRP6-E3E4/DKK1c complex that will be discussed below, the three crystal structures presented here represent five E1E2/E3E4 dual propeller structures from distinct crystallization conditions and from completely different crystal packing environments. It is striking that, when superimposed, all five dual propeller structures have essentially identical mainchain traces (Fig. 3a,b). The relative orientations among all four domains (YWTD–EGF–YWTD–EGF) are fixed in all five E1E2/E3E4 dual propeller structures (Fig. 3a,b). This strongly suggests that the two propellers in both LRP6-E1E2 and E3E4 are structurally coupled and can be considered as relatively rigid structural blocks. This rigidity between neighboring YWTD–EGF pairs may be largely attributable to the bridging EGF domains in E1 and E3 that interact with YWTD propellers on both sides. There are various direct inter-propeller interactions for YWTD propeller pairs E1–E2 and E3–E4, but residues involved in these interactions are not conserved between E1–E2 and E3–E4 interfaces and may play auxiliary roles in stabilizing LRP6-E1E2 and E3E4 structural blocks.
Figure 3.
Rigidity of the two LRP6-ECD structural blocks and hinge between these two blocks. (a) Superposition of two LRP6-E1E2 copies in the asymmetric unit of the LRP6-E1E2 crystal lattice. (b) Superposition of LRP6-E3E4 and LRP6-E1E2. LRP6-E3E4 and E1E2 are shown in green and cyan, respectively. (c) EM analysis of human LRP6-E1E4. (d) A model for the two structural blocks of the LRP6 extracellular domain and the hinge between these two blocks.
To determine if the orientation between these two structural blocks is also fixed, we visualized LRP6-E1E4, which contains all four LRP6 propeller–EGF pairs, using electron microscopy (EM) of negatively-stained LRP6-E1E4 samples. EM images indicated that LRP6-E1E4 can be visualized in many different shapes that are apparently not from a uniform conformation (Fig. 3c). We propose that there is likely a hinge between LRP6-E2 and E3. This notion is consistent with the previous observation that LRP6-E3 cannot be secreted when overexpressed in mammalian cells on its own or in combination with E1E2, but its secretion is vastly enhanced when expressed together with LRP6-E4, indicating a lack of co-folding effect between LRP6-E2 and E3 (ref. 33). It is important that both copies of the second EGF domain (EGF2) are well ordered in the identical position relative to the second β-propeller in our LRP6-E1E2 crystal structure (Fig. 3a). There are only two conserved linker residues (Pro630-Glu631) between the C-terminus of the well-ordered EGF2 domain and the beginning of globularly folded propeller 3, both of which are located on bottom surfaces of the LRP6-E1E2 and E3E4 blocks. The hinge flexibility provided by such a short linker is very limited. For example, the bottom surfaces of LRP6-E1E2 and E3E4 may approach each other, but the top surfaces of propellers from the same LRP6 molecule, which may provide binding surfaces for various LRP6 partners, cannot face each other (Fig. 3d).
Crystal structure of LRP6-E3E4 in complex with DKK1c
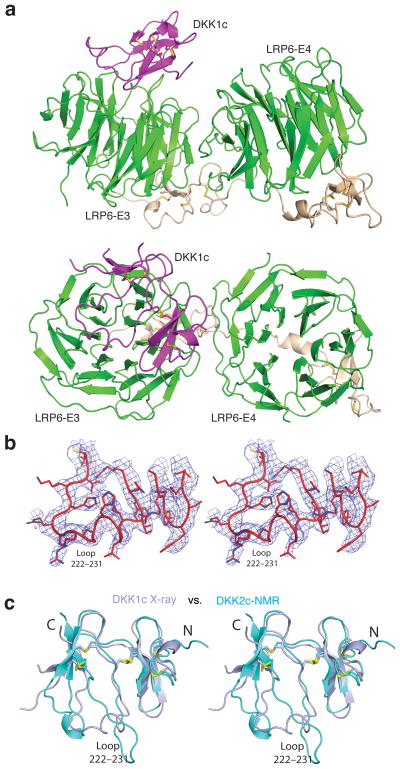
We next separately purified LRP6-E3E4 and the C-terminal domain of DKK1 (DKK1c). The two proteins were then mixed together with DKK1c in stoichiometric excess, and the complex containing both proteins, designated as the LRP6-E3E4/DKK1c complex, was purified using size-exclusion chromatography (SEC) in preparation for crystallization. The crystal structure was eventually determined by molecular replacement and refined at 3.1 Å resolution (Fig. 4a,b). There are two LRP6-E3E4 and one DKK1c molecules in the asymmetric unit. The LRP6-E3E4 structure is largely unchanged by the binding of DKK1c (Supplementary Fig. 2). The structural topology of DKK1c, including the formation of five disulfide bonds, is the same as the unbound DKK2c NMR structure previously reported34. However, there are significant conformational differences in DKK1c compared with the unbound DKK2c NMR structure. Instead of having six β-strands in DKK2c, there are only four β-strands for DKK1c with two each residing on two sides of a relatively flat structure (Fig. 4c). There is one long loop between these two β-sheets, largely flexible in the DKK2c NMR structure, which is crucial for the binding of both LRP6-E3 copies. Both of these two LRP6-E3 propellers use their top surface to recognize the two sides of DKK1c in our crystal lattice.
Figure 4.
Crystal structure of the LRP6-E3E4/DKK1c complex. (a) An overall view of the complex in two orientations roughly related by a horizontal 90° rotation. Only the interface A is shown here (see text). DKK1c is in magenta. (b) Stereo view of a representative portion of the Fo-Fc electron density map (contoured at 2.0 σ), showing the DKK1c loop 222–231 region. The map was calculated using phases from LRP6-E3E4 only. (c) Comparison of DKK1c and DKK2c structures. Crystal structure of DKK1 in the LRP6-E3E4/DKK1 complex (in light blue) and the NMR structure of DKK2c (in cyan) are superimposed.
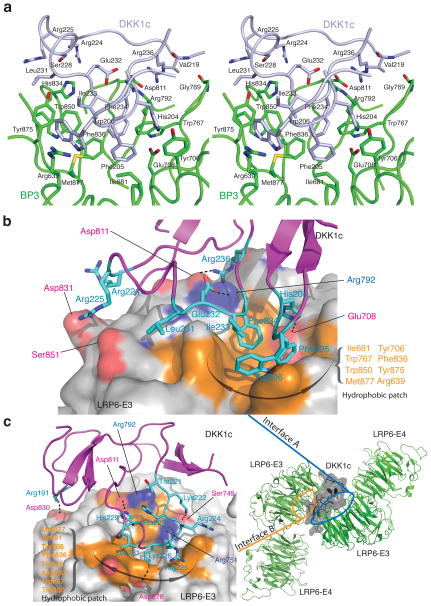
Intriguingly, one of these two DKK1c surfaces involved in LRP6 binding is the proposed binding site for Kremen (see below), a protein that plays a role in the endocytosis and turnover of LRP5/6 (ref. 20,35). Since the physiological relevance of this interface (interface B) remains to be tested, here we emphasize only the other interface (interface A, Fig. 4, 5a,b) that is supported by our functional assay (see below), and that is consistent with the previous biochemical data35.
Figure 5.
Specific interactions in the two LRP6-E3/DKK1c interfaces. (a) Stereo view of LRP6-E3E4/DKK1 interface A. DKK1c is shown in light blue. Residues involved in the interactions are labeled and shown in green (LRP6-E3E4) and light blue (DKK1c) stick models. (b) A surface representation of the LRP6–DKK1c interaction (interface A only). LRP6-E3 is shown in surface illustration. LRP6 residues involved in the interface are shown on the solid surface, whereas DKK1 residues are shown in stick models. Hydrophobic LRP6 and DKK1c residues directly involved in hydrophobic interactions are also labeled in orange and cyan, respectively. (c) The second LRP6–DKK1 interface (interface B) observed in the crystal lattice. (Right) Overall structure of LRP6-E3E4/DKK1c complex in the crystal lattice. Two LRP6-E3E4 are arranged roughly in an anti-parallel orientation. DKK1c is shown in transparent surface illustration. (Left) Detailed interactions on the interface B. LRP6-E3 is shown in surface illustration and its interface residues are shown as in (b). The physiological relevance of the interface B remains to be tested.
LRP6-E3E4/DKK1c interfaces
In both interfaces (interfaces A and B), the long loop of DKK1c (residues 222–231) forms the major binding site. Both E3 YWTD propellers in the asymmetric unit dock on opposite sides of this DKK1 loop, using their top surfaces containing a hydrophobic patch that is formed by residues Phe836, Trp850, Trp767, Tyr875, Met877, Tyr706 and Ile681. This hydrophobic patch forms the core interactions in both interfaces. It should be noted that residues in this hydrophobic patch are conserved among LRP5/6 propellers 1, 2 and 3, but not propeller 4 (Supplementary Fig. 1). This provides an explanation for why LRP6-E4 does not interact with DKK1c in our structure.
Interface A buries an surface of 1946 Å2, which is typical for specific protein-protein interactions36. In interface A, the hydrophobic patch on the top surface of LRP6-E3 interacts with DKK1c Phe205 and Trp206 in the short loop between the first two β-strand in DKK1c, and three other hydrophobic side-chains in the DKK1c long loop (Leu231, Ile233 and Phe234) (Fig. 5a,b). This hydrophobic interaction is buttressed by several salt bridges and hydrogen bonds between LRP6-E3 and DKK1c: LRP6 Asp811 with DKK1 Arg236, LRP6 Glu708 with DKK1 His204, LRP6 Arg792 with DKK1 Glu232, and LRP6 His834 with DKK1 Ser228. In addition, the sidechains of DKK1 Arg224 and Arg225 are at the tip of the loop and can interact with LRP6-E3 top surfaces on both sides. Arg236 and His204 have been implicated in LRP6–DKK1 interactions in a previous study35.
In interface B, most interactions are made by a segment in the DKK1c long loop on the top surface of LRP6-E3 propeller (Fig. 5c). This interface buries a surface area of 1877 Å2. Here the LRP6-E3 top surface hydrophobic patch containing Phe836, Trp850, Tyr875 and Met877 is covered by a helix-like structure formed by DKK1 residues 226–231 (KGSHGL) with well-fit complementary surfaces (Fig. 5c). The Lys226 side-chain points straight into the hole formed by E3 propeller and interacts with LRP6 Asp878. This interaction is enhanced by four hydrogen bonds in this region: LRP6 Asp811 – DKK1 His229, LRP6 Ser749 – DKK1 Lys222, LRP6 Arg792 – Thr221, and LRP6 Arg751 – DKK1 His223. In addition to interactions in this long loop region, there are a number of other interactions, including a salt bridge between LRP6 Asp830 and DKK1 Arg191 that is located near the other end of the interface B. It should be noted that DKK1 residues Arg191, Ser192 and Lys226 were implicated in Kremen binding35.
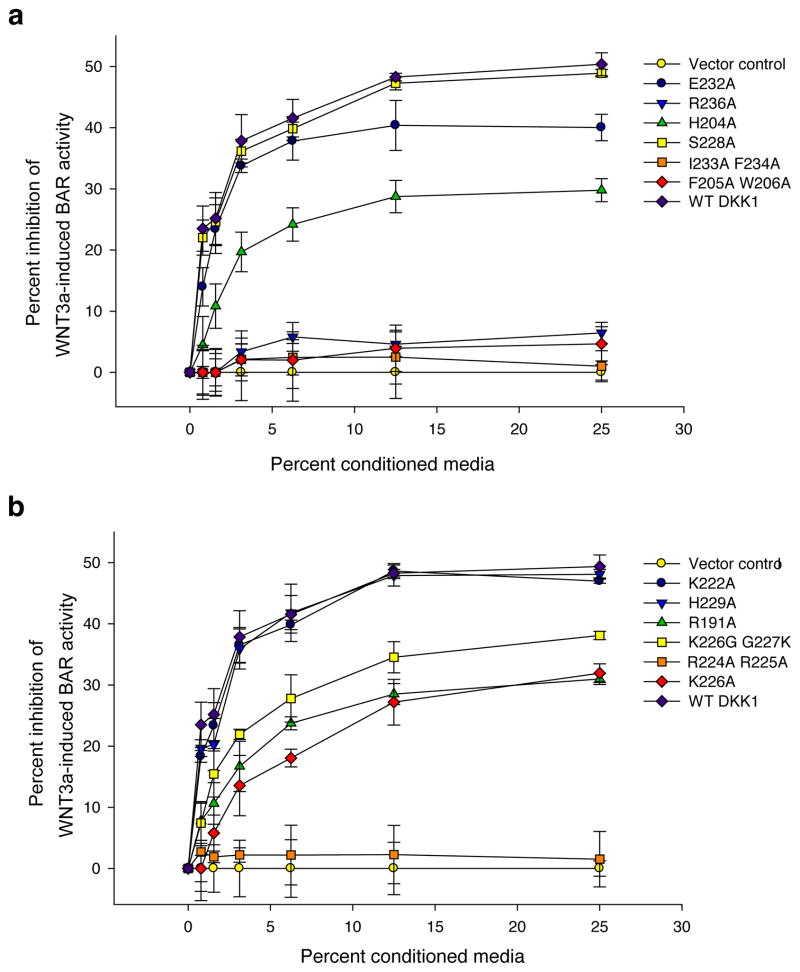
Mutagenesis analysis of DKK1 residues in the LRP6–DKK1 interface
To validate the physiological relevance of our crystal structures and to evaluate the significance of the residues involved in the interaction between LRP6 and DKK1, we mutated DKK1 at positions within the observed LRP6–DKK1 interfaces. Specifically, we converted different amino acids into the neutral amino acid Alanine, and then used a robust luciferase reporter assay for Wnt/β-catenin signaling to monitor whether the mutations altered the ability of DKK1 to repress Wnt/β-catenin signaling in cells cultured in vitro. Mutation of interface A residues had strong effects (Fig. 6a). Mutation of DKK1c hydrophobic residues (Ile233 Phe234, Phe205 Trp206), as well as Arg236, completely abolished the ability of DKK1 to inhibit Wnt/β-catenin signaling, whereas H204A and E232A showed reduced activity (Fig. 6a). This is consistent with previous results that mutations in mouse DKK1 corresponding to human DKK1 H204E and R236E abolish repression of Wnt/β-catenin signaling by DKK1 (ref. 35). In comparison, mutations in interface B had much milder effects (Fig. 6b). The only mutation involving interface B that completely abolished DKK1 activity was the R224A R225A double mutation. In this case, Arg224 and Arg225 should be counted for both interfaces A and B, as they can interact with both LRP6 copies in the crystal structure. In summary, missense mutations on both DKK1c surfaces can suppress the ability of DKK1 to antagonize Wnt/β-catenin signaling. Together, these results are consistent with the binding interfaces revealed by our crystal structure and support their physiological relevance.
Figure 6.
DKK1 residues in DKK1–LRP5/6 interface are important for its inhibitory effects on Wnt/β-catenin signaling. Conditioned media was generated for each of the indicated DKK1 constructs and each was tested for its ability to inhibit Wnt3A mediatedβ-catenin signaling in RKO cells as measured by a β-catenin activated reporter (BAR). (a) mutations on interface A; (b) mutations on interface B. Assays were performed in triplicate and error bars represent standard deviation. Reporter data were normalized to expression of the corresponding DKK1 WT or mutant protein as determined by desitometric analysis of a western blot (Supplementary Fig. 6).
Biochemical characterization of the LRP6–DKK1c interaction
To further characterize the interaction between DKK1c and LRP6, we determined the affinity of DKK1c for LRP6-E1E2 and LRP6-E3E4 using an Alpha Technology binding assay (Supplementary Fig. 3). Purified DKK1c interacts with both LRP6-E1E2 and LRP6-E3E4 fragments with dissociation constants (Kd values) of 70 and 50 nM, respectively, comparable with previously reported corresponding Kd values for the full-length DKK1 (64 and 21 nM, respectively)17. This result and our mutagenesis data (Fig. 6) are consistent with the conclusion that DKK1c is necessary and sufficient for the LRP6–DKK1 interaction and Wnt inhibition19.
We next sought to explore the stoichiometry of Dkk1c and LRP6 in solution by size-exclusion chromatography and dynamic light scattering analysis. The LRP6-E3E4/DKK1c complex existed mostly in a 1:1 molar ratio in solution (Supplementary Fig. 4), which suggests that the LRP6-E3E4/DKK1c interaction via interface B is not stably formed in solution. In contrast, the presence of DKK1c can significantly induce LRP6-E1E4 oligomerization (Supplementary Fig. 4), which is consistent with the possibility of DKK1c containing two LRP6 binding surfaces. It remains to be tested whether the LRP6–DKK1 interface B we observed is physiologically relevant. This is an intriguing issue since the presence of Wnt3a quickly induces plasma membrane-associated LRP6 aggregates37, in which the local LRP6 concentration could be high enough for the LRP6–DKK1 interaction via interface B.
Discussion
The Wnt/β-catenin signaling pathway is involved in many biological processes and is regulated by both extracellular and intracellular signals. It is generally accepted that the initiation of signal transduction at the plasma membrane involves formation of a ternary complex between the secreted Wnt ligand and the integral membrane proteins Frizzled and LRP5/6. While members of the Frizzled family act as receptors for Wnts, LRP5/6 is an essential Wnt co-receptor that serves as a critical point of signal integration by interacting with multiple partners, including Dickkopf (DKK), sclerostin (SOST), and R-spondin in addition to Wnts11,29,38,39. While Wnt and R-spondin activate the Wnt/β-catenin pathway, DKK and SOST are known to be potent inhibitors of this pathway. The LRP5/6 extracellular domain (ECD) contains four “YWTD-type β-propeller – EGF like domain” pairs, followed by three LA domains (Fig. 1a). The LA domains are expected to be structurally uncoupled, according to structural studies of LA domains in LDLR16,40, and are dispensable for Wnt/β-catenin signaling17. It is important to know if these propeller–EGF domains function independently like “strings and beads”, or are structurally coupled, thereby allowing for allosteric regulation.
LRP5/6-ECD contains two rigid “propeller–EGF” structural blocks
It is striking that in the three crystal structures presented here, all five LRP6-E1E2, E3E4 pairs have almost identical main-chain structures, despite being in a distinct chemical environment in their respective crystal lattices. This argues strongly that the LRP6-E1E2 and E3E4 pairs are structurally coupled and form relatively rigid structure blocks, in which the top surfaces of the coupled propellers are roughly next to each other. Our EM analysis indicates that the relative orientation of the E1E2 and E3E4 blocks is not fixed. However, the short linker connecting these two structural blocks is located on the bottom surfaces of both blocks, and one of the two residues in the linker is a proline that makes the main-chain less flexible than most of other residues. This not only allows the bottom surfaces of the two blocks to approach each other, but also prevents the top surfaces from approaching each other (Fig. 3d). The protein SOST has been shown to interact with the top surface of the E1 propeller, and to inhibit Wnt signaling by competing with Wnt for binding to LRP5/6-E1 (ref. 17,34,41–45). At least some of the Wnt proteins are expected to bind to the propeller top surfaces as well since DKK1 was shown to compete with Wnt3a and Wnt9b for binding to LRP6 (ref. 17). Given that most Wnt proteins and the extracellular domain of Frizzled are much smaller than the E1E2 and E3E4 blocks (~90 Å in the long dimension) of LRP5/6 in this restrained inter-block orientation, one can expect that the top surface(s) of the LRP5/6 extracellular domain needs to be close to the membrane when forming the Fzd–Wnt–LRP ternary complex. It is plausible that Wnt signaling activities require appropriate spatial arrangement of Fzd, Wnt and LRP5/6, and the dynamic properties of LRP5/6 structure could be a key factor.
Would such structural coupling affect Wnt signaling? One potential example is a LRP6-R611C missense mutation that was found in patients with coronary artery disease. LRP6 Arg611 is located near the tip of extruding EGF2 domain and may play a role in stabilizing the position of this EGF domain by forming a salt bridge with Glu477 in the E2 propeller domain (Supplementary Fig. 5a). R611C may relax the relative orientation between YWTD and EGF domains in LRP6-E2, which may in turn affect the orientation or structural dynamics between the two LRP6 structural blocks. Alternatively, R611C mutation may perturb the disulfide bond formation and therefore the folding of the EGF2 domain by providing an additional cysteine residue.
Structural basis of the LRP5/6–DKK1 interaction
Our LRP6-E3E4/DKK1c crystal structure reveals two LRP6/DKK1c interfaces. Interface A uses a hydrophobic interaction as the core for binding and its existence and physiological relevance is supported by current and previous mutagenesis data. Based on the key interactions identified by our crystal structure, it is apparent that LRP6-E4 cannot bind to DKK1 as it lacks this hydrophobic patch on the top surface (Fig. 2, Supplementary Fig. 1). Does DKK1c interact with LRP6-E1, or E2 or with both? Since LRP6-E1, but not LRP6-E2, has a top surface very similar to that of LRP6-E3 (Fig. 2c), LRP6-E3 and E1 may form two discrete DKK1c binding sites with very similar binding modes, whereas LRP6-E2 may not be compatible with DKK1c binding. This structural analysis is consistent with the LRP5 mutations found in HMB patients. These mutations are in the top surface of LRP5-E1 propeller, and have been shown to disrupt the LRP5–DKK1 interaction32 (Fig. 1b).
Our structures provide explanations for the activities of LRP5 mutations associated with high bone mass (HBM). For example, the LRP5(G171V) mutation is associated with HBM and displays reduced binding to DKK1 (ref. 46). In our crystal structure, the mainchain amine of Val219 of DKK1c forms a hydrogen bond with the mainchain carbonyl of LRP6-E3 residue Gly769 (Supplementary Fig. 5b). LRP6 G768V mutation (corresponding to G171V in LRP5-E1) would change the conformation of a β-turn formed by Gly768 and Gly769 and may disrupt this hydrogen bond. Assuming LRP5-E1 and LRP6-E3 interact with DKK1 in the same way, G171V may weaken the LRP5–DKK1 interaction for the same reason.
It is intriguing that we observed a second LRP6–DKK1 interface (interface B), which overlaps with a previously proposed Kremen-binding surface35. Our mutagenesis data on interface B (Fig. 6b) could be partially explained by the lack of Kremen binding activities, but the potential involvement of interface B in the LRP5/6–DKK1 interaction in the cell cannot be excluded. Importantly, if LRP6 can interact with both surfaces of DKK1c, LRP6 can also compete with Kremen binding, and thus the resulting output would depend on the local concentrations of LRP5/6, DKK1 and Kremen. How DKKs interact with LRP5/6 and regulate Wnt signaling in vivo will require further investigations.
Implications for Wnt signaling and drug discovery
Antibodies or inhibitors that disrupt the LRP5/6–DKK1 interactions are potentially useful for osteoporosis and other diseases29,30. The top surfaces of LRP6-E1 and E3 propellers contain hydrophobic pockets that are amenable for developing small molecule inhibitors (Fig. 5b), especially since most of these residues are from the loop regions of propeller domains and thus have a certain degree of conformational flexibility to generate deeper pockets for small molecule drugs. In addition, our structural model of LRP6 extracellular domain indicates that the LRP6 ligand binding domains may be regulated allosterically. Therefore, we predict that it is possible to develop allosteric Wnt pathway regulators by targeting at the LRP5/6 extracellular domain.
It has been previously shown that DKK1 competes with Wnt for LRP5/6 binding17. In addition, it has been shown that Wnt1 and Wnt9b bind to LRP6-E1E2, while Wnt3a binds to E3E4, and that E1 and E3 may be the major ligand binding sites17,47,48. Because of the structural rigidity, to allow for the competition by small DKK proteins, Wnt proteins may bind to the top surface of E1 and E3 as well. Since the hydrophobic patches on the top surfaces of LRP6 E1 and E3 are the LRP6–DKK1 interaction cores, we predict that these two hydrophobic patches may form part of the Wnt binding sites. We expect these and other predictions based on structural analyses to advance the discovery of drugs targeting the Wnt/β-catenin pathway.
ONLINE METHODS
Protein expression and purification
Extracellular domains of human LRP6, including LRP6-E1E2 (20–629), LRP6-E3E4 (630–1244) and LRP6-E1E4 (20–1244), were overexpressed using a pAcGP67A vector in a BaculoGold secreted baculovirus/Hi5 insect cell system. Human DKK1 C-terminal Cys-rich domain DKK1c (183–266) was cloned and expressed as a thioredoxin/His6-fusion protein in E. coli BL-21 strain. LRP6 proteins were purified using Ni-NTA resin, HiTrap Q anion exchange, and Superdex-200 size-exclusion columns (GE Life Sciences). Human DKK1c proteins were purified using Ni-NTA resin. After the removal of the thioredoxin-tag, DKK1c was further purified with HiTrap S cation exchange, and a Superdex-75 size-exclusion column. Five-fold excess of DKK1c was mixed with corresponding LRP6 proteins and further purified by a Superdex-200 size-exclusion column. The proteins (LRP6-E1E2 or LRP6-E3E4) and protein complex (LRP6-E3E4/DKK1c) were concentrated to 5 m g ml−1, 10 mg ml−1 and 5 mg ml−1, respectively, for crystallization.
Crystallization and data collection
Crystals of human LRP6-E1E2 were grown at 20 °C by hanging drop vapor diffusion. Equal volume of protein sample was mixed with the crystallization solution (80mM sodium citrate pH 5.5, 20–21% PEG3350 (w/v), 40 mM KSCN). Single crystals were transferred into crystallization solution with 16% glycerol (v/v), and flash frozen in liquid nitrogen. Crystals of human LRP6-E3E4 were grown at 20 °C from hanging drops with a well solution containing 100 mM N-(2-Acetamido) Iminodiacetic Acid (ADA) pH 6.5, 100 mM MgCl2, 12% PEG6000 (w/v). Single crystals were transferred to cryo-protection solution (crystallization buffer with 20% glycerol (v/v)) and flash frozen in liquid nitrogen. Crystals of human LRP6-E3E4/DKK1c complex were grown at 20 °C from hanging drops with a well solution containing 20mM citric acid, 80 mM Bis-tris propane, pH 8.8, 19–20% PEG3350 (w/v). Single crystals were soaked in cryo-buffer (crystallization buffer with 25% glycerol (v/v)) overnight and frozen in liquid nitrogen. All datasets used in this work were collected at ALS beamlines BL8.2.1 and BL8.2.2 (LRP6-E1E2, λ=0.9774Å; LRP6-E3E4, λ=0.9790Å; LRP6-E3E4/DKK1c, λ=0.999Å) and processed using the software Mosflm/CCP4 (ref. 49) or HKL2000 (ref. 50).
Structure determination and refinement
All three structures were determined by molecular replacement with the program PHASER51, using the crystal structure of LDL receptor YWTD domain14 (PDB 1IJQ) as the initial search model. Solutions for individual YWTD domains were found sequentially. After several cycles of refinement using REFMAC5 (ref. 52), EGF-like domains (and DKK1c in the case of the LRP6-E3E4/DKK1c complex structure) were manually built using Coot53. Crystallographic refinement was carried out with REFMAC5 with TLS parameters. For the LRP6-E1E2 structure, the last residue well ordered in identical position in both LRP6-E1E2 copies is Val629, the last residue in the EGF2 domain. LRP6 residue Pro630 is visible in only one copy. In the LRP6-E3E4 structures, the first residue that is well ordered in the same position in all three LRP6-E3E4 copies is Ala632, which is immediately before the first β-strand of YWTD propeller 3. Ramachandran statistics for the refined coordinates (residues in favored region (%), number of outliers): LRP6-E1E2 (98.2, 0), LRP6-E3E4 (98, 2) and LRP6-E3E4/DKK1c (98.5, 3). The final refinement statistics are summarized in Table 1.
EM imaging
TEM grids coated with continuous carbon films were glow-discharged in air for 20 s. 6 μl of LRP-E1E4 was loaded onto the carbon side for 2 minutes, and blotted from the side. Then 10 μl of 2% uranyl acetate (w/v) was loaded onto the carbon side for 2 minutes, and blotted from the side. The sample was dried in air. Images were obtained by using a Tecnai F20 electron microscope (FEI, OR) at 200 keV with a 30 μm objective aperture. Images were taken at 97,000 magnification and about −1 μm defocus and recorded on a Gatan 2K×2K CCD camera with an effective pixel size of 0.11 nm.
Cell culture
HEK293T (CRL-11268) and RKO (CRL-2577) cells were obtained from ATCC and were cultured in DMEM supplemented with 10% FBS (Invitrogen, CA) and 1% Pen/Strep (Invitrogen, CA).
Conditioned media
WNT3A conditioned media was harvested from WNT3A expressing L-cells as previously described54. DKK1 conditioned medias were harvested from HEK293T cells transiently transfect with DKK1 expression constructs. Briefly, HEK293T cells were seeded in 6-well plates and transfected with 1.5 mg of the appropriate FLAG-tagged DKK1 expression construct using Lipofectamine 2000 as directed by the manufacturer (Invitrogen, CA). DKK1 conditioned medias were harvested 24 hours post transfection.
Western blot
Conditioned medias were combined with LDS sample buffer and a final concentration of 50mM DTT. Samples were run on NuPAGE 4–12% Bis-Tris gels and transferred to nitrocellulose. Blots were stained with Ponceau S and imaged for protein loading comparison (Supplementary Fig. 6). Blots were probed with anti-FLAG antibody (Sigma, MO), followed by HRP-conjugated secondary antibody (Jackson, PA) and developed with ECL detection kit (Pierce, IL).
BAR reporter assay
Detailed information on the β-catenin activated reporter (BAR) has been previously described55. RKO cells stably expressing the BAR reporter (RKO/BAR) were generated as previously described54. RKO/BAR luciferase cells were also infected with a lentivirus carrying Renilla luciferase driven by a constitutive EF1alpha promoter which was used for assay normalization. RKO/BAR cells were seeded in 96-well plates at 30,000 cells per well. 24 hours after seeding, the media was replaced with a single dose of WNT3A conditioned media combined with increasing amounts of control, DKK1, or mutant DKK1 conditioned medias and incubated for an additional 24 hours. Media was then aspirated and cells were lysed with 25μl of 1X passive lysis buffer (Promega, WI). 10μL of lysate was assayed for both Firely (BAR reporter) and Renilla (normalization) luciferase activity using the dual luciferase assay (Promega, WI) and an Envision multilabel plate reader (PerkinElmer, MA).
Alpha binding assay
The measurement was based on the “Determining Kd with an Alpha assay” protocol from PerkinElmer. Streptavidin Donor beads and Glutathione Acceptor beads (from PerkinElmer, MA) were used in this assay. Biotinylation of LRP6-E1E2 and E3E4 proteins were performed with ChromaLink Biotin Protein Labeling Kit (Solulink, CA). Gradients of untagged DKK1c (0 to 10 μM) were used to titrate the interaction between biotinylated LRP6 proteins and GST-DKK1c. The concentrations of biotinylated LRP6 proteins and GST-DKK1c used were 4 nM and 10 nM, respectively. The concentration for both donor and acceptor beads was 20μg mL−1. The assay was done in a solution containing 1 × PBS buffer (pH7.5) with 0.5 % BSA. Data were analyzed with a one-site competition model using nonlinear regression with the software GRAPHPAD PRISM (GraphPad, CA).
Supplementary Material
Acknowledgments
We are grateful to the staff at ALS beamlines BL 8.2.1 and 8.2.2 for assistance with synchrotron data collection. We thank Dr. Guojun Bu (Washington University) for human LRP6 cDNA. This work was supported by NIH grants CA90351 to W.X. R.T.M. is an investigator of the HHMI. T.B. was supported by a Training Grant from NIH/NIAMS (T32AR056969).
Footnotes
Accession codes
Protein Data Bank: Diffraction data and coordinates of LRP6-E1E2, LRP6-E3E4 and the LRP6-E3E4/DKK1c complex structures are deposited under accession codes 3S94, 3S8Z and 3S8V, respectively.
Competing Financial interests
R.T.M. is a cofounder of, and consultant with, FATE Therapeutics. The other authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Z.C., T.B., R.T.M., L.W. and W.X. participated in the experimental design and wrote the paper. Z.C. did protein purification, crystallization, crystal structural determination, Alpha binding assay, and SEC, DLS analysis. T.B. did BAR assay. Z.W. developed protein expression systems. L.W. did EM analysis. S.M. provided technical supports.
References
- 1.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 2.Moon RT, Bowerman B, Boutros M, Perrimon N. The promise and perils of Wnt signaling through beta-catenin. Science. 2002;296:1644–6. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–68. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niehrs C, Shen J. Regulation of Lrp6 phosphorylation. Cell Mol Life Sci. 2010;67:2551–62. doi: 10.1007/s00018-010-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehrli M, et al. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–30. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 8.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–8. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 9.Tamai K, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–5. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 10.Semenov MV, et al. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–61. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 11.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–77. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 12.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–6. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 13.Mao B, et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–5. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 14.Jeon H, et al. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- 15.Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;283:837–62. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 16.Rudenko G, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–8. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 17.Bourhis E, et al. Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind L, Koonin EV. A colipase fold in the carboxy-terminal domain of the Wnt antagonists--the Dickkopfs. Curr Biol. 1998;8:R477–8. doi: 10.1016/s0960-9822(98)70309-4. [DOI] [PubMed] [Google Scholar]
- 19.Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–10. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao B, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–7. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 21.Ellwanger K, et al. Targeted disruption of the Wnt regulator Kremen induces limb defects and high bone density. Mol Cell Biol. 2008;28:4875–82. doi: 10.1128/MCB.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien AJ, et al. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–8. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinzone JJ, et al. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood. 2009;113:517–25. doi: 10.1182/blood-2008-03-145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N, et al. A potential role for Dkk-1 in the pathogenesis of osteosarcoma predicts novel diagnostic and treatment strategies. Br J Cancer. 2007;97:1552–9. doi: 10.1038/sj.bjc.6604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–23. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 26.Boyden LM, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–21. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 27.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–9. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani A, et al. LRP6 mutationin a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–82. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams BO, Insogna KL. Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone. J Bone Miner Res. 2009;24:171–8. doi: 10.1359/jbmr.081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron R, Rawadi G, Roman-Roman S. Wnt signaling: a key regulator of bone mass. Curr Top Dev Biol. 2006;76:103–27. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- 31.Yaccoby S, et al. Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood. 2007;109:2106–11. doi: 10.1182/blood-2006-09-047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–55. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CC, Pearson C, Bu G. Cooperative folding and ligand-binding properties of LRP6 beta-propeller domains. J Biol Chem. 2009;284:15299–307. doi: 10.1074/jbc.M807285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, et al. Structural insight into the mechanisms of Wnt signaling antagonism by Dkk. J Biol Chem. 2008;283:23364–70. doi: 10.1074/jbc.M802375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, et al. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J Biol Chem. 2008;283:23371–5. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilic J, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–22. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 38.Johnson ML, Summerfield DT. Parameters of LRP5 from a structural and molecular perspective. Crit Rev Eukaryot Gene Expr. 2005;15:229–42. doi: 10.1615/critreveukargeneexpr.v15.i3.50. [DOI] [PubMed] [Google Scholar]
- 39.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–81. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 40.Jeon H, Blacklow SC. Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem. 2005;74:535–62. doi: 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 41.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280:26770–5. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 42.Li X, et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–7. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 43.Ellies DL, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone MinerRes. 2006;21:1738–49. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 44.Balemans W, et al. The binding between sclerostin and LRP5 is altered by DKK1 and by high-bone mass LRP5 mutations. Calcif Tissue Int. 2008;82:445–53. doi: 10.1007/s00223-008-9130-9. [DOI] [PubMed] [Google Scholar]
- 45.Weidauer SE, et al. NMR structure of the Wnt modulator protein Sclerostin. Biochem Biophys Res Commun. 2009;380:160–5. doi: 10.1016/j.bbrc.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 46.Murrills RJ, et al. A cell-based Dkk1 binding assay reveals roles for extracellular domains of LRP5 in Dkk1 interaction and highlights differences between wild-type and the high bone mass mutant LRP5(G171V) J Cell Biochem. 2009;108:1066–75. doi: 10.1002/jcb.22335. [DOI] [PubMed] [Google Scholar]
- 47.Ettenberg SA, et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci U S A. 2010;107:15473–8. doi: 10.1073/pnas.1007428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Y, et al. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. Macromolecular Crystallography, part A. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–52. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 55.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. doi: 10.1007/978-1-59745-249-6_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.