Abstract
Transcription factors and their binding sites have been proposed as primary targets of evolutionary adaptation because changes to single transcription factors can lead to far-reaching changes in gene expression patterns. Nevertheless, there is very little concrete evidence for such evolutionary changes. Industrial wine yeast strains, of the species Saccharomyces cerevisiae, are a geno- and phenotypically diverse group of organisms that have adapted to the ecological niches of industrial winemaking environments and have been selected to produce specific styles of wine. Variation in transcriptional regulation among wine yeast strains may be responsible for many of the observed differences and specific adaptations to different fermentative conditions in the context of commercial winemaking. We analyzed gene expression profiles of wine yeast strains to assess the impact of transcription factor expression on metabolic networks. The data provide new insights into the molecular basis of variations in gene expression in industrial strains and their consequent effects on metabolic networks important to wine fermentation. We show that the metabolic phenotype of a strain can be shifted in a relatively predictable manner by changing expression levels of individual transcription factors, opening opportunities to modify transcription networks to achieve desirable outcomes.
SACCHAROMYCES cerevisiae is the yeast species most widely used in the fermentation industry (oenology, bread making, and brewing). Most genetic studies of S. cerevisiae have been carried out using a handful of strains (Mortimer et al. 1957; Mortimer and Johnston 1986) that were selected for their ease of use under laboratory conditions.
By contrast, industrial yeast strains are geno- and phenotypically highly diverse (Frezier and Dubourdieu 1992; Schütz and Gafner1994; Rossouw et al. 2009), having adapted to the ecological niches provided by industrial or semi-industrial environments. In the wine industry a large number of such strains are commercially produced, most of which were originally isolated from spontaneous wine fermentations (Johnston et al. 2000). Although the original or natural ecological niche of S. cerevisiae is subject to conjecture, industrial environments have undoubtedly sculpted the recent evolution of the strains currently used in industry, offering an excellent opportunity for comparative studies to investigate evolutionary relationships and the molecular mechanisms underlying phenotypic differentiation.
Wine yeast strains were primarily selected for their ability to completely ferment (to ferment to dryness) very high levels (>200 g/liter) of sugars in a largely anaerobic environment. Beyond this fundamental trait, strains have been selected for specific and diverse purposes, for example to support the production of different styles of wine or to produce different aroma profiles. These strains therefore represent a wide range of phenotypic traits, which is a reflection of significant genetic diversity.
A number of studies have focused on evolutionary adaptations of wine yeast strains. It has been suggested that the diploid status of most wine yeast strains may confer an advantage in terms of rapid adaptation to variable external environments and provide a way to increase the dosage of genes important for fermentation (Bakalinsky and Snow 1990; Salmon 1997). Furthermore, subtelomeric chromosomal regions are subject to duplications and rearrangements via ectopic exchanges (Bidenne et al. 1992; Rachidi et al. 1999). Another reported mode of evolution of Saccharomyces is the formation of interspecific hybrids. The resulting genome plasticity promotes faster adaptation in response to environmental changes (Puig and Perez-Ortin 2000; Libkind et al. 2011) by providing the genetic diversity upon which natural selection operates.
Adaptations of these strains to the specific oenological environment and their selection for specific biotechnological purposes are also reflected in global transcriptomic, proteomic, and metabolomic profiles. Studies of wine yeast strains have correlated differences in fermentation phenotypes to gene expression, protein levels, and metabolic regulation (Rossouw et al. 2008, 2009, 2010). These studies focused on the aroma-relevant exometabolome as produced by different wine yeast strains, since this metabolome largely determines the aromatic perception of fruitiness and complexity of wines, and is therefore of particular interest to winemakers.
It has been proposed that some of the primary evolutionary targets of strain diversification are transcription factors and their binding sites (Dermitzakis and Clark 2002). Data show that although S. cerevisiae and S. mikatae have similar genome sequences, they are significantly different in their transcription-factor binding profiles (Borneman et al., 2007a,b). It has been hypothesized that the extensive binding site differences observed between the different species reflect rapid specialization of Saccharomyces for distinct ecological environments (Borneman et al. 2007a,b).
For this study, the production of volatile aroma compounds was correlated to previously established transcriptional profiles of five different wine yeast strains under simulated winemaking conditions. We were able to identify transcription factors (TFs) whose expression profiles may contribute to the different metabolism-related phenotypes observed in different strains. In particular, we assessed whether the metabolic phenotype of one strain could be engineered to more closely resemble that of another strain by adjusting the expression of key transcription factors. This would support the hypothesis that changes in expression of specific transcription factors are responsible for the evolutionary adaptation of different Saccharomyces strains. The identification of such key TFs promises targeted improvement of fermentation performance (Hou et al. 2009).
Methods
Strains, media, and culture conditions
The yeast strains used in this study are listed in Table1. All are diploid Saccharomyces cerevisiae strains used in industrial wine fermentations. Yeast cells (inoculated from single, characterized colonies) were cultivated at 30° in YPD synthetic media 1% yeast extract (Biolab, South Africa), 2% peptone (Fluka, Germany), 2% glucose (Sigma, Germany). Solid medium was supplemented with 2% agar (Biolab).
Table 1 . Strains used in this study.
| Strain | Source |
|---|---|
| VIN13 | Anchor Yeast, South Africa |
| BM45 | Lallemand, Inc., Montréal, Canada |
| DV10 | Lallemand, Inc., Montréal, Canada |
| SOK2-VIN13 | This study |
| RAP1-VIN13 | This study |
Fermentation media
Fermentation experiments were carried out with synthetic must MS300, which approximates a natural must as previously described (Bely et al. 1990). The medium contained 125 g/liter glucose and 125 g/liter fructose, and the pH was adjusted to 3.3 with NaOH.
Fermentation conditions
All fermentations were carried out under microaerobic conditions in 100-ml glass bottles (containing 80 ml of the medium) sealed with rubber stoppers with a CO2 outlet. All fermentations were carried out in triplicate, i.e., independent biological repeats. The fermentation temperature was approximately 22° and no continuous stirring was performed during the course of the fermentation. Fermentation bottles were inoculated with YPD cultures in the logarithmic growth phase (around OD600 = 1) to an OD600 of 0.1 (i.e., a final cell density of approximately 106 cfu/ml). The cells from the YPD precultures were briefly centrifuged and resuspended in MS300 to avoid carryover of YPD to the fermentation media. The fermentations followed a time course of 14 days and the bottles were weighed daily to assess the progress of fermentation. Samples of the fermentation media and cells were taken at days 2, 5, and 14 as representative of the exponential, early logarithmic, and late logarithmic growth phases, respectively.
Growth measurement
Cell proliferation (i.e., growth) was determined spectrophotometrically (PowerwaveX, Bio-Tek Instruments) by measuring the optical density (at 600 nm) of 200-µl samples of the suspensions over the 14-day experimental period.
Analytical methods
High-performance liquid chromatography (HPLC):
Culture supernatants were obtained from the cell-free upper layers of the fermentation media. For the purposes of glucose determination and carbon recovery, culture supernatants and starting media were analyzed by HPLC on an AMINEX HPX-87H ion exchange column using 5 mM H2SO4 as the mobile phase. Agilent RID and UV detectors were used in tandem for peak detection and quantification. Analysis was carried out using the HPChemstation software package.
Gas chromatograph–flame ionization detector (GC-FID):
Each 5-ml sample of synthetic must taken during fermentation was spiked with an internal standard of 4-methyl-2-pentanol to a final concentration of 10 mg/liter. To each of these samples 1 ml of solvent (diethyl ether) was added and the tubes sonicated for 5 min. The top layer in each tube was separated by centrifugation at 3000 rpm for 5 min and the extract analyzed. Three microliters of each sample was injected into the GC. All extractions were done in triplicate.
The analysis of volatile compounds was carried out on a Hewlett Packard 5890 Series II GC coupled to an HP 7673 auto-sampler and injector and an HP 3396A integrator. The column used was a Lab Alliance organic-coated, fused silica capillary with dimensions of 60 m × 0.32 mm internal diameter with a 0.5-μm coating thickness. The injector temperature was set to 200°, the split ratio to 20:1, and the flow rate to 15 ml/min, with hydrogen used as the carrier gas for a flame ionization detector held at 250°. The oven temperature was increased from 35° to 230° at a ramp of 3°/min.
Internal standards (Merck, Cape Town) were used to calibrate the machine for each of the compounds measured.
Microarray analysis:
Sampling of cells from fermentation and total RNA extraction was performed as described by Abbott et al. (2007). Samples were taken from independent fermentations in triplicate on days 2, 5, and 14. For a complete description of the hybridization conditions refer to Rossouw et al. (2008). Transcript data can be downloaded from the GEO repository under the following accession numbers: GSE11651 (for the original VIN13, BM45, EC1118, 285, and DV10 data sets analyzed in Rossouw et al. 2009) and GSE26929 (for the SOK2-overexpressing strain and VIN13 control data sets).
Transcriptomics data analysis:
The microarray data were background corrected and normalized with robust multichip average (Irizarry et al. 2003) and the resultant log2 transformed data were mean centered for each probe set. Determination of differential gene expression between experimental parameters was conducted using SAM (significance analysis of microarrays) version 2 (Tusher et al. 2001). The two-class, unpaired setting was used and genes with a Q-value <0.5 (P < 0.0005) and a fold change greater than 2 (positive or negative) were taken into consideration as differentially expressed genes.
The sequences for each of the individual probes of the Affymetrix Yeast 2.0 Genechip were mapped to the yeast genome by the use of blastn (Altschul et al. 1990). A Perl program was written to perform the following tasks: (1) 100% identity matches (over the full length of the probe) were extracted from the blastn results; (2) the probes were subsequently assembled into probe sets and the resultant probe set to gene relationships modeled as a graph; (3) ambiguous probe sets, i.e., those that were found to map to more than one gene (node degree >1), were removed from the input gene list for the subsequent random forest analyses.
Random forest analysis (Breiman 2001) was carried out on the normalized and mean centered expression data by the use of the randomForest R package (Liaw and Wiener 2002). A random forest classification model was created using the strains as classes, regardless of time point. Fifteen thousand trees were generated in the creation of the model with 73 randomly selected variables (probe sets) used at each split. The out-of-bag (OOB) estimate of error rate was 4.65%. The mean decrease of accuracy measure of variable importance was extracted from the random forest model and used to rank the contribution of all probe sets according to their ability to discriminate between different strains. The probe sets occurring within the 200 most important variables from the random forest model described above were selected for further in depth analysis and evaluation.
Gene expression profiles were clustered using the short time series expression miner (STEM; Ernst and Bar-Joseph 2006).
Multivariate data analysis:
The patterns within the different sets of data were investigated by principal-component analysis (PCA; Qlucore Omics Explorer v. 2.2). PCA is a bilinear modeling method, which gives a visually interpretable overview of the main information in large, multidimensional data sets. By plotting the principal components it is possible to view statistical relationships between different variables in complex data sets and detect and interpret sample groupings, similarities, or differences, as well as the relationships between the different variables (Mardia et al. 1979).
Univariate statistics and visualization:
The levels of aroma compounds from target strains and transcription factor overexpression strains were compared to their respective control strains and the statistical significance of the changes evaluated with a t-test at a 95% confidence interval. To better visualize the statistical relationships in the data set the following algorithm was implemented in Perl: a mathematical graph was created with a node for each control strain (VIN13 or BM45). Subsequently, those compounds that showed a statistically significant difference from the control in either the target or the overexpression strain were added as a node to the graph and an edge created to the control strain node. For each strain showing a significant change a node was added to the graph and an edge created between it and the previously mentioned compound node. Fold change was calculated for each compound in each strain as a simple ratio between the compound level in the strain and that of its control. If the ratio was less than one its negative reciprocal was taken. This fold change information as well as descriptive information for each node was then written into an annotation file. Cytoscape v. 2.8.1 (Smoot et al. 2011) was used to visualize the resulting graph and annotation. The nodes were shaded according to fold change on a red (positive) or blue (negative) color scale.
Overexpression constructs and transformation:
The two plasmids constructed for use in this study are pDM-PhR-RAP1 (genotype, 2μ LEU2 TEF1P PhR322 TEF1T PGKP RAP1 PGKT) and pDM-PhR-SOK2 (genotype, 2μ LEU2 TEF1P PhR322 TEF1T PGKP SOK2 PGKT). Primers used for amplification of transcription factor encoding genes are listed in Supporting Information, Table S1. Standard procedures for the isolation of DNA were used throughout this study (Ausubel et al. 1994). Standard DNA techniques were also carried out as described by Sambrook et al. (1989). All enzymes for cloning, restriction digest, and ligation reactions were obtained from Roche Diagnostics (Randburg, South Africa) and used according to supplier specifications. Sequencing of all plasmids was carried out on an ABI PRISM automated sequencer. All plasmids contain the dominant marker PhR conferring phleomicin resistance (PhR) and were transformed into host VIN13 and BM45 cells via electroporation (Wenzel et al. 1992; Lilly et al. 2006).
Quantitative real-time PCR analysis (QRT-PCR):
RNA extractions from fermenting yeasts were carried out as per the microarray analyses. Primer design for QRT-PCR analysis was performed using the Primer Express software v. 3 (Applied Biosystems) and reagents were purchased from KAPA Biosystems. Spectral data were captured by the 7500 cycler (Applied Biosystems). Data analyses were conducted using Signal Detection Software (SDS) v. 1.3.1. (Applied Biosystems) to determine the corresponding Ct values and PCR efficiencies, respectively, for the samples analyzed (Ramakers et al. 2003). The genes selected for QRT-PCR, as well as the primer sequences used for amplification are described in Table S2.
Transcriptomic analysis of overexpressing strains:
Fermentations of the SOK2-overexpressing strain as well as the VIN13 control were carried out in triplicate in synthetic must as described previously. Samples for transcriptomic analysis were taken from three independent biological repeats at day 2 of fermentation, during the exponential growth phase. The microarray data can be viewed at the GEO repository under the accession number (GSE26929).
Results
Transcription factor enrichment
In our previous work (Rossouw et al. 2008), the transcriptome of five distinct industrial wine yeast strains was analyzed at three time points in synthetic wine must fermentations, day 2 (exponential growth phase), day 5 (early stationary growth phase), and day 14 (late stationary-growth phase). Strains were also monitored for sugar utilization and production of ethanol, glycerol, and 32 volatile aroma compounds (Rossouw et al. 2008, 2009).
Normalized expression values for the different strains and time points were analyzed by random forest analysis (Breiman 2001), and the top 200 strain discriminatory genes were ranked according to their ability to differentiate between the different strains. These genes were subsequently subjected to transcription factor enrichment as described by Teixeira et al. (2006) to identify the main regulatory structures present in the data. Transcription factors that reportedly regulate most of the highly discriminatory genes from the random forest outputs were thus identified and ranked according to the percentage of genes identified by the random forest, which are regulated by these transcription factors. Enrichment of transcription factors was performed on the total set of 200 genes, as well as on a smaller subset of 42 genes from the random forest output, which are thought to be involved in metabolism based on GO functional annotations. From Table 2 it is clear that a few key transcription factors may account for the majority of genes responsible for the differential transcriptional response between strains.
Table 2 . Top 10 hits for transcription factor enrichment analysis of random forest outputs (% of total) for strain discriminatory genes in the total gene list and in the metabolism-specific subset.
| All genes | % | Metabolic genes | % |
|---|---|---|---|
| Ste12 | 39 | Ste12p | 45 |
| Sfp1p | 36 | Sok2p | 29 |
| Yap1p | 33 | Rap1p | 26 |
| Rap1p | 26 | Yap6p | 24 |
| Aft1p | 26 | Cin5p | 24 |
| Sok2p | 25 | Phd1p | 19 |
| Msn2p | 24 | Skn7p | 17 |
| Met4p | 24 | Tec1p | 17 |
| Msn4p | 18 | Nrg1p | 17 |
| Rpn4p | 18 | Ino4p | 17 |
Most of the identified transcription factors are involved in the synchronization of stress responses, the regulation of carbon utilization and the modulation of cell membrane and cell wall properties. Genes in these categories can be directly linked to the major changes that yeast experience during fermentation and presumably also reflect the evolutionary framework of domesticated strains.
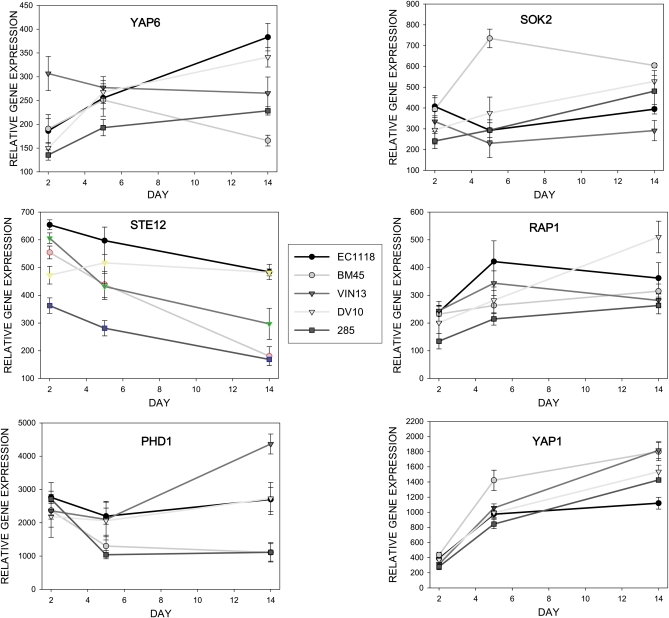
The transcriptome data were screened to identify the transcription factors in Table 2 that showed differences in either expression level and/or expression pattern between different strains over time. Some of the TF genes did show significant differences in expression levels between one or more strains at particular time points, but overall expression trends and patterns over time were similar. Importantly, six of the transcription factor-encoding genes and notably some of the top-scoring candidates of the TF enrichment, namely YAP1, YAP6, SOK2, PHD1, STE12, and RAP1, did show significant differences between strains in terms of relative transcript abundance and expression patterns over time (Figure 1).
Figure 1 .
Expression patterns of six genes encoding key transcription factors based on transcription factor enrichment of 200 top-scoring strain-discriminatory genes from random forest analysis. The expression values are derived from microarray experiments and are the average of three biological repeats ±SD.
Interestingly, strains with similar physiological properties regarding metabolite profiles and cell wall properties as described in Rossouw et al. (2009) (e.g.., EC1118 and DV10, as well as BM45 and 285) also presented similar profiles regarding the expression patterns of these six transcriptional regulators. These transcription factors play important roles in cellular metabolism and regulation, although their specific functions are not fully characterized, and information regarding regulatory networks and specific targets is limited. Yap1p is induced in response to oxidative stress conditions (Okazaki et al. 2007) and is believed to regulate the expression of several genes involved in protein mannosylation as well as the invasive growth response (Haugen et al. 2004; Thorsen et al. 2007). Yap6p is involved in a variety of stress-related programs, including the response to DNA damage and oxidative, osmotic, and toxic metal stresses (Tan et al. 2008). Three other key transcription factor encoding genes in the enrichment analysis, namely SOK2, PHD1, and STE12, show highly variable expression patterns between strains (Figure 1). Their protein products are all involved in pseudohyphal growth and regulation of key mannoproteins such as Flo11p (Gimeno and Fink 1994; Pan and Heitman 2000), as well as a host of other metabolic processes. Finally, Rap1p is a multipurpose DNA-binding protein that functions in transcriptional activation, silencing, and replication in yeast. Genes containing Rap1p binding sites include genes encoding proteins involved in amino acid biosynthesis and regulation of carbon metabolism (Yarragudi et al. 2007).
Overexpression of selected transcription factors
To determine whether the different expression patterns of these key regulators could be reconciled with the metabolic and phenotypic differences observed between the strains, we selected two of these genes, namely SOK2 and RAP1, for overexpression analysis. The SOK2 gene was cloned from the BM45 strain and overexpressed in VIN13, while the RAP1 gene was cloned from DV10 and overexpressed in BM45. Our goal was to elevate the expression levels of these transcription factors in the overexpression strains to more closely match the expression levels observed in the “donor” strains.
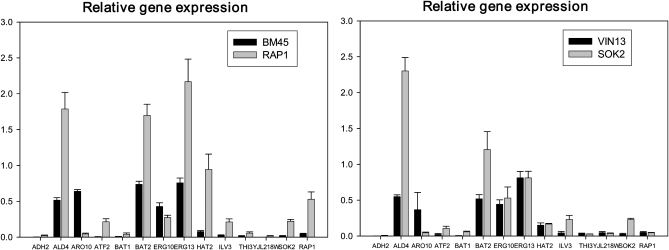
Figure 2 clearly shows that the expression levels of SOK2 and RAP1 in the transformed strains were successfully and significantly increased in comparison to their respective controls. To assess whether the overexpression of these factors had an impact on genes under their control, several known or suggested target genes of Sok1p and of Rap1p (Table S3) were selected for expression analysis using real-time PCR, while two genes, ERG10 and THI3, were included as negative controls.
Figure 2 .
Relative gene expression (normalized to PDA1 expression) of RAP1, SOK2, and selected target genes. Values are the average of three biological repeats ±SD.
Both negative controls (THI3 and ERG10) showed no change in expression for the transformants, while most of the known or suggested target genes of the two transcription factors, such as ERG13, BAT2, and ALD4, were increased in expression (Figure 2). Of these suggested targets, only ARO10 did not show any increase in both the RAP1 and SOK2 overexpression strains. Considering that the identification of target genes in databases is not always based on direct biological evidence (Li et al. 2008), these data provide strong evidence that the transformed strains show expression patterns that indeed reflect increased levels of the two transcription factors.
Fermentation properties of the overexpressing strains
The three original strains (DV10, VIN13, and BM45), as well as the two transformants were inoculated into synthetic wine must and the fermentations monitored over the 14-day fermentation period. All fermentations completed to dryness and the levels of ethanol and glycerol production were similar for the two transformed strains and their respective controls (data not shown).
The impact of changes in transcription factor expression levels on the wine aroma-relevant metabolite profile produced by the different strains was assessed. For this purpose, the concentrations of 22 exometabolites were measured at days 2, 5, and 14 of fermentation, in keeping with our original sampling scheme (Rossouw et al. 2008). The results are summarized in Table S4, Table S5, Table S6, and Figure 3.
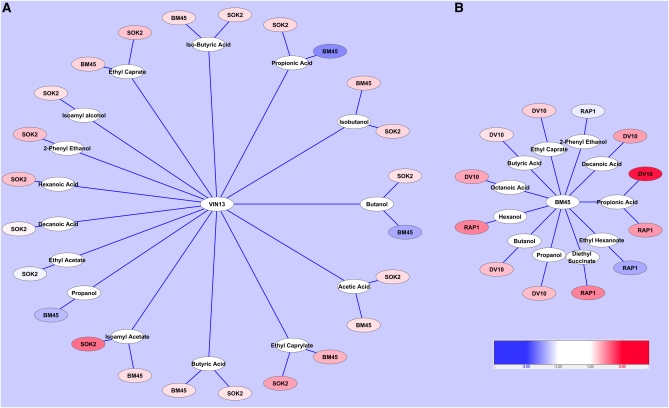
Figure 3 .
Statistically significant changes in aroma compounds among control, target, and transformed strains on day 14 of fermentation. (A) The levels of aroma compounds from both the target strain (BM45) and the SOK2-overexpression strain that were shown to be statistically significantly different from the control (VIN13). (B) The levels of aroma compounds from both the target strain (DV10) and the RAP1-overexpression strain that were shown to be statistically significantly different from the control (BM45). The degree of fold change is represented by red (positive) and blue (negative) color scales.
Clearly, significant differences in the production of volatile aroma compounds at all three stages of fermentation when transformed and untransformed parental strains are compared. The differences were most pronounced for the SOK2 transformant, but significant differences were also evident for the RAP1-overexpressing strain. By the end of fermentation, more than half of the aroma compounds measured were present at substantially different concentrations in the SOK2-overexpressing strain in comparison to the parental VIN13 strain, similar to the BM45 target strain. In the case of the RAP1 transformant, four compounds were significantly increased, and two compounds decreased with reference to the control BM45 strain (Table S6 and Figure 3). For certain volatiles (such as propanol and isoamyl alcohol) the increased concentrations observed in the transformed strains exceeds that of the target strains. Levels of overexpression of the transcription factors in our experiments are not controlled in a precise manner and therefore are not identical to the levels in the original target strains. The impact of overexpression on individual metabolite levels is thus likely to differ (being either more or less) from the exact concentrations determined for the original strains.
Importantly, for the SOK2-overexpressing strain, most of the specific metabolic changes as shown in Figure 3 can be directly accounted for by the observed differences in gene expression as determined by transcriptomic analysis. Although we did not assess the transcriptional response of the RAP1-overexpressing strain, changes in metabolite levels in this strain also correlate well with known targets of Rap1p and the enzymatic activities of these enzymes. For example, one of the target genes, ERG13 (Kasahara et al. 2007), is involved in the production of diethyl succinate, which is present at much higher concentrations at the end of fermentation in the transformed strain compared to the BM45 reference strain (Table S6 and Figure 3).
Transcriptomic analysis of a SOK2-overexpressing strain
Samples from the SOK2 overexpression fermentations were taken for transcriptomic analysis at day 2 of fermentation, during the exponential growth phase. Close to 1000 transcripts were found to be significantly differentially expressed with a fold change of >2 or <−2. Of these, 258 transcripts were upregulated and 677 downregulated. In terms of alignment with the real-time data, the trends for the 13 transcripts quantified in the real-time analysis were similar to the data derived from the transcriptome analysis, but for ILV3, ALD4, and BAT2, where the significant increases in expression evident in the real-time data (Figure 2) were not reflected in the microarray data. This difference may be explained by different SOK2 expression levels in the two experiments, i.e., a sixfold increase in the real-time experiments vs. a twofold increase in the microarray data. It is well established that such differences are commonly seen when 2µ-based multiple copy plasmids are used to amplify gene expression and that many targets of transcription factors are responsive to the precise concentration of the activator (Sauer and Jäckle 1991; Ni et al. 2009; Zheng et al. 2010).
Of the differentially expressed transcripts (>2- or <−2-fold), 20% were targets of Sok2p as previously described in the literature (Borneman et al. 2006; Borneman et al. 2007a,b; Horak et al. 2002; Lee et al. 2002). The remaining 80% of differentially expressed genes may be accounted for by secondary effects of the overexpression or indeed reflect unidentified downstream targets of Sok2p.
When comparing the SOK2-overexpressing strain with the VIN13 control, the upregulated genes showed enrichment for the GO categories of metabolism, specifically amino acid metabolism (Table S7). This aligns with the known metabolic regulation of Sok2p. In the case of the downregulated genes, GO processes such as autophagy and energy reserve metabolic processes were the most strongly represented (Table S8).
In the context of the aroma profile changes seen in the transformed strains, gene expression differences in fermentation pathways and pathways related to amino acid metabolism are the most important as amino acids are the precursors for the higher alcohols and esters produced during alcoholic fermentation. Table S9 shows the fold changes for genes in these pathways for fold changes >1.5 or <−1.5. A major increase in expression (fold change >4) is evident for ATF2 (a known target of Sok2p; Workman et al. 2006). The Atf2p enzyme is responsible for the production of a number of volatile esters from their corresponding alcohols, such as ethyl acetate, isoamyl acetate, and phenylethyl acetate (Vestrepen et al. 2003). Isoamyl acetate concentrations in the overexpression strain were significantly higher at all time points considered (Table S4, Table S5, and Table S6), corroborating the effect of elevated gene expression on the amount of a metabolite produced. Likewise, activation of ALD6 by Sok2p (Borneman et al. 2006; Chua et al. 2006) could explain the increase in acetic acid concentrations (Table S4, Table S5, and Table S6) in the SOK2-overexpressing strain as acetate is the direct product of the reaction catalyzed by the aldehyde dehydrogenase isomer encoded by ALD6 (Saint-Prix et al. 2004). Increased expression levels of three ILV genes (1, 2, and 5) involved in branched-chain amino acid metabolism (Holmberg and Petersen 1988) may account for the dramatic increase in the two end-products of this pathway, namely isobutanol and isobutyric acid. Increased expression of ADH5 and ADH4 in particular also account for the higher concentrations of several higher alcohols and esters (such as 2-phenylethanol) as these enzymes carry out key dehydrogenation reactions in the Ehrlich pathway (Dickinson et al. 2002).
Multivariate and univariate analysis
Our original question pertained to whether the metabolic phenotype of one strain could be shifted in the direction of another by adjusting the expression of a key transcription factor. This would suggest that changes in the regulation/expression of specific transcription factors could be responsible for major phenotypic divergence and adaptation of different Saccharomyces species or different strains within a species. To address this issue we followed both a multivariate approach (PCA) and created a statistical graph to visualize the overall structure of the volatile metabolite data set in a qualitative and quantitative manner.
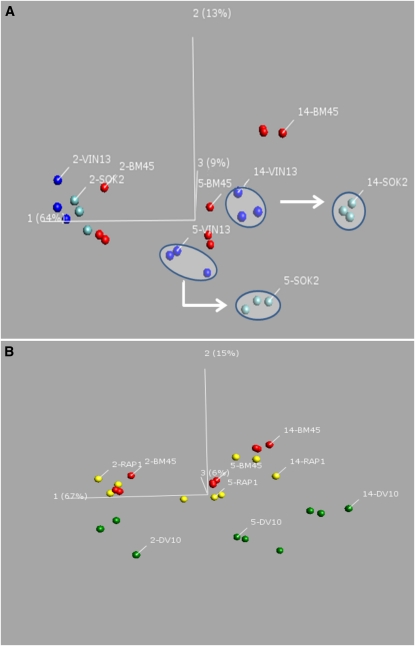
Figure 3 clearly shows that the vast majority of metabolites in the transcription factor overexpression strain have shifted in the same direction as the target strain, either matching closely or in some cases overshooting the target. Only a few compounds shift in an opposite direction to that of the target strain. The PCA analysis in Figure 4 shows the overall shift in the metabolic profiles at each time point in fermentations performed with each of the five strains (DV10, VIN13, BM45, SOK2-VIN13, and RAP1-VIN13). On day 2 of fermentation the differences between the sample groupings of the SOK2-overexpressing VIN13 and the reference VIN13 strain is still small. The same is true of the RAP1-overexpressing strain and its BM45 control strain. However, by day 5 of fermentation the two transformed industrial strains form clearly distinct clusters that are separated from their control samples along the first three principal components. The same is true for day 14, when the distances between distinct sample groupings are even greater for the first two principal components.
Figure 4 .
Principal component analysis of aroma compound concentrations in strains overexpressing individual transcription factors as compared to the corresponding untransformed parental as well as to the strain with naturally higher levels of expression of the same transcription factor. (A) A PC1 vs. PC2 vs. PC3 plot of the VIN13 SOK2-overexpression strain (light blue), the VIN13 control strain (dark blue), and the BM45 target strain (red). Component 1 accounts for 64%, component 2 for 13%, and component 3 for 9% of model variation. (B) The BM45 RAP1-overexpressing strain (yellow), control BM45 strain (red), and target DV10 strain (green) are shown. In this case component 1 accounts for 67% of model variation, component 2 for 15%, and component 3 for 6% of model variation. Samples are labeled according to timepoint (day 2, 5, or 14) and strain.
As can be seen in Figure 4B, the overall exometabolite composition of the RAP1-overexpressing strain has shifted from the BM45 control strain in the direction of the DV10 target cluster for days 5 and 14 of fermentation. Similarly, SOK2-overexpressing samples shift from the VIN13 control cluster (Figure 4A), in the direction of the target BM45 cluster, even shifting beyond the target cluster in days 5 and 14.
Discussion
The adjustment of key transcription factor expression levels in a wine yeast strain can indeed alter metabolism on a large scale. More specifically, we were able to moderate metabolism in a qualitatively reasonably defined manner by engineering the expression levels of transcription factors identified by the analysis of high-quality comparative gene expression data. This was achieved despite the complexity of the regulation of aroma compound metabolism, which is affected by many other parameters, such as the prevailing redox balance, the concentration of intermediates, and the flux through upstream and downstream pathways, which affects the rates and directionality of many promiscuous enzymes that catalyze the reactions of higher alcohol and ester synthesis.
The data clearly support the hypothesis that microevolution, which has provided us with the plethora of industrial Saccharomyces strains known today, could use transcription factor moderation and/or binding site alteration to effect a large-scale rewiring of metabolic and regulatory circuits in the cell. The possibility thus exists to modify or enhance industrial wine yeasts in a holistic manner by carefully selecting and modifying high-level master regulatory systems, instead of instituting numerous single gene changes at the effector level.
Supplementary Material
Acknowledgments
We thank Jo McBride and the Cape Town Centre for Proteomic and Genomic Research for the microarray analysis and the staff and students at the Institute for Wine Biotechnology for their support and assistance in numerous areas. Funding for the research presented in this article was provided by the National Research Foundation of South Africa and research funding organization of the South African wine industry, Winetech.
Literature Cited
- Abbott D. A., Knijnenburg T. A., De Poorter L. M., Reinders M. J., Pronk J. T., et al. , 2007. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEM. Yeast Res. 7: 819–833 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., 1994. Current Protocols In Molecular Biology. John Wiley, New York [Google Scholar]
- Bakalinsky A. T., Snow R., 1990. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6: 367–382 [DOI] [PubMed] [Google Scholar]
- Bely L., Sablayrolles J., Barre P., 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Vitic. 40: 319–324 [Google Scholar]
- Bidenne C., Blondin B., Dequin S., Vezinhet F., 1992. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr. Genet. 22: 1–7 [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., et al. , 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Zhang Z. D., Rozowsky J., Seringhaus M. R., Gerstein M., et al. , 2007a. Transcription factor bindind site identification in yeast: a comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct. Integr. Genomics 4: 335–345 [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., et al. , 2007b. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Breiman L., 2001. Random forests. Mach. Learn. 45: 5–22 [Google Scholar]
- Chua G., Morris Q. D., Sopko R., Robinson M. D., Ryan O., et al. , 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 103: 12045–12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E. T., Clark A. G., 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Eshantha L., Salgado J., Hewlins M. J. E., 2002. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278: 8028–8034 [DOI] [PubMed] [Google Scholar]
- Ernst J., Bar-Joseph Z., 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezier V., Dubourdieu D., 1992. Ecology of yeast strain Saccharomyces cerevisiae during spontaneous fermentation in a Bordeaux winery. Am. J. Enol. Vitic. 43: 375–380 [Google Scholar]
- Gimeno C. J., Fink G. R., 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14: 2100–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen A. C., Kelley R., Collins J. B., Tucker C. J., Deng C., et al. , 2004. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5: R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg S., Petersen J. G., 1988. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 13: 207–217 [DOI] [PubMed] [Google Scholar]
- Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., et al. , 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16: 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Cao X., Wang M., Lu M., 2009. Effect of overexpression of transcription factors on the fermentation properties of Saccharomyces cerevisiae industrial strains. Lett. Appl. Microbiol. 49: 14–19 [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., et al. , 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J. R., Baccari C., Mortimer R. K., 2000. Genotypic characterization of strains of commercial wine yeasts by tetrad analysis. Res. Microbiol. 151: 583–590 [DOI] [PubMed] [Google Scholar]
- Kasahara K., Ohtsuki K., Ki S., Aoyama K., Takahashi H., et al. , 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 6686–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., et al. , 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Li X., Macartur S., Bourgon R., Nix D., Pollard D. A., et al. , 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M., 2002. Classification and Regression by randomForest. R News 2: 18–22 [Google Scholar]
- Libkind D., Hittinger C. T., Valério E., Gonçalves C., Dover J., et al. , 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 108: 14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M., Bauer F. F., Styger G., Lambrechts M. G., Pretorius I. S., 2006. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEM. Yeast Res. 6: 726–743 [DOI] [PubMed] [Google Scholar]
- Mardia K. V., Kent J. T., Bibby J. H., 1979. Multivariate Analysis. Academic Press, London [Google Scholar]
- Mortimer R., Johnston J., 1986. Geneology of principal strains of the Yeast Genetics Stock Center. Genetics 113: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R., Lerner R., Barr J., 1957. Ultraviolet-induced biochemical mutants of Saccharomyces cerevisiae. U.S. Afon.Energy. Comm. Doc. UCRL 3746: 1–10 [Google Scholar]
- Ni L., Bruce C., Hart C., Leigh-Bell J., Gelperin D., et al. , 2009. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 23: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S., Tachibana T., Naganuma A., Mano N., Kuge S., 2007. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol. Cell 27: 675–688 [DOI] [PubMed] [Google Scholar]
- Pan X., Heitman J., 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell–cell adhesion. Mol. Cell. Biol. 20: 8364–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Perez-Ortin J. E., 2000. Stress response and expression patterns in wine fermentations of yeast genes induced at the diauxic shift. Yeast 16: 139–148 [DOI] [PubMed] [Google Scholar]
- Rachidi N., Barre P., Blondin B., 1999. Multiple Ty-mediated chromosomal translocation lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol. Gen. Genet. 261: 841–850 [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F., 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rossouw D., Naes T., Bauer F. F., 2008. Linking gene regulation and the exo-metabolome: A comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9: 530–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D., Olivares-Hernandes R., Nielsen J., Bauer F. F., 2009. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl. Environ. Microbiol. 75: 6600–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D., Van Den Dool A. H., Jacobson D., Bauer F. F., 2010. Comparative transcriptomic and proteomic profiling of industrial wine yeast strains. Appl. Environ. Microbiol. 10.1128/AEM.00586–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Prix F., Bönquist L., Dequin S., 2004. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+ -dependent Aldp6p and Aldp5p isoforms play a major role in acetate formation. Microbiology 150: 2209–2220 [DOI] [PubMed] [Google Scholar]
- Salmon J. M., 1997. Enological fermentation kinetics of an isogenic ploidy series derived form an industrial Saccharomyces cerevisiae strain. J. Ferment. Bioeng. 83: 253–260 [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual, Ed. 2 Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Sauer F., Jäckle H., 1991. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature 10: 563–566 [DOI] [PubMed] [Google Scholar]
- Schütz M., Gafner J., 1994. Dynamics of the yeast strain population during spontaneous alcoholic fermentation determined by CHEF gel electrophoresis. Lett. Appl. Microbiol. 19: 253–257 [Google Scholar]
- Smoot M., Ono K., Ruscheinski J., Wang P.-L., Ideker T., 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Feizi H., Luo C., Fan S. H., Ravasi T., et al. , 2008. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc. Natl. Acad. Sci. USA 105: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., et al. , 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34: D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M., Lagniel G., Kristiansson E., Junot C., Nerman O., et al. , 2007. Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol. Genomics 30: 35–43 [DOI] [PubMed] [Google Scholar]
- Tusher C. G., Tibshirani R., Chu G., 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestrepen K. J., Van Laere S. D., Vanderhaegen B. M., Derdelinckx G., Dufour J. P., et al. , 2003. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl. Environ. Microbiol. 69: 5228–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel T. J., Migliazza A., Steensma H. Y., Van Den Berg J. A., 1992. Efficient selection of phleomycin-resistant Saccharomyces cerevisiae transformants. Yeast 8: 667–668 [DOI] [PubMed] [Google Scholar]
- Workman C. T., Mak H. C., Mccuine S., Tagne J. B., Agarwal M., et al. , 2006. A systems approach to mapping DNA damage response pathways. Science 312: 1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A., Parfrey L. W., Morse R. H., 2007. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 35: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhao H., Mancera E., Steinmetz L. M., Snyder M., 2010. Genetic analysis of variation in transcription factor binding in yeast. Nature 464: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.