Abstract
Recent studies suggest that growth inhibition by 1,25-dihydroxyvitamin D3 represents an innovative approach to ovarian cancer therapy. To understand the molecular mechanism of 1,25-dihydroxyvitamin D3 action, we profiled the hormone-induced changes in the transcriptome of ovarian cancer cells using microarray technology. More than 200 genes were identified to be regulated by 1,25-dihydroxyvitamin D3. Reverse transcription-PCR analyses confirmed the regulation of a group of apoptosis-related genes, including the up-regulation of the decoy receptor that inhibits tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) action, TRAIL receptor 4, and the down-regulation of Fas, the receptor that mediates the action of Fas ligand. The regulation was further confirmed at the protein level. Consistent with the regulation of the death receptors, pretreatment with 1,25-dihydroxyvitamin D3 decreased apoptosis induced by TRAIL and Fas ligand. Because persistent 1,25-dihydroxyvitamin D3 treatment has been shown to induce apoptosis in ovarian cancer, the hormone appears to exert a dual effect on the death of ovarian cancer cells. Knockdown of TRAIL receptor 4 by RNA interference or ectopic expression of Fas relieved the suppressive effect of 1,25-dihydroxyvitamin D3, showing that molecular manipulation of death receptors is a viable approach to overcome the protective effect of 1,25-dihydroxyvitamin D3 on the apoptosis of ovarian cancer. These strategies may allow ovarian cancer patients to benefit from therapy with both 1,25-dihydroxyvitamin D3 and ligands for death receptors, such as TRAIL, shown to selectively induce apoptosis in cancer but not normal cells.
Vitamin D3 (VD)2 is a steroid hormone best known for its role in calcium homeostasis. Its deficiency causes rickets in children and osteomalacia in adults. VD also regulates the proliferation and differentiation of normal and malignant cells of many tissue types. Because sunlight controls the first step of VD synthesis, namely, the photoconversion of 7-dehydrocholesterol to vitamin D3 (1), the hormone is considered a “sun” medicine that is effective for the treatment and/or prevention of type II rickets, osteoporosis, autoimmune disorders, as well as epithelial cancers of many types.
Ovarian cancer (OCa) is a fatal disease with an overall 5-year survival rate of about 40%. OCa mortality and incidence rates are lower in countries within 20 degrees of the equator (2) where there is a high amount of sunlight. In the United States, women between the ages of 45 and 54 living in the North have 5 times the OCa mortality rate of women living in Southern states (3). The inverse correlation between sunlight exposure and OCa mortality indicates that decreased synthesis of VD may contribute to OCa initiation and/or progression. This idea has been further substantiated by recent studies (4–7) showing that 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3), the active form of VD, inhibits the growth of multiple OCa cell lines (4–6) and OVCAR3 tumor xenografts in nude mice (7).
Effects of VD are mediated through the vitamin D receptor (8), which is a member of the steroid/thyroid receptor superfamily of ligand-regulated transcription factors. In response to the activation by 1,25-(OH)2D3, the receptor recruits multiple co-activators, including members of the p160 SRC family (9), which are associated with histone acetyltransferase activity, and the DRIP complex (10), which serves as a mediator between the vitamin D receptor and RNA polymerase II complex. Recent studies (11) have suggested that the anti-tumor activity of VD is mediated through its effect on gene transcription. In OCa cells, we have shown that the transcriptional up-regulation of GADD45 is essential for the 1,25-(OH)2D3-induced cell cycle arrest at the G2/M checkpoint (4). 1,25-(OH)2D3 also increases p27 protein stability in OCa cells through down-regulation of Skp2 and cyclin E mRNA to induce cell cycle arrest at the G1/S checkpoint (6). These studies suggest that the action of 1,25-(OH)2D3 through the nuclear activation of vitamin D receptor is important for the anti-tumor activity of the hormone in OCa.
Because the anti-tumor activity of 1,25-(OH)2D3 is mediated through regulation of gene expression, a more complete understanding of the mechanism underlying 1,25-(OH)2D3 action in OCa cells necessitates identification of changes in gene expression induced by the hormone. In the present study, we profile transcriptional changes in OVCAR3 cells induced by 1,25-(OH)2D3 using multiple independent microarray analyses. We concentrated our subsequent analysis to a few apoptosis-related genes from this list. Our studies reveal that 1,25-(OH)2D3 regulates the expression of death receptors and protects cancer cells from apoptosis induced by death ligands. The findings suggest that a better understanding of both beneficial and adverse effects of 1,25-(OH)2D3 in OCa cells is necessary for the development of effective OCa therapeutic strategies that utilize active VD compounds, either alone or in combination with other apoptosis-inducing agents.
MATERIALS AND METHODS
Chemical Reagents, Antibodies, and Cell Cultures
1,25-(OH)2D3 was purchased from Calbiochem (La Jolla, CA). TRAIL, biological active Fas ligand, staurosporine, calcium ionophore A23187, and etoposide were from Sigma. The antibodies anti-TRAIL (BD Pharmingen), anti-TRAIL receptor 4 (TRAIL-R4), anti-Fas (Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-TRAIL-R2 (Oncogene Research Products, San Diego, CA), and anti-caspase-7 (Cell Signaling Technology Inc., Beverly, MA) were purchased commercially.
All cell lines used in the study have been described previously (6). OVCAR3 cells were cultured in RPMI 1640 medium supplemented with 15% fetal bovine serum, 2 mm l-glutamine, penicillin (50 units/ml), streptomycin (50 µg/ml), 10 mm HEPES, 1 mm sodium pyruvate, 4.5 g/liter glucose, 1.5 g/liter sodium bicarbonate, and 10 µg/ml bovine insulin. Other cell lines were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% fetal bovine serum. For 1,25-(OH)2D3 treatment, the hormone was dissolved in ethanol, diluted to the desired concentration in culturing medium, and added to the cells. For longer treatments, the medium was replaced with fresh medium containing 1,25-(OH)2D3 or vehicle every third day.
Microarray Analyses and Data Statistics
For microarray analyses, OVCAR3 cells at about 70% confluence were treated with 10−7 m 1,25-(OH)2D3 for 0, 8, 24, or 72 h. 1,25-(OH)2D3 was added at different times to allow the treated cells to be harvested at the same time. Ethanol was included as vehicle controls, and all cells were exposed to the same amount of ethanol for the same length of time. Total RNA was extracted using TRIzol reagent (Invitrogen) and further purified using Qiagen RNeasy columns. An initial experiment was performed using the U95Av2 chip from Affymetrix Inc. (Santa Clara, CA). From this experiment it was determined that GADD45 expression changed in response to VD (4). Three subsequent experiments were performed using the U133A chips (Affymetrix Inc.). In these experiments the response of the cells to 1,25-(OH)2D3 was determined by Northern blotting analysis of GADD45 before using the isolated RNA for microarray analysis.
The RNA samples were processed for hybridization to microarrays by the Microarray Core Facility of the H. Lee Moffitt Cancer Center. Briefly, RNA was converted to double-stranded cDNA using an oligo(dT)24 primer containing a T7 promoter sequence. The resulting double-stranded cDNA was transcribed into biotin-labeled cRNA using T7 RNA polymerase. This cRNA was hybridized to GeneChip probe arrays. Unbound RNA was washed from the chips and the remaining biotinylated RNA was stained and the chips scanned. Scanned chip images were analyzed using microarray suite software (MAS 5.0) from Affymetrix. In total, four independent experiments were carried out.
Raw data were processed using Irizarry and Speed’s Robust Multichip Analysis. After normalization, significance analysis of microarray (SAM) analysis (12) was performed by comparing gene expression profiles at time points of 8, 24, and 72 h to those of vehicle control (0 time point). Significant genes were clustered using GeneSpring software 6.0 (Silicon Genetics, Redwood City, CA). This resulted in groupings based on the general pattern of expression changes with time following 1,25-(OH)2D3 addition. The final list of genes was then generated by evaluating each probeset from the SAM analysis for consistency. A gene was considered to be consistently regulated if the same gene expression profile was obtained in all three experiments performed with the U133A chips and detected by all probesets. Gene identification was based on the sequence of the probes used on the arrays (13).
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
RT-PCR was conducted according to the protocol of Invitrogen with minor modifications. Briefly, 1 µg of pooled RNA from three independent experiments was reverse transcribed with the oligo(dT)20 primer in a total reaction volume of 20 µl. The amplification of target genes was carried out using 1 µl of reverse transcription product in a total reaction volume of 25 µl. PCR was performed using a GeneAmp PCR System 2700 (Applied Biosystem, Inc., Foster City, CA) for about 25 cycles with each cycle consisting of denaturing at 94 °C for 30 s followed by annealing for 45 s at various temperatures (supplemental Table S1) and elongation for 1 min at 72 °C. PCR products were visualized by electrophoresis on 1.5% agarose gels. Primers were synthesized by Invitrogen and the sequences are listed in supplemental Table S1.
Immunoblotting Analysis, Cell Growth, and Apoptotic Assays
For immunoblotting analyses, cellular extracts were separated on SDS-PAGE gels and blotted onto nitrocellulose membranes. Antibodies were used at 1:500 to 1:1000 dilutions. Proteins were detected using ECL as described (6).
To assay cell growth, methylthiazole tetrazolium (MTT) assays were performed as described (6). For each data point, eight samples were analyzed in parallel. Absorption at 595 nm (A595 nm) was measured on a MRX microplate reader (DYNEX Technologies, Chantilly, VA).
The apoptotic index of cells treated with death ligands and 1,25-(OH)2D3 was determined by flow cytometry following staining with Annexin V-fluorescein isothiocyanate and propidium iodide according to the manufacturer’s protocol (Annexin V kit, Santa Cruz Biotechnology Inc.). Flow cytometry was performed in a BD Biosciences FACS-Calibur flow cytometer. The data from 10,000 cells per time point were acquired and analyzed using CellQuest software.
The apoptotic index of transfected cells was determined as previously described (6). Transfected cells were fixed in 3.7% formaldehyde/phosphate-buffered saline and stained with 4′6′-diamidino-2-phenylindole. The apoptotic index of green fluorescence protein (GFP)-positive cells was determined by scoring 400 GFP-positive cells in randomly selected microscopic fields for chromatin condensation and apoptotic body formation.
Caspase Assays
The activity of caspase-3 was determined by the amount of fluorescent AMC liberated from the cleavage of the peptide substrate, Ac-DEVD-AMC, specific for caspase-3, using an assay kit from BD Biosciences. AMC was measured using a spectrofluorometer at excitation of 380 nm and emission of 420 nm. Caspase-7 activity was similarly determined by the amount of free fluorescent AFC released from substrate, Ac-DEVD-AFC, by caspase-7 immunoprecipitates following the protocol of the assay kit from Cell Signaling (Beverly, MA). AFC fluorescence was measured at excitation of 400 nm and emission of 505 nm.
Plasmid, siRNA, and Transfection
TRAIL-R4 specific siRNA was synthesized from Upstate Inc. (Chicago, IL). The sense and antisense sequences were 5′-GGACAUGCAAAGGAAACAAtt-3′ and 5′-UUGUUUCCUUUGCAUGUCCtt-3′, respectively. OVCAR3 cells were transfected with either pCMV-Fas and pEGFP or TRAIL-R4 siRNA using Lipofectamine or Oligofectamine according to the manufacturer’s protocol (Invitrogen). pCMV plasmid and nonspecific siRNA were used as controls. Cells were pretreated with 10−7 m 1,25-(OH)2D3 or vehicle for 3 days before the transfection, then 6 h post-transfection, cells were replaced in fresh medium containing 1,25-(OH)2D3 or vehicle. Forty-eight h later, cells were processed for immunoblotting analyses or treated with TRAIL or Fas ligand for apoptosis assays.
RESULTS
Identification of 1,25-(OH)2D3-regulated Genes by Microarray in OVCAR3 Cells
To profile the changes in the pattern of gene expression in human OCa cells induced by 1,25-(OH)2D3, OVCAR3 cells were treated with 10−7 m 1,25-(OH)2D3 for 0, 8, 24, or 72 h in four independent experiments. Total RNA was isolated, reverse transcribed, and subjected to microarray analyses. The first analysis was performed with the U95Av2 Affymetrix chips, which identified GADD45 among many others as being up-regulated. Subsequent analysis with conventional approaches confirmed this and identified GADD45 as a primary target gene for 1,25-(OH)2D3 (4). To ensure the reproducibility of the microarray data, three independent analyses were subsequently performed with the U133A Affymetrix chips, which contain 22,283 probe sets and can be used to analyze the expression level of ~15,000 known human genes. For quality control, RNA samples were first tested by Northern blots for up-regulation of GADD45 to ensure that the cells from which the RNA was isolated had responded to the treatment (data not shown). Samples from cells treated with the hormone for 3 days were included to ensure that genes that are regulated by persistent 1,25-(OH)2D3 treatment might be identified. These genes could mediate the anti-tumor activity of the hormone even though they may not be primary target genes.
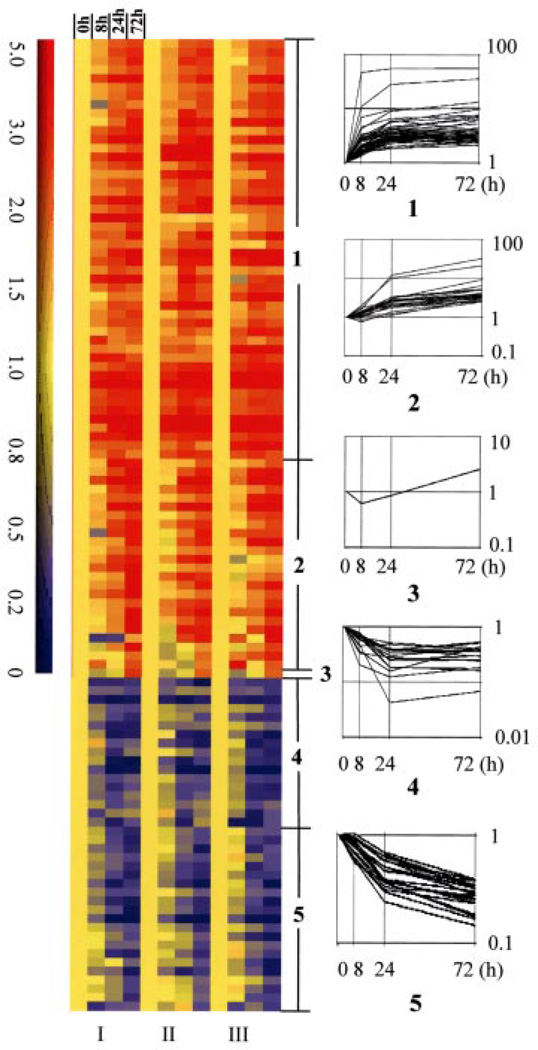
The raw data from the three microarray studies with U133A were first analyzed with SAM (12), a statistical method based on the conventional t test but developed specifically for microarray analysis. Using the 5% false discovery rate, we found that 710 probesets representing about 500 genes were modulated by 1,25-(OH)2D3. We reduced this list by requiring that a gene must change by at least 2.5-fold at one point during the time course of the experiment. The selected genes were clustered into groups using GeneSpring software, which generated five clusters (Fig. 1), of which, clusters 1 to 3 were up-regulated and clusters 4 and 5 were down-regulated. Genes in clusters 1 and 4 were induced or repressed at 8- or 24-h treatment and prolonged treatment did not change the fold regulation dramatically. Examples are GADD45 and 1,25-(OH)2D3 24-hydroxylase genes, which both contain vitamin D response elements in their genome (4, 14). Genes in clusters 2 and 5 were steadily up- or down-regulated and their fold regulation kept increasing as the time of treatment was extended. The cyclin A1 gene fell into cluster 3, which was repressed significantly at 8 h treatment but became induced at 72 h (2.58-fold) of 1,25-(OH)2D3 treatment. The final gene list contains only those genes whose pattern of expression was consistent in all three experiments and across all probesets that detect the same gene. This reduced the list to 58 genes that appeared to be up-regulated (TABLE ONE) and 38 genes that appeared to be down-regulated (TABLE TWO). Many of these genes were also identified in the previous experiment using U95Av2 chips (TABLES ONE and TWO).
FIGURE 1. Gene expression profiles in OVCAR3 cells after 1,25-(OH)2D3 treatment.
RNA isolated from OVCAR3 cells treated with 1,25-(OH)2D3 for 0, 8, 24, or 72 h was subjected to microarray analyses. Three independent analyses (I–III) were performed in Affymetrix U133A chips. A total of 96 genes with less than 5% false detective rate in SAM analysis (delta = 0.653) and at least 2.5-fold changes were selected for clustering analysis (GeneSpring software, Silicon Genetics). Genes are presented in rows. The time of 1,25-(OH)2D3 treatment and the number of array analyses are indicated above and below the column. Up- and down-regulated genes are shown with a colorimetric scale to indicate the relative expression levels. Numbers 1–5 show five different clusters identified by gene tree clustering.
TABLE ONE. Genes up-regulated by 1,25-(OH)2D3 (total: 58).
Genes are grouped by gene molecular functions. Mean -fold changes and S.D. are presented.
| GenBank | Gene name | -Fold change (mean ± S.D.) | qa | U95 | ||
|---|---|---|---|---|---|---|
| 8 h | 24 h | 72 h | ||||
| % | ||||||
| Cell cycle and apoptosis | ||||||
| NM_001924 | GADD45b | 2.77 ± 0.09 | 3.71 ± 0.13 | 4.02 ± 0.15 | 2.6 | √ |
| NM_003914 | Cyclin A1b | −1.63 ± 0.11 | −1.17 ± 0.21 | 2.58 ± 0.18 | 1.1 | √ |
| NM_003810 | TRAIL | 2.76 ± 0.02 | 2.69 ± 0.10 | 2.31 ± 0.17 | 1.5 | √ |
| NM_003840 | TRAIL-R4 | 1.85 ± 0.13 | 2.83 ± 0.44 | 3.00 ± 0.40 | 4.6 | √ |
| Oncogene and tumor suppressors | ||||||
| NM_004675 | Ras homolog gene family member I (ARHI) | 1.78 ± 0.44 | 2.43 ± 0.11 | 6.51 ± 0.25 | 1.2 | √ |
| NM_022337 | RAB38, Ras oncogene family | −1.01 ± 0.20 | 2.01 ± 0.28 | 2.70 ± 0.19 | 1.6 | N/Ac |
| NM_005252 | Fos | 6.79 ± 0.64 | 9.42 ± 0.23 | 10.61 ± 0.08 | 4.1 | √ |
| NM_004442 | EphB2 | −1.05 ± 0.30 | 2.98 ± 0.50 | 9.38 ± 0.47 | 1.6 | √ |
| NM_006408 | Anterior gradient 2 homolog | 2.77 ± 0.12 | 4.11 ± 0.27 | 7.00 ± 0.21 | 0.6 | N/A |
| Enzymes | ||||||
| NM_000782 | 24-Hydroxylaseb | 46.8 ± 0.05 | 54.94 ± 0.24 | 55.91 ± 0.19 | 0.6 | √ |
| NM_002774 | Kallikrein 6 | 2.60 ± 0.09 | 5.56 ± 0.14 | 7.21 ± 0.18 | 0.6 | √ |
| NM_005308 | G protein-coupled receptor kinase 5 (GPRK5)b | 4.17 ± 0.25 | 8.80 ± 0.27 | 13.34 ± 0.10 | 1.1 | √ |
| NM_007207 | Dual specificity phosphatase 10 (DUSP10)b | 2.92 ± 0.11 | 4.23 ± 0.08 | 3.67 ± 0.08 | 0.6 | √ |
| NM_015170 | Sulfatase 1 | 1.29 ± 0.14 | 2.64 ± 0.15 | 3.65 ± 0.16 | 0.9 | √ |
| NM_000402 | Glucose-6-phosphate dehydrogenase (G6PD)b | 1.41 ± 0.12 | 3.23 ± 0.11 | 5.01 ± 0.20 | 0.6 | √ |
| NM_003784 | Serine proteinase inhibitor clade B, 7 | 1.99 ± 0.55 | 10.11 ± 0.20 | 21.30 ± 0.08 | 0.6 | N/A |
| NM_016931 | NADPH oxidase 4 (NOX4) | 1.27 ± 0.35 | 2.63 ± 0.25 | 3.21 ± 0.14 | 4.6 | N/A |
| NM_002168 | Mitochondrial isocitrate dehydrogenase 2 (NADP+) | 1.05 ± 0.12 | 1.92 ± 0.13 | 2.50 ± 0.19 | 1.1 | √ |
| NM_002849 | Protein-tyrosine phosphatase, receptor type, Rb | 1.50 ± 0.02 | 1.95 ± 0.27 | 2.66 ± 0.32 | 0.6 | N/A |
| NM_012232 | RNA polymerase I and transcript release factor | 1.20 ± 0.16 | 2.22 ± 0.24 | 2.98 ± 0.08 | 1.1 | N/A |
| NM_001056 | Sulfotransferase family, cytosolic, 1C, member 1 | 1.32 ± 0.42 | 2.48 ± 0.28 | 2.75 ± 0.09 | 4.6 | N/A |
| NM_000904 | NAD(P)H dehydrogenase, quinone 2 | 1.49 ± 0.16 | 2.24 ± 0.09 | 2.95 ± 0.17 | 4.6 | N/A |
| NM_006741 | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | 1.53 ± 0.24 | 3.03 ± 0.27 | 2.80 ± 0.11 | 4.6 | N/A |
| NM_001977 | Glutamyl aminopeptidase (aminopeptidase A) | 1.88 ± 0.27 | 3.41 ± 0.03 | 2.73 ± 0.18 | 1.5 | N/Cd |
| NM_006105 | Rap guanine nucleotide exchange factor (GEF) 3 | 2.58 ± 0.17 | 3.52 ± 0.05 | 2.69 ± 0.25 | 2.7 | N/A |
| NM_001993 | Coagulation factor III | 1.61 ± 0.18 | 1.81 ± 0.18 | 2.51 ± 0.21 | 4.6 | N/A |
| Signal transducers, growth factors, and receptors | ||||||
| NM_004626 | Wingless-type MMTV integration site, 11 (WNT11) | 1.60 ± 0.15 | 12.15 ± 0.24 | 32.85 ± 0.18 | 0.6 | √ |
| NM_000728 | Calcitonin-related polypeptide β | 3.13 ± 0.17 | 4.60 ± 0.27 | 6.40 ± 0.17 | 2.6 | √ |
| NM_001401 | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor 2 (Edg2) | 2.00 ± 0.31 | 3.99 ± 0.43 | 5.28 ± 0.15 | 0.6 | √ |
| M37435 | Human macrophage-specific colony-stimulating factor | 1.43 ± 0.09 | 2.41 ± 0.06 | 2.50 ± 0.15 | 0.6 | √ |
| Transcription factors | ||||||
| NM_021784 | Forkhead box A2 | 1.66 ± 0.23 | 3.52 ± 0.25 | 3.24 ± 0.17 | 1.5 | N/A |
| NM_005257 | GATA-6 | 3.37 ± 0.09 | 3.87 ± 0.17 | 4.31 ± 0.13 | 4.6 | √ |
| Immunity proteins, structural proteins, cell adhesion molecules, and cytoskeleton | ||||||
| NM_004345 | Cathelicidin antimicrobial peptide | 10.72 ± 0.32 | 28.19 ± 0.10 | 35.89 ± 0.10 | 0.6 | N/A |
| NM_001845 | Collagen, type IV, α1 | 1.85 ± 0.13 | 3.16 ± 0.12 | 3.08 ± 0.09 | 1.5 | √ |
| NM_001846 | Collagen, type IV, α2 | 1.46 ± 0.09 | 2.25 ± 0.03 | 2.78 ± 0.11 | 1.2 | √ |
| NM_001003407 | Actin binding LIM protein 1 (ABLIM) | 1.38 ± 0.01 | 3.17 ± 0.25 | 3.23 ± 0.21 | 0.9 | N/A |
| AL832563 | Neural cell adhesion molecule 1 | 1.70 ± 0.21 | 2.64 ± 0.21 | 3.23 ± 0.17 | 4.0 | N/A |
| NM_000582 | Nephropontinb | 2.12 ± 0.34 | 2.74 ± 0.02 | 6.11 ± 0.06 | 3.3 | N/A |
| NM_005554 | Keratin 6A | 1.02 ± 0.37 | 1.96 ± 0.32 | 4.27 ± 0.14 | 0.6 | √ |
| NM_001407 | Cadherin, EGF LAG seven-pass G-type receptor 3 | −1.18 ± 0.64 | 1.11 ± 0.88 | 3.05 ± 0.14 | 3.3 | N/A |
| NM_005261 | GTP-binding protein overexpressed in skeletal muscle (GEM)b | 1.36 ± 0.08 | 1.97 ± 0.20 | 3.16 ± 0.21 | 2.6 | √ |
| NM_000138 | Fibrillin 1 (FBN1) | 1.16 ± 0.45 | 3.33 ± 0.52 | 5.38 ± 0.27 | 4.6 | √ |
| Others | ||||||
| XM_378901 | Filaggrin gene | 1.31 ± 0.04 | 2.42 ± 0.23 | 5.08 ± 0.12 | 0.6 | √ |
| NM_002888 | Retinoic acid receptor responder 1 (RARRES1)b | 3.04 ± 0.23 | 5.71 ± 0.21 | 9.73 ± 0.13 | 0.6 | √ |
| AK092245 | Clone 141H5 contains parts of a novel chordin-like protein | 1.46 ± 0.05 | 2.95 ± 0.06 | 3.81 ± 0.00 | 0.6 | √ |
| NM_017724 | Leucine-rich repeat interacting protein 2 | 1.53 ± 0.09 | 2.65 ± 0.26 | 3.28 ± 0.13 | 1.5 | N/A |
| NM_018431 | Docking protein 5 | 2.08 ± 0.07 | 3.19 ± 0.03 | 3.26 ± 0.14 | 0.6 | √ |
| AF191019 | Estradiol-induced (E2IG4) | 2.63 ± 0.17 | 3.68 ± 0.02 | 3.44 ± 0.16 | 0.6 | N/A |
| NM_017734 | Palmdelphin | 4.52 ± 0.12 | 6.51 ± 0.06 | 7.55 ± 0.28 | 0.6 | N/A |
| NM_030915 | Likely ortholog of mouse limb-bud and heart gene | 1.96 ± 0.13 | 2.77 ± 0.07 | 3.08 ± 0.13 | 0.6 | N/A |
| D84109 | RBP-MS/type 3 | 1.55 ± 0.09 | 1.79 ± 0.25 | 2.63 ± 0.10 | 0.3 | √ |
| NM_020182 | Transmembrane prostate androgen-induced RNA (TMEPAI) | 1.75 ± 0.57 | 6.08 ± 0.60 | 3.98 ± 0.10 | 4.6 | N/A |
| AL049370 | cDNA DKFZp586D0918 | 1.07 ± 0.28 | 1.21 ± 0.21 | 2.51 ± 0.12 | 0.6 | N/C |
| NM_134264 | WD repeat and SOCS box-containing 1 | 2.56 ± 0.25 | 1.82 ± 0.10 | 2.10 ± 0.37 | 0.6 | N/A |
| NM_000039 | Apolipoprotein A-I | 1.30 ± 0.86 | 2.75 ± 0.26 | 3.96 ± 0.18 | 0.9 | N/A |
| NM_138444 | Potassium channel tetramerization domain containing 12 | 2.53 ± 0.16 | 3.50 ± 0.10 | 3.78 ± 0.14 | 0.6 | N/A |
| NM_003982 | Solute carrier family 7 (cationic amino acid transporter, y + system), member 7 | 1.21 ± 0.13 | 2.14 ± 0.22 | 3.57 ± 0.42 | 4.0 | N/A |
| NM_002343 | Lactotransferrin | 1.17 ± 0.09 | 2.83 ± 0.87 | 3.37 ± 0.44 | 2.2 | N/A |
q value refers to the possibility of a false positive for a specific gene by SAM analysis.
Genes previously reported to be 1,25-(OH)2D3-regulated in microarray analyses of other cell types.
N/A, not applicable.
N/C, -fold change less than 2.50.
TABLE TWO. Genes down-regulated by 1,25-(OH)2D3 (total: 38).
Genes are grouped by gene molecular functions. Mean -fold changes and S.D. are presented.
| GenBank | Gene name | -Fold change (mean ± S.D.) | qa | U95 | ||
|---|---|---|---|---|---|---|
| 8 h | 24 h | 72 h | ||||
| % | ||||||
| Cell cycle and apoptosis | ||||||
| NM_014059 | Response gene to complement 32 (RGC 32) | −4.90 ± 0.13 | −8.14 ± 0.38 | −5.47 ± 0.08 | 0.6 | N/Ab |
| NM_000043 | Fas | −1.54 ± 0.14 | −1.96 ± 0.27 | −2.50 ± 0.21 | 0.6 | √ |
| Oncogene and tumor suppressors | ||||||
| NM_005564 | Lipocalin 2 (LCN2) | −1.04 ± 0.13 | −1.59 ± 0.23 | −3.05 ± 0.21 | 0.6 | N/A |
| NM_002089 | GRO-β | −1.68 ± 0.20 | −3.92 ± 0.10 | −3.35 ± 0.25 | 0.6 | √ |
| NM_002090 | GRO-γ | −1.69 ± 0.19 | −3.32 ± 0.16 | −2.54 ± 0.36 | 2.2 | √ |
| NM_007177 | TU3A | −1.64 ± 0.10 | −3.35 ± 0.17 | −5.97 ± 0.25 | 0.6 | N/A |
| Enzymes | ||||||
| NM_001001567 | Phosphodiesterase 9A | −1.69 ± 0.21 | −4.14 ± 0.33 | −6.82 ± 0.94 | 0.6 | √ |
| NM_001116 | Adenylate cyclase 9 | −1.47 ± 0.12 | −3.01 ± 0.42 | −3.60 ± 0.07 | 0.6 | √ |
| NM_000295 | Serine proteinase inhibitor, clade A, member 1 | −1.46 ± 0.26 | −1.68 ± 0.23 | −3.62 ± 0.33 | 3.3 | N/A |
| Signal transducers, growth factors, and receptors | ||||||
| NM_004591 | Small inducible cytokine subfamily A (Cys-Cys), 20 (SCYA20) | −2.09 ± 0.39 | −23.64 ± 0.31 | −14.59 ± 0.77 | 0.6 | N/A |
| NM_002993 | Small inducible cytokine subfamily B (Cys-X-Cys), 6 (SCYB6) | −1.53 ± 0.24 | −2.88 ± 0.12 | −2.82 ± 0.15 | 1.2 | N/A |
| NM_000627 | Latent transforming growth factor β-binding protein 1 | −1.41 ± 0.22 | −5.36 ± 0.20 | −6.27 ± 0.38 | 0.6 | N/A |
| NM_001999 | Fibrillin 2 | 1.03 ± 0.09 | −1.44 ± 0.09 | −2.53 ± 0.07 | 0.6 | √ |
| NM_001955 | Endothelin 1 | −2.79 ± 0.41 | −6.39 ± 0.21 | −.28 ± 0.20 | 4.0 | N/A |
| Transcription factors | ||||||
| NM_003317 | Thyroid transcription factor 1 | −1.28 ± 0.12 | −2.10 ± 0.15 | −2.64 ± 0.14 | 2.6 | √ |
| NM_004235 | Kruppel-like factor 4 (gut) | −1.29 ± 0.11 | −1.87 ± 0.15 | −2.08 ± 0.15 | 4.0 | N/A |
| Immunity proteins | ||||||
| NM_002371 | Mal, T-cell differentiation protein | −1.25 ± 0.05 | −2.68 ± 0.20 | −3.80 ± 0.22 | 0.6 | N/A |
| NM_000600 | Interleukin 6 | −1.67 ± 0.06 | −2.70 ± 0.34 | −2.46 ± 0.17 | 0.9 | N/A |
| NM_000584 | Interleukin 8 | −1.36 ± 0.04 | −2.50 ± 0.19 | −1.98 ± 0.17 | 0.6 | √ |
| NM_003641 | Interferon-induced transmembrane protein 1 | −1.03 ± 0.02 | −1.71 ± 0.37 | −2.93 ± 0.29 | 0.6 | N/A |
| NM_024626 | Immune costimulatory protein B7-H4 | −1.10 ± 0.14 | −2.59 ± 0.42 | −4.20 ± 0.33 | 2.2 | N/A |
| NM_152854 | CD40 | −2.51 ± 0.41 | −1.50 ± 0.51 | −2.90 ± 0.15 | 1.2 | N/A |
| Structural proteins, cell adhesion molecules, and cytoskeleton | ||||||
| NM_015515 | Keratin 23 | −1.60 ± 0.15 | −3.19 ± 0.06 | −4.24 ± 0.32 | 0.6 | N/A |
| Others | ||||||
| NM_018286 | Hypothetical protein FLJ10970 | −1.69 ± 0.22 | −2.49 ± 0.45 | −3.46 ± 0.21 | 1.1 | N/A |
| NM_024626 | Immune costimulatory protein B7-H4 | −1.10 ± 0.14 | −2.59 ± 0.42 | −4.20 ± 0.33 | 2.2 | N/A |
| NM_002964 | S100 calcium-binding protein A8 | −1.42 ± 0.22 | −2.73 ± 0.29 | −5.73 ± 0.28 | 0.6 | N/A |
| NM_183373 | Hypothetical protein FLJ21175 | −1.26 ± 0.13 | −1.97 ± 0.12 | −3.08 ± 0.29 | 0.6 | N/A |
| NM_182487 | Olfactomedin-like 2A | −1.77 ± 0.21 | −2.52 ± 0.39 | −2.78 ± 0.30 | 0.6 | √ |
| NM_030792 | Hypothetical protein PP1665 | −1.14 ± 0.20 | −2.14 ± 0.05 | −2.86 ± 0.14 | 0.6 | N/A |
| NM_006403 | Enhancer of filamentation 1 | −3.05 ± 0.12 | −3.57 ± 0.16 | −4.14 ± 0.55 | 0.6 | √ |
| NM_002965 | S100 calcium-binding protein A9 | −1.23 ± 0.35 | −1.95 ± 0.50 | −3.42 ± 0.51 | 0.9 | N/A |
| NM_018192 | Leprecan-like 1 | −1.59 ± 0.45 | −5.45 ± 0.39 | −5.69 ± 0.62 | 1.5 | N/A |
| CR599608 | Full-length cDNA clone CS0DJ006YH17 of T cells | −2.26 ± 0.19 | −2.08 ± 0.07 | −2.65 ± 0.15 | 3.3 | N/A |
| NM_014746 | Ring finger protein 144 | −1.38 ± 0.06 | −2.68 ± 0.35 | −2.80 ± 0.50 | 4.6 | N/A |
| AK075399 | cDNA PSEC0089 fis | −1.22 ± 0.26 | −2.67 ± 0.38 | −3.92 ± 0.35 | 4.0 | N/A |
| NM_014178 | Syntaxin binding protein 6 | −1.29 ± 0.28 | −4.12 ± 0.24 | −3.96 ± 0.19 | 4.6 | N/A |
| NM_020349 | Ankyrin repeat domain 2 | −1.78 ± 0.22 | −2.73 ± 0.57 | −5.47 ± 0.36 | 2.2 | N/A |
| BC011980 | Clone IMAGE:3860421 | −1.51 ± 0.58 | −2.65 ± 0.87 | −1.89 ± 0.48 | 3.3 | N/A |
q value refers to the possibility of a false positive for a specific gene by SAM analysis.
N/A, not applicable.
As shown in TABLES ONE and TWO, 1,25-(OH)2D3-regulated genes represent molecules involved in almost every aspect of cellular functions, suggesting that 1,25-(OH)2D3 may have widespread effects on OCa biology beyond the growth inhibition demonstrated in our previous studies (4, 6). Except for 1,25-(OH)2D3 24-hydroxylase that was increased by 55.91-fold, most of the genes were regulated modestly, possibly illustrating the fact that chronic treatment is required to induce a significant change in cell growth or suggesting that small changes in many genes can lead to significant changes in the growth properties of a cell. The vast majority of the genes have not been previously reported to be regulated by 1,25-(OH)2D3. They thus represent potential novel target genes for the hormone. Several of the identified genes, such as c-FOS, an oncogene with growth inhibitory activity, were previously shown to be regulated by 1,25-(OH)2D3 (15, 16). On the other hand, several known 1,25-(OH)2D3 target genes identified in other cell types, like vitamin D receptor (17), P21 (18), and insulin-like growth factor-binding protein-3 (19), did not show significant changes in this particular cell type. Furthermore, the transcript of cyclin E (6), SKP2 (6), and hTERT (20) in OVCAR3 cells was shown to be decreased by 1,25-(OH)2D3 in our previous PCR studies, which was not detected in the current array analyses. The exact reason is unclear but may be because of the fact that the level of their mRNA expression is so low that it cannot be distinguished from the average noise level of hybridization on the chip. For example, we specifically examined hTERT signal (probeset 207199_at) on U133 chips after hybridization and found that it was fairly low. Consistently, we have been unable to detect hTERT mRNA in OVCAR3 cells by Northern blot analyses and all our published data were obtained from RT-PCR.
Validation of the Regulation of Apoptosis-related Genes by 1,25-(OH)2D3
Our recent studies have shown that 1,25-(OH)2D3 regulates apoptosis in OVCAR3 cells (20). To identify genes that may mediate the apoptotic response, we incorporated apoptosis as a parameter in GeneSpring software and found 13 apoptosis-related genes that were significantly regulated as judged by SAM analysis using a 5% false discovery rate. This includes 5 up-regulated genes and 8 down-regulated genes (TABLE THREE). Because fold change is an arbitrary parameter, we did not include this criterion when selecting apoptotic genes.
TABLE THREE. Apoptosis-related genes regulated by 1,25-(OH)2D3.
| GenBank | Gene name | -Fold change (mean ± S.D.) | qa | ||
|---|---|---|---|---|---|
| 8 h | 24 h | 72 h | |||
| % | |||||
| Up-regulated | |||||
| NM_001924 | GADD45 | 2.77 ± 0.09 | 3.71 ± 0.13 | 4.02 ± 0.15 | 2.6 |
| NM_003810 | TRAIL | 1.91 ± 0.16 | 1.61 ± 0.25 | 1.83 ± 0.16 | 1.5 |
| NM_005157 | ABL1 | 1.47 ± 0.08 | 1.61 ± 0.04 | 1.63 ± 0.08 | 0.9 |
| NM_003840 | TRAIL-R4 | 1.85 ± 0.13 | 2.83 ± 0.44 | 3.00 ± 0.40 | 4.6 |
| NM_000660 | Transforming growth factor-β1 | 1.27 ± 0.20 | 1.58 ± 0.23 | 1.66 ± 0.04 | 4.8 |
| Down-regulated | |||||
| NM_000043 | Fas | −1.54 ± 0.14 | −1.96 ± 0.27 | −2.50 ± 0.21 | 0.6 |
| NM_001227 | Caspase-7 | −1.19 ± 0.09 | −1.32 ± 0.01 | −1.46 ± 0.09 | 4.6 |
| NM_003842 | TRAIL-R2 | 1.00 ± 0.20 | −1.22 ± 0.06 | −1.30 ± 0.22 | 4.2 |
| NM_014456 | PDCD4 | −1.38 ± 0.26 | −1.30 ± 0.09 | −1.33 ± 0.38 | 1.2 |
| NM_006595 | API5-like 1 (API5L1) | −1.09 ± 0.18 | −1.11 ± 0.08 | −1.25 ± 0.08 | 4.6 |
| NM_001316 | Cellular apoptosis susceptibility protein (CSE1) | −1.04 ± 0.01 | −1.09 ± 0.04 | −1.28 ± 0.09 | 0.6 |
| NM_004330 | BNIP2 | −1.03 ± 0.04 | −1.22 ± 0.12 | −1.18 ± 0.09 | 1.5 |
| NM_04964 | HDAC1 | −1.12 ± 0.05 | −1.16 ± 0.09 | −1.23 ± 0.01 | 1.5 |
q value refers to the possibility of a false positive for a specific gene by SAM analysis.
To confirm the regulation of the apoptosis-related genes by 1,25-(OH)2D3, RT-PCR analysis was performed with pooled RNA from three independent preparations. PCR analysis confirmed the regulation by 1,25-(OH)2D3 of all 12 genes (Fig. 2). We used GADD45 as a positive control in these experiments.
FIGURE 2. Verification of the regulation by 1,25-(OH)2D3 at mRNA level of apoptosis-related genes identified in microarray analyses.
OVCAR3 cells were treated for the indicated times, and RNA was prepared and subjected to RT-PCR analyses with primers specific for different genes. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control was included in each PCR analysis and representative data are shown. PDCD4, programmed cell death 4; CSE1, cellular apoptosis susceptibility 1; BNIP2, BCL2 adenovirus E1B 19-kDa interacting protein 2; HDAC1, histone deacetylase 1; AP5IL1, API5-like 1; TGF-β, transforming growth factor-β.
From the list of apoptotic genes, it is striking that multiple genes working on the extrinsic apoptotic pathway mediated through death receptors were regulated by 1,25-(OH)2D3. Fas, TRAIL receptor 2 (TRAIL-R2), and caspase-7 were down-regulated, whereas TRAIL and TRAIL-R4, a decoy receptor that suppresses the apoptosis response to TRAIL, were up-regulated. It thus appears that 1,25-(OH)2D3 may regulate the extrinsic apoptotic pathway in OCa cells.
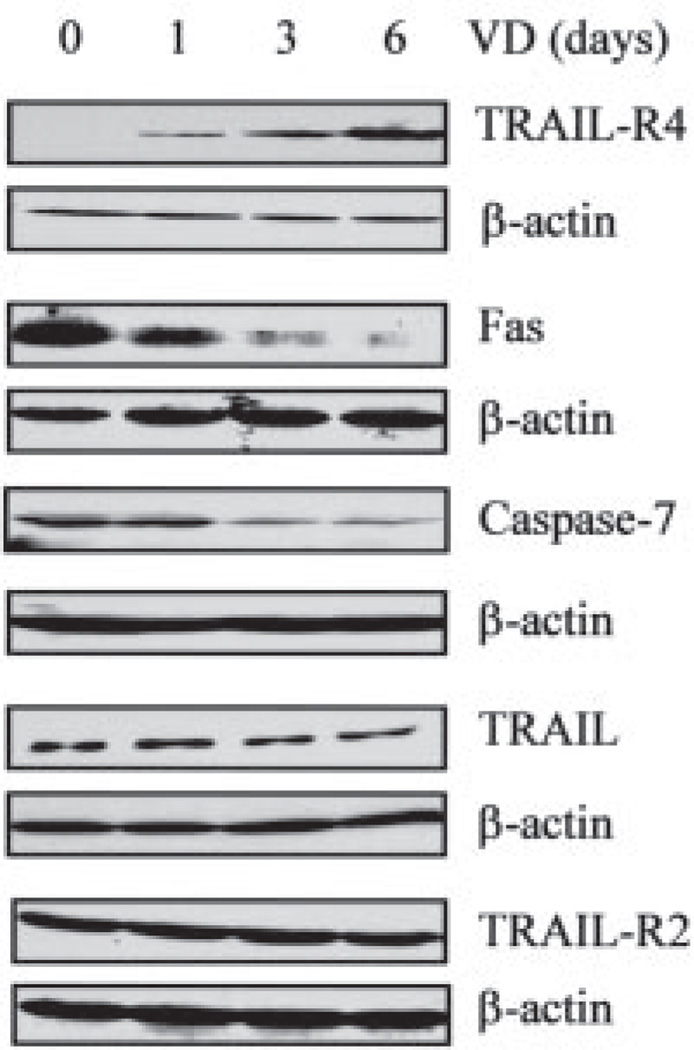
Before exploring ways to confirm the above prediction, we went further to determine whether the regulation of these genes by 1,25-(OH)2D3 was manifested at the protein level (Fig. 3). The immunoblotting analyses showed that TRAIL-R4 was induced, whereas Fas and caspase-7 proteins were repressed, which was consistent with the regulation seen at the RNA level. Conversely, the TRAIL protein appeared to be down-regulated and the protein level of TRAIL-R2 did not change. This demonstrates that regulation at the RNA level is not always manifested at the protein level. Overall, protein analyses support the prediction that 1,25-(OH)2D3 may suppress the extrinsic apoptotic pathway mediated through death receptors.
FIGURE 3. Regulation at the protein level of apoptotic genes along the death receptor pathway by 1,25-(OH)2D3.
OVCAR3 cells were treated with 10−7 m 1,25-(OH)2D3 (VD) for the indicated times and the levels of TRAIL-R4, Fas, caspase-7, TRAIL, and TRAIL-R2 protein expression were determined by immunoblotting. β-Actin blot was included to show even loading.
1,25-(OH)2D3 Suppresses Death Receptor-mediated Apoptosis in OVCAR3 Cells
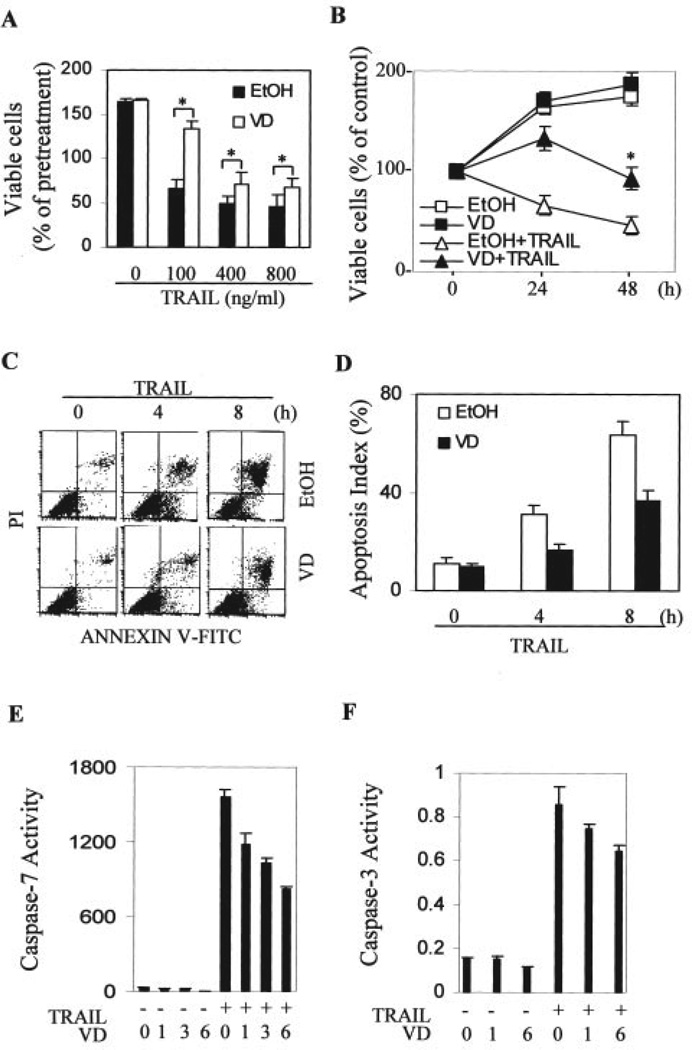
We next directly tested whether 1,25-(OH)2D3 suppressed the extrinsic apoptotic pathway mediated through the death receptors. First, we tested the effects of 1,25-(OH)2D3 on TRAIL-induced cytotoxicity in OVCAR3 cells. As shown in Fig. 4, pretreatment with 1,25-(OH)2D3 suppressed the decrease in cell numbers induced by TRAIL at various concentrations (Fig. 4A) or by 100 ng/ml at different time points (Fig. 4B). More direct assessment of apoptosis markers reveals that the effects on cell loss are because of changes in the amount of induced apoptosis (Fig. 4, C–F). Annexin V-positive cells are reduced by pretreatment with 1,25-(OH)2D3. In addition, the activation of caspase-7 (Fig. 4E) and caspase-3 (Fig. 4F) by TRAIL was suppressed by the addition of 1,25-(OH)2D3. Caspase-7 was activated more dramatically by TRAIL (about 54-fold) than caspase-3 (about 5-fold), suggesting that caspase-7 may be preferentially involved in apoptosis induced by TRAIL. Caspase 7 is decreased by 1,25-(OH)2D3 treatment, which may explain why 1,25-(OH)2D3 treatment has a more dramatic effect on caspase 7 than on caspase 3. In addition, it seemed that the longer the 1,25-(OH)2D3 pretreatment, the stronger was the resulting suppression.
FIGURE 4. Suppression of TRAIL-induced apoptosis by 1,25-(OH)2D3 in OVCAR3 cells.
A, cells were pretreated with 1,25-(OH)2D3 (VD) or vehicle (EtOH) for 3 days. Equal numbers of treated cells were plated into 96-well plates and further treated with the indicated concentrations of TRAIL for 24 h. MTT assays were performed and absorption at 595 nm (A595 nm) was determined. Eight samples were analyzed in parallel for each data point and the experiments were reproduced twice. Percentages of viable cells were calculated relative to the A595 values of cells before TRAIL treatment (pretreated cells), which was set as 100%. *, p < 0.01 when compared with the corresponding values from cells pretreated with vehicle. B, cells were pretreated similarly as in A and subsequently treated with or without 100 ng/ml TRAIL for the indicated times before MTT assays were performed. Percentages of viable cells were calculated by dividing A595 values with the corresponding value before treatment for each group. *, p < 0.01 when compared with the group treated with TRAIL following vehicle. C and D, cells were pretreated similarly as in A followed by secondary treatment with 100 ng/ml TRAIL for the indicated times. Apoptosis of treated cells was determined by flow cytometry after Annexin V staining. A representative profile of the flow cytometry analysis is shown in C. The apoptotic index from two independent analyses is shown in D. E, effect of 1,25-(OH)2D3 on TRAIL-induced caspase-7 activation. Cells were treated with 10−7 m 1,25-(OH)2D3 or EtOH for 0, 1, 3, or 6 days followed by treatment with 100 ng/ml TRAIL for 4 h. Capase-7 activity was determined as described under “Materials and Methods.” F, effect of 1,25-(OH)2D3 on TRAIL-induced caspase-3 activation. Cells were treated as in E and caspase-3 activity was measured. FITC, fluorescein isothiocyanate.
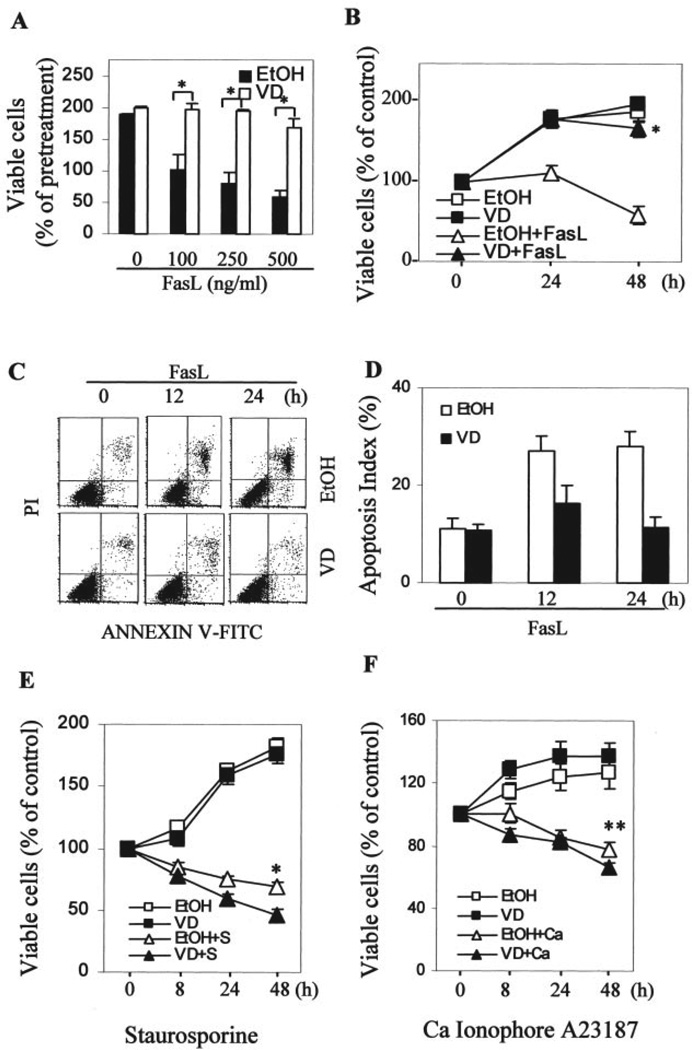
The fact that Fas was down-regulated by 1,25-(OH)2D3 suggests that protective action of 1,25-(OH)2D3 may not be limited to apoptosis induced by TRAIL. Therefore, we also tested the effect of 1,25-(OH)2D3 on Fas ligand-induced cytotoxicity in OVCAR3 cells. Similar to the above described TRAIL data, pretreatment with 1,25-(OH)2D3 prevented the decrease in cell number induced by Fas ligand at various concentrations (Fig. 5A) or by 250 ng/ml Fas ligand at different time points (Fig. 5B). Similar apoptotic analyses also confirmed that the protective hormonal effect on Fas ligand-induced toxicity in growth assays was because of the suppression of Fas ligand-induced apoptosis (Fig. 5, C and D).
FIGURE 5. Selective suppression of Fas ligand (FasL)-induced apoptosis by 1,25-(OH)2D3 in OVCAR3 cells.
A–D, cells were treated and MTT (panels A and B) and apoptotic (panels C and D) assays were performed as described in the legend to Fig. 4, panels A–D. The difference is that the cells were treated with FasL instead of TRAIL. For panel A, cells were treated for 48 h and, for panels B–D, cells were treated with 250 ng/ml FasL. A and B, *, p < 0.01 when compared with the corresponding values obtained from cells pretreated with vehicle. E and F, cells pretreated with 10−7 m 1,25-(OH)2D3 or vehicle for 3 days were treated with 1 µm staurosporin or 0.5 µm calcium ionophore A23187 for the indicated times. MTT assays were performed, and percentages of viable cells were calculated by dividing A595 values with the corresponding value before treatment. *, p < 0.01 when compared with cells treated with vehicle followed by staurosporin. **, p < 0.05 when compared with cells treated with vehicle followed by A23187. FITC, fluorescein isothiocyanate.
To determine whether the suppressive effect of 1,25-(OH)2D3 is universal to caspase-dependent apoptosis or restricted to the extrinsic death receptor pathway, we examined the effect of 1,25-(OH)2D3 on cytotoxicity induced by different stimuli that are known to induce cell death through caspase-dependent apoptosis. This includes the protein kinase C inhibitor staurosporine and the calcium ionophore A23187. We observed that 1,25-(OH)2D3 did not suppress staurosporine- (Fig. 5E) or A23187 (Fig. 5F) -induced apoptosis. Instead, 1,25-(OH)2D3 pretreatment possibly enhanced staurosporine- or calcium ionophore A23187-induced apoptosis. This suggests that the protective effect is likely to be specific to the extrinsic apoptotic pathway mediated through death receptors.
Cell Specificity of the Protective Effect of 1,25-(OH)2D3 on TRAIL-induced Apoptosis
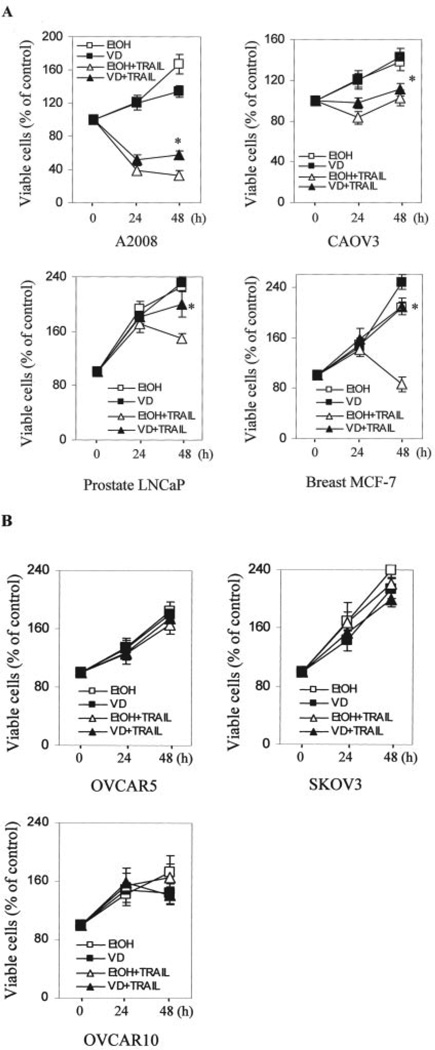
To determine whether the protective effect of 1,25-(OH)2D3 against TRAIL-induced apoptosis is restricted to OVCAR3 cells, additional VD-sensitive OCa cells were sequentially treated with 1,25-(OH)2D3 and TRAIL. As shown in Fig. 6, panel A, 1,25-(OH)2D3 exhibited detectable protection against TRAIL in the VD-sensitive cell lines 2008 and CAOV3. The protective effect was stronger in LNCaP prostate and MCF-7 breast cancer cells (Fig. 6A). Thus, the protective effects of 1,25-(OH)2D3 against TRAIL-induced apoptosis are shared by VD-sensitive OCa cells as well as some breast and prostate cancer cells.
FIGURE 6. Suppression of TRAIL-induced apoptosis by 1,25-(OH)2D3 in multiple cancer cell lines.
A and B, cells pretreated similarly as in Fig. 4A were subsequently treated with or without 100 ng/ml TRAIL for the indicated times before MTT assays were performed with the exception that LNCaP cells were treated with 800 ng/ml TRAIL. Percentages of viable cells were calculated by dividing A595 values with the corresponding value before treatment. *, p < 0.01 when compared with cells treated with TRAIL following vehicle.
Some ovarian cancer cell lines are resistant to VD-induced growth suppression (6). To determine how these VD-resistant cells respond to TRAIL-induced cytotoxicity as well as the protection by 1,25-(OH)2D3, we analyzed the response of OVCAR10, SKOV3, and OVCAR5 cells in cell growth assays (Fig. 6B). As expected, 1,25-(OH)2D3 did not affect their growth, confirming their resistance to 1,25-(OH)2D3. Interestingly, VD-resistant cells appear to be also resistant to TRAIL-induced cytotoxicity even without any 1,25-(OH)2D3 treatment.
Strategies to Overcome the Adverse Effect of 1,25-(OH)2D3 on Apoptosis Mediated through Death Receptors
Because VD-sensitive OCa cells are also sensitive to TRAIL, the combinational treatment may represent an innovative strategy for OCa therapy. Therefore, surmounting the interference of TRAIL action by 1,25-(OH)2D3 may be of clinical significance. Reversal of the up-regulation of TRAIL-R4 may eliminate the adverse effect of 1,25-(OH)2D3. Conversely, restored expression of the down-regulated Fas may also reverse the effects of 1,25-(OH)2D3 on Fas ligand-induced apoptosis.
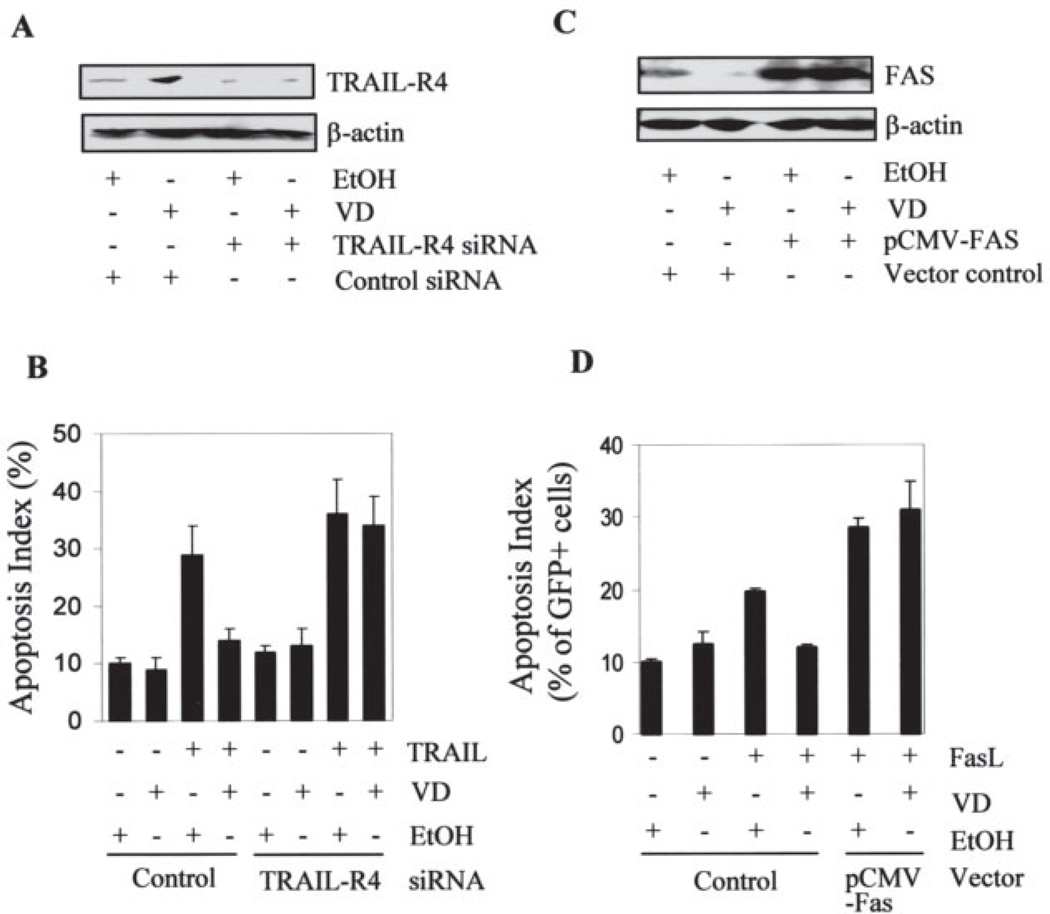
To test whether this adverse effect of 1,25-(OH)2D3 could be eliminated through manipulation of TRAIL-R4, we transiently transfected OVCAR3 cells with TRAIL-R4 siRNA to knockdown the expression of TRAIL-R4. As shown in Fig. 7, panel A, TRAIL-R4 siRNA decreased the expression of TRAIL-R4 and its induction by 1,25-(OH)2D3. Consistent with decreased TRAIL-R4 expression, apoptosis induced by TRAIL was increased in the cells transfected with siRNA, and 1,25-(OH)2D3 pretreatment no longer protected the cells from TRAIL-induced apoptosis (Fig. 7B).
FIGURE 7. Suppression of the death receptor-mediated apoptosis by 1,25-(OH)2D3 is eliminated by molecular manipulation of TRAIL-R4 and Fas.
OVCAR3 cells pretreated with 10−7 m 1,25-(OH)2D3 or vehicle were transfected with TRAIL-R4-specific siRNA (A and B) or pCMV-Fas together with pEGFP plasmids (C and D). Six h post-transfection, cells were placed in fresh medium containing 1,25-(OH)2D3 or vehicle. 48 h later, cells were harvested and immunoblotting analysis was performed to determine TRAIL-R4 or Fas protein levels (panels A and C). For apoptosis analyses, cells were treated with 100 ng/ml TRAIL for 4 h (panel B) or 250 ng/ml Fas ligand for 12 h before fixation (panel D). TRAIL-induced apoptosis was determined by flow cytometry after Annexin-V staining (B). Fas ligand-induced apoptosis was scored by counting the percentage of GFP-positive cells undergoing nuclear condensation (D).
Conversely, we used an expression vector to increase the expression of Fas in 1,25-(OH)2D3-treated cells. As shown in Fig. 7C, the transfected cells expressed Fas at a much higher level than control cells and the down-regulation of Fas protein is no longer detectable because of dominance of the ectopic expression. After scoring the apoptotic index of the transfected cells (GFP-positive cells), it became clear that Fas overexpression increased Fas ligand-induced apoptosis and diminished the protection by 1,25-(OH)2D3 (Fig. 7D). These analyses show that molecular manipulation of death receptors is an effective strategy that may allow OCa patients to benefit from combined therapy with both 1,25-(OH)2D3 and the ligands for death receptors.
DISCUSSION
The present study is the first to identify VD-targeted genes in OCa cells by large-scale profiling using microarray and represents an important step in our effort to understand the molecular mechanisms underlying VD action in OCa. Multiple aspects of the data are significant. First, a large number of genes were found to be regulated by 1,25-(OH)2D3. Most of these genes are reported here for the first time as 1,25-(OH)2D3-regulated. This large response provides an initial indication that cellular responses to VD treatment can be complex. Second, our identification of these genes is based on the analysis of multiple independent experiments. This gives us the ability to identify genes like TRAIL-R4, which was regulated modestly by 1,25-(OH)2D3. However, this regulation was confirmed at both the RNA and protein levels and caused a functional consequence in OCa. Third, and more importantly, we were able to validate the functional significance of the microarray data in OCa biology by demonstrating, with detailed molecular analyses, that the regulation of Fas and TRAIL receptors, detected by microarray, could be translated into the actual suppression of death ligand-induced apoptosis. These studies reveal an undesired effect of VD action in human cancer cells and provide a second indication that 1,25-(OH)2D3 induces a complex biological response, showing the necessity to understand both beneficial and adverse effects of VD compounds for its successful application to the clinical management of human cancers. Finally, our studies show that manipulation of death receptors is a viable approach to eliminate this adverse effect of 1,25-(OH)2D3 on the extrinsic apoptotic pathway by the combined application of both 1,25-(OH)2D3 and death ligands for OCa therapy.
Microarray analyses on 1,25-(OH)2D3-regulated gene expression have been performed in prostate cancer (21), breast cancer (22), colon cancer (23, 24), and squamous carcinoma (25, 26) cells (supplemental Table S2). Except the enzyme 24-hydroxylase, which processes 1,25-(OH)2D3, no other gene has been identified by more than three groups and only a few genes were commonly identified by two different groups. As pointed out by others (21, 25), this probably reflects the fact that the action of 1,25-(OH)2D3 in different cell lines involves different sets of genes. This emphasizes the necessity for study of VD-regulated genes in each cancer type on an individual basis.
TABLES ONE and TWO contain genes encoding proteins involved in cell growth immunity, motility, cell-cell adhesion, and cell-extracellular matrix interactions. This implies that 1,25-(OH)2D3 may influence many aspects of OCa cell homeostasis and could suppress OCa invasion and metastasis by regulating cellular processes in more ways than simple growth inhibition. Nonetheless, there are several genes on the list that have previously been associated with the growth or progression of OCa. For example, Edg2 has been implicated in growth inhibition (27). Edg2 is one of the receptors for lysophosphatic acid, a potential biomarker for human OCa (28). The induction of Edg2 by 1,25-(OH)2D3 is likely to be directly involved in the hormone-induced suppression of OCa cell growth. The expression of CXC chemokines including interleukin-8, small inducible cytokine subfamily B6 (SCYB6), Gro-β and -γ are down-regulated by 1,25-(OH)2D3. Interleukin-8 is a well documented stimulator of OCa growth and motility (29). Down-regulation of interleukin-8 has been reported to contribute to the growth suppression of 1,25-(OH)2D3 in skin cells (30). It is possible that it also plays a role in OCa. Gro-α was recently shown to promote tumor growth, transformation, metastasis, and angiogenesis (31), and the function of Gro-β and -γ was assumed to be similar to Gro-α (32, 33).
Our published studies suggest that cell death is an important mechanism of 1,25-(OH)2D3-induced growth suppression of OCa cells in vitro (20) and in nude mice (7). Consistent with this observation was the fact that multiple apoptosis-related genes were found to be VD-regulated in the current microarray analysis (TABLE THREE). VD up-regulated transforming growth factor-β and ABL1, known pro-apoptotic molecules (34, 35), as well as GADD45, which is associated with apoptosis (36, 37). Among down-regulated genes, API5-like 1 (API5L1) has been shown to be anti-apoptotic (38) and histone deacetylase 1 (HDAC1) controls the acetylation status of proapoptotic proteins such as p53 (39). Inhibition of this HDAC activity induces apoptosis in cancer cells (40). Counterintuitively, some genes that are implicated in promoting apoptosis, such as programmed cell death 4 (41, 42), cellular apoptosis susceptibility 1 (43), and BCL2 adenovirus E1B 19-kDa interacting protein 2 (44), were down-regulated by 1,25-(OH)2D3 (20). It remains to be determined if these changes occur at the protein level and what role these changes play in OCa cells.
Anti-apoptotic actions of 1,25-(OH)2D3 have been described in keratinocytes (45, 46), osteoblasts (47), lymphocytes (48), thyrocytes (49), and osteosarcoma cells (50). The functions are consistent with the positive physiological role of 1,25-(OH)2D3 in regulating the survival and differentiation of cells of skin, bone, and the immune system. However, it is striking that 1,25-(OH)2D3 induces apoptosis in multiple human epithelial cancer cell types (51–53) but suppresses death receptor-mediated apoptosis. With respect to this point, it is important to point out that, although 3-day treatment with 1,25-(OH)2D3 suppressed death receptor-mediated apoptosis, treatment with the hormone for longer than 6 days induced apoptosis by gradually decreasing the level of hTERT expression (20). Besides, our data suggests that the 1,25-(OH)2D3 induced suppression of apoptosis appears to be restricted to the extrinsic pathway because vitamin D exerted a synergistic effect with chemical agents such as staurosporine (Fig. 5). Thus, therapy with active VD compounds alone or in combination with other chemical agents remains an attractive approach for the treatment of human OCa.
TRAIL is known to selectively induce apoptosis in transformed cells but not in normal cells. This unique property makes it a very attractive therapeutic agent for human cancers. The relief of the suppressive effect of 1,25-(OH)2D3 on TRAIL-induced apoptosis by siRNA for TRAIL-R4 suggests that it may be beneficial to combine TRAIL-R4 siRNA, TRAIL, and 1,25-(OH)2D3 for treatment of OCa. This is particularly important because 1,25-(OH)2D3-sensitive OCa cells appear to be also sensitive to TRAIL (Fig. 6). Because siRNA is being actively pursued as a novel approach for cancer therapy and suppression of TRAIL-induced apoptosis also occurs in prostate and breast cancer cells, the combination therapeutic concept proposed here for OCa is likely to be applicable to other 1,25-(OH)2D3-sensitive human cancers as well.
Supplementary Material
Acknowledgments
We thank Paul Byvoet for critical reading of the manuscript. Microarray and flow cytometry were performed in the Microarray and the Flow Cytometry core facilities at H. Lee Moffitt Cancer Center and Research Institute.
Footnotes
This work was supported by New Investigator Award DAMD17-01-1-0731 from U. S. Department of Defense Ovarian Cancer Program and NCI, National Institutes of Health Grant 1R01 CA111334 (to W. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
The abbreviations used are: VD, vitamin D3; 1,25-(OH)2D3, 1,25-dihydroxyvitamin D3; OCa, ovarian cancer; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; TRAIL-R2, TRAIL receptor 2; TRAIL-R4, TRAIL receptor 4; FasL, Fas ligand; SAM, significance analysis of microarray; RT, reverse transcription; MTT, methylthiazole tetrazolium; GFP, green fluorescence protein; EGFP, enhanced green fluorescence protein; siRNA, small interference RNA; AMC, 7-amino-4-methylcoumarin.
REFERENCES
- 1.Webb AR, Kline L, Holick MF. J. Clin. Endocrinol. Metab. 1988;67:373–378. doi: 10.1210/jcem-67-2-373. [DOI] [PubMed] [Google Scholar]
- 2.Muir C, Waterhouse J, Mack T, Powell J, Whelan S. Cancer Incidence in Five Continents, International Agency for Research on Cancer Control (IARC), Scientific Publication No. 88. Vol. V. Lyon, France: IARC; 1987. [Google Scholar]
- 3.Devesa SS, Grauman MA, Blot WJ, Pennello GA, Hoover RN, Fraumeni JF., Jr . Atlas of Cancer Mortality in the United States. Bethesda, MD: National Institutes of Health Publication No. 99-4564, NCI, National Institutes of Health; 1999. [Google Scholar]
- 4.Jiang F, Li P, Fornace AJ, Jr, Nicosia SV, Bai W. J. Biol. Chem. 2003;278:48030–48040. doi: 10.1074/jbc.M308430200. [DOI] [PubMed] [Google Scholar]
- 5.Miettinen S, Ahonen MH, Lou YR, Manninen T, Tuohimaa P, Syvala H, Ylikomi T. Int. J. Cancer. 2004;108:367–373. doi: 10.1002/ijc.11520. [DOI] [PubMed] [Google Scholar]
- 6.Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. J. Biol. Chem. 2004;279:25260–25267. doi: 10.1074/jbc.M311052200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Jiang F, Li P, Li C, Ma Q, Nicosia SV, Bai W. Clin. Cancer Res. 2005;11:323–328. [PubMed] [Google Scholar]
- 8.Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, Pike JW, Shine J, O’Malley BW. Proc. Natl. Acad. Sci. U. S. A. 1988;85:3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 10.Rachez C, Freedman LP. Gene (Amst.) 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 11.Hedlund TE, Moffatt KA, Miller GJ. Endocrinology. 1996;137:1554–1561. doi: 10.1210/endo.137.5.8612485. [DOI] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbig J, Sprinkle R, Enkemann SA. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou A, Elgort MG, Allegretto EA. J. Biol. Chem. 1997;272:19027–19034. doi: 10.1074/jbc.272.30.19027. [DOI] [PubMed] [Google Scholar]
- 15.Johansen C, Kragballe K, Henningsen J, Westergaard M, Kristiansen K, Iversen L. J. Investig. Dermatol. 2003;120:561–570. doi: 10.1046/j.1523-1747.2003.12095.x. [DOI] [PubMed] [Google Scholar]
- 16.Maurer U, Jehan F, Englert C, Hubinger G, Weidmann E, DeLuca HF, Bergmann L. J. Biol. Chem. 2001;276:3727–3732. doi: 10.1074/jbc.M005292200. [DOI] [PubMed] [Google Scholar]
- 17.Ross TK, Darwish HM, DeLuca HF. Vitam. Horm. 1994;49:281–326. doi: 10.1016/s0083-6729(08)61149-8. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 19.Boyle BJ, Zhao XY, Cohen P, Feldman D. J. Urol. 2001;165:1319–1324. [PubMed] [Google Scholar]
- 20.Jiang F, Bao J, Li P, Nicosia SV, Bai W. J. Biol. Chem. 2004;279:53213–53221. doi: 10.1074/jbc.M410395200. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan AV, Shinghal R, Raghavachari N, Brooks JD, Peehl DM, Feldman D. Prostate. 2004;59:243–251. doi: 10.1002/pros.20006. [DOI] [PubMed] [Google Scholar]
- 22.Swami S, Raghavachari N, Muller UR, Bao YP, Feldman D. Breast Cancer Res. Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- 23.Wood RJ, Tchack L, Angelo G, Pratt RE, Sonna LA. Physiol. Genomics. 2004;17:122–129. doi: 10.1152/physiolgenomics.00002.2003. [DOI] [PubMed] [Google Scholar]
- 24.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon- Cardo C, Munoz A. Cancer Res. 2003;63:7799–7806. [PubMed] [Google Scholar]
- 25.Lin R, Nagai Y, Sladek R, Bastien Y, Ho J, Petrecca K, Sotiropoulou G, Diamandis EP, Hudson TJ, White JH. Mol. Endocrinol. 2002;16:1243–1256. doi: 10.1210/mend.16.6.0874. [DOI] [PubMed] [Google Scholar]
- 26.Akutsu N, Lin R, Bastien Y, Bestawros A, Enepekides DJ, Black MJ, White JH. Mol. Endocrinol. 2001;15:1127–1139. doi: 10.1210/mend.15.7.0655. [DOI] [PubMed] [Google Scholar]
- 27.Fang X, Gaudette D, Furui T, Mao M, Estrella V, Eder A, Pustilnik T, Sasagawa T, Lapushin R, Yu S, Jaffe RB, Wiener JR, Erickson JR, Mills GB. Ann. N. Y. Acad. Sci. 2000;905:188–208. doi: 10.1111/j.1749-6632.2000.tb06550.x. [DOI] [PubMed] [Google Scholar]
- 28.Shen Z, Wu M, Elson P, Kennedy AW, Belinson J, Casey G, Xu Y. Gynecol. Oncol. 2001;83:25–30. doi: 10.1006/gyno.2001.6357. [DOI] [PubMed] [Google Scholar]
- 29.So J, Navari J, Wang FQ, Fishman DA. Gynecol. Oncol. 2004;95:314–322. doi: 10.1016/j.ygyno.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Gurlek A, Pittelkow MR, Kumar R. Endocr. Rev. 2002;23:763–786. doi: 10.1210/er.2001-0044. [DOI] [PubMed] [Google Scholar]
- 31.Loukinova E, Dong G, Enamorado-Ayalya I, Thomas GR, Chen Z, Schreiber H, Van Waes C. Oncogene. 2000;19:3477–3486. doi: 10.1038/sj.onc.1203687. [DOI] [PubMed] [Google Scholar]
- 32.O’Donovan N, Galvin M, Morgan JG. Cytogenet. Cell Genet. 1999;84:39–42. doi: 10.1159/000015209. [DOI] [PubMed] [Google Scholar]
- 33.Haskill S, Peace A, Morris J, Sporn SA, Anisowicz A, Lee SW, Smith T, Martin G, Ralph P, Sager R. Proc. Natl. Acad. Sci. U. S. A. 1990;87:7732–7736. doi: 10.1073/pnas.87.19.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havrilesky LJ, Hurteau JA, Whitaker RS, Elbendary A, Wu S, Rodriguez GC, Bast RC, Jr, Berchuck A. Cancer Res. 1995;55:944–948. [PubMed] [Google Scholar]
- 35.Barila D, Rufini A, Condo I, Ventura N, Dorey K, Superti-Furga G, Testi R. Mol. Cell. Biol. 2003;23:2790–2799. doi: 10.1128/MCB.23.8.2790-2799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, Haber DA. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 37.Baudet C, Chevalier G, Chassevent A, Canova C, Filmon R, Larra F, Brachet P, Wion D. J. Neurosci. Res. 1996;46:540–550. doi: 10.1002/(SICI)1097-4547(19961201)46:5<540::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Gianfrancesco F, Esposito T, Ciccodicola A, D’Esposito M, Mazzarella R, D’Urso M, Forabosco A. Cytogenet. Cell Genet. 1999;84:164–166. doi: 10.1159/000015247. [DOI] [PubMed] [Google Scholar]
- 39.Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. J. Biol. Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T, Yokozaki H, Kuniyasu H, Hayashi K, Naka K, Ono S, Ishikawa T, Tahara E, Yasui W. Int. J. Cancer. 2000;88:992–997. doi: 10.1002/1097-0215(20001215)88:6<992::aid-ijc24>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 41.Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H, Matsuhashi S, Colburn NH. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14037–14042. doi: 10.1073/pnas.96.24.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lankat-Buttgereit B, Gregel C, Knolle A, Hasilik A, Arnold R, Goke R. Mol. Cell. Endocrinol. 2004;214:149–153. doi: 10.1016/j.mce.2003.10.058. [DOI] [PubMed] [Google Scholar]
- 43.Behrens P, Brinkmann U, Wellmann A. Apoptosis. 2003;8:39–44. doi: 10.1023/a:1021644918117. [DOI] [PubMed] [Google Scholar]
- 44.Belcredito S, Vegeto E, Brusadelli A, Ghisletti S, Mussi P, Ciana P, Maggi A. Brain Res. Brain Res. Rev. 2001;37:335–342. doi: 10.1016/s0165-0173(01)00138-2. [DOI] [PubMed] [Google Scholar]
- 45.Diker-Cohen T, Koren R, Liberman UA, Ravid A. Ann. N. Y. Acad. Sci. 2003;1010:350–353. doi: 10.1196/annals.1299.064. [DOI] [PubMed] [Google Scholar]
- 46.De Haes P, Garmyn M, Degreef H, Vantieghem K, Bouillon R, Segaert S. J. Cell. Biochem. 2003;89:663–673. doi: 10.1002/jcb.10540. [DOI] [PubMed] [Google Scholar]
- 47.Duque G, El Abdaimi K, Henderson JE, Lomri A, Kremer R. Bone. 2004;35:57–64. doi: 10.1016/j.bone.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Cippitelli M, Fionda C, Di Bona D, Di Rosa F, Lupo A, Piccoli M, Frati L, Santoni A. J. Immunol. 2002;168:1154–1166. doi: 10.4049/jimmunol.168.3.1154. [DOI] [PubMed] [Google Scholar]
- 49.Wang SH, Koenig RJ, Giordano TJ, Myc A, Thompson NW, Baker JR., Jr Endocrinology. 1999;140:1649–1656. doi: 10.1210/endo.140.4.6659. [DOI] [PubMed] [Google Scholar]
- 50.Hansen CM, Hansen D, Holm PK, Binderup L. J. Steroid Biochem. Mol. Biol. 2001;77:1–11. doi: 10.1016/s0960-0760(01)00033-4. [DOI] [PubMed] [Google Scholar]
- 51.Guzey M, Kitada S, Reed JC. Mol. Cancer Ther. 2002;1:667–677. [PubMed] [Google Scholar]
- 52.Weitsman GE, Ravid A, Liberman UA, Koren R. Ann. N. Y. Acad. Sci. 2003;1010:437–440. doi: 10.1196/annals.1299.079. [DOI] [PubMed] [Google Scholar]
- 53.Stewart LV, Weigel NL. Exp. Biol. Med. 2004;229:277–284. doi: 10.1177/153537020422900401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.