Abstract
Aging is the major biomedical challenge of this century. The percentage of elderly people, and consequently the incidence of age-related diseases such as heart disease, cancer, and neurodegenerative diseases, is projected to increase considerably in the coming decades. Findings from model organisms have revealed that aging is a surprisingly plastic process that can be manipulated by both genetic and environmental factors. Here we review a broad range of findings in model organisms, from environmental to genetic manipulations of aging, with a focus on those with underlying gene-environment interactions with potential for drug discovery and development. One well-studied dietary manipulation of aging is caloric restriction, which consists of restricting the food intake of organisms without triggering malnutrition and has been shown to retard aging in model organisms. Caloric restriction is already being used as a paradigm for developing compounds that mimic its life-extension effects and might therefore have therapeutic value. The potential for further advances in this field is immense; hundreds of genes in several pathways have recently emerged as regulators of aging and caloric restriction in model organisms. Some of these genes, such as IGF1R and FOXO3, have also been associated with human longevity in genetic association studies. The parallel emergence of network approaches offers prospects to develop multitarget drugs and combinatorial therapies. Understanding how the environment modulates aging-related genes may lead to human applications and disease therapies through diet, lifestyle, or pharmacological interventions. Unlocking the capacity to manipulate human aging would result in unprecedented health benefits.
I. Introduction
Aging, the inevitable and irreversible process of loss of viability and increase in vulnerability, is shaping modern society and medicine. In some European countries, such as Italy, Spain, and the United Kingdom, it is estimated that by 2050, the proportion of people older than 60 will rise from roughly 20% to almost 40% (Weiss, 2002). Because age is a risk factor for most human diseases, ranging from arthritis to life-threatening diseases such as type 2 diabetes and most types of cancer, this “graying” of the population is arguably the major biological and biomedical challenge of the 21st century. Age-related diseases such as heart disease, cancer, and neurodegenerative diseases are already among the leading causes of death in industrialized countries, and their prevalence will inevitably increase (Olshansky et al., 2006; Butler et al., 2008). By 2050, there may be four times more patients with Alzheimer's disease. On top of an aging population, the obesity epidemic affecting several modern societies, such as the United States and the United Kingdom, further emphasizes the need to develop interventions that minimize the effects of the metabolic syndrome and impact of diseases such as type 2 diabetes (Wang et al., 2007).
With an aging population, there is a great and urgent need to develop approaches and therapies targeting the aging process and age-related diseases (Butler et al., 2008). Delaying the process of aging, even slightly, would have profound social, medical and economic benefits (Olshansky et al., 2006; Butler et al., 2008). For example, slowing aging by a mere 7 years would cut mortality of age-related diseases by half at every age. Therefore, the potential benefits from research on the basic biology and genetics of aging are unparalleled in terms of improving quality of life and health. Although much debate remains regarding the molecular causes of aging, findings from model organisms show that aging is surprisingly plastic and can be manipulated by both genetic and environmental factors (Finch and Ruvkun, 2001; Kenyon, 2010). In principle, therefore, it is possible to manipulate human aging. Unlocking this capacity to manipulate aging in people would result in unprecedented human health benefits, and it opens new opportunities for industry.
The remarkable discoveries of the past 2 decades showing that single genes can regulate aging in model organisms demonstrate that aging can be genetically manipulated (Finch and Ruvkun, 2001; Kenyon, 2010). Hundreds of genes that modulate longevity have now been identified in model organisms (de Magalhães et al., 2009a). In some cases (e.g., in worms), mutations in single genes can extend lifespan by almost 10-fold (Ayyadevara et al., 2008). Nonetheless, aging is a complex process that derives not from single genes but from the interactions of multiple genes with each other and with the environment. Evidence from animal systems shows a major impact of the environment on aging, yet environmental manipulations of aging act through genes and proteins, usually by triggering signaling pathways and modulating gene expression. In fact, some genes have been shown in model organisms to have varying effects on lifespan depending on diet (Heikkinen et al., 2009). Genes that can regulate aging in model organisms cannot be directly applied to humans through genetic manipulations for numerous legal, ethical, and technical reasons. If we could understand how the environment modulates these aging-related genes, we might be able to create antiaging therapies applicable to humans, potentially through diet, lifestyle, and even pharmacological interventions. Therefore, understanding genome-environment interactions in the context of aging can be a powerful approach to identify attractive targets for drug design.
In this review, we give an overview of the major environmental factors that modulate aging in animals, in particular those with underlying gene-environment interactions with potential for improving human health and drug discovery. Moreover, we provide a snapshot of the relevance of these to human biology and to antiaging applications in diet, industry, pharmacy, and healthcare.
II. Environmental Manipulations of Aging in Animals
There are multiple examples of environmental manipulations of aging in animal systems. A well known example of this is temperature; in many poikilotherms, environmental temperature has been known for decades to modulate life history, including lifespan. Invertebrate model organisms such as fruit flies (Drosophila melanogaster) and nematode worms (Caenorhabditis elegans) exhibit longer lifespan (up to a certain threshold) at lower temperatures (Lamb, 1968; Klass, 1977). Such effects have been observed in other species such as fish (for review, see Conti, 2008). Although obviously not easy to apply to humans, it is interesting to note that mice genetically engineered to have a lower body temperature are long-lived (Conti et al., 2006; Conti, 2008; Carrillo and Flouris, 2011).
Organisms adapt to their environment and, particularly during development, can alter their phenotypes in response to environmental cues, a process known as phenotypic plasticity. Often, as organisms adapt to their environment, they adapt their developmental programs in ways that affect adult lifespan. The classic example comes from the caste systems found in social insects such as ants and bees. Queens, who are fed a different diet, can considerably outlive workers (Finch, 1990; Rueppell et al., 2007). Another example in alternative life history strategies is the dauer pathway of C. elegans. In low-food or crowded conditions, worms enter a form of developmental arrest called dauer, once believed to forestall aging completely, that considerably extends lifespan (Klass and Hirsh, 1976). Larvae of the sea slug (Phestilla sibogae), in contrast, require a particular environmental cue (a food source) in order to metamorphose, and, because postlarval lifespan is unaffected by how long it takes for the larvae to metamorphose, lifespan can be considerably extended by artificially preventing the larvae from coming in contact with its metamorphic stimulus (Miller and Hadfield, 1990). These animals clearly illustrate how environmental conditions can result in developmental plasticity that has profound effects on aging and longevity.
More recently, work has focused on another nematode worm, Strongyloides ratti, that exhibits alternative life histories. S. ratti is a parasite, the free-living form is very short-lived (∼5 days), yet once inside a host, female worms can live for more than a year (Gardner et al., 2006). These remarkable (>50-fold) differences in lifespan from the same genome are, as far as we are aware, the largest lifespan difference caused by the environment. It should be noted, however, that the two forms of S. ratti are quite different morphologically and physiologically, and so identifying the specific mechanisms involved in life-extension is difficult.
One last example is the Australian redback spider (Latrodectus hasselti), in which male spiders do not eat as adults and are thus short-lived. Their development, reproduction, and aging can be modulated, however, by the presence of female spiders: male spiders reared in the absence of female spiders develop slowly and maintain a high body condition, whereas male spiders reared in the presence of female spiders develop rapidly and have a shorter lifespan. It is possible that the male spiders sense pheromones from female spiders, which trigger the male spiders' development and consequently degeneration and death (Kasumovic et al., 2009). Environmental sensing involving the olfactory system and, at least in invertebrates, specific sensory neurons has been shown to be part of the cascade linking the environment to longevity (Libert and Pletcher, 2007).
Although there are many other examples, these animals illustrate how environmental conditions can lead to extreme alterations in aging and longevity. Their relevance to humans may appear at first glance to be minimal. For instance, epidemiological evidence suggests that, if anything, single men have on average a shorter lifespan (Kaplan and Kronick, 2006), so parallels between the redback spider and humans are unlikely to exist. Remarkably, however, the recent investigation in model organisms of some of the genetic and molecular pathways involved in environmental manipulations of aging, particularly in the dauer formation in worms, indicates these may lead to human applications and therapies. Among the first genes shown to regulate longevity was daf-2, identified by Kenyon et al. (1993) by focusing on genes that regulate the dauer pathway (“daf” comes from abnormal dauer formation). Worms with daf-2 mutations live more than twice as long as normal. In recent years, some of the aging-related genes identified in worms have been shown to have mammalian homologs that modulate longevity and delay age-related diseases in mice, in particular as part of the insulin/insulin-like growth factor (IGF12)/growth hormone (GH) pathway (Bartke, 2005), and variants in these genes have even been associated with human longevity, such as the daf-2 homolog IGF1R (Suh et al., 2008). Therefore, there is great potential for human homologs of genes shown to modulate aging in model organisms to represent pharmaceutical targets with human applications.
III. Diet, Health, and Aging
The previous examples of how diet can modulate aging (e.g., social insects and the dauer pathway) are extreme cases not observed in humans. There is evidence, however, that the environment, and diet in particular, can influence aging trajectories in humans. Such environmental influences can be observed from an early age with long-lasting effects. Early nutrition can affect late-life diseases, such as cardiovascular disease (Barker and Osmond, 1986) and mortality (Gluckman et al., 2008; Hanson and Gluckman, 2008). Likewise, infections in early life can increase inflammatory levels and, together with diet, contribute to late-life diseases (Finch, 2010). The specific genes and mechanisms involved are largely unknown, but these epidemiological studies clearly demonstrate that early life environment can affect aging, and these effects are most likely mediated by gene-environment interactions.
There is a vast amount of literature showing the dietary influences on health, longevity, and aging. In mammals, and humans in particular, there has been a great interest in identifying what constitutes a healthy diet, and numerous studies have focused on the health and longevity benefits of specific dietary components. From epidemiological studies to studies in model organisms—including longevity studies (for a review, see Lebel et al., 2011)—thousands of compounds and diets have been studied with varying degrees of success. Although it is important to understand how variations in diet and how specific dietary components affect health and longevity, it is crucial to point out that understanding how to manipulate the basic process of aging (even slightly) will bring more health benefits than any dietary manipulation or lifestyle studied to date (Olshansky et al., 2006; Butler et al., 2008). As such, herein, we focus on interventions that may retard the aging process.
By far the most widely studied dietary manipulation of aging is caloric restriction (CR), also called dietary restriction. CR consists of restricting the food intake of organisms normally fed ad libitum without triggering malnutrition and is the only dietary intervention shown, to date, to increase longevity and modulate the process of aging in several model organisms (Bishop and Guarente, 2007; Fontana et al., 2010; Spindler, 2010). Even in mammals, such as mice and rats, CR can extend longevity by up to 50%, delay physiological aging, and postpone or diminish the morbidity of most age-related diseases (Masoro, 2005). Ongoing studies in rhesus monkeys suggest that CR can lower the incidence of aging-related deaths in primates (Colman et al., 2009).
Although effects vary across species and strains, evidence from rodents suggests that CR can retard the aging process in mammals and hence delay the appearance and onset of the major age-related pathological conditions, including immune diseases, neurological diseases, diabetes, stroke, metabolic and cardiovascular diseases, as well as cancer (Weindruch and Walford, 1988; Masoro, 2005; Fontana et al., 2010). Studies in rhesus monkeys suggest that CR can reduce body fat and inflammation in primates, in addition to delaying the onset of age-related diseases (Colman et al., 2009). It is noteworthy that results from flies and mice suggest that CR exerts its beneficial effects, including reduced mortality, even when started late in life (Mair et al., 2003; Spindler, 2005). The optimum level of restriction for a given species is not known, however, and in mice seems to depend on genetic background (Liao et al., 2010).
Despite its health and longevity benefits, CR in mammals is associated with negative functional consequences and side-effects such as a reduction in fecundity, muscle mass, and wound healing capacity as well as increased susceptibility to infections (Dirks and Leeuwenburgh, 2006; Fontana et al., 2010). Clearly these side effects are problematic for establishing such dietary regimens in our current society as a means to retard the rate of aging. Therefore, developing CR mimetics, drugs or foods that reproduce the actions of CR without its side effects, is of immense scientific, social, and commercial interest (Ingram et al., 2006).
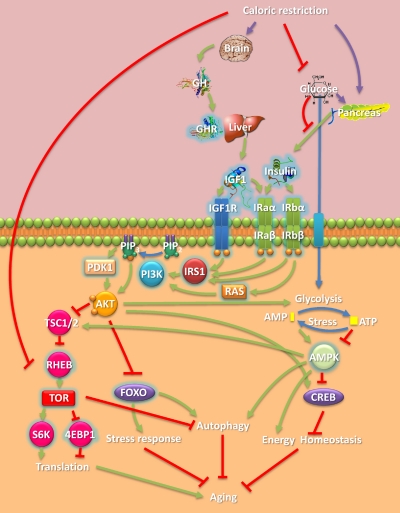
Although the mechanisms by which CR extends lifespan remain a subject of debate, and CR responses are complex, neuroendocrine adaptations and ultimately cellular responses to the environment seem to be crucial. One emerging hypothesis is that CR exerts its effects through hormonal changes that affect cells and induce a survival response (Sinclair, 2005). It is noteworthy that the mechanisms by which CR modulates aging have been shown in model organisms to be mediated by genes and, although not fully understood, signaling pathways (Fig. 1). The ability of individual genes and pathways to affect CR is a major area of research with many potential human applications (Bishop and Guarente, 2007).
Fig. 1.
Overview of CR-associated signaling and some of its key players. CR acts on the hypothalamus, which controls the secretion of GH from the pituitary. CR also lowers glucose levels and diminishes secretion of insulin from the pancreas. GH acting on the liver causes the release to the plasma of IGF1. IGF1 binds to IGF1R or insulin receptor a (IRaα-IRaβ) and triggers its autophosphorylation, which in turn serves as an anchor for recruiting various downstream effectors. Phosphotidylinositol-3-kinase (PI3K) is either activated via direct interaction with, for example, the insulin-like substrate 1 (IRS1) or RAS. PI3K catalyzes the phosphorylation of phosphatidyl-inositols, such as the conversion of phosphatidyl inositol 4,5-bisphosphate (PIP2) to phosphatidyl inositol 3,4,5-trisphosphate (PIP3). PIP3 serves as a binding site for phosphoinositide-dependent protein kinase (PDK1), which activates the serine/threonine protein kinase AKT. AKT phosphorylates tuberous sclerosis protein 1 and 2 (TSC1/2) as well as FOXO transcription factors and cAMP response element-binding (CREB). TSC1/2 phosphorylation by AKT inhibits the Ras homolog enriched in brain (RHEB). RHEB stimulates the phosphorylation of the ribosomal protein S6 kinase (S6K1) and 4E-BP1 through activation of TOR. TOR's activation of S6K1 and inhibition of 4E-BP1 enhances translation. FOXO phosphorylation prevents its nuclear translocation and activation of its stress response target genes. Various cellular forms of stress and AKT activity lead to the depletion of ATP and elevation of AMP. AMPK positively regulates TSC2 and FOXO, negatively regulates CREB, and enforces energy homeostasis. FOXO and AMPK promote autophagy, whereas TOR suppresses it. Under CR, GH levels decline and therefore, via this pathway, TOR signaling decreases (CR also suppresses TOR signaling through other mechanisms) whereas AMPK and some FOXO factors are activated, in turn decreasing translation while increasing stress responses and autophagy, which seem to be some of the mechanisms by which CR retards aging. Human homologs of genes directly linked to CR life-extending effects in model organisms are highlighted with a blue halo.
Given the heterogeneity of human populations, it is unlikely that CR will have the marked longevity benefits in humans that it has in rodents, in particular for those individuals already on a relatively healthy diet (de Grey, 2005; Phelan and Rose, 2005). In fact, CR in wild-derived mice, which, unlike typical laboratory strains, are genetically heterogeneous, does not alter average lifespan even if it increases maximum lifespan (Harper et al., 2006). This suggests variable responses to CR depending on genotype, possibly making CR beneficial to some individuals but not to others. Nonetheless, the widespread effects of CR in multiple diseases across species suggest that some benefits can be expected for some people (Fontana et al., 2010), in particular given the modern obesity epidemic and growing incidence of metabolic diseases such as type 2 diabetes. One preliminary study in humans suggested benefits of CR on cardiovascular disease (Fontana et al., 2004), and biomarkers of CR, such as body temperature and insulin levels, have been associated with human longevity (Roth et al., 2002). In men and women undergoing CR for an average of six years, CR has been reported to lower body temperature (Soare et al., 2011). Moreover, the Okinawan population in Japan, by avoiding high-calorie sugars, saturated fats, and processed foods and instead consuming more vegetables and fruits, seems to have undergone a mild form of CR for decades that could have contributed to the lower risk of age-related chronic diseases and mortality among older Okinawans compared with elderly people in the rest of Japan (Willcox et al., 2006).
It seems that organisms from yeast to mammals have evolved genetic programs to cope with periods of starvation that can also postpone aging and age-related diseases, but how can we take advantage of those mechanisms to improve human health? Because assaying the longevity effects of CR in humans is practically impossible, studying its molecular mechanisms in lower life forms could be beneficial to humans through the identification of candidate genes, pathways and molecular mechanisms. Although CR will not be suitable for everyone, targeting its mechanisms and developing CR mimetics may lead to drug development for a number of age-related and metabolic diseases.
IV. Genome-Environment Interactions as Targets for Dietary Interventions and Drug Discovery
“…[It's] possible that we could change a human gene and double our life span.”—Cynthia Kenyon (Duncan, 2004)
According to the GenAge database of aging-related genes (http://genomics.senescence.info/genes/), more than 700 genes have been identified that regulate lifespan in model organisms (de Magalhães et al., 2009a). Many of these genes and their associated pathways—such as the insulin/IGF1/GH pathway—have been shown to affect longevity across different model organisms (Kenyon, 2010). Therefore, at least some mechanisms of aging are evolutionarily conserved and may have potential therapeutic applications (Baur et al., 2006). For example, evidence suggests the use of lowered IGF signaling (e.g., by targeting IGF receptors) to treat certain age-related diseases such as cancer (Pollak et al., 2004), Alzheimer's disease (Cohen et al., 2009), and autoimmune diseases (Smith, 2010). Moreover, a number of genes and pathways associated with longevity and CR are part of nutrient-sensing pathways that also regulate growth and development, including the insulin/IGF1/GH pathway (Narasimhan et al., 2009; Stanfel et al., 2009). Many of these genes modulate the response to environmental signals, such as food availability, and act in signaling pathways that if understood can be targeted (Fig. 1). The genetic regulation of aging is therefore an emerging field with multiple applications in the human nutrition, cosmetic, and pharmaceutical industries.
In addition to genes associated with aging, research has focused on identifying genes associated with the life-extending effects of CR. One method is to identify genes that decrease or cancel out the life-extending effects of CR when mutated (Gems et al., 2002; Bishop and Guarente, 2007). More than 100 such genes have been identified in model organisms (D. Wuttke, C. Vora, J. P. de Magalhães, unpublished observations). The growth hormone receptor (GHR) is the only gene so far identified in mammals that mediates CR lifespan effects (Bonkowski et al., 2006); most CR-related genes have been identified in lower life forms, such as yeast, flies, and worms (http://genomics.senescence.info/diet/). Nonetheless, most of these genes have mammalian homologs and represent potential targets for interventions to improve human health and/or optimize a healthy diet (Fig. 1). Potential pharmacological interventions may be developed by targeting these genes and pathways; examples of this approach are discussed below.
Applied research based on aging-related pathways is dominated by CR mimetics, and some developments have already captured widespread attention (Ingram and Roth, 2011). In yeast, the NAD+-dependent class III of histone deacetylase enzymes called sirtuins have been reported to mediate the life-extending effects of CR. Specifically, Sir2 overexpression extended lifespan in yeast and CR failed to extend the lifespan of Sir2 mutants (Lin et al., 2000). By screening compounds that activate Sir2, Howitz et al. (2003) identified a number of molecules, including plant polyphenols such as resveratrol. Because resveratrol is usually found in plants and in particularly high concentrations in red wine, it was argued that these findings may have implications for health care and for establishing healthy lifestyles (Guerrero et al., 2009). Feeding resveratrol to yeast, flies, worms, and fishes results in life extension (for review, see Baur, 2010). In mammals, resveratrol was reported to activate the closest mammalian homolog of Sir2, SIRT1, and survivorship of mice on a high-fat diet increased if supplemented with resveratrol (Baur et al., 2006). CR up-regulated the protein level of SIRT1 in several rat tissues, also culturing cells in serum of CR rats up-regulated SIRT1 protein levels; insulin and IGF1 have been shown to reduce SIRT1 protein levels (Cohen et al., 2004).
Based on the work described above, a number of laboratories and companies, including Sirtris (http://www.sirtrispharma.com/), cofounded by David Sinclair, have since focused on identifying compounds that modulate the levels or activity of sirtuins. One screen for environmental chemicals that inhibit sirtuins found that dihydrocoumarin, a flavoring agent found in food and cosmetics, inhibited SIRT1 and increased apoptosis (Olaharski et al., 2005). Likewise, one study assayed a panel of 18 drugs commonly used in clinical practice for SIRT1 expression and identified three compounds (l-thyroxin, sodium nitroprusside, and, surprisingly, insulin) to be activators of SIRT1 (Engel and Mahlknecht, 2008). Although the relevance of these results remains to be established (see below for concerns about resveratrol and SIRT1), these approaches demonstrate how knowledge of aging-related genes can be helpful in terms of optimizing diet for health and on assessing long-term use of drugs.
As detailed ahead, testing compounds directly for effects on human aging is not possible; companies often focus on age-related diseases for which drug development can be assessed and validated for therapeutic effects. Consequently, Sirtris has taken a leading role in translating research on sirtuins to multiple therapeutic areas, particularly in terms of activating SIRT1 as a therapy for type 2 diabetes (Lavu et al., 2008). Indeed, more potent small molecule SIRT1 activators than resveratrol identified via high-throughput screening have been reported to improve insulin sensitivity and lower plasma glucose in obese mice (Milne et al., 2007). At the time of writing, clinical trials for sirtuin activators are also being tested for muscular atrophy, cancer, psoriasis, and sepsis (http://clinicaltrials.gov/ct2/show/NCT01154101; http://clinicaltrials.gov/ct2/show/NCT00920803; http://clinicaltrials.gov/ct2/show/NCT01262911; http://clinicaltrials.gov/ct2/show/NCT00920556; http://clinicaltrials.gov/ct2/show/NCT00964340). The work on sirtuins is an example of how it is possible to move from genes acting on longevity toward developing pharmacological products targeting age-related diseases.
It is important to note that resveratrol and even the health benefits of SIRT1 activation have more recently come under attack. Resveratrol does not extend the lifespan of mice fed a normal diet (Pearson et al., 2008; Miller et al., 2011), and the effects of SIRT1 on mammalian aging and CR are controversial. It has been reported that resveratrol does not directly activate SIRT1 (Pacholec et al., 2010), although more recent results from Sirtris argue otherwise (Dai et al., 2010), and even if the mechanism is indirect, there is evidence that resveratrol has SIRT1-dependent effects in mammalian cells (Baur, 2010). Crucially, although SIRT1 overexpression in mice has been reported to have some health benefits, such as protecting against some types of cancer, it does not extend longevity (Herranz et al., 2010). Moreover, contrary to Sirtris' work, studies in rats suggest that SIRT1 inhibition may be a potential therapy for type 2 diabetes (Erion et al., 2009).
Even if sirtuins and resveratrol do not live up to their expectations, this research is pioneering in terms of genome-environment interactions and nutritional manipulations of aging. These studies also show the path from basic discovery on the biology of aging to potential antiaging and pharmacological interventions and can therefore be applied to other genes and pathways. The lessons learned from the pitfalls of SIRT1 and resveratrol research can also help others to translate basic research on the biology of aging to the clinic, such as avoiding the use of short-lived rodent strains (e.g., by using unhealthy diets), which may lead to findings that only apply to a subset of individuals.
As mentioned above, a number of genes regulating longevity also control growth and development. Some of these, such as the insulin/IGF1/GH pathway, have been suggested to play a role in the mechanisms of CR (Fig. 1). An emerging critical player is the target of rapamycin (TOR) signaling pathway, which involves both nutrient sensing and regulation of growth. Several genes in the TOR pathway, and the TOR gene itself, regulate longevity in flies (Kapahi et al., 2004) and both longevity and dauer diapause in worms (Jia et al., 2004). Strikingly, not only have genetic manipulations of the TOR gene extended lifespan in yeast and worms (Stanfel et al., 2009) but also feeding rapamycin (which inhibits TOR and is also known as sirolimus) to middle-aged mice significantly (9–14%) increased lifespan (Harrison et al., 2009). Whether rapamycin is extending lifespan by delaying of aging or by affecting a specific disease, such as cancer, remains unclear. More recent studies show that starting rapamycin administration earlier in life does not result in a significantly greater increase in lifespan (10–18%) than that obtained in middle-aged mice (Miller et al., 2011).
Rapamycin has serious side effects, particularly as an immunosuppressor, and thus it is not suitable as an antiaging drug. As in sirtuins, however, these studies highlight the road from basic discovery on the biology of aging to antiaging interventions. Further studies of the TOR pathway and of repressors more specific of its downstream signaling pathway are ongoing. Whether rapamycin produces a change in another parameter related to energy uptake or utilization is unknown, and determining which of its effects modulate lifespan is an important unsolved question. Like resveratrol, TOR has attracted considerable attention from the pharmaceutical industry, particularly in the context of cancer (Meric-Bernstam and Gonzalez-Angulo, 2009).
Other candidate CR mimetics are also being explored, including 2-deoxyglucose and the diabetes drug metformin, which inhibit glycolysis, and many others are in the pipeline (Ingram et al., 2006; Ingram and Roth, 2011). Metformin, which activates the nutrient and energy sensor AMP-activated protein kinase (AMPK) previously associated with lifespan and CR in worms (Kenyon, 2010) extends lifespan of murine disease models (Anisimov et al., 2008), yet failed to extend the lifespan of normal rats (Smith et al., 2010). One study reported that metformin slightly increased lifespan in female mice but decreased lifespan in male mice (Anisimov et al., 2010). Another recent study suggested beneficial effects of metformin in a mouse model of Alzheimer's disease independent of AMPK activation (Kickstein et al., 2010). For some aging-related pathways, therapies may focus on specific diseases or even on cosmetic applications.
Knowledge of genetic and molecular pathways related to aging and its modulation can also be translated into predictions on health effects of dietary components (Müller and Kersten, 2003). Therefore, in addition to pharmaceuticals, another marketplace for basic aging research involves supplements, which avoids the need for clinical trials. Indeed, companies are now focusing on nutritional supplements that target genes/pathways involved in aging. One example is Genescient (http://www.genescient.com/), a biotechnology company; its strategy involves choosing supplements that affect pathways that may be important in long-lived flies as assayed from gene expression analyses (Rose et al., 2010).
V. Functional Genomics of Aging, Target Prioritization, and Future Prospects
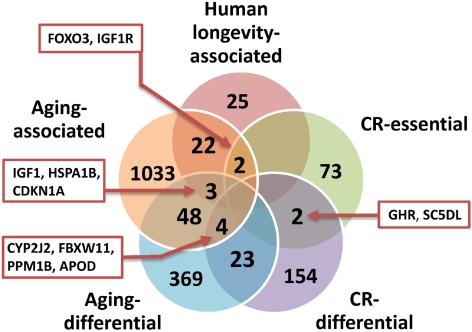
The ability to modulate SIRT1 and the recent findings from rapamycin offer a glimpse of what can be achieved by focusing on aging-related genes. A single gene that regulates aging can have a profound impact on health and several age-related diseases. Given how CR delays the onset of multiple age-related diseases, such as type 2 diabetes and cancer, manipulation of CR pathways might have applications at least in disease prevention and possibly even in a clinical setting. However, in addition to sirtuins and TOR, there are already hundreds of genes associated with aging and CR in model organisms, and this provides an excellent opportunity for target discovery (Fig. 2). Besides, we have only begun to study the genetics of aging and thus such genes represent only the “tip of the iceberg.” Therefore, there is a huge potential to discover new therapeutic targets among aging-related genes and those that regulate the effects of diet on lifespan.
Fig. 2.
Overview and overlap of genes related to aging, human longevity, and CR. Shown are the intersections between human orthologs of genes identified via genetic manipulation experiments in model organisms (aging-associated), genes that disrupt or cancel life-extending effects of CR when mutated in model organisms (CR-essential), aging differentially expressed genes in mammals (aging-differential), CR-differentially expressed genes in mammals (CR-differential) and genes associated with human longevity in at least one epidemiological study (human longevity-associated). All data obtained from the GenAge database (http://genomics.senescence.info/genes/), except for the CR-associated genes, which come from data sets assembled by the authors from the literature, available on request.
A. Functional Genomics of Aging and Longevity
In addition to genetic manipulation experiments to identify genes of interest, another approach involves identifying genes specifically up-regulated or down-regulated in a particular experimental setting. One study found longevity effects when genes differentially expressed in long-lived worms were mutated (Murphy et al., 2003). Likewise, another study associated the up-regulation of one gene, the eukaryotic translation initiation factor 4E binding protein (4E-BP), with CR effects in mitochondrial activity and life-extension in flies (Zid et al., 2009), although it should be noted that null mutations of 4E-BP decrease lifespan in normal flies (Tettweiler et al., 2005). Another study in flies focused on the temporal transcriptional responses caused by feeding and identified nutrient-responsive genes, many of which under the control of the forkhead box transcription factor dFOXO (Gershman et al., 2007). We have analyzed aging gene expression data to identify common molecular signatures of mammalian aging. Some of the genes overexpressed with age seem to be a response to aging, in that they have been previously found to have protective functions (de Magalhães et al., 2009b). As such, these genes may help organisms manage aging and could be targets for manipulation. Likewise, gene expression analysis of CR has been conducted to identify associated genes (Lee et al., 1999, 2000). A number of molecular signatures have emerged from such studies that could be useful to identify candidate processes and pathways that affect aging, biomarkers (see below), and candidate regulators (Anderson and Weindruch, 2010; Hong et al., 2010).
Gene expression profiles of aging and CR may serve as biomarkers for testing drug effects on inducing “youthful levels” or CR-like gene expression patterns, respectively, without doing full lifespan experiments (Spindler, 2006). SIRT1 activators have been evaluated this way by Sirtris (Smith et al., 2009). Because some mouse models respond differently to CR, studies have focused on identifying differential responses that can be associated with the mechanisms of CR and/or used as biomarkers (Bartke et al., 2008). More recent work has also focused on identifying genes that influence the life-extending effects of CR by classic genetics using differences in lifespan in different mouse recombinant inbred strains (Liao et al., 2010).
In addition to aging- and CR-related genes, another source of candidate genes and pathways for drug design are human longevity-associated genes (Barzilai and Shuldiner, 2001; Browner et al., 2004; Kenyon, 2010). Dozens of genes have now been associated with human longevity (de Magalhães et al., 2009a), although only a handful of genes have been shown to have consistent effects across populations.
Many longevity-associated genes are related to specific diseases and have deleterious genotypes for which incidence decreases with age, resulting in the alternative allele unrelated to disease to become more prevalent in older individuals. However, some evidence suggests that a few longevity-conferring genes, such as the cholesteryl ester transfer protein (CETP), have alleles with protective effects against diseases (Bergman et al., 2007). Therefore, extreme longevity is not due simply to a lack of disease-associated alleles, but some alleles seem to protect against multiple age-related diseases. CETP is a promising target for drug discovery because its alleles have been associated not only with longevity but also with a lower risk of specific age-related diseases, such as cardiovascular diseases, cognitive decline, and dementia (including Alzheimer's disease) (Barzilai et al., 2003; Sanders et al., 2010). In fact, CETP inhibitors (such as torcetrapib) that can raise HDL cholesterol have already been developed, and it is hoped these can be used as preventive therapy for heart disease (Kontush et al., 2008). The identification of drugs that mimic the effects of longevity genes, and of alleles associated with exceptional human longevity in particular (e.g., centenarians), is thus an appealing area of research. One current difficulty is that for many of these longevity genes, we do not know the biochemistry behind the longevity effects or the functional consequences of the different alleles; therefore, we do not know whether we should aim to activate or inhibit these genes. Some promising candidates have emerged, however.
One of the first genes associated with human longevity was apolipoprotein E (APOE), which is involved in lipid metabolism and cholesterol transport. Variants of APOE have been associated with age-related diseases such as cardiac disease and Alzheimer's disease (Schächter et al., 1994). APOE has gathered considerable interest because its polymorphisms are associated with response to therapy in patients with Alzheimer's disease (Evans and McLeod, 2003). Besides, one recent study showed that APOE isoforms differentially regulate clearance of amyloid-β from the brain (Castellano et al., 2011). Consequently, some human longevity genes are already under the spotlight of academia and industry, and this number is bound to increase in the near future as high-throughput sequencing becomes widespread, facilitating genome-wide association studies of longevity (de Magalhães et al., 2010). Moreover, human longevity genes can give clues as to which pathways are associated with healthy human aging, which in turn can be targeted by drugs.
B. Prioritizing Targets for Drug Discovery and Network Approaches
Genome analyses from CR, aging, and human longevity genes provide biological targets for drug discovery. Screening natural products, existing drugs, and chemical libraries for molecules that affect “druggable” targets associated with aging may lead to compounds of therapeutic value. Given the hundreds of genes associated with aging and CR, however, it is important to identify the most promising targets. Integrating information from different datasets can help prioritize candidates (Fig. 2). It is interesting to note the two genes shown in model organisms to be related with aging, associated with human longevity, and essential to CR effects: IGF1R and FOXO3 (Fig. 2). IGFR1 is part of the insulin/IGF1/GH pathway, the down-regulation of which has been associated with life-extension in several model systems and, as mentioned above, is already a target of pharmacological interventions. The FOXO transcription factor FOXO3 is a homolog of dFOXO and of daf-16, in which mutations suppress the life-extending effects of daf-2 (Kenyon et al., 1993). FOXO transcription factors are, in fact, part of the same insulin/IGF1/GH pathway (Fig. 1) that modulates lifespan across organisms (Kenyon, 2010). A strong association between FOXO3 and human longevity has been reported (Willcox et al., 2008) and subsequently validated in other populations (for review, see Kenyon, 2010). FOXO3 was also associated with insulin levels and prevalence of cancer, heart disease, and type 2 diabetes (Willcox et al., 2008). Further work is necessary to understand the modulation of FOXO3 and its molecular mechanisms affecting longevity, but it is a promising target for drug development.
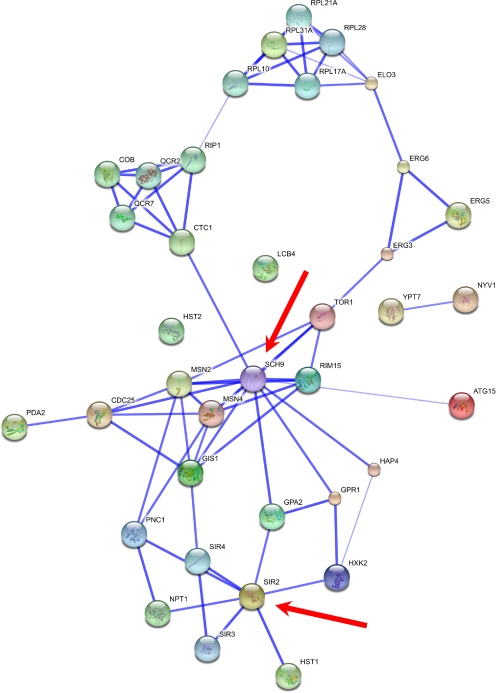
To facilitate target gene prioritization, a number of additional approaches may be employed. For example, in silico studies of transcriptional regulation can allow the identification of upstream regulators (for review, see de Magalhães et al., 2010). Furthermore, an emerging approach to study the complex interactions between the multiple components of biological systems is network biology (Barabási et al., 2011). Given the complexity of aging, network approaches may be particularly suited to identify crucial regulators of its modulation by the environment. For instance, knowing the protein-protein interaction network of candidate proteins allows the identification of hubs, proteins with a large number of interactions, which tend to be more biologically relevant (Fig. 3). Together with other biological (e.g., kinases and receptors are often seen as promising drug targets), medical, and strategic considerations already used for target selection in drug discovery (for review, see Knowles and Gromo, 2003), the integrated knowledge of aging-related pathways can help identify suitable targets for drug discovery. In addition, the advent of large-scale databases of compounds and drugs, such as DrugBank (Wishart et al., 2008), STITCH (Kuhn et al., 2008), and the Connectivity Map (Lamb et al., 2006), paves the way to cross-linking longevity/CR-associated genes with drug databases to identify candidate molecules for effects on aging.
Fig. 3.
Network of CR-related proteins from yeast. Some proteins, such as Sch9 and Sir2 (indicated by red arrows), have a high number of interacting partners (hubs), whereas others have no interactions. Such analyses could be used to identify candidate regulatory hubs as well as promising new candidate genes that interact with known CR-related proteins. Sir2 mammalian homologs are the focus of considerable research (see text), whereas Sch9's mammalian homolog AKT2 is important in insulin signaling and glucose transport. Figure created using STRING (http://string-db.org/).
Advances in the integration of biological (including aging-related) datasets are paralleled by advances in data integration and network analyses in nutrition and pharmacology (Hopkins, 2008). Biological systems are intrinsically complex; for example, CR signaling involves nonlinear pathways, feedback loops, and compensatory mechanisms (Fig. 1). Multitarget drugs and combinatorial therapies may therefore be more successful than single-target drugs. A network-based view of drug discovery is emerging to account for the complexity of human biology (Schadt et al., 2009; Erler and Linding, 2010). Network approaches allows drug developers to take advantage of the large volumes of “-omic” datasets being generated and exploit, rather than dismiss, the intricacy of biology, disease, and drug responses to develop new therapies (for review, see Cho et al., 2006; Schadt et al., 2009). Moreover, focusing on drugs that target multiple proteins, rather than ligands that act on individual targets, has advantages in terms of efficacy and toxicity (Hopkins, 2008). Employing combinations of compounds to target multiple pathways and avoid compensatory mechanisms is another approach, one already used in cancer therapies (Meric-Bernstam and Gonzalez-Angulo, 2009), and in the context of aging is being explored by companies such as Genescient.
Current progress in genomics, high-throughput methods, informatics, and systems biology should help to develop network approaches that test target combinations resulting in the emerging paradigm of network pharmacology (Keith et al., 2005; Hopkins, 2008). Systematic drug-design strategies directed against multiple targets hold much promise in the field of aging (Csermely et al., 2005), although challenges remain in developing accurate computer models of relevant pathways and suitable in vitro and in vivo models for testing. In the same vein, progress in personalized medicine and in predicting individual responses (e.g., using SNPs) to the environment (including diet, lifestyle, and drugs), will be key to maximizing environmental interventions that improve health and counteract aging. Therefore, network approaches to both aging and pharmacology are promising future avenues (Simkó et al., 2009).
C. Translation to Extend Human Healthspan
Although a number of genes and even a few drugs have emerged as candidates for targeting the aging process pharmacologically, several problems are associated with translation to human aging. In principle, human clinical trials on aging cannot be performed. One major problem is that aging cannot be quantified, and even a trial running for several years would struggle to identify endpoints. Lifespan or survival could be quantified, as well as health biomarkers such as low blood pressure, insulin sensitivity, inflammatory markers, glucose metabolism, etc., but these may or may not reflect alterations in the aging process.
Another issue is whether long-term interventions are practical in humans (Kirkland and Peterson, 2009). Interventions such as CR mimetics may only be effective in humans if applied for many years, in which case their safety and side effects would have to be demonstrated. For example, aspirin has beneficial anti-inflammatory and antithrombotic properties and can slightly extend lifespan in male, but not female, mice (Strong et al., 2008). Human epidemiological studies suggest its long-term use can reduce the risk of certain types of cancer and cardiovascular disease (Strong et al., 2008; Rothwell et al., 2011), yet it also increases risk of gastrointestinal bleeding (Derry and Loke, 2000). Therefore, it will be crucial to understand the long-term effects of compounds in trials associated with antiaging drugs.
Many aging-related genes have pleiotropic effects, and so targeting them may have beneficial effects in one disease yet may be detrimental for another age-related disease. As indicated above, the only gene shown to be essential for CR in mammals is GHR. Mouse knockouts are also long-lived (Bonkowski et al., 2006). Therefore, inhibitors of GHR may be used to decrease insulin/IGF1/GH signaling and may be deemed to be of potential therapeutic value. Recent data from subjects with GHR deficiency showed decreased mortality from cancer and type 2 diabetes, but cardiac disease mortality appears to be increased. Overall mortality does not seem to be different (Guevara-Aguirre et al., 2011).
One possibility, as touched upon above, is to conduct clinical trials for specific age-related diseases to obtain approval for particular drugs from regulatory agencies (e.g., U.S. Food and Drug Administration). On the basis of findings in model organisms, people could engage in long-term use of approved drugs indicated for specific diseases. Supplements can also be marketed for long-term effects, though all these can be seen as backdoor approaches. In addition, short-term studies in humans may be feasible for disease, followed by individual analysis of other age-associated endpoints (Kirkland and Peterson, 2009). Another potential area to translate findings from the bench to the bedside is focusing on dysfunction and frailty in the elderly (Kirkland and Peterson, 2009). This would imply clinical studies in elderly patients, which has its own problems (Evans, 2011), but established frailty indicators could be used as endpoints (Kirkland and Peterson, 2009).
Overall, demonstrating that a particular intervention is affecting human aging, as done in model organisms, is virtually impossible. Interventions, including drugs, emerging from basic research on aging will probably target specific age-related pathological conditions and/or dysfunction. Subsequent studies of health biomarkers and multiple age-related diseases may reveal broader effects. Success in animal models or short-term human studies may be sufficient to convince potential patients of the usefulness of particular dietary supplements or approaches, as exemplified by those voluntarily undergoing CR (http://www.crsociety.org/), which can serve as basis for further studies (Soare et al., 2011).
D. Future Prospects in Epigenetics and Aging
One field of immense untapped potential is epigenetics. Initially defined by Conrad Waddington as “the interaction of genes with their environment, which bring the phenotype into being,” epigenetics represents an extra layer of instructions not encoded in the primary DNA sequence. The role of this extra layer in the regulation of genome-environment interactions is beginning to emerge. Epigenetics involve chemical modifications of DNA nucleotide residues (such as cytosine methylation) and associated proteins such as histones that alter the DNA's structure and function and can activate or repress genes (Goldberg et al., 2007).
Epigenetic modifications can be the result of stress or diet (Mathers, 2006; Fraga and Esteller, 2007; Sedivy et al., 2008). Although precise targets for epigenetic modifications during aging or CR are unknown, this is an area with great potential, because epigenetic-driven changes in gene expression as a result of diet or lifestyle are thought to contribute to lifelong health (Mathers, 2006). In fact, histone deacetylases, like sirtuins, modify epigenetic patterns (Fraga and Esteller, 2007).
Merry et al. (2008) showed in rats that dietary supplementation with α-lipoic acid, although by itself unable to extend lifespan, allowed animals switched from CR to ad libitum feeding at 12 months of age to maintain the survival trajectory and extended longevity characteristic of CR; conversely, animals switching from ad libitum to CR did not exhibit extended longevity. In contrast, switching rats not fed lipoic acid between ad libitum feeding and CR also switched the survival trajectory. These results suggest that supplementation with lipoic acid induced a “memory effect” on the animals, locking them into the survival trajectory of the feeding regimen before the switch (Merry et al., 2008). It is noteworthy that animals fed lipoic acid and switched from CR to ad libitum feeding gained weight, just like animals not fed lipoic acid, and thus it seems that although the longevity effects of CR were preserved, other effects of CR were not. Because lipoic acid can induce hyperacetylation of histones and potentially acts as a histone deacetylase inhibitor, the hypothesis that epigenetic mechanisms are involved in this memory effect that specifically influences the life-extending effects of CR is attractive (Merry et al., 2008), although it is also possible that changes in energy uptake or utilization could be involved, and further work is needed to elucidate the underlying mechanisms.
Most approaches outlined thus far rely on manipulation of gene activities by diet or drugs to identify genes in signaling cascades or pathways associated with aging or its manipulation. Yet difficulties may surface because our knowledge of these pathways is still incomplete. Epigenetic modifications can modify hundreds or thousands of genes, so targeting a given epigenetic protein or modification may be a more powerful approach, although much work remains for this strategy to be feasible. Therefore, the case for an epigenetic link between nutrition and longevity is strong, even if the specific epigenetic role of nutrients in modulating aging remains unknown (Niculescu and Lupu, 2011).
VI. Concluding remarks
Aging is the major driving factor of disease in the 21st century. Manipulation of aging-related genes by diet, lifestyle, and pharmaceuticals could dramatically improve human health and could be used to develop drugs against age-related diseases such as cancer, heart disease, type 2 diabetes, obesity, and neurodegenerative diseases. The hundreds of aging-related genes and genes related to CR already identified offer enormous opportunities for target discovery (Fig. 2). Although aging-related genes cannot be modified in humans, understanding how these can be manipulated by diet or pharmaceuticals can have a profound impact on health. In other words, work on the genetics of aging allows the identification of novel genomic targets for drug development, opening the door for aging pharmacogenomics.
Marred by decades of “quackery” (including grafting testicles from young animals into men), the science of aging has come a long way in gaining respectability (Stipp, 2010). Already more than 20 companies worldwide are focusing specifically on the aging process (http://whoswho.senescence.info/corp.php), in addition to “big pharma,” with aging-oriented research and development projects. Although this number is modest, it shows the growing potential of a field that is bound to increase. In 2008, GlaxoSmithKline purchased Sirtris for $720 million (Sipp, 2008), a huge amount for a company with no clinical data; presumably the purchase was based on the extraordinary potential suggested by a compound capable of delaying aging. Even though questions have been raised about their efficiency, resveratrol and other drugs targeting SIRT1 showcase how a gene initially identified as a regulator of aging in yeast can be used as a pharmaceutical target for multiple human diseases. It demonstrates confidence in the field and in the idea that aging is not immutable. The recent problems raised concerning SIRT1 and resveratrol research also serve as a cautionary tale of the hurdles in translation of laboratory discoveries to the clinic.
We now know of hundreds of genes that regulate aging in model organisms, dozens associated with longevity in humans, and hundreds differentially expressed with age. This vast amount of information yields increased power for personalized and stratified medicine, for identifying biomarkers of aging, and for drug development to extend lifespan and ameliorate age-related diseases. Overall, it gives us a blueprint (albeit still imperfect) of how aging is controlled that we can use to potentially manipulate the basic aging process, whatever its underlying molecular mechanisms may be. Moreover, our knowledge of nutrient-sensing pathways that mediate the effects of CR has greatly increased in recent years, opening new opportunities for drug discovery and ultimately for perhaps developing an antiaging pill that retards aging with minimal side effects.
In conclusion, we now know of many target genes that either individually or collectively could be used for screening molecules (nutritional compounds and drugs) that may modulate aging. Even if proving that a particular diet or drug can delay aging is not feasible from a scientific and regulatory perspective, there is a huge potential to identify molecules that ameliorate age-related diseases and/or dysfunction. This represents a tremendous opportunity for companies working in nutrition and pharmacology in a field on an upward trajectory.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council [Grant BB/H008497/1] (to J.P.M.); the Ellison Medical Foundation (to J.P.M.); a Marie Curie International Reintegration Grant within EC-FP7 (to J.P.M.); the Erasmus program (to D.W., M.P.); and the Bundesministerium für Bildung und Forschung (to D.W.). We thank everyone at the GABBA Annual Symposium in Porto, Portugal, June 2009, for discussions that spurred this work and all participants at the Lifestyle and Ageing Conference in Pisa, Italy, October 2010, for fruitful discussions on these topics. We also thank Brian Merry for valuable discussions and comments on a draft of the manuscript. We also thank Joana Costa for helping type the manuscript.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: de Magalhães, Wuttke, Wood, Plank, and Vora.
This article is available online at http://pharmrev.aspetjournals.org.
- 4E-BP
- eukaryotic translation initiation factor 4E binding protein
- AMPK
- AMP-activated protein kinase
- APOE
- apolipoprotein E
- CETP
- cholesteryl ester transfer protein
- CR
- caloric restriction
- daf
- abnormal dauer formation
- GH
- growth hormone
- GHR
- growth hormone receptor
- IGF
- insulin-like growth factor
- IGF1R
- insulin-like growth factor I receptor
- SIRT1
- sirtuin 1
- TOR
- target of rapamycin.
References
- Anderson RM, Weindruch R. (2010) Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol Metab 21:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Berstein LM, Egormin PA, Piskunova TS, Popovich IG, Zabezhinski MA, Tyndyk ML, Yurova MV, Kovalenko IG, Poroshina TE, et al. (2008) Metformin slows down aging and extends life span of female SHR mice. Cell Cycle 7:2769–2773 [DOI] [PubMed] [Google Scholar]
- Anisimov VN, Piskunova TS, Popovich IG, Zabezhinski MA, Tyndyk ML, Egormin PA, Yurova MV, Rosenfeld SV, Semenchenko AV, Kovalenko IG, et al. (2010) Gender differences in metformin effect on aging, life span and spontaneous tumorigenesis in 129/Sv mice. Aging (Albany NY) 2:945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. (2008) Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell 7:13–22 [DOI] [PubMed] [Google Scholar]
- Barabási AL, Gulbahce N, Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C. (1986) Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1:1077–1081 [DOI] [PubMed] [Google Scholar]
- Bartke A. (2005) Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology 146:3718–3723 [DOI] [PubMed] [Google Scholar]
- Bartke A, Bonkowski M, Masternak M. (2008) How diet interacts with longevity genes. Hormones (Athens) 7:17–23 [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. (2003) Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA 290:2030–2040 [DOI] [PubMed] [Google Scholar]
- Barzilai N, Shuldiner AR. (2001) Searching for human longevity genes: the future history of gerontology in the post-genomic era. J Gerontol A Biol Sci Med Sci 56:M83–M87 [DOI] [PubMed] [Google Scholar]
- Baur JA. (2010) Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev 131:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. (2007) Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS Comput Biol 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. (2007) Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 8:835–844 [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. (2006) Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci USA 103:7901–7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browner WS, Kahn AJ, Ziv E, Reiner AP, Oshima J, Cawthon RM, Hsueh WC, Cummings SR. (2004) The genetics of human longevity. Am J Med 117:851–860 [DOI] [PubMed] [Google Scholar]
- Butler RN, Miller RA, Perry D, Carnes BA, Williams TF, Cassel C, Brody J, Bernard MA, Partridge L, Kirkwood T, et al. (2008) New model of health promotion and disease prevention for the 21st century. BMJ 337:a399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo AE, Flouris AD. (2011) Caloric restriction and longevity: effects of reduced body temperature. Ageing Res Rev 10:153–162 [DOI] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. (2011) Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med 3:89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CR, Labow M, Reinhardt M, van Oostrum J, Peitsch MC. (2006) The application of systems biology to drug discovery. Curr Opin Chem Biol 10:294–302 [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. (2009) Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139:1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305:390–392 [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. (2009) Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325:201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti B. (2008) Considerations on temperature, longevity and aging. Cell Mol Life Sci 65:1626–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti B, Sanchez-Alavez M, Winsky-Sommerer R, Morale MC, Lucero J, Brownell S, Fabre V, Huitron-Resendiz S, Henriksen S, Zorrilla EP, et al. (2006) Transgenic mice with a reduced core body temperature have an increased life span. Science 314:825–828 [DOI] [PubMed] [Google Scholar]
- Csermely P, Agoston V, Pongor S. (2005) The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci 26:178–182 [DOI] [PubMed] [Google Scholar]
- Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, et al. (2010) SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem 285:32695–32703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Grey AD. (2005) The unfortunate influence of the weather on the rate of ageing: why human caloric restriction or its emulation may only extend life expectancy by 2–3 years. Gerontology 51:73–82 [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM. (2009a) The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell 8:65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Curado J, Church GM. (2009b) Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25:875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Finch CE, Janssens G. (2010) Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res Rev 9:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry S, Loke YK. (2000) Risk of gastrointestinal haemorrhage with long term use of aspirin: meta-analysis. BMJ 321:1183–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks AJ, Leeuwenburgh C. (2006) Caloric restriction in humans: potential pitfalls and health concerns. Mech Ageing Dev 127:1–7 [DOI] [PubMed] [Google Scholar]
- Duncan DE. (2004) Discover dialogue: biologist Cynthia Kenyon. The idea that aging is something that's not a given is a new paradigm. Discover Magazine 25:16–19 [Google Scholar]
- Engel N, Mahlknecht U. (2008) Aging and anti-aging: unexpected side effects of everyday medication through sirtuin1 modulation. Int J Mol Med 21:223–232 [PubMed] [Google Scholar]
- Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, et al. (2009) SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci USA 106:11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Linding R. (2010) Network-based drugs and biomarkers. J Pathol 220:290–296 [DOI] [PubMed] [Google Scholar]
- Evans WE, McLeod HL. (2003) Pharmacogenomics: drug disposition, drug targets, and side effects. N Engl J Med 348:538–549 [DOI] [PubMed] [Google Scholar]
- Evans WJ. (2011) Drug discovery and development for ageing: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci 366:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. (1990) Longevity, Senescence, and the Genome, The University of Chicago Press, Chicago [Google Scholar]
- Finch CE. (2010) Evolution in health and medicine Sackler colloquium: evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA 107 (Suppl 1):1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G. (2001) The genetics of aging. Annu Rev Genomics Hum Genet 2:435–462 [DOI] [PubMed] [Google Scholar]
- Fontana L, Meyer TE, Klein S, Holloszy JO. (2004) Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci USA 101:6659–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. (2010) Extending healthy life span–from yeast to humans. Science 328:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. (2007) Epigenetics and aging: the targets and the marks. Trends Genet 23:413–418 [DOI] [PubMed] [Google Scholar]
- Gardner MP, Gems D, Viney ME. (2006) Extraordinary plasticity in aging in Strongyloides ratti implies a gene-regulatory mechanism of lifespan evolution. Aging Cell 5:315–323 [DOI] [PubMed] [Google Scholar]
- Gems D, Pletcher S, Partridge L. (2002) Interpreting interactions between treatments that slow aging. Aging Cell 1:1–9 [DOI] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS. (2007) High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics 29:24–34 [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL. (2008) Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359:61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128:635–638 [DOI] [PubMed] [Google Scholar]
- Guerrero RF, García-Parrilla MC, Puertas B, Cantos-Villar E. (2009) Wine, resveratrol and health: a review. Nat Prod Commun 4:635–658 [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, et al. (2011) Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med 3:70ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. (2008) Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol 102:90–93 [DOI] [PubMed] [Google Scholar]
- Harper JM, Leathers CW, Austad SN. (2006) Does caloric restriction extend life in wild mice? Aging Cell 5:441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460:392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen S, Argmann C, Feige JN, Koutnikova H, Champy MF, Dali-Youcef N, Schadt EE, Laakso M, Auwerx J. (2009) The Pro12Ala PPARgamma2 variant determines metabolism at the gene-environment interface. Cell Metab 9:88–98 [DOI] [PubMed] [Google Scholar]
- Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. (2010) Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SE, Heo HS, Kim DH, Kim MS, Kim CH, Lee J, Yoo MA, Yu BP, Leeuwenburgh C, Chung HY. (2010) Revealing system-level correlations between aging and calorie restriction using a mouse transcriptome. Age (Dordr) 32:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL. (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4:682–690 [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191–196 [DOI] [PubMed] [Google Scholar]
- Ingram DK, Roth GS. (2011) Glycolytic inhibition as a strategy for developing calorie restriction mimetics. Exp Gerontol 46:148–154 [DOI] [PubMed] [Google Scholar]
- Ingram DK, Zhu M, Mamczarz J, Zou S, Lane MA, Roth GS, deCabo R. (2006) Calorie restriction mimetics: an emerging research field. Aging Cell 5:97–108 [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. (2004) The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131:3897–3906 [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. (2004) Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14:885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan RM, Kronick RG. (2006) Marital status and longevity in the United States population. J Epidemiol Community Health 60:760–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasumovic MM, Brooks RC, Andrade MC. (2009) Body condition but not dietary restriction prolongs lifespan in a semelparous capital breeder. Biol Lett 5:636–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR. (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4:71–78 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366:461–464 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. (2010) The genetics of ageing. Nature 464:504–512 [DOI] [PubMed] [Google Scholar]
- Kickstein E, Krauss S, Thornhill P, Rutschow D, Zeller R, Sharkey J, Williamson R, Fuchs M, Köhler A, Glossmann H, et al. (2010) Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc Natl Acad Sci USA 107:21830–21835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JL, Peterson C. (2009) Healthspan, translation, and new outcomes for animal studies of aging. J Gerontol A Biol Sci Med Sci 64:209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M, Hirsh D. (1976) Non-ageing developmental variant of Caenorhabditis elegans. Nature 260:523–525 [DOI] [PubMed] [Google Scholar]
- Klass MR. (1977) Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev 6:413–429 [DOI] [PubMed] [Google Scholar]
- Knowles J, Gromo G. (2003) A guide to drug discovery: target selection in drug discovery. Nat Rev Drug Discov 2:63–69 [DOI] [PubMed] [Google Scholar]
- Kontush A, Guérin M, Chapman MJ. (2008) Spotlight on HDL-raising therapies: insights from the torcetrapib trials. Nat Clin Pract Cardiovasc Med 5:329–336 [DOI] [PubMed] [Google Scholar]
- Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. (2008) STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 36:D684–D688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313:1929–1935 [DOI] [PubMed] [Google Scholar]
- Lamb MJ. (1968) Temperature and lifespan in Drosophila. Nature 220:808–809 [DOI] [PubMed] [Google Scholar]
- Lavu S, Boss O, Elliott PJ, Lambert PD. (2008) Sirtuins–novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov 7:841–853 [DOI] [PubMed] [Google Scholar]
- Lebel M, Picard F, Ferland G, Gaudreau P. (2011) Drugs, nutrients, and phytoactive principles improving the health span of rodent models of human age-related diseases. J Gerontol A Biol Sci Med Sci http://dx.doi.org/10.1093/gerona/glr038 [DOI] [PubMed]
- Lee CK, Klopp RG, Weindruch R, Prolla TA. (1999) Gene expression profile of aging and its retardation by caloric restriction. Science 285:1390–1393 [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. (2000) Gene-expression profile of the ageing brain in mice. Nat Genet 25:294–297 [DOI] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. (2010) Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9:92–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Pletcher SD. (2007) Modulation of longevity by environmental sensing. Cell 131:1231–1234 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128 [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L. (2003) Demography of dietary restriction and death in Drosophila. Science 301:1731–1733 [DOI] [PubMed] [Google Scholar]
- Masoro EJ. (2005) Overview of caloric restriction and ageing. Mech Ageing Dev 126:913–922 [DOI] [PubMed] [Google Scholar]
- Mathers JC. (2006) Nutritional modulation of ageing: genomic and epigenetic approaches. Mech Ageing Dev 127:584–589 [DOI] [PubMed] [Google Scholar]
- Meric-Bernstam F, Gonzalez-Angulo AM. (2009) Targeting the mTOR signaling network for cancer therapy. J Clin Oncol 27:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry BJ, Kirk AJ, Goyns MH. (2008) Dietary lipoic acid supplementation can mimic or block the effect of dietary restriction on life span. Mech Ageing Dev 129:341–348 [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. (2011) Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 66:191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Hadfield MG. (1990) Developmental arrest during larval life and life-span extension in a marine mollusc. Science 248:356–358 [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Kersten S. (2003) Nutrigenomics: goals and strategies. Nat Rev Genet 4:315–322 [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283 [DOI] [PubMed] [Google Scholar]
- Narasimhan SD, Yen K, Tissenbaum HA. (2009) Converging pathways in lifespan regulation. Curr Biol 19:R657–R666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Lupu DS. (2011) Nutritional influence on epigenetics and effects on longevity. Curr Opin Clin Nutr Metab Care 14:35–40 [DOI] [PubMed] [Google Scholar]
- Olaharski AJ, Rine J, Marshall BL, Babiarz J, Zhang L, Verdin E, Smith MT. (2005) The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet 1:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshansky SJ, Perry D, Miller RA, Butler RN. (2006) In pursuit of the longevity dividend. Scientist 20:28–35 [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, et al. (2010) SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem 285:8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, et al. (2008) Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab 8:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan JP, Rose MR. (2005) Why dietary restriction substantially increases longevity in animal models but won't in humans. Ageing Res Rev 4:339–350 [DOI] [PubMed] [Google Scholar]
- Pollak MN, Schernhammer ES, Hankinson SE. (2004) Insulin-like growth factors and neoplasia. Nat Rev Cancer 4:505–518 [DOI] [PubMed] [Google Scholar]
- Rose MR, Long AD, Mueller LD, Rizza CL, Matsagas KC, Greer LF, Villeponteau B. (2010) Evolutionary nutrigenomics, in The Future of Aging (Fahy GM, West MD, Coles LS, Harris SB. eds) pp 357–366, Springer, Heidelberg [Google Scholar]
- Roth GS, Lane MA, Ingram DK, Mattison JA, Elahi D, Tobin JD, Muller D, Metter EJ. (2002) Biomarkers of caloric restriction may predict longevity in humans. Science 297:811. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. (2011) Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet 377:31–41 [DOI] [PubMed] [Google Scholar]
- Rueppell O, Bachelier C, Fondrk MK, Page RE., Jr (2007) Regulation of life history determines lifespan of worker honey bees (Apis mellifera L.). Exp Gerontol 42:1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AE, Wang C, Katz M, Derby CA, Barzilai N, Ozelius L, Lipton RB. (2010) Association of a functional polymorphism in the cholesteryl ester transfer protein (CETP) gene with memory decline and incidence of dementia. JAMA 303:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. (1994) Genetic associations with human longevity at the APOE and ACE loci. Nat Genet 6:29–32 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Friend SH, Shaywitz DA. (2009) A network view of disease and compound screening. Nat Rev Drug Discov 8:286–295 [DOI] [PubMed] [Google Scholar]
- Sedivy JM, Banumathy G, Adams PD. (2008) Aging by epigenetics–a consequence of chromatin damage? Exp Cell Res 314:1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkó GI, Gyurkó D, Veres DV, Nánási T, Csermely P. (2009) Network strategies to understand the aging process and help age-related drug design. Genome Med 1:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA. (2005) Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev 126:987–1002 [DOI] [PubMed] [Google Scholar]
- Sipp D. (2008) GSK moves on Sirtris. Nat Biotechnol 26:595. [DOI] [PubMed] [Google Scholar]
- Smith DL, Jr, Elam CF, Jr, Mattison JA, Lane MA, Roth GS, Ingram DK, Allison DB. (2010) Metformin supplementation and life span in Fischer-344 rats. J Gerontol A Biol Sci Med Sci 65:468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kenney RD, Gagne DJ, Frushour BP, Ladd W, Galonek HL, Israelian K, Song J, Razvadauskaite G, Lynch AV, et al. (2009) Small molecule activators of SIRT1 replicate signaling pathways triggered by calorie restriction in vivo. BMC Syst Biol 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ. (2010) Insulin-like growth factor-I regulation of immune function: a potential therapeutic target in autoimmune diseases? Pharmacol Rev 62:199–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soare A, Cangemi R, Omodei D, Holloszy JO, Fontana L. (2011) Long-term calorie restriction, but not endurance exercise, lowers core body temperature in humans. Aging (Albany NY) 3:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler SR. (2005) Rapid and reversible induction of the longevity, anticancer and genomic effects of caloric restriction. Mech Ageing Dev 126:960–966 [DOI] [PubMed] [Google Scholar]
- Spindler SR. (2006) Use of microarray biomarkers to identify longevity therapeutics. Aging Cell 5:39–50 [DOI] [PubMed] [Google Scholar]
- Spindler SR. (2010) Caloric restriction: from soup to nuts. Ageing Res Rev 9:324–353 [DOI] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. (2009) The TOR pathway comes of age. Biochim Biophys Acta 1790:1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp D. (2010) The Youth Pill: Scientists at the Brink of an Anti-Aging Revolution, Penguin Books, London [Google Scholar]
- Strong R, Miller RA, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, et al. (2008) Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell 7:641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. (2008) Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA 105:3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. (2005) Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev 19:1840–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Colditz GA, Kuntz KM. (2007) Forecasting the obesity epidemic in the aging U.S. population. Obesity (Silver Spring) 15:2855–2865 [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL. (1988) The Retardation of Aging and Disease by Dietary Restriction, C. C. Thomas, Springfield [Google Scholar]
- Weiss G. (2002) Europe wakes up to aging. Sci Aging Knowledge Environ 2002:ns10. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. (2008) FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA 105:13987–13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox DC, Willcox BJ, Todoriki H, Curb JD, Suzuki M. (2006) Caloric restriction and human longevity: what can we learn from the Okinawans? Biogerontology 7:173–177 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36:D901–D906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]