Abstract
In the human hematopoietic system, aging is associated with decreased bone marrow cellularity, decreased adaptive immune system function, and increased incidence of anemia and other hematological disorders and malignancies. Recent studies in mice suggest that changes within the hematopoietic stem cell (HSC) population during aging contribute significantly to the manifestation of these age-associated hematopoietic pathologies. Though the mouse HSC population has been shown to change both quantitatively and functionally with age, changes in the human HSC and progenitor cell populations during aging have been incompletely characterized. To elucidate the properties of an aged human hematopoietic system that may predispose to age-associated hematopoietic dysfunction, we evaluated immunophenotypic HSC and other hematopoietic progenitor populations from healthy, hematologically normal young and elderly human bone marrow samples. We found that aged immunophenotypic human HSC increase in frequency, are less quiescent, and exhibit myeloid-biased differentiation potential compared with young HSC. Gene expression profiling revealed that aged immunophenotypic human HSC transcriptionally up-regulate genes associated with cell cycle, myeloid lineage specification, and myeloid malignancies. These age-associated alterations in the frequency, developmental potential, and gene expression profile of human HSC are similar to those changes observed in mouse HSC, suggesting that hematopoietic aging is an evolutionarily conserved process.
Keywords: lineage potential, hematopoiesis, microarray, leukemia
Hematopoiesis is initiated by hematopoietic stem cells (HSC) that can self-renew and progressively differentiate into a hierarchy of committed progenitors that ultimately give rise to mature blood cells (1). Though the mechanisms of aging in the hematopoietic system are comprised of a combination of cell-intrinsic and -extrinsic causes that ultimately alter the generation and function of mature blood lineages, there is increasing evidence that implicates alterations within the HSC population as one of the mechanisms behind hematopoietic aging. We and others have found that as mice age, their HSC numbers increase, but competitive repopulation ability is reduced, suggesting a decrease in mouse HSC function with age (2–7). Additionally, elderly mouse HSC exhibit a marked decrease in lymphopoiesis and increase in myelopoiesis (2, 6). In the mouse, we and others have identified distinct clonal subtypes of HSC that differentially respond to external cytokine stimuli and exhibit lineage bias upon transfer to irradiated hosts (8–13). The majority of HSC from elderly mice are myeloid biased, whereas most HSC from young mice are balanced in lymphopoiesis and myelopoiesis (8–13).
In humans, age-associated hematopoietic changes include decreased bone marrow cellularity (14), declines in lymphopoiesis (7, 15, 16), red cell abnormalities such as anemia (17), and increases in the incidence of myelodysplastic syndromes, myeloproliferative disorders, and myeloid malignancies (18). In human bone marrow, the HSC population is highly enriched within the Lin−CD34+CD38−CD90+CD45RA− population (19–24). Previous studies addressing the age-associated changes in human HSC have relied on indirect evaluations of stem and progenitor populations and have therefore been less quantitative than studies of mouse HSC aging. The proliferative capacity of human HSC appears to decline from fetal liver, to cord blood, to adult bone marrow (25), and some indirect evidence suggests reductions in stem cell reserves with age (26, 27), particularly in the context of anemia (28). Clinically, in the setting of bone marrow transplantation, increasing donor age correlates with increasing transplant-related mortality (29). Few studies have evaluated the frequency of bone marrow progenitor populations directly: one study found that the number of CD34+ bone marrow cells decreases with age (30), whereas another study found that the frequency of CD34+CD38− bone marrow cells increases with age (31). However, CD34+ and CD34+CD38− bone marrow populations are both heterogeneous, only a small fraction of which are HSC.
Therefore, to more precisely identify changes within the stem cell compartments that contribute to human hematopoietic aging, we evaluated the putative human HSC compartment (immunophenotypically defined as Lin−CD34+CD38−CD90+CD45RA−) from healthy, young (20–35 y of age) human bone marrow samples (hereafter referred to as young HSC) and healthy elderly (65+ y of age) human bone marrow samples (hereafter referred to as elderly HSC). We characterized, by flow cytometry, the frequency, distribution, and cell-cycle profile of immunophenotypic HSC and other hematopoietic progenitor populations, and we found that aged immunophenotypic human HSC are increased in frequency and are less quiescent than young HSC. We sorted immunophenotypic human HSC from young and elderly bone marrow samples and tested their ability to proliferate and differentiate in vitro and in vivo. Both young and elderly immunophenotypic human HSC were able to generate lymphoid and myeloid progeny in culture, but elderly HSC exhibited significantly myeloid-biased differentiation potential compared with young HSC under equal conditions. Elderly immunophenotypic human HSC xenotransplanted into immunodeficient mice did not engraft or generate lymphoid progeny as efficiently as young human HSC. We also performed gene expression profiling of sorted young and elderly immunophenotypic human HSC. Our results suggest that a number of mechanisms behind the phenotype of hematopoietic aging are transcriptionally regulated at the level of diverse subsets of HSC.
Results
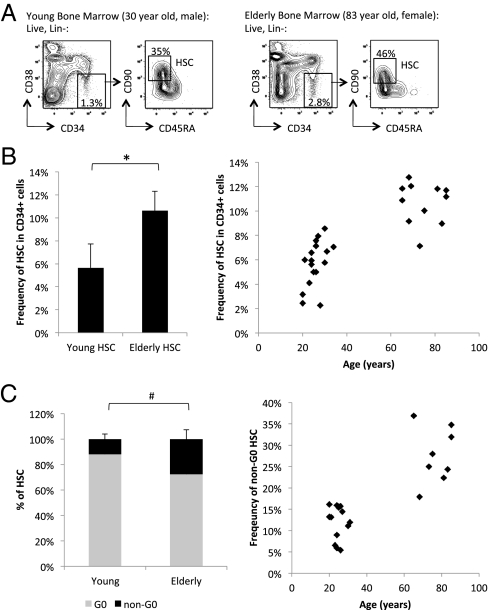
To directly characterize human HSC during aging, we first quantified by flow cytometry the frequency of immunophenotypic human HSC (Lin−CD34+CD38−CD90+CD45RA−) in hematologically normal young and elderly bone marrow samples (Fig. 1A). We found that elderly bone marrow contain increased frequency of HSC within the CD34+ population (Fig. 1B) as well as within the bone marrow mononuclear fraction (SI Appendix, Fig. S1), consistent with a recent finding in a small number of samples showing an age-associated increase in frequency of HSC (32). Quantification of the exact number of HSC was not possible because of the inherent variability in the technique of human bone marrow aspiration. We also observed, on average, an age-associated increase, albeit not statistically significant, in the frequency of multipotent progenitors (MPP; Lin−CD34+CD38−CD90−CD45RA−) (33) (SI Appendix, Fig. S2). We further investigated the age-associated expansion of human HSC by evaluating the cell-cycle status of young and elderly HSC using RNA (Pyronin Y) and DNA (Hoechst 33342) dyes (Fig. 1C and SI Appendix, Fig. S3). We found that a greater percentage of young HSC are Pyronin Y low, and likely in the quiescent G0 phase of the cell cycle, compared with elderly HSC, of which there is a greater percentage that are Pyronin Y high, and likely in nonquiescent G1, S, or G2 phases (Fig. 1C and SI Appendix, Fig. S3 A and B).
Fig 1.
Increased frequency of HSC in normal elderly bone marrow compared to young. (A) Gating strategy and flow cytometric profile of HSC (Lin−, CD34+, CD38−, CD90+, CD45RA−) in representative hematopoietically normal young (Left) and elderly (Right) bone marrow samples. The left panels for each sample are gated on lineage negative (Lin−) live cells, and the right panels are gated on Lin−CD34+CD38− live cells. (B) Summary of HSC as frequency of total Lin−CD34+ population from multiple young (n = 13) and elderly (n = 11) bone marrow samples. *P < 10−7. (C) Summary of quiescent G0 (Hoechst 33342low, Pyronin Ylow, correlating with 2N DNA and low levels of RNA) and non-G0 (Pyronin Yhigh, correlating with 2N to 4N DNA and higher levels of RNA) HSC frequency out of total HSC from multiple young (n = 13) and elderly (n = 8). # P < 0.013. Error bars represent standard deviation.
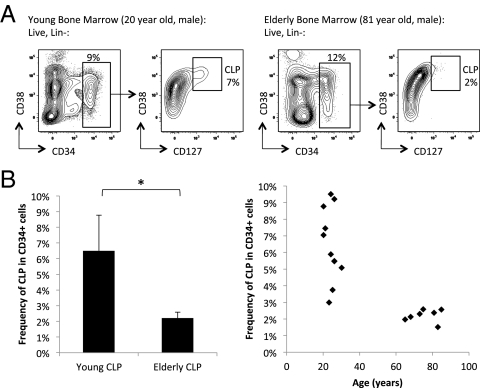
Because HSC differentiate into mature blood cells via a succession of committed progenitors, we next examined these populations in young and elderly bone marrow samples to determine whether age-associated increases in human HSC frequency corresponded to changes in frequency of human myeloid and lymphoid progenitors. We did not detect any differences in the frequency of the immunophenotypic common myeloid progenitors (CMP; Lin−CD34+CD38+CD123+CD45RA−), granulocyte-macrophage progenitors (GMP; Lin−CD34+CD38+CD123+CD45RA+), and megakaryocyte-erythrocyte progenitors (MEP; Lin−CD34+CD38+CD123−CD45RA−) (34) from young and elderly bone marrow (SI Appendix, Fig. S4 A–C). However, elderly bone marrow did exhibit a relative decrease in the frequency of common lymphoid progenitors (CLP; Lin−CD34+CD38+CD127+; Fig. 2 and SI Appendix, Fig. S5). Therefore, whereas the myeloid progenitor populations appear to be maintained with age, lymphoid progenitors decline.
Fig 2.
Decreased frequency of CLP in normal elderly bone marrow compared to young. (A) Gating strategy and flow cytometric profile of CLP (Lin−, CD34+, CD127+) in representative hematopoietically normal young (Left) and elderly (Right) bone marrow samples. The left panels for each sample are gated on lineage negative (Lin−) live cells, and the right panels are gated on Lin−CD34+ live cells. (B) Summary of CLP as frequency of total Lin−CD34+ population from multiple normal young (n = 10) and elderly (n = 7) bone marrow samples. *P < 2.0 × 10−4.
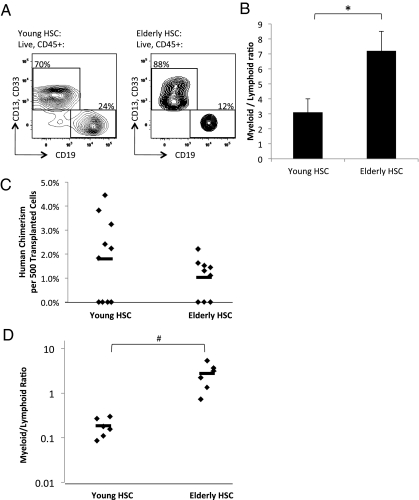
To determine the changes in the developmental potential of aged human HSC, we analyzed the ability of young and elderly HSC to generate myeloid and lymphoid progeny in vitro, and engraft and differentiate in vivo. We used FACS to isolate young and elderly immunophenotypic human HSC. FACS-purified HSC were cocultured with AC6.2.1 cells, which can act as a surrogate for normal bone marrow stroma in supporting both myeloid and B-lymphoid differentiation of human HSC (24). We determined by flow cytometry the percentage of B cells (CD19+) and myeloid cells (CD33+ and/or CD13+) generated by young and elderly human HSC (Fig. 3A). Young HSC, compared with elderly HSC, cultured for 14 d on AC6.2.1 cells yielded greater numbers, albeit not statistically significant, of human CD45+ cells per HSC, suggesting greater plating efficiency (SI Appendix, Fig. S6A). Experiments using single HSC from either young or elderly bone marrow samples plated on AC6.2.1 did not yield quantifiable colonies at any appreciable frequency. Notably, we found that elderly HSC exhibit significantly diminished capacity to give rise to lymphoid B lineage cells, resulting in an increased proportion of myeloid cells being generated per HSC cultured on AC6.2.1, and an increased myeloid-to-lymphoid ratio (Fig. 3B). The decreased efficiency in the generation of lymphoid B lineage cells by elderly HSC may be a mechanism behind the observed myeloid-biased behavior of elderly HSC. Additionally, this reduction in lymphopoiesis potential may account in part for the decreased plating efficiency of elderly HSC, because the number of myeloid cells generated in AC6.2.1 culture per HSC is similar between young and elderly HSC (SI Appendix, Fig. S6A). We did not observe significant differences in the ability of young and elderly HSC to form myeloid and erythroid colonies in methylcellulose culture (SI Appendix, Fig. S6B), further suggesting that myeloerythroid differentiation capacity is preserved in elderly HSC.
Fig 3.
Diminished lymphoid versus myeloid differentiation capacity of HSC from normal elderly bone marrow compared to young bone marrow. (A) Gating strategy and flow cytometric profile of representative progeny derived from FACS-purifed HSC from hematologically normal young (Left) and elderly (Right) bone marrow samples cultured for 14 days on AC6.2.1 stromal cell line. The panels for each sample are gated on human CD45+ live cells. (B) Summary of CD13+ and/or CD33+ myeloid versus CD19+ B-lymphoid distribution generated from 50−200 HSC from multiple normal young (n = 8) and elderly (n = 6) bone marrow samples cocultured with AC6.2.1 stromal cell line. *P < 0.011. (C) Summary of bone marrow engraftment as measured by percent human chimerism per 500 transplanted HSC from unique young (n = 10) and elderly (n = 9) bone marrow samples. Each diamond represents an individual mouse transplanted with HSC from unique human bone marrow samples, and the bar indicates the average. On average, mice transplanted with young human HSC developed approximately twofold more human chimerism than mice transplanted with elderly human HSC, but this difference was not statistically significant due to the numbers of mice transplanted with young (n = 4) or elderly (n = 3) human HSC, which did not have any detectable chimerism. However, the approximately twofold difference in percent human chimerism between mice successfully engrafted with either young (n = 6) or elderly (n = 6) human HSC is statistically significant (P < 0.02). (D) Summary of human CD13+ and/or CD33+ myeloid versus CD19+ B-lymphoid distribution from bone marrow of mice successfully engrafted with young (n = 6) or elderly (n = 6) human HSC. Each diamond represents an individual mouse transplanted with HSC from a unique individual and the bar indicates the average. On average, the myeloid-to-lymphoid ratio was increased in the mice transplanted with elderly HSC by 14-fold. #P < 0.004. Error bars represent standard deviation.
We also isolated immunophenotypic human HSC from 10 young and nine elderly bone marrow samples, and transplanted each of them i.v. into an immunodeficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) pup. At 16 wk posttransplant, we found that six of 10 mice (60%) transplanted with young HSC, and six of nine mice (66%) transplanted with elderly HSC, contained human CD45+ cells in the bone marrow. Mice successfully engrafted with elderly human HSC, compared with young human HSC, had lower human chimerism per HSC transplanted (Fig. 3C). Additionally, bone marrow of mice engrafted with elderly HSC, compared with young HSC, contained a higher myeloid-to-lymphoid progeny (CD33+ and/or CD13+ to CD19+) ratio (Fig. 3D), suggesting that xenotransplanted elderly HSC, compared with young HSC, generate myeloid progeny more efficiently than lymphoid progeny. The data may reflect an inherent myeloid bias within the elderly compared with young HSC population, and the process of lineage specification in human hematopoiesis likely begins in diverse stem cells. Spleens from engrafted mice contained human CD45+CD3+ T cells, but their frequencies were too low to identify any significant differences, and bone marrow from engrafted mice contained human glycophorin A+ erythroid cells and human CD41/61+ platelets (SI Appendix, Fig. S7).
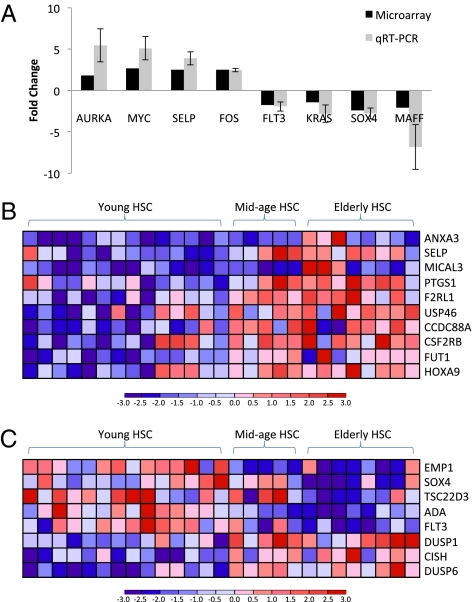
To identify changes in gene expression that may underlie the differences in frequency and developmental potential of young and elderly HSC, we used FACS to sort HSC from 14 young and eight elderly bone marrow samples for transcriptional profiling (>95% purity; SI Appendix, Fig. S8). We also obtained gene expression data for five midage (42–61 y of age) bone marrow samples. Using the significance analysis of microarray (SAM) algorithm (35), we identified 285 age-regulated genes in human HSC [false discovery rate (FDR) < 30%; SI Appendix, Table S1]. We confirmed the changes in expression of a subset of genes in sorted HSC from four young and four elderly independent samples using quantitative RT-PCR (Fig. 4A). Using Ingenuity Pathways Analysis (IPA) software, we found that this set of age-regulated genes significantly enriched for transcriptional networks and biological functions associated with cell-cycle, cell growth and proliferation, and hematopoietic development (SI Appendix, Fig. S9 A–C). We also found that elderly HSC up-regulate genes associated with signaling pathways, including ERK/MAPK and GM-CSF signaling, that are involved in the expansion and proliferation of hematopoietic stem and progenitor compartments (SI Appendix, Fig. S9D). Interestingly, biological functions and pathways associated with DNA repair and cell death are concurrently enriched (SI Appendix, Fig. S9C). Additionally, we found elderly HSC up-regulate several genes that have been implicated in human hematopoietic myeloid malignancies, including Aurka, Fos, Hoxa9, Myc, and Trim13 (Fig.4A and SI Appendix, Table S2). In contrast, young HSC express approximately twofold-higher levels of Maff (Fig. 4A and SI Appendix, Table S2), which is involved primarily in translocations found in lymphoid leukemias more commonly seen in young patients. We also found, using IPA, that the age-regulated genes in HSC are significantly enriched for those that are involved in acute myeloid leukemia signaling (SI Appendix, Fig. S9D). In addition, we found that elderly HSC primarily up-regulate genes that specify myeloerythroid fate and function (Fig. 4B), and down-regulate genes associated with lymphopoiesis (Fig. 4C). These findings that the age-associated increased expression of myeloid-specification gene SELP and decreased expression of lymphoid-specification genes FLT3 and SOX4 in human HSC have also been observed in mouse HSC (2, 6, 36). The increase in transcription of myeloid lineage-specific genes in the elderly human HSC population correlates with our in vitro and in vivo results, suggesting that the lineage biases of aged immunophenotypic human HSC is in part transcriptionally defined.
Fig 4.
Gene expression profiling of HSC from normal elderly and young bone marrow reveals transcriptional differences reflecting myeloid lineage-bias of elderly HSC. (A) Validation of microarray data by quantitative RT-PCR: average fold-change in the expression of selected genes as determined by microarray analysis (14 young and 8 elderly HSC samples) and quantitative RT-PCR (4 young and 4 elderly HSC samples; independent of samples analyzed by microarrays). Error bars represent standard deviation. Heat maps reflecting expression levels of (B) myeloid-specific and (C) lymphoid-specific age-regulated genes that are significantly differentially expressed between young and elderly HSC.
Discussion
In this study, we have characterized the age-associated effects on human hematopoiesis at the level of the stem cell. In aged individuals, we have found that immunophenotypic human HSC, similar to mouse HSC, are increased in frequency. Because the process of bone marrow aspiration often allows for the inclusion of a small amount of peripheral blood in the acquired sample, and because the CD34+ population is found primarily in the bone marrow, we calculated the frequency of immunophenotypic human HSC, as a percentage of CD34+ cells, as the best proxy for the frequency of HSC in bone marrow. The increased variability in the frequency of immunophenotypic human HSC as a percentage of total bone marrow mononuclear cells is potentially due in part to differing amounts of peripheral blood cells within the acquired sample.
In addition, we have found that elderly immunophenotypic human HSC have poorer engraftment efficiency and are relatively more myeloid-biased than young HSC. The relative decrease in frequency of CLP in human bone marrow combined with the relative loss of B-cell potential and corresponding increase of myeloid potential of immunophenotypic HSC both in vitro and in vivo suggest that lineage bias of the HSC population changes with age. We were unable to measure significant differences in other lineages due to low chimerism; more robust in vitro and in vivo models for quantitative assessment of T-cell, erythroid, and megakaryocytic potentials of human HSCs will be needed to more thoroughly assess the lineage potential of human HSCs and confirm our hypothesis. Efficient functional assays of the engraftment and lineage potential of single human HSC would be the most ideal method to characterize aging human HSC, because the immunophenotypic human HSC population we have analyzed is likely heterogeneous, containing distinct subsets of HSC and potentially non-HSCs. Nevertheless, by examining a highly purified population that is significantly enriched for bone marrow-derived human HSC, we have begun to identify subtle but important age-associated properties of the HSC population, in terms of its frequency and developmental potential, which would have been missed if one looked at more heterogeneous hematopoietic progenitor pools.
Our data indicate that human hematopoietic aging, similar to mouse hematopoietic aging, is associated with changes in human HSC gene expression that reflect the quantitative and functional alterations seen in elderly HSC. The increase in elderly HSC frequency may be partly due to increased frequency of HSC in active cell-cycle phases. Elderly HSC up-regulate genes associated with cell-cycle, cell growth and proliferation, and hematopoietic development, corresponding well with the increased proportion of elderly HSC that we observed to likely be in more active cell-cycle phases. However, this increase in HSC frequency does not necessarily translate into improved HSC function, supported by our finding that elderly HSC do not have as high engraftment efficiency in our xenotransplantation model.
We and others have proposed that stem cells, due to their inherent self-renewing potential and longevity, are ideal reservoirs for DNA damage over the life of the organism, and can therefore accumulate the multiple genetic/epigenetic events required for a normal cell to become a cancer cell (37–40). Our gene expression data shows that elderly HSC up-regulate genes associated with DNA repair and cell death, possibly indicating that elderly HSC have activated cell-cycle checkpoints in response to DNA damage. Even though more elderly HSC may be in the process of proliferating, we speculate that they may also be arrested at cell-cycle checkpoints, perhaps due to the presence of DNA damage, for which there is evidence in elderly mouse HSC (40, 41). This, in turn, may lead to increased recruitment of quiescent HSC into the cell cycle to maintain adequate levels of functional HSC at a given time.
In this study, we also showed that the decline in lymphopoiesis with age can be traced to the behavior of the stem cell, and the transcriptional up-regulation of genes that specify myeloid-lineage differentiation likely underlies the myeloid skewing observed in elderly HSC. This change in developmental potential observed within the HSC population during aging could be due to (i) all HSC changing from balanced myeloid-lymphoid potential to myeloid-biased with age or (ii) intrinsically myeloid-biased HSC outcompeting balanced-potential HSC during aging. In mice, small numbers of myeloid-biased HSC can be found among balanced-potential HSC in young 2-mo-old mice (8, 11, 12), suggesting that the young mouse HSC pool may contain lineage-biased clones that compete for niches and expansion signals. The young human HSC population may similarly contain clones of lineage-biased cells, the selection of which, during aging, results in the predominance of myeloid-biased HSC in the elderly. This myeloid-biased skewing of elderly HSC lineage potential that we have observed may be one mechanism behind the increased frequency of myeloid disorders and malignancies in elderly people. Another factor that may contribute to the increased incidence of myeloid malignancies with age is that elderly human HSC up-regulate genes implicated in myeloid malignancies, such as Aurka, Fos, Hoxa9, Myc, and Trim13. Although these genes likely play roles in the normal maintenance and functions of HSC, we speculate that the increased transcription of these genes may potentially facilitate translocations to these loci and thereby malignant transformation in elderly bone marrow. HSC in elderly bone marrow, having accumulated a lifetime of genomic insults and being transcriptionally as well as functionally myeloid biased, may also be more likely to contribute to the development myeloid, as opposed to lymphoid, diseases.
Changes within the human HSC population during aging could also be influenced by alterations in the interactions between HSC and their aging niches. In the mouse, there is evidence to suggest that the young and elderly mouse HSC populations respond differently to cytokines such as IL-7 and TGF-β (9, 12). In young and elderly human HSC, we observed differential expression of cytokine receptors and pathways, including enrichment of the ERK/MAPK signaling and GM-CSF signaling pathways in elderly HSC, which may be physiological responses by different subtypes of lineage-biased human HSC to the aging hematopoietic environment. In particular, the increase in elderly human HSC frequency and their myeloid bias may reflect the hematopoietic system's attempt to maintain homeostasis, ensuring adequate functional mature progeny. We speculate that the inability of elderly human HSC to maintain homeostasis contributes to age-associated cytopenias, including anemias and dysplasias.
Notably, the results from our functional analysis and gene expression profiling of young and elderly human HSC significantly parallel the data we and others have generated on young and elderly mouse HSC (2, 6, 36). Both aged human and mouse HSC are increased in frequency, and they are transcriptionally and functionally myeloid-biased in their differentiation potential. Not surprisingly, the set of differentially expressed genes between young and elderly human HSC shares overlap with the set of differentially expressed genes between young and elderly mouse HSC. These similarities strongly suggest that the biological processes that cause the hematopoietic aging phenotype are similar between human and mouse, and that mouse hematopoietic aging is a reasonable model of human hematopoietic aging.
Our data directly implicate the human HSC and their age-associated alterations in the frequency, function, and gene expression as vital contributors to the aging in the human hematopoietic system. Further studies will address the pathways involved in the aging of the healthy human HSC population and also characterize HSC from age-associated hematopoietic diseases to better understand the processes involved in changing healthy elderly HSC into diseased elderly HSC.
Methods
Human Samples.
Normal young human bone marrow mononuclear cells were purchased from AllCells, Inc. Normal young and elderly human bone marrow samples were obtained from hematologically normal donors at the Stanford Medical Center with informed consent, according to an institutional review board (IRB)-approved protocol (Stanford IRB nos. 5112 and 10831). We analyzed a total of 15 elderly (ages 65–85), 28 young (ages 20–31), and 5 midage (ages 42–61) normal bone marrow samples. Peripheral blood complete blood count values, from samples for which data are available, can be found in SI Appendix, Table S3. Mononuclear cells from bone marrow samples were prepared using Ficoll-Paque PLUS (GE Healthcare) and either analyzed/sorted fresh or cryopreserved in 90% FBS, 10% DMSO in liquid nitrogen. There is inherent variability in the technique of human bone marrow aspiration, during which peripheral blood may be aspirated along with bone marrow; therefore, the mononuclear cell fraction obtained from each aspiration may contain a small but unpredictable percentage of peripheral blood mononuclear cells in addition to bone marrow mononuclear cells. Estimates of bone marrow cellularity were determined by examination of bone marrow core biopsies. Human CD34-positive cells were enriched using magnetic beads (Miltenyi Biotec; Stemcell Technologies).
Flow Cytometry Analysis and Cell Sorting.
The following panel of antibodies (Caltag/Invitrogen and BD Biosciences) was used for analysis and sorting of human hematopoietic stem and progenitor populations: PE-Cy5–conjugated anti-human lineage markers (anti-CD2, RPA-2.10; anti-CD3, S4.1; anti-CD4, S3.5; anti-CD7, CD7-6B7; anti-CD8, 3B5; anti-CD10, 5-1B4; anti-CD11b, ICRF44; anti-CD14, TU.K.4; anti-CD19, SJ25-C1; anti-CD20, 13.6E12; anti-CD56, B159; anti-GPA, GA-R2), PB-conjugated anti-CD45RA, MEM56; PE-Cy7–conjugated anti-CD38, HIT2; FITC-conjugated or Alexa Fluor 700-conjugated anti-CD90 (Thy-1), 5E10; PE-conjugated or FITC-conjugated anti-CD123; PE-conjugated anti-CD127, hIL-7R-M21.
The following panel of antibodies was used for analysis of differentiated human hematopoietic populations and human engraftment/chimerism: PB-conjugated CD45, HI30; APC-conjugated anti-CD34, 8G12; Alexa Fluor 750-conjugated CD3, S4.1; Alexa Fluor 700-conjugated CD19, SJ25-C1; PE-conjugated CD13, TK1; PE-conjugated CD33, P67.6; PE-Cy5–conjugated GPA, GA-R2; FITC-conjugated CD41a, HIP8. The following panel of antibodies (eBiosciences) was used to identify mouse leukocytes and red blood cells, respectively: Alexa Fluor 488- or PE-Cy7–conjugated CD45.1, A20.1.7; PE-Cy5– or PE-Cy7–conjugated Ter119. Single-cell suspensions were prepared using standard methods from bone marrow of transplanted mice. Red blood cells were lysed using ACK buffer.
For analyses and sorting, except otherwise noted below, cells were stained with the appropriate antibody combinations for 30–60 min on ice, and dead cells were excluded by propidium iodide staining. Gating strategy used to separate CD90+ from CD90−, CD45RA+ from CD45RA−, CD123+ from CD123−, and CD127+ from CD127− populations was fluorescence minus one.
For analysis of the HSC cell cycle, cells were resuspended at a concentration of 106 cells/mL of HBSS medium (10% FCS, 20 mM Hepes, 1 g/L glucose, and 50 μg/mL verapamil) and incubated for 30 min at 37 °C with 20 μg/mL Hoechst 33342 (Invitrogen); 1 μg/mL Pyronin Y (Sigma-Aldrich) was added and cells were incubated for another 10 min at 37 °C, stained with the appropriate antibody combination for identification of HSC for 30 min on ice, washed, and analyzed. Gating strategy used to identify quiescent G0 (Hoechst 33342low, Pyronin Ylow, correlating with 2N DNA and low levels of RNA) and non-G0 (Pyronin Yhigh, correlating with 2N–4N DNA and higher levels of RNA) populations was first delineated based on Lin−CD34+ population from the same samples (SI Appendix, Fig. S3B), and identical gates were used to identify G0 and non-G0 subsets within the HSC population (SI Appendix, Fig. S3A).
Cells were analyzed and sorted using a FACSAria II cytometer (BD Biosciences). A total of 50–300 HSC were sorted for in vitro assays, 500–2,000 HSC were sorted for in vivo assays, and ∼1,000–10,000 HSC were sorted for RNA purification. Analysis of flow cytometry data were performed using FlowJo Software (Treestar).
In Vitro Assays: Methylcellulose and AC6.2.1 Coculture.
Methylcellulose colony formation was assayed by sorting 300 cells into individual wells of a six-well plate, each containing 3 mL of complete methylcellulose (Methocult GF+ H4435; Stemcell Technologies). Plates were incubated for 12–14 d at 37 °C, and colonies then scored based on morphology.
To analyze the myeloid and B-cell lymphoid potential of HSC (20, 21, 24), 50–200 HSC were sorted into individual wells of a 96-well plate containing semiconfluent AC6.2.1 stromal cells (42) in Iscove's modified Dulbecco's medium, GlutaMAX, penicillin/streptomycin, nonessential amino acids, and sodium pyruvate. Plates were incubated for 14 d at 37 °C in 5% oxygen, and cells then analyzed by flow cytometry.
Mouse Transplantation.
NSG mice obtained originally from the Jackson Laboratory were bred in a specific pathogen-free environment according to a protocol approved by the Stanford Administrative Panel on Laboratory Animal Care. P0–P2 newborn pups were preconditioned with 100 rads of γ-irradiation up to 24 h before transplantation (33, 43). A total of 500–2,000 FACS-purified HSC were resuspended in PBS containing 2% FCS and transplanted i.v. via the anterior facial vein using a 30- or 31-gauge needle.
Statistical Analysis.
Student t test was performed using Excel (Microsoft).
RNA Purification, Amplification, and Microarray Analysis.
Total RNA was extracted using TRIzol (Invitrogen) or Ambion RNA Isolation Kit (Applied Biosystems by Life Technologies) according to the manufacturer's protocols and treated with DNase I (Qiagen). All RNA samples were quantified with the RiboGreen RNA Quantitation Kit (Molecular Probes), subjected to reverse transcription, two consecutive rounds of linear amplification, and production and fragmentation of biotinylated cRNA (Affymetrix). Fifteen micrograms of cRNA from each sample was hybridized to Affymetrix HG U133 Plus 2.0 microarrays. Hybridization and scanning were performed according to the manufacturer's instructions (Affymetrix). Raw data from all samples are available from the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32719).
Raw data were normalized using the standard robust multichip average algorithm, together with 21,701 Affymetrix U133 Plus 2.0 human microarrays downloaded from GEO, according to methods previously described (44). Probe sets were identified to be present, and their associated transcripts expressed in elderly or young HSC if the mean of the normalized values of the probe sets of either group was greater than the threshold value calculated using the StepMiner algorithm, as previously described (45). The normalized data from probe sets that were determined to be present were then used in SAM (35) and Ingenuity Pathways Analysis (Ingenuity Systems). The categorization of genes into lymphoid and myeloid groupings was done based on evaluation of relevant literature as well as available gene expression profiling data of human and mouse lymphoid and myeloid progenitors and their differentiated progeny. Heat maps were generated using HeatMapViewer (GenePattern; Broad Institute).
Supplementary Material
Acknowledgments
The authors thank Renee Mehra for administrative and logistical support; Ravi Majeti, Christopher Park, Matthew Inlay, and Charles Chan for helpful advice and discussions; Theresa Storm and Libuse Jerabek for excellent laboratory management; Ken Cheung for statistical advice; and the Stanford Functional Genomics Facility for array processing services. Support for this work was provided by the Stanford Medical Scientist Training Program (W.W.P.), a grant from the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation (to D.S.), and National Institute of Aging Grant R01AG029124 (to S.L.S. and I.L.W).
Footnotes
Conflict of interest statement: W.J.M. is on the board of and owns stock and options in Stemedica Cell Technologies, Inc. I.L.W. is on the board of StemCells, Inc., and owns stock in Amgen, Inc.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32719).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116110108/-/DCSupplemental.
References
- 1.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers SM, et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim M, Moon HB, Spangrude GJ. Major age-related changes of mouse hematopoietic stem/progenitor cells. Ann N Y Acad Sci. 2003;996:195–208. doi: 10.1111/j.1749-6632.2003.tb03247.x. [DOI] [PubMed] [Google Scholar]
- 4.Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- 5.Pearce DJ, Anjos-Afonso F, Ridler CM, Eddaoudi A, Bonnet D. Age-dependent increase in side population distribution within hematopoiesis: Implications for our understanding of the mechanism of aging. Stem Cells. 2007;25:828–835. doi: 10.1634/stemcells.2006-0405. [DOI] [PubMed] [Google Scholar]
- 6.Rossi DJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: Aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111:5553–5561. doi: 10.1182/blood-2007-11-123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103:4111–4118. doi: 10.1182/blood-2003-10-3448. [DOI] [PubMed] [Google Scholar]
- 10.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Beerman I, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci USA. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogawa T, Kitagawa M, Hirokawa K. Age-related changes of human bone marrow: A histometric estimation of proliferative cells, apoptotic cells, T cells, B cells and macrophages. Mech Ageing Dev. 2000;117(1–3):57–68. doi: 10.1016/s0047-6374(00)00137-8. [DOI] [PubMed] [Google Scholar]
- 15.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5(2):133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 16.Miller JP, Allman D. The decline in B lymphopoiesis in aged mice reflects loss of very early B-lineage precursors. J Immunol. 2003;171:2326–2330. doi: 10.4049/jimmunol.171.5.2326. [DOI] [PubMed] [Google Scholar]
- 17.Guralnik JM, Eisenstaedt RS, Ferrucci L, Klein HG, Woodman RC. Prevalence of anemia in persons 65 years and older in the United States: Evidence for a high rate of unexplained anemia. Blood. 2004;104:2263–2268. doi: 10.1182/blood-2004-05-1812. [DOI] [PubMed] [Google Scholar]
- 18.Lichtman MA, Rowe JM. The relationship of patient age to the pathobiology of the clonal myeloid diseases. Semin Oncol. 2004;31(2):185–197. doi: 10.1053/j.seminoncol.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci USA. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Péault B, Weissman IL, Buckle AM, Tsukamoto A, Baum C. Thy-1-expressing CD34+ human cells express multiple hematopoietic potentialities in vitro and in SCID-hu mice. Nouv Rev Fr Hematol. 1993;35(1):91–93. [PubMed] [Google Scholar]
- 22.McCune JM, et al. The SCID-hu mouse: Murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- 23.Negrin RS, et al. Transplantation of highly purified CD34+Thy-1+ hematopoietic stem cells in patients with metastatic breast cancer. Biol Blood Marrow Transplant. 2000;6:262–271. doi: 10.1016/s1083-8791(00)70008-5. [DOI] [PubMed] [Google Scholar]
- 24.Murray L, et al. Analysis of human hematopoietic stem cell populations. Blood Cells. 1994;20:364–369. discussion 369–370. [PubMed] [Google Scholar]
- 25.Lansdorp PM, Dragowska W, Mayani H. Ontogeny-related changes in proliferative potential of human hematopoietic cells. J Exp Med. 1993;178:787–791. doi: 10.1084/jem.178.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gale RE, Fielding AK, Harrison CN, Linch DC. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol. 1997;98:512–519. doi: 10.1046/j.1365-2141.1997.2573078.x. [DOI] [PubMed] [Google Scholar]
- 27.Marley SB, et al. Evidence for a continuous decline in haemopoietic cell function from birth: Application to evaluating bone marrow failure in children. Br J Haematol. 1999;106(1):162–166. doi: 10.1046/j.1365-2141.1999.01477.x. [DOI] [PubMed] [Google Scholar]
- 28.Lipschitz DA, Mitchell CO, Thompson C. The anemia of senescence. Am J Hematol. 1981;11(1):47–54. doi: 10.1002/ajh.2830110106. [DOI] [PubMed] [Google Scholar]
- 29.Kollman C, et al. Donor characteristics as risk factors in recipients after transplantation of bone marrow from unrelated donors: The effect of donor age. Blood. 2001;98:2043–2051. doi: 10.1182/blood.v98.7.2043. [DOI] [PubMed] [Google Scholar]
- 30.Waterstrat A, et al. In: Telomeres and Telomerase in Ageing, Disease, and Cancer. Rudolph KL, editor. Berlin: Springer; 2008. pp. 111–140. [Google Scholar]
- 31.Taraldsrud E, et al. Age and stress related phenotypical changes in bone marrow CD34+ cells. Scand J Clin Lab Invest. 2009;69(1):79–84. doi: 10.1080/00365510802419447. [DOI] [PubMed] [Google Scholar]
- 32.Beerman I, Maloney WJ, Weissmann IL, Rossi DJ. Stem cells and the aging hematopoietic system. Curr Opin Immunol. 2010;22:500–506. doi: 10.1016/j.coi.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci USA. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers SM, et al. Hematopoietic fingerprints: An expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nalapareddy K, Jiang H, Guachalla Gutierrez LM, Rudolph KL. Determining the influence of telomere dysfunction and DNA damage on stem and progenitor cell aging: What markers can we use? Exp Gerontol. 2008;43:998–1004. doi: 10.1016/j.exger.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Weissman IL. Normal and neoplastic stem cells. Novartis Found Symp. 2005;265:35–50. discussion 50–54, 92–97. [PubMed] [Google Scholar]
- 39.Lieber MR, Karanjawala ZE. Ageing, repetitive genomes and DNA damage. Nat Rev Mol Cell Biol. 2004;5(1):69–75. doi: 10.1038/nrm1281. [DOI] [PubMed] [Google Scholar]
- 40.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 41.Rossi DJ, et al. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6:2371–2376. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- 42.Whitlock CA, Tidmarsh GF, Muller-Sieburg C, Weissman IL. Bone marrow stromal cell lines with lymphopoietic activity express high levels of a pre-B neoplasia-associated molecule. Cell. 1987;48:1009–1021. doi: 10.1016/0092-8674(87)90709-4. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa F, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor gamma chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahoo D, et al. MiDReG: A method of mining developmentally regulated genes using Boolean implications. Proc Natl Acad Sci USA. 2010;107:5732–5737. doi: 10.1073/pnas.0913635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahoo D, Dill DL, Gentles AJ, Tibshirani R, Plevritis SK. Boolean implication networks derived from large scale, whole genome microarray datasets. Genome Biol. 2008;9:R157. doi: 10.1186/gb-2008-9-10-r157. 1–R157.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.