Abstract
Although the proteins comprising many signaling systems are known, less is known about their numbers per cell. Existing measurements often vary by more than 10-fold. Here, we devised improved quantification methods to measure protein abundances in the Saccharomyces cerevisiae pheromone response pathway, an archetypical signaling system. These methods limited variation between independent measurements of protein abundance to a factor of two. We used these measurements together with quantitative models to identify and investigate behaviors of the pheromone response system sensitive to precise abundances. The difference between the maximum and basal signaling output (dynamic range) of the pheromone response MAPK cascade was strongly sensitive to the abundance of Ste5, the MAPK scaffold protein, and absolute system output depended on the amount of Fus3, the MAPK. Additional analysis and experiment suggest that scaffold abundance sets a tradeoff between maximum system output and system dynamic range, a prediction supported by recent experiments.
Keywords: quantitative immunoblotting, single-cell fluorescence quantification, genetic algorithmic model optimization
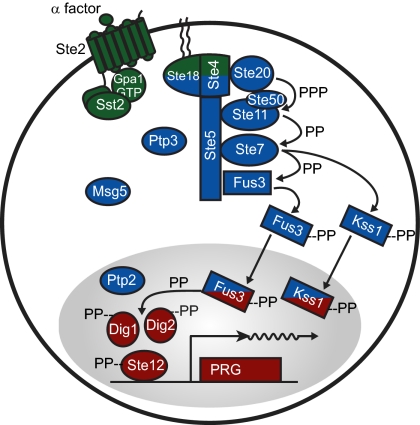
The Saccharomyces cerevisiae pheromone response system has been useful for understanding eukaryotic cell signaling (1–3). In haploid cells, the pheromone response system detects mating pheromone in the extracellular environment secreted by yeast of the opposite mating type. The system triggers a number of responses, including induction of gene expression, arrest in cell cycle progression, changes in morphology, and eventually, mating. The molecules and interactions by which the system operates are relatively well-understood (Fig. 1).
Fig. 1.
The yeast pheromone response system represented as three subsystems. The receptor/G protein (green), MAPK cascade (blue), and gene expression (red) subsystems. System input in MATa cells is α-factor, secreted by MATα yeast cells. α-Factor binds the G protein-coupled receptor Ste2, causing release of the Gβ-Gγ dimer Ste4-Ste18 from the inhibitory Gα subunit Gpa1. Gβ-Gγ then recruits the scaffold protein Ste5 to the cell membrane and bridges an interaction between the scaffold Ste5 and the kinase Ste20. Ste5 binds three sequentially activated kinases of a MAPK cascade. Ste20 then phosphorylates three sites on the MAPKKK Ste11. Phosphorylated Ste11 then phosphorylates two sites on a MAPKK Ste7, which in turn, phosphorylates two sites on each of two MAPKs, Fus3 and Kss1. Both active MAPKs then phosphorylate transcriptional regulators (Ste12, Dig1, and Dig2) and thereby, induce pheromone responsive gene expression. Active Fus3 also phosphorylates additional substrates that promote morphological changes and arrest the cell cycle. A number of phosphatases (Msg5, Ptp2, and Ptp3) inactivate the MAPKs.
Changes in the number of molecules per cell (hereafter called abundances) of signaling proteins can alter quantitative signaling behaviors. For example, in the pheromone response system, overexpression of Fus3 increases pheromone-induced sensitivity to cell cycle arrest (4), and overexpression of Gpa1 decreases pheromone-induced transcription (5). Similarly, changes in the abundances of MAPK cascade scaffold proteins (Ste5 in the yeast pheromone response system) (Fig. 1) can alter system behavior. In models, when scaffold is limiting, increasing scaffold abundance first increases system output and then, eventually sequesters the associated protein kinases onto separate scaffolds, diminishing the number of fully assembled complexes and system output (6). This peak and decline behavior has been experimentally shown for Ste5 in this yeast system (7) and the Kinase Suppressor of Ras (KSR) scaffold protein in Ras signaling in Xenopus oocytes (8).
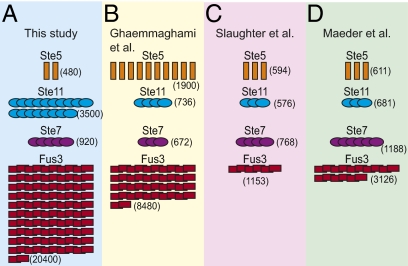
Previous investigators reported focused and genome-wide inventories of pheromone system protein abundances (9–13). These measurements differed by up to 12-fold (Fig. 2) (14, 15). We thought some discrepancies might be because of differences and systematic biases in quantification methods (Discussion). We developed improved measurement methods and used these methods to quantify system proteins. We used the measurements together with quantitative models of the system, and we examined consequences of these numbers and their remaining uncertainties for system behaviors.
Fig. 2.
Reported measures of protein abundances vary greatly across studies. Graphical representation of numbers of MAPK cascade proteins as measured by immunoblotting in (A) this study (Table S1) and (B) the work by Ghaemmaghami et al. (9) and fluorescent correlation spectroscopy in (C) the work by Slaughter et al. (12) and (D) the work by Maeder et al. (11). Each protein icon represents 200 molecules/cell.
Results
Accurate Measurement of Yeast Pheromone System Protein Abundances.
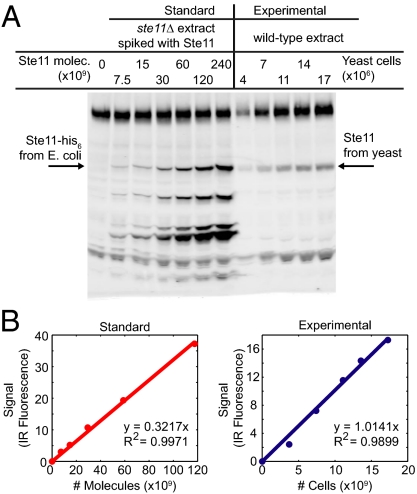
To measure pheromone system protein abundances (Fig. 3 and Fig. S1), we developed and deployed improvements that eliminated several sources of measurement error in standard quantitative immunoblotting protocols (9, 16, 17). Four improvements merit special mention. First, we prepared protein extracts using a chemical lysis procedure that gave increased and more consistent protein recovery than other methods (Fig. S2) (9, 18). Second, we used a calibration standard for each measured protein. This change was critical, because proteins differed greatly in electrophoretic transfer from gel and retention on the membrane (SI Materials and Methods and Fig. S2). Third, we quantified antigen-bound primary antibody with a secondary antibody linked to an infrared fluorophore (not an enzyme), thereby making signal intensity linear within a large dynamic range (19). Fourth, we ran (on each quantification gel) a dilution series of cell extract and a dilution series of the calibration standard for the quantified protein (Fig. 3), thereby reducing error in quantification by interpolation. To gain insight into the remaining variation, we used the improved methods to conduct four to nine independent measurements of independent cultures.
Fig. 3.
Improved protein quantification through careful immunoblotting. (A) Fluorescence scanner image of a representative Ste11 immunoblot. We diluted the Ste11-His6 protein standard into a protein extract from a ste11Δ strain and loaded the protein standards on a gel alongside protein extract from a WT strain (corresponding to the indicated number of cells). We probed the membrane with primary antibodies against Ste11 and secondary antibodies linked to a fluorophore. (B) Plot of total fluorescence intensities of the Ste11 bands above. We fit lines to standard (Left) and experimental (Right) data and used the slopes of the lines to calculate 3,152 molecules/cell of Ste11 in this experiment.
For different proteins, measured average abundances ranged from under 40 to over 20,000 per cell (Fig. 2A and Table S1). Measurements showed significant gel to gel and sample to sample variation, with SDs between 6% and 33%, corresponding to 1.2- to 2-fold variation [(mean + SD)/(mean − SD)] across all measured proteins. Multiple independent measurements for each protein reduced the SE of the estimated mean values to ∼8% of mean abundances (Table S1).
The least abundant essential system protein, the MAPK scaffold Ste5, was present at ∼480 molecules/cell. Ste5 was, thus, 2- to 43-fold less abundant than the different kinases that it binds (Ste11, Ste7, Fus3, and Kss1) (Fig. 2A). The pheromone response system shares kinase components with two other signaling systems: the filamentous growth (FG) and high osmolarity glycerol response (HOG) systems (20). In all three systems, MAPK (Fus3, Kss1, or Hog1) abundance was greater than MAPKKK (Ste11) abundance, which in turn, was greater than MAPKK abundance (Ste7 or Pbs2) (Table S1).
We determined the effect of system induction by exposing cells to high pheromone (1 μM) for 15 min and then measuring abundance. Five proteins (Gpa1, Fus3, Ste12, Msg5, and Far1) showed stimulation-dependent increases above the measurement error by factors ranging from 1.3- to 2.1-fold (Table S1). The observed increases were expected: pheromone stimulation diminishes Far1 degradation (21) and increases transcription of the GPA1, FUS3, STE12, MSG5, and FAR1 genes (22).
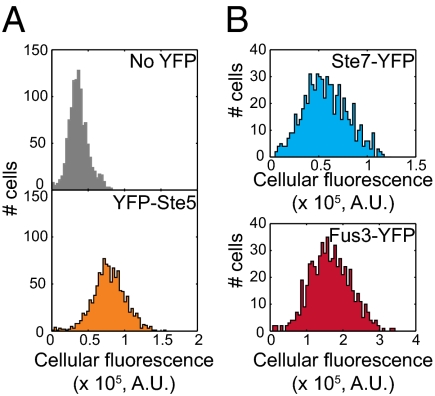
To cross-calibrate these numbers and measure their cell to cell variation, we quantified YFP-tagged proteins in single cells. We showed previously (23) that quantification based on fluorescent proteins can be accurate given knowledge of the light-collecting biases in experimental equipment, the rate of dilution caused by cell growth, the ratio of steady state expression of tagged protein to native protein, the rate of maturation of the fluorophore, and the rate of degradation of the fused protein (lack of correction for fluorophore maturation and degradation results in underquantification). Fluorescence measurements showed that cells averaged (±SEM) 434 ± 34 molecules of Ste5, consistent with the immunoblotting measurement of 484 ± 61. To compare cell to cell variation for other system proteins, we quantified fluorescence for four additional protein fusions: Fus3-YFP, Ste7-YFP, Dig1-YFP, and CFP-Ste12. Coefficients of variation (CVs) for total fluorescence were 28–40% for the five proteins (Fig. 4B and Table S2). Notably, higher abundance proteins did not exhibit higher cell to cell variation as previously reported for yeast proteins in general (24, 25). We did not calculate single cell abundance for these proteins, because we had not measured their degradation rates, with the exception of Ste5 (23). However, fluorescence of cells expressing Fus3-YFP was only approximately twofold higher than fluorescence of cells expressing YFP-Ste5 (Fig. 4), suggesting that degradation of the Fus3-YFP fusion may be high (Results and Discussion).
Fig. 4.
We captured images and used Cell-ID to calculate total fluorescence cells containing YFP-tagged proteins as described in ref. 23 and SI Materials and Methods. (A) YFP-Ste5 protein measurements. The population distributions of fluorescence are shown for strains with one STE5 gene encoding untagged (Upper) or YFP-tagged (Lower) protein. The cell to cell coefficients of variation were 36.2% and 28.4%, respectively, for 1,095 and 1,110 cells. After applying corrections for protein maturation, degradation, photobleaching, autofluorescence, and probability of photon detection (23) (SI Materials and Methods), we estimated that cells had, on average, 434 molecules of YFP-Ste5 per cell. (B) Cell to cell variation in fluorescence of cells containing Ste7-YFP (Upper) or Fus3-YFP (Lower). The coefficients of variation were 35.6% and 30.4%, respectively.
Fig. 2 illustrates some differences between our current measurements and previous reports (Table S1). The most significant differences are a lower Ste5 abundance compared with an earlier whole-genome immunoblotting study (9), a higher Fus3 abundance compared with two studies that relied on fluorescence measurements (11, 12), and a higher value for Ste11 compared with all three studies. In the case of Fus3, we suspect that the underestimation of Fus3 abundance might be caused by omission of correction for fluorophore maturation and Fus3 degradation rate.
Computational Investigation of MAPK Cascade Dynamic Range.
We sought to identify and investigate properties of the system that might be sensitive to precise protein abundances (and the remaining variability in the measurements). To search for behaviors that might be sensitive to low scaffold abundance and particular abundances of other system components, we developed a quantitative model of the MAPK cascade, YeastMAPKCascade (YMC).
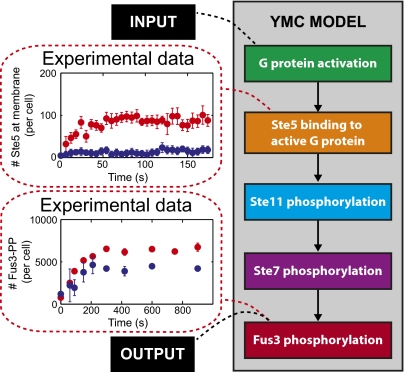
We generated YMC from the Yeast Pheromone Model repository (YPM; http://YeastPheromoneModel.org) (26), a wiki-based repository detailing the molecular components and reactions of the pheromone response system. YPM also contains a synonymous embedded model of the pheromone system encoded in the BioNetGen language (27). We manually simplified the full pheromone response system model encoded in the YPM to eliminate molecules and reaction rules that were not directly involved in the MAPK cascade (Fig. 5, SI Materials and Methods, and Fig. S3). We designated the system output of YMC to be the amount of activated (doubly phosphorylated) Fus3 (Fus3-PP). To couple model activation to simulated extracellular pheromone levels, we included two first-order reactions with rate constants that varied as a function of pheromone concentration: one rate constant for G-protein activation (by dissociation) and a second rate constant for G-protein deactivation (by reassociation). The pheromone response system has basal activity in the absence of pheromone (3, 7), which ensures expression of key system proteins, including Ste2 (the receptor) and Fus3, in unstimulated cells (22). To reproduce basal activity, which had not been attempted in a number of previous modeling studies of this system (6, 28–31), we set a nonzero minimum for the G-protein dissociation rate in the absence of pheromone.
Fig. 5.
Constraining a mechanistic signaling system model through dynamic experimental data. A block diagram for the YMC. Model input is the dose-dependent rates of G-protein activation and deactivation, which characterize the effects of pheromone treatment on the activity of the G protein. Model parameters are constrained by experimental measurements of Ste5 translocation to the plasma membrane (formation of a complex between Ste5 and the active G protein) and experimental measurements of the model output, Fus3 phosphorylation, in response to subsaturating (blue) and saturating (red) pheromone. The full model is available in Fig. S3 and SI Materials and Methods.
The resulting model, YMC, included 41 parameters: 32 reaction rate constants, 7 protein abundances, the mean cell volume, and the rate of increase in mean volume (Table S3). We used YMC to simulate the response of the system to three different levels of pheromone: no pheromone (to establish prestimulation steady state system activity), low pheromone (0.1 nM), and saturating pheromone (either 50 or 100 nM) (SI Materials and Methods). Because YMC did not contain MAPK-dependent feedbacks, we compared simulation results with experimental data obtained in the presence of an inhibitor of the Fus3 MAPK (SI Materials and Methods).
The YPM contains and documents known and estimated values for 12 of 41 parameters, including the six abundances measured and reported here. We estimated the other parameters by using a genetic algorithm (32) to identify parameter sets that produced a dynamic behavior that matched experimental measurements of Ste5 membrane translocation and Fus3 phosphorylation (discussed below). Genetic algorithms can often find parameter sets that give good agreement between computed and measured behaviors when others fail, and thus, they are commonly used to optimize parameters in biochemical models (33). Just as important, genetic algorithms can be used to generate multiple, different parameter sets that each result in agreement between computed and measured behaviors (34).
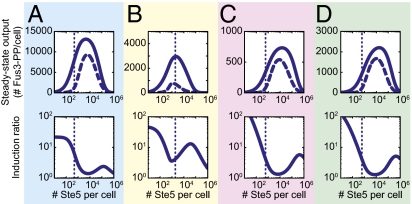
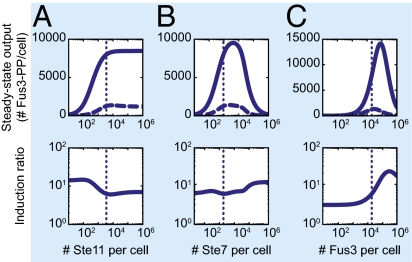
We, thus, used a genetic algorithm (GA) to produce multiple sets of parameter values (SI Materials and Methods) and named the model defined by the optimal set of parameter values YMCv1 (YeastMAPKcascade_v1) (Fig. S4A). We then examined the effect of changed abundances on system output. To do this examination, we varied levels of Ste5, Ste11, Ste7, and Fus3 from 10 to 106 molecules/cell (a number that could not be realized experimentally) and computed system behavior. As Ste5 abundance increased, the steady state-induced output (Fus3-PP) first increased greatly, peaked, and then, eventually declined (Fig. 6A). This peak in system output at intermediate Ste5 abundance was expected from the modeling work of Levchenko et al. (6) and the experimental observations by Chapman and Asthagiri (7). We next studied the expected impact of varying Ste11, Ste7, and Fus3 abundances within the same range. As Ste11 increased, the computed steady state YMCv1 output increased and then, leveled off (Fig. 7A), because a sufficient amount of Ste5-bound Ste11 formed to enable maximal phosphorylation of Ste7. In contrast, with increasing Ste7 or Fus3, system output increased, peaked, and then, decreased (Fig. 7 B and C). This dependence of system output on Ste7 and Fus3 abundances was qualitatively similar to the dependence on Ste5 abundance, because Ste7 and Fus3 form a complex; additionally, the YMC model stipulates that this complex cannot bind Ste5, consistent with experimental observations (35).
Fig. 6.
Regardless of measured system protein abundances, Ste5 scaffold abundance sets a system-level tradeoff between maximum signaling and dynamic range. We simulated the steady state system output and dynamic range using the (A) YMCv1, (B) YMC_Gmgm, (C) YMC_Sltr, and (D) YMC_Mdr models. Upper shows steady state system output (Fus3-PP per cell) in the absence of pheromone (dashed line) and in response to saturating amounts of pheromone (solid line) for a range of Ste5 abundances. Lower shows induction ratio calculated by dividing the steady state system output with saturating pheromone by the output with no pheromone. Dotted lines in all plots indicate measured Ste5 abundance.
Fig. 7.
Ste7, Ste11, and Fus3 abundances do not set a tradeoff between system output and dynamic range. We simulated the steady state system output and dynamic range of YMCv1 vs. (A) Ste11, (B) Ste7, and (C) Fus3 abundances. Upper shows steady state system output (Fus3-PP per cell) in the absence of pheromone (dashed line) and in response to saturating amounts of pheromone (solid line) for a range of kinase abundances (Upper). Lower shows induction ratio calculated from the steady state system output. Dotted lines in all plots indicate measured abundances.
We inspected the above results for the effect of abundance on basal, uninduced system output. Basal output was strongly affected by changes in the abundance of Ste5 but only weakly affected by changes in levels of other system components (Figs. 6A and 7). We next examined system dynamic range as measured by induction ratio (the ratio of induced to basal output) in response to differences in protein abundances. Induction ratio was also highly sensitive to changes in Ste5 abundance (Fig. 6A). Strikingly, the induction ratio was lowest near the Ste5 abundance predicted to produce the highest system output. Stated differently, the abundance of Ste5 established a tradeoff between total system output and dynamic range, and changes in the abundance of Ste5 changed the tradeoff. Ste5 abundance also showed a tradeoff using a different measure of dynamic range commonly used in electrical engineering: absolute difference between the basal and induced output or swing (36) (Fig. S5A). By contrast, neither induction ratio nor swing showed a tradeoff between total system output and dynamic range on varying the abundances of Ste11, Ste7, and Fus3 (Fig. 7 and Fig. S5A). However, as predicted at very high Ste5 abundances at which the protein is no longer limiting (7), changes in Ste7 abundance affected signaling (SI Materials and Methods), consistent with the notion that Ste7 (920 molecules/cell) becomes limiting under these circumstances.
We then quantified effects of Ste5 abundance in living cells. We constructed otherwise isogenic derivatives of the reference strain that replaced the WT STE5 locus with between one and seven integrated copies of YFP-Ste5 and that carried a PPRM1-CFP reporter (2). The YFP-Ste5 derivative enabled absolute single cell quantification of the changes in Ste5 abundance caused by the genetic manipulation. Qualitative results were as expected and previously reported (7); with increasing Ste5 abundance, both basal and induced activity increased. As our model predicted, overexpression of Ste5 using seven genomic copies of the Ste5 gene increased basal system activity (7.5-fold ± 4.3-fold) more than it increased induced system activity (1.3-fold ± 0.1-fold). Because of the effect on basal output, increasing Ste5 abundance by a factor of seven reduced the measured induction ratio by a factor of 5.9 ± 3.8 (Fig. S6). This great increase in basal output in response to small absolute increases in WT Ste5 number (from 1× Ste5 to 2× Ste5) (Fig. S6) is consistent with Ste5 being limiting.
Additional sensitivity analysis of all model parameters, including abundances, revealed that the only other parameter that produced a strong tradeoff between pathway output and dynamic range was cell volume (SI Materials and Methods and Table S3). The tradeoff observed for cell volume was not surprising given that changes in cell volume affected all protein concentrations in the model, including the concentration of the lowest abundance protein Ste5.
We investigated the extent to which these behaviors depended on particular parameter sets. To do this investigation, we used four different well-separated and well-fitting parameter sets, also generated by the GA from the measured abundances and time course data, to generate four distinct alternate models (YMCv2–5) (SI Materials and Methods). For each parameter set, Ste5 abundance set a tradeoff between system output and dynamic range, whereas the abundances of Ste11, Ste7, and Fus3 did not (Fig. S5 B–E).
Finally, we tested the dependence of the model's behavior on these measured abundances. To do this test, we used the GA to optimize three additional versions of YMC based on reports from Ghaemmaghami et al. (9), Slaughter et al. (12), and Maeder et al. (11) (Fig. 2). We named these models after the first authors of the different studies: YMC_Ghaemmaghami (YMC_Gmgm), YMC_Slaughter (YMC_Sltr), and YMC_Maeder (YMC_Mdr) (Fig. S4 and Table S3). We also created a model, YMC_optimized (YMC_opt), in which the GA estimated best fit protein abundances in addition to all of the other model parameters. Notably, all four models showed lower absolute system output than YMCv1, which we attributed to the undercounting (or for YMC_opt, underestimation) of the protein kinase Fus3 (Table S3). However, all four altered abundance models displayed some tradeoff between total system output and dynamic range that depended on Ste5 abundance but not changes in the abundance of Ste11, Ste7, or Fus3 (Fig. 6 B–D and Fig. S7).
Discussion
Accurate Measurements of Protein Abundances.
We developed an improved immunoblotting protocol and took advantage of previous work that enabled accurate single cell quantification of fluorescent fusion proteins (23). We used these methods to quantify abundances of key components of the pheromone response system. The immunoblotting and single cell measurements have complementary strengths. Quantitative immunoblotting does not depend on genetic modifications to cells and does not require knowledge of protein degradation rates. Quantification of signal from genetically encoded fluorescent fusion proteins does not require antibodies for each protein and yields information about differences in protein abundance among individual cells and for individual cells over time. The methods also have different limitations. Making specific antibodies for immunoblotting measurements takes significant effort (37). Fluorescent protein-based methods require determining that the fluorescent protein tag does not alter the population abundance correction for fluorophore maturation and correcting for protein degradation rate, which is typically calculated from comparison with abundance determined by immunoblotting (23). Such data and corrections were not available for previous studies (11, 12, 16).
Some of these measured abundances, particularly for the Ste11 and Fus3 protein kinases, are larger than those abundances reported (9–13), and their consequences include higher absolute system output. These discrepancies might, in principle, arise from differences in growth conditions and strain background. However, we suspect that significant differences arise from biases in the measurement methods (SI Materials and Methods has a comparison of immunoblotting methods). Our work establishes plausible causes for different protein counts. For example, because the first proteome-wide immunoblotting study (9) used a single protein standard with a tandem affinity purification (TAP) tag to calibrate measurements of thousands of different TAP-tagged yeast proteins, protein-specific differences in membrane transfer and retention (Fig. S2) could have resulted in widespread undercounting. Our immunoblotting measurements revealed a much larger range of abundances than those measurements that we took by fluorescence methods. In particular, although we measured ∼43-fold more Fus3 than Ste5 by immunoblotting (Fig. 2A and Table S1), Fus3-YFP cells were only two times as bright as YFP-Ste5 cells (Fig. 4), and previously published fluorescent correlation spectroscopy studies also suggested a more modest two- to fivefold excess of Fus3 over Ste5 (11, 12). We expect undercounting in fluorescence quantification when the fusion protein degradation rate is rapid relative to the fluorophore maturation rate or when the fluorescent moiety itself destabilizes the entire fusion. Consistent with this idea, Ste11 is degraded after system induction (38); our results suggest that Fus3 may be degraded as well. Although we took care to try to imagine, identify, and reduce sources of experimental bias, the resulting (up to twofold) variation in measurements among individual experiments (Table S1) defines significant remaining experimental uncertainty.
Scaffold Abundance Sets System Dynamic Range.
Previous modeling studies of scaffold-based signaling systems predicted that scaffold abundance helped determine maximal signaling output (6, 39). However, these studies did not consider the effect of scaffold concentration on basal system activity and thus, did not explore the impact of changing scaffold levels on system dynamic range. Only a few published models of the pheromone response system have included nonzero system activity in the absence of pheromone, and these efforts did not investigate its importance (10, 40).
By accounting for basal activity, we found that the abundance of the scaffold Ste5, but not other MAPK cascade components, strongly affected the dynamic range of the system in response to saturating pheromone inputs. Furthermore, we observed a computed tradeoff between system output and dynamic range as a function of scaffold abundance. We showed that this tradeoff was insensitive to all different tested sets of parameter values, including different sets of measured protein abundances, but that different values for the number of protein kinases affected predicted system output. We then confirmed the tradeoff by careful measurement in living cells (Fig. S6). Two experimental studies are qualitatively consistent with these findings, where cells overexpressing Ste5 produce higher basal system output, higher induced system output, and lower dynamic range than WT cells (7, 41).
The models suggested that differences in protein abundances should impact the quantitative performance tradeoffs. For example, the Ste5 scaffold abundance (480) measured here suggested that the system compromises between total system output and high-induction ratio, whereas the abundances measured in ref. 9 predict that the system favors total output at the expense of induction ratio (Fig. 6 A and B). Direct experiments in living cells were consistent with our conclusion (Fig. S6). Similarly, the high abundance for the Fus3 MAPK that we find may allow high absolute system output, even with relatively low scaffold numbers.
Our results show that scaffold abundance defines a set point for a particular tradeoff between system output and dynamic range, and they suggest that different abundances of scaffold proteins in other systems may set different tradeoffs. Although the abundance of Ste5 in S. cerevisiae does not change in response to pheromone stimulation (this work and refs. 11 and 12), these results also raise the possibility that scaffold abundance in other systems might change during signaling events to allow changes in system performance.
In S. cerevisiae, induced system output must be high enough to elicit appropriate physiological changes in response to pheromone treatment (for example, to overcome opposing phosphatases, trigger cell cycle arrest, and activate Ste12) (42). Conversely, in uninduced cells, basal output must be high enough to maintain synthesis of system proteins with expression that depends on the activity of the transcription factor Ste12 (for example, Ste2 and Fus3) but low enough to avoid triggering growth arrest (22). The large dynamic range of the system seems consistent with our recent findings that a large output range increases the precision with which a signaling system can respond differently to different amounts of input (3). During the history of S. cerevisiae (43), the pheromone system is likely to have been under natural and human-guided selection; the current relatively low scaffold abundance might represent an evolutionary solution that optimized performance within these constraints.
Materials and Methods
Yeast Strains and Growth Conditions.
All S. cerevisiae strains were derivatives of W303a (44). We used ACLY379 (2), a MATa bar1 derivative, as the reference for all measurements. We performed nucleic acid and yeast manipulations with standard procedures (45). To create strains for diluting protein standards, we deleted genes from ACLY379 using PCR-based gene disruption as described (46). For strains with YFP- or CFP-tagged proteins, we replaced the chromosomal copy of the gene with an fluorescent protein-fusion gene under the control of the native upstream regulatory region. We grew cells at 30 °C to midlog phase in synthetic defined (SD) medium containing appropriate auxotrophic nutrient mixtures (BSM formulations, BIO-101; Qbiogene) with yeast nitrogen base (Difco; Becton Dickinson) and 2% glucose. We constructed strains with multiple copies of the YFP-STE5 gene as detailed in SI Materials and Methods.
Quantitative Immunoblotting.
We identified and eliminated many sources of quantitative error in commonly used Western blotting procedures. We outline the protocol briefly here and detail it in SI Materials and Methods. We lysed cells and extracted total protein as described (18) (Fig. S2A). We prepared calibration standards for each protein of interest by serially diluting known amounts of purified, bacterially expressed His6-tagged protein into protein extracts from yeast cells deleted for the gene of interest. Each gel had one lane with extract from a deletion strain, six lanes with twofold serial dilutions of protein standard, and at least three lanes with different quantities of lysate from untreated and α-factor–treated reference cells (Fig. S1). We performed gel electrophoresis and protein transfer on a membrane, blotted the gel, and probed the membrane with antibodies (SI Materials and Methods). We used the primary antibodies listed in Table S1 (37) and secondary antibodies covalently linked to infrared fluorophores.
Measurement of Fluorescent Protein Fusions and Reporter Gene Output.
We performed optical microscopic cytometry as described elsewhere (3, 23) and in SI Materials and Methods. For cell handling, image capture, image analysis, and data processing, we used the open-source software packages Cell-ID and PAW (23, 47).
Computational Modeling.
We developed the MAPK cascade model from a model of the entire yeast pheromone response system from the YeastPheromoneModel (YPM) repository (http://YeastPheromoneModel.org) (26). Briefly, we simplified YPM to include only species and reactions that comprise the core signal transduction through the MAPK cascade (Fig. S3). Notably, we omitted Ste11 and Ste7 degradation to avoid introducing additional complexity and unknown parameters. We also omitted Ste5 oligomerization; although Ste5 oligomerization is important for signal transduction (48), the exact stoichiometry and mechanistic consequences of oligomerization are not known. Inclusion of Ste5 dimerization would have increased the number of species in the model from 236 to over 20,000, dramatically slowing down simulation and hindering parameter estimation. We wrote MATLAB scripts that used a genetic algorithm (Genetic Algorithm and Direct Search Toolbox, v2.1, R2007a; MathWorks) to perform parameter optimization and execute BioNetGen (version 2.0.46) (27) simulations of the models (SI Materials and Methods). We varied abundances over the wide range of 10–106 molecules to explore the full potential of the pathway architecture rather than limiting ourselves to abundances that could easily be achieved experimentally.
Supplementary Material
Acknowledgments
We thank Mary Maxon for antibodies and Tina Chin for help with constructing the expression plasmids. We are grateful to members of the Molecular Sciences Institute and the Colman-Lerner, Endy, and Brent laboratories for comments and discussions on the manuscript. This work was supported by National Institutes of Health from National Human Genome Research Institute for the Center for Quantitative Genome Function Grant P50 HG002370 (to R.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004042108/-/DCSupplemental.
References
- 1.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 2.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 3.Yu RC, et al. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456:755–761. doi: 10.1038/nature07513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elion EA, Grisafi PL, Fink GR. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 5.Guo M, et al. The yeast G protein alpha subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol Cell. 2003;12:517–524. doi: 10.1016/s1097-2765(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 6.Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman SA, Asthagiri AR. Quantitative effect of scaffold abundance on signal propagation. Mol Syst Biol. 2009;5:313. doi: 10.1038/msb.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cacace AM, et al. Identification of constitutive and ras-inducible phosphorylation sites of KSR: Implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 10.Hao N, Yildirim N, Wang Y, Elston TC, Dohlman HG. Regulators of G protein signaling and transient activation of signaling: Experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J Biol Chem. 2003;278:46506–46515. doi: 10.1074/jbc.M308432200. [DOI] [PubMed] [Google Scholar]
- 11.Maeder CI, et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 12.Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc Natl Acad Sci USA. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: Specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark CD, Palzkill T, Botstein D. Systematic mutagenesis of the yeast mating pheromone receptor third intracellular loop. J Biol Chem. 1994;269:8831–8841. [PubMed] [Google Scholar]
- 15.Raths SK, Naider F, Becker JM. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem. 1988;263:17333–17341. [PubMed] [Google Scholar]
- 16.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 17.Coligan JE. Current Protocols in Protein Science. New York: Wiley; 1996. [Google Scholar]
- 18.Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingrich JC, Davis DR, Nguyen Q. Multiplex detection and quantitation of proteins on western blots using fluorescent probes. Biotechniques. 2000;29:636–642. doi: 10.2144/00293pf02. [DOI] [PubMed] [Google Scholar]
- 20.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blondel M, et al. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 2000;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts CJ, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 23.Gordon A, et al. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2007;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- 24.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 25.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 26.Thomson TM. PhD thesis. Cambridge, MA: Massachusetts Institute of Technology; 2008. Models and analysis of yeast mating response: Tools for model building, from documentation to time-dependent stimulation. [Google Scholar]
- 27.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioNetGen: Software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 28.Yi TM, Kitano H, Simon MI. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc Natl Acad Sci USA. 2003;100:10764–10769. doi: 10.1073/pnas.1834247100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kofahl B, Klipp E. Modelling the dynamics of the yeast pheromone pathway. Yeast. 2004;21:831–850. doi: 10.1002/yea.1122. [DOI] [PubMed] [Google Scholar]
- 30.Shao D, Zheng W, Qiu W, Ouyang Q, Tang C. Dynamic studies of scaffold-dependent mating pathway in yeast. Biophys J. 2006;91:3986–4001. doi: 10.1529/biophysj.106.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilioti Z, et al. Oscillatory phosphorylation of yeast Fus3 MAP kinase controls periodic gene expression and morphogenesis. Curr Biol. 2008;18:1700–1706. doi: 10.1016/j.cub.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koza JR. Genetic Programming: On the Programming of Computers by Means of Natural Selection. Cambridge, MA: MIT Press; 1992. [Google Scholar]
- 33.Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: A comparison of global optimization methods. Genome Res. 2003;13:2467–2474. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwacke JH, Voit EO. The potential for signal integration and processing in interacting MAP kinase cascades. J Theor Biol. 2007;246:604–620. doi: 10.1016/j.jtbi.2006.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusari AB, Molina DM, Sabbagh W, Jr, Lau CS, Bardwell L. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horowitz P, Hill W. The Art of Electronics. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- 37.Pincus D, Benjamin K, Burbulis I, Tsong AE, Resnekov O. Reagents for investigating MAPK signalling in model yeast species. Yeast. 2010;27:423–430. doi: 10.1002/yea.1758. [DOI] [PubMed] [Google Scholar]
- 38.Esch RK, Errede B. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc Natl Acad Sci USA. 2002;99:9160–9165. doi: 10.1073/pnas.142034399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burack WR, Shaw AS. Signal transduction: Hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 40.Yildirim N, Hao N, Dohlman HG, Elston TC. Mathematical modeling of RGS and G-protein regulation in yeast. Methods Enzymol. 2004;389:383–398. doi: 10.1016/S0076-6879(04)89023-2. [DOI] [PubMed] [Google Scholar]
- 41.Garrenton LS, et al. Nucleus-specific and cell cycle-regulated degradation of mitogen-activated protein kinase scaffold protein Ste5 contributes to the control of signaling competence. Mol Cell Biol. 2009;29:582–601. doi: 10.1128/MCB.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 43.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 44.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 45.Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular and Cell Biology. New York: Wiley; 2002. [Google Scholar]
- 46.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Brun R, Couet O, Vandroni C, Zanarini O. PAW Physics Analysis Workstation. CERN Program Library Entry Q121. Geneva: CERN Geneva; 1989. [Google Scholar]
- 48.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.