Abstract
The binding and polymerization of RecA protein to DNA is required for recombination, which is an essential function of life. We study the pressure and temperature dependence of RecA binding to single-stranded DNA in the presence of adenosine 5'-[γ-thio]triphosphate (ATP[γ-S]), in a temperature regulated high pressure cell using fluorescence anisotropy. Measurements were possible at temperatures between 5–60 °C and pressures up to 300 MPa. Experiments were performed on Escherichia coli RecA and RecA from a thermophilic bacteria, Thermus thermophilus. For E. coli RecA at a given temperature, binding is a monotonically decreasing and reversible function of pressure. At atmospheric pressure, E. coli RecA binding decreases monotonically up to 42 °C, where a sharp transition to the unbound state indicates irreversible heat inactivation. T. thermophilus showed no such transition within the temperature range of our apparatus. Furthermore, we find that binding occurs for a wider range of pressure and temperature for T. thermophilus compared to E. coli RecA, suggesting a correlation between thermophilicity and barophilicity. We use a two-state model of RecA binding/unbinding to extract the associated thermodynamic parameters. For E. coli, we find that the binding/unbinding phase boundary is hyperbolic. Our results of the binding of RecA from E. coli and T. thermophilus show adaptation to pressure and temperature at the single protein level.
Keywords: pressure–temperature phase diagram, DNA binding protein, protein stability, protein functionality
Homologous recombination is an essential mechanism for genomic stability and for generating genetic variations. In eukaryotic cells, RecA protein or its variants are required for recombination (1). In prokaryotes, RecA protein is responsible for DNA repair and plays an important role in the SOS response (2). All organisms have inherited RecA homologues such as Rad51 in humans. Besides being an essential protein, RecA is one of the most studied and sequenced protein. Due to its universal existence in almost all life forms, RecA has been used as a phylogenetic marker for bacteriophage, bacteria, archea, and eukaryotes (3).
In the presence of ATP, a RecA molecule binds to three nucleotides of a ssDNA and further polymerizes to form a helical filament (4). RecA depolymerization occurs with the hydrolysis of ATP to form ADP and releases energy in the process. In the presence of a nonhydrolizable form of ATP, adenosine 5'-[γ-thio]triphosphate (ATP[γ-S]), RecA binding to ssDNA is much stronger compared to that with ATP. In the presence of ATP[γ-S], RecA can polymerize on ssDNA, but the rate of depolymerization is significantly reduced. ATP[γ-S] disrupts the natural dynamic instability. In the presence of ATP, the dynamic instability is thought to act as a type of kinetic proofreading (5).
Many archea and bacteria can live and grow in extreme conditions such as high pressure, temperature, and pH (6–8). For example, Serratia marcescens, a psychrophilic bacteria, can live at -20 °C while Pyrococcus CH1, a hyperthermophilic archea, can grow at 110 °C and 120 MPa (9, 10). The proliferation of these bacterial extremophiles raises an important question as to how proteins can sustain such harsh conditions because proteins are only stable in a range of thermodynamic conditions such as pressure and temperature (11–16).
A thermodynamic limit on the stability of proteins also puts a limit on the functionality of proteins (15, 17, 18) and hence the existence of living systems. Essential proteins must have adapted to the species habitat (19–25). Adaptation means the preservation of the functionality of proteins. The adaptation of proteins to high pressures and temperatures is usually attributed to various factors such as increased hydrophobicity of the protein residues, uncorrelated domain structures, and extremely slow unfolding (26). It is not clear if thermostability is correlated with increased barostability.
In this paper, we study and compare the pressure and temperature dependence of the equilibrium RecA binding to ssDNA in vitro in the presence of ATP[γ-S], using the method of fluorescence anisotropy, for a mesophilic bacteria Escherichia coli (EC) and a thermophilic bacteria Thermus thermophilus (TT). The methods presented here can also be applied to study other DNA binding proteins.
Materials and Methods
Fluorescence Anisotropy.
Fluorescence anisotropy is a common technique to measure binding (27, 28). It has been applied to investigate RecA binding to DNA as a function of DNA sequence and structure (29).
When a fluorescent molecule is excited with linearly polarized light, the emitted light will have the same polarization. If the fluorescent molecule rotates while it is excited, then light will be emitted with a rotated polarization. Measurement of emitted light intensity in the vertical Iv and horizontal Ih orientations can therefore detect the average rotation. The fluorescence anisotropy A is defined as
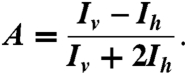 |
[1] |
The anisotropy of a spherical molecule is given by
 |
[2] |
where A0 is the intrinsic anisotropy, τ is the lifetime of the fluorophore, θ is the rotational correlation time of the molecule, η is the viscosity of water, and V is the molecular volume (30). An increased binding of RecA to ssDNA increases the rotational correlation time of the molecule and hence leads to an increase in the anisotropy.
In our experiments, we used ssDNA of 39 thymine nucleotides, which does not form any secondary structure or base stacking. It has a persistence length of approximately six nucleotides (31). Each RecA molecule binds to three bases and hence a completely covered ssDNA in our case will have 13 RecA bound to it. We find that the typical value of the anisotropy of the bare DNA, ADNA, is 0.05 or less. At atmospheric pressure, if the RecA concentration is high enough, then 13 RecA molecules can completely cover the DNA giving a large anisotropy, which is typically A13 = 0.25 (29). The measured value of A13 in our experiments is substantial compared to the theoretical maximum of A ≈ 0.4 (27). A is a function of Ih/Iv and does not depend on the total fluorescence intensity. Although the effects of pressure, temperature, and pH may alter fluorescein’s total fluorescence intensity, these effects do not alter the accuracy of the anisotropy.
Optics and High-Pressure Measurement Setup.
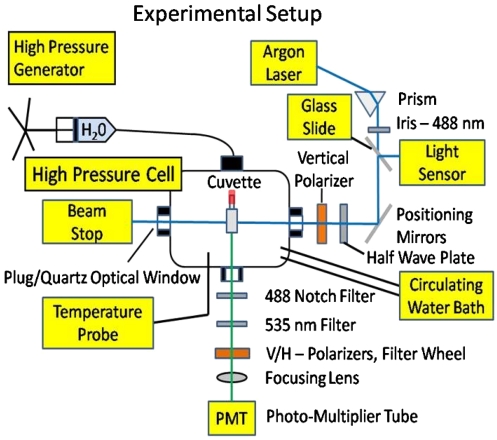
A schematic of the experimental setup is shown in Fig. 1. Anisotropy measurements were done with a temperature regulated high pressure cell (ISS) containing three quartz windows at right angles. A piston compresses water up to 300 MPa. Water transmits the pressure to a 400-μL square cuvette sealed with a flexible rubber cap (Spectrocell). The sample cuvette is excited directly with the 488-nm line of an argon laser that is vertically polarized with a linear polarizer (NT47-216 Edmunds Optics). The emitted fluorescent light is collected at a right angle to the incident laser beam. It passes through a collimating lens, followed by an emission filter for fluorescein (HQ535/50 Chroma), and then through a notch filter (zet-488 Chroma) to block scattered laser light. Next, it passes through a motorized filter wheel (FW103S Thor Labs) holding linear polarizers for the vertical or horizontal directions. The light is then focused onto a sensitive photomultiplier tube (H7421-40 Hamamatsu) with an f = 100-mm lens.
Fig. 1.
Experimental setup for temperature-regulated fluorescence anisotropy measurements at high pressures.
Biochemistry.
We experimented with standard TMD buffer (Tris) consisting of 25 mM Tris·HCl to pH 7.5 at 25 °C, 10 mM MgCl2, and 1 mM DTT. We also experimented with HMD buffer (Hepes) consisting of 25 mM Hepes/KOH to pH 7.5 at 25 °C, 10 mM MgCl2, and 1 mM DTT. We used a concentration of 1 nM ssDNA (39T) with a 5′ modification of 6-carboxyfluorescein (Genelink). The concentration of ATP[γ-S] (Enzo Life Sciences, Inc.) was always 0.1 mM. EC RecA protein was obtained from USB Affymetrix, and TT RecA protein was obtained from New England Biolabs. The concentration of EC RecA and TT RecA for ATP[γ-S] experiments were both 0.5 μM for comparison. Although binding of EC RecA to ssDNA was instantaneous at ambient conditions, it took several hours for TT RecA to cover the ssDNA in TMD buffer. For TT RecA, we kept the solution at 40 °C overnight before we began measurements.
Results
Dependence of Binding on Pressure at a Constant Temperature.
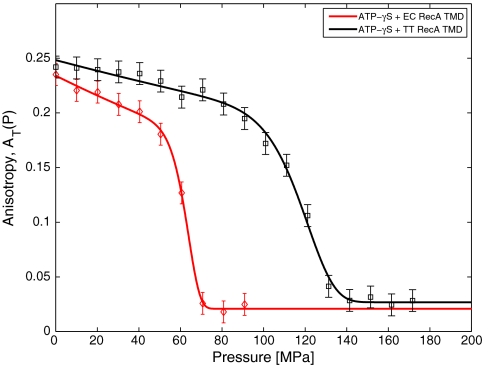
We measure the anisotropy AT(P) as a function of pressure P for fixed temperatures, T. In Fig. 2, we show AT(P) at T = 35 °C for EC RecA + ATP[γ-S], and TT RecA + ATP[γ-S]. We find that AT(P) is always a monotonically decreasing function of pressure. The anisotropy shows a sharp change at high pressure like a first-order phase transition.
Fig. 2.
Comparison of the pressure dependence of the anisotropy AT(P) at T = 35 °C for EC RecA + ATP[γ-S] (♦) and TT RecA + ATP[γ-S] (□). The anisotropy is a monotonically decreasing function of pressure. Symbols are the data from experiments and solid lines are the fit through the data. The number of counts for each state point was fixed and the error bars were taken to be identical to those determined at a single point.
Dependence of Binding on Temperature at a Constant Pressure.
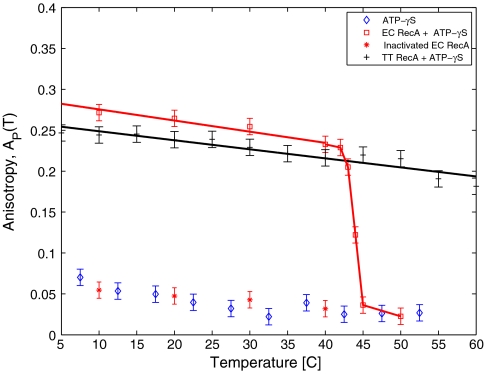
To investigate the temperature dependence of binding at a constant pressure, we measured AP(T) for P = 0.1 MPa (1 atm) as shown in Fig. 3. For EC RecA + ATP[γ-S], AP(T) decreases linearly followed by a sharp transition at T ≈ 42 °C, where AP(T) drops to the value for bare DNA, ADNA. The temperature dependence of AP(T) in case of TT RecA shows a similar linear decrease consistent with Eq. 2 but does not show a sharp transition in the range of the temperatures allowed by our apparatus. In order to investigate the reversibility of the transition in the case of EC RecA, we increased the temperature to 50° then decreased the temperature back to 10 °C slowly. We find that AP(T) does not recover back over the timescale of 200 min (Fig. 3), suggesting that the transition is irreversible. However, we did notice a partial recovery in AP(T) after several days.
Fig. 3.
Temperature dependence of anisotropy AP(T) at atmospheric pressure. For EC RecA + ATP[γ-S] (□), AP(T) gradually decreases with T, followed up by a sharp transition to the anisotropy of bare DNA at T ≈ 42 °C. Upon further lowering the temperature in steps (*), the anisotropy does not increase to its original value, suggesting that heat inactivation is irreversible. The experiment run without RecA and just ATP[γ-S] is shown for comparison to the heat inactivation, indicating that no RecA is bound. For TT RecA + ATP[γ-S] (+), the anisotropy decreases gradually from low temperatures up to 60 °C. Much higher temperatures are required to observe the hypothetical sharp transition in anisotropy.
The irreversibility of binding at high T suggests that domain of RecA responsible for binding to ssDNA denatures irreversibly at high temperatures. We expect that the binding domain stability should be highly correlated with the global stability of a protein. RecA should therefore exhibit regions of binding in (P,T) plane similar to the global stability. Besides high-temperature denaturation, many proteins denature at low temperatures and high pressures. This is known as cold denaturation 32. In the case of EC RecA, AP(T) shows nonmonotonic behavior for P > 20 MPa, and hence suggests cold and heat unbinding at high pressures (see Fig. 4A).
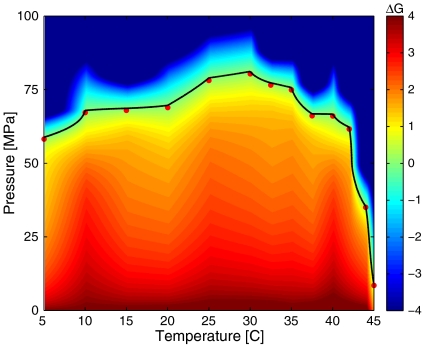
Fig. 4.
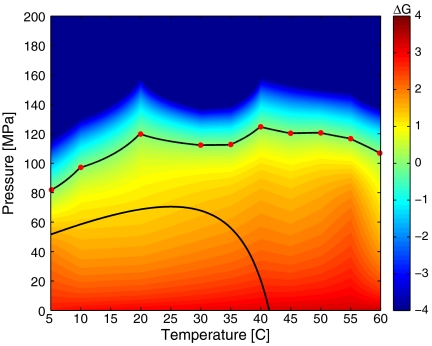
ΔG(P,T) in units of kBT298 for EC RecA + ATP[γ-S] + TMD buffer computed for the experimental measurements using the two-state model. The pressures and temperatures along ΔG = 0 are indicated as the solid black curve with the experimentally measured values indicated as red circles.
The (P,T) Binding Phase Diagrams with ATP[γ-S].
To obtain the phase diagram of binding, we measure the pressure–temperature dependence of anisotropy A(P,T) with ATP[γ-S] and two common buffers, TMD (Tris) and HMD (Hepes). We further use a two-state model of binding/unbinding to extract the free energy difference between the bound and unbound states from our anisotropy measurements. Within a linear approximation of anisotropy per RecA molecule bound, we can define the fractional occupancy ϕ(P,T) of RecA on ssDNA
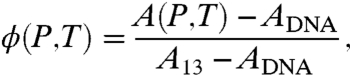 |
[3] |
where A13 is the anisotropy of the system when 13 RecA bind to ssDNA and ADNA is when none are bound. The fractional occupancy ϕ(P,T) can be written in terms of the free energies of the bound state GB and unbound state GU as
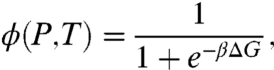 |
[4] |
where ΔG = GU - GB. Hence ΔG(P,T) can be written as
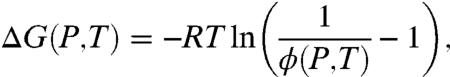 |
[5] |
where R is the molar gas constant. Within this approximation, the phase boundary ΔG(P,T) = 0 has a very simple interpretation, the pressure–temperature curve on which the average occupancy of RecA on ssDNA is one-half. The free energy for intermediate state points is obtained using linear interpolation of the anisotropy values from the experimental measurements.
In Fig. 4, we show ΔG(P,T) for EC RecA with ATP[γ-S] in TMD buffer. A surface fit for the data in Fig. 4 is shown in Fig. 5. Analogous figures for EC RecA in HMD buffer are shown in Fig. 6 and Fig. 7. The experimentally measured pressures and temperatures corresponding to half occupancy are indicated by the red circles. For both of the buffers, the pressure–temperature phase diagram of binding resembles that of protein structural stability with some subtle differences. Usually, pressure values for denaturation of proteins reported in experiments are much higher than what we find for unbinding, suggesting that perhaps the central DNA binding domain (4) is more sensitive to pressure and temperature than the overall structural stability of RecA. EC RecA shows both heat and cold unbinding with ATP[γ-S] in both buffers. The cold unbinding is possibly due to cold denaturation (32) of RecA or a cold degradation of the DNA binding domain of RecA. Along with the measurement, we show the fitted values of ΔG(P,T) using Hawley’s expansion of free energy change (see Thermodynamic Analysis).
Fig. 5.
ΔG(P,T) in units of kBT298 for EC RecA + ATP[γ-S] + TMD for the two-state model fitted to Hawley’s expansion. The fitted phase boundary ΔG = 0 is shown as the solid black curve.
Fig. 6.
ΔG(P,T) in units of kBT298 for EC RecA + ATP[γ-S] + HMD buffer computed for the experimental measurements using the two-state model. The pressures and temperatures along ΔG = 0 are indicated as the solid black curve with the experimentally measured values indicated as red circles.
Fig. 7.
ΔG(P,T) in units of kBT298 for EC RecA + ATP[γ-S] + HMD buffer for the two-state model fitted to Hawley’s expansion is shown. The fitted phase boundary ΔG = 0 is shown as the solid black curve.
A comparison of Fig. 4 and Fig. 6 shows that the phase boundary of binding/unbinding of RecA to ssDNA shows slight differences in the two buffers, suggesting that the temperature-dependent pH changes in case of TMD has rather little effect on binding. We further find that the phase diagram extends up to 45 °C with HMD versus 42 °C with TMD.
The temperature derivative of pressure along the phase boundary is related to the change in entropy ΔS and the change in volume ΔV between bound and unbound state as
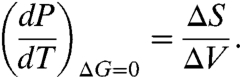 |
[6] |
Hence in the case of EC RecA, high-temperature unbinding is characterized by ΔS > 0 and ΔV < 0. The low-temperature unbinding is characterized by ΔS < 0 and ΔV < 0. Our observations agree with the entropy and volume changes of hydrophobic polymers upon heat and cold denaturation (15, 33). The entropy change between the two states goes to zero when dP/dT goes to zero (15, 34), a point separating heat and cold unbinding regimes in the (P,T) plane.
Comparison with Extremophilic RecA Protein.
Our studies on the EC RecA binding suggest that the functionality of RecA is adapted to the temperature range in which EC grows. We measured the pressure dependence of binding for TT RecA over a range of temperatures allowed by our apparatus 5–60 °C. Above 60 °C, the o rings of the pressure cell no longer seal. ΔG(P,T) as a function of temperature and pressure for TT RecA is shown in Fig. 8. The ΔG = 0 curve is rather flat for the range of temperatures we can access with our apparatus. The shape of the ΔG = 0 curve suggests that TT RecA binding to ssDNA is possible above 110 MPa, the pressure at the deepest known depth of the ocean, whereas EC RecA cannot. It is no surprise that a strain of TT can thrive near deep sea hydrothermal vents without any effect on the growth (35). Our experiments suggest that RecA from different bacteria are adapted to the thermodynamic conditions in which they grow.
Fig. 8.
The measured ΔG(P,T) in units of kBT298 for TT RecA + ATP[γ-S] + TMD buffer. The TT RecA phase boundary, ΔG = 0, is the upper solid black curve. For a comparison of phase diagrams, we also plot the ΔG = 0 curve for EC RecA + ATP[γ-S] + TMD buffer (lower black curve).
The pH Dependence of Binding.
Although TMD buffer is the standard buffer for RecA research, its properties are not always best suited to temperature-dependent measurements due to its large temperature coefficient of pH. We measured that the pH of TMD buffer varies linearly with temperature. The measured temperature coefficient was -0.027 ± 0.001 pH/°C. Moreover, TMD is a good buffer only in the pH range 7.5–9 and varies by an entire pH unit between 5 and 45 °C. Thus high temperatures are outside the range of the buffering capacity of TMD. To see if this effect was significant, we repeated our phase diagram measurements with HMD. HMD has a smaller temperature coefficient of pH change -0.014 pH °C and the working range of pH 6.8–8.2.
We also measured unbinding as a function of pressure for EC and TT RecA at T = 35°C for a pH range 5–9. We find that the dependence of the pressure P1/2 [pressure at ϕ(P,T) = 0.5] on pH is rather flat and can be fit with a line. For the P1/2 of EC RecA, 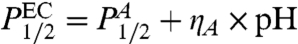 , where the values of the parameters are
, where the values of the parameters are 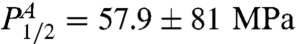 ηA = 0.51 ± 1.25 MPa/pH unit. For TT RecA, the dependence was similarly flat with
ηA = 0.51 ± 1.25 MPa/pH unit. For TT RecA, the dependence was similarly flat with  , where the parameters are
, where the parameters are  ηB = -4.3 ± 1.7 MPa/pH unit. Both
ηB = -4.3 ± 1.7 MPa/pH unit. Both  and
and 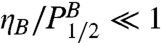 . Thus, the pH effect on binding is weak. We found that the fluorescence anisotropy measurements break down systematically above pH 9 or below pH 4, where DNA screening (pH 9) is affected or fluorescence is quenched (pH 4).
. Thus, the pH effect on binding is weak. We found that the fluorescence anisotropy measurements break down systematically above pH 9 or below pH 4, where DNA screening (pH 9) is affected or fluorescence is quenched (pH 4).
Stability and Reversibility.
We have performed various experiments that bear on the reproducibility of our data and the thermodynamic reversibility of the RecA + ssDNA system. It is shown that pH or the type of protein may influence reversibility in pressure (36, 37). Measurements of phase diagrams were made by first equilibrating the system at a given temperature and atmospheric pressure then going up in pressure in a step of 10 MPa to the maximum pressure. The subsequent equilibration of the system at different pressures is very fast (usually less than 2 min) as evident from the stable values of the anisotropy. After reaching the maximum pressure of measurement, which takes about 20 min, the pressure was set back to atmospheric pressure for a new temperature measurement. For EC RecA, we find that anisotropy and hence the binding is reversible with cycling pressure between 0.1 and 300 MPa at a given temperature. For TT RecA, the binding recovery timescale varied around several hours.
Thermodynamic Analysis.
In this section, we use the free energy difference calculated from the anisotropy measurements (Eq. 4) to extract thermodynamic parameters and the shape of the Gibbs free energy surface in (P,T) plane using Hawley’s expansion of ΔG (35). The Gibbs free energy change between the unbound and bound states is defined as
| [7] |
The free energy difference can be expanded as Taylor series in terms of pressure and temperature to second order.
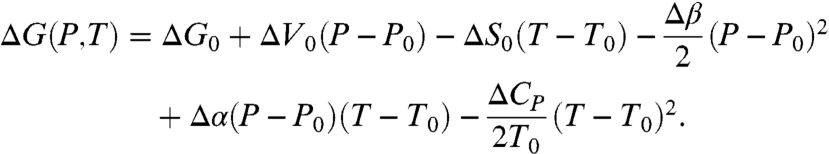 |
[8] |
We take T0 = 298 K and P0 = 0.1 MPa as standard conditions. It is assumed that the thermodynamic parameters appearing in Eq. 8 have no additional pressure or temperature dependence but otherwise take on different values in the unbound and bound state. Δα = αU - αB, Δβ = βU - βB, and so forth. This expansion involves terms including changes in standard free energy ΔG0, entropy ΔS0, volume ΔV0, specific heat ΔCP, compressibility Δβ, and thermal expansion Δα. Δα and Δβ are specially defined as
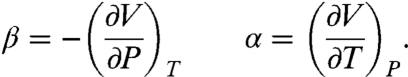 |
[9] |
 |
[10] |
The thermodynamic coefficients are determined by several rounds of least squares fitting, first for the overdetermined set of coefficients around ΔG = 0, and then again against the entire dataset to determine the scale of parameters. The values between the HMD and TMD coefficients are comparable.
The geometry of the quadratic curve depends on the ratio r = B2/(4AC). For r < 1, Eq. 10 describes an ellipse, r = 1 is a parabola, and r > 1 is a hyperbola. In terms of the thermodynamic parameters, r is calculated as
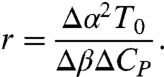 |
[11] |
The extracted parameters for EC RecA for TMD buffer and HMD buffer are listed in Table 1.
Table 1.
The extracted parameters for EC RecA
| Parameter | Units | ATP[γ-S]-TMD | ATP[γ-S]-HMD |
| Δβ | mL/(MPa·mol) | 0.057 ± 0.615 | 0.0038±0.3249 |
| Δα | mL/(K·mol) | 9.97 (99) | 7.75 (51) |
| ΔCp | kJ/(K·mol) | 10.6 (16) | 4.7 (11) |
| ΔV0 | mL/mol | −225.9 (23) | −178 (15) |
| ΔV0 | Å3/molecule | −28.9 (29) | −22.7 (19) |
| ΔS0 | kJ/(K·mol) | 0.658 (43) | 0.492 (39) |
| ΔG0 | kJ/mol | 15.75 (98) | 13.4 (13) |
| r | L | 49± 525 | 1,000 ± 87,000 |
Discussion
We have investigated the effects of pressure and temperature on the equilibrium binding of RecA protein to ssDNA E. coli and T. thermophilus RecA proteins in the presence of ATP[γ-S].
At a given temperature, the binding is a monotonically decreasing function of pressure for both TT and EC RecA under all conditions measured. At high pressures, the drop in binding is sharp. For EC RecA at constant temperature, binding is reversible in pressure. For EC RecA, binding is not reversible at high temperatures above 42 °C suggesting aggregation. The cutoff in the region of binding at high temperature correlates with the measured drop in growth rate of EC at 42 °C (38). The pressure-temperature phase diagram of binding of RecA from TT shows a wider region of binding in the (P,T) plane. This suggests that a thermophilic bacteria such as the one studied here could also be viable at high pressures. Indeed, strains of TT have been found to thrive near thermal vents in the ocean where both the pressure and temperature are very large (34).
The binding of RecA protein from EC shows a hyperbolic phase diagram in the (P,T) plane near the transition to the commonly found elliptic phase diagrams found for the stability of various proteins (35, 39, 40). For TT, the region of the phase diagram above T > 60 °C was not accessible, so we could not calculate the shape of the entire phase boundary like EC and extract thermodynamic parameters. We note that the global denaturation of a protein versus the functional disruption of a domain may have different (P,T) dependencies. Our method does not distinguish between local unfolding of the DNA binding domain and global unfolding of the protein. To distinguish between these two, another method such as circular dichroism is necessary (35).
The unbinding of RecA at pressures around 50–60 MPa at 37 °C also sheds light on the SOS response in EC measured at high pressures (41). Because RecA tends to depolymerize at high pressures, the conventional UV irradiation pathway (2) would not be applicable to the high pressure SOS response.
Our RecA measurements are applicable to RecA from any species. In future studies, RecA could be purified from other species (42). Other extremophiles of interest in clude Serratia marcescens, which grows at -20 °C, or Photobacterium profundum, which lives at 70 MPa. A phase-diagrams comparison of RecA from such organisms would be of great interest. We hypothesize that the region of binding for each extremophilic RecA would span the pressure and temperature range where bacteria survive, like for EC and TT.
We have shown that pressure and temperature adaptation occurs even at the single molecule level for the essential recombination protein RecA. Bioinformatic studies may shed some light on which variations in RecA proteins confer adaptation to extreme environments.
Acknowledgments.
We thank S. Leibler, A. Buguin, Y. Maeda, J. Chuang, and R. Bar-Ziv for helpful discussions and advice. This work is supported by National Science Foundation PHY-0848815. P.K. acknowledges the support of National Academies Keck Future Initiatives Award.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roca R, Cox M. RecA protein: Structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 2.Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005;3:1174–1176. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu D, et al. Stalking the fourth domain in metagenomic data: Searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS One. 2011;6:e18011. doi: 10.1371/journal.pone.0018011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Story RM, Weber IT, Steitz TA. The structure of the EC RecA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 5.Tlusty T, Bar-Ziv R, Libchaber AJ. High-fidelity DNA sensing by protein binding fluctuations. Phys Rev Lett. 2004;93:258103. doi: 10.1103/PhysRevLett.93.258103. [DOI] [PubMed] [Google Scholar]
- 6.Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–112. [Google Scholar]
- 7.Kato C, et al. Extremely barophilic bacteria isolated from the Mariana Trench, Challenger Deep, at a depth of 11,000 meters. Appl Environ Microbiol. 1998;64:1510–1513. doi: 10.1128/aem.64.4.1510-1513.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deguchi S, et al. Microbial growth at hyperaccelerations up to 403,627 g. Proc Natl Acad Sci USA. 2011;19:7997–8002. doi: 10.1073/pnas.1018027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivkina EM, Friedmann EI, McKay CP, Gilichinsky DA. Metabolic activity of permafrost bacteria below the freezing point. Appl Environ Microbiol. 2000;66:3230–3233. doi: 10.1128/aem.66.8.3230-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X, et al. Pyrococcus CH1, an obligate piezophilic hyperthermophile extending the upper pressure temperature limits for life. ISME J. 2009;3:873–876. doi: 10.1038/ismej.2009.21. [DOI] [PubMed] [Google Scholar]
- 11.Bridgeman PW. The coagulation of albumen by pressure. J Biol Chem. 1914;19:511–512. [Google Scholar]
- 12.Kauzmann W. Thermodynamics of unfolding. Nature. 1987;325:763–764. [Google Scholar]
- 13.Dill KA. Theory for the folding and stability of globular proteins. Biochemistry. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 14.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 15.Buldyrev SV, Kumar P, Debenedetti PG, Rossky PJ, Stanley HE. Water-like solvation thermodynamics in a spherically symmetric solvent model with two characteristic lengths. Proc Natl Acad Sci USA. 2007;104:20177–20182. doi: 10.1073/pnas.0708427104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buldyrev SV, Kumar P, Sastry S, Stanley HE, Weiner S. Hydrophobic collapse and cold denaturation. J Phys Condens Matter. 2010;22:284109. doi: 10.1088/0953-8984/22/28/284109. [DOI] [PubMed] [Google Scholar]
- 17.Gross M, Jaenicke R. Proteins under pressure, the influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur J Biochem. 1994;2:617–630. doi: 10.1111/j.1432-1033.1994.tb18774.x. [DOI] [PubMed] [Google Scholar]
- 18.Mozhaev VV, Heremans K, Frak J, Masson P, Balny C. High pressure effects on protein structure and function. Proteins. 1996;24:81–91. doi: 10.1002/(SICI)1097-0134(199601)24:1<81::AID-PROT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19.Sterner R, Liebl W. Thermophilic adaptation of proteins. Crit Rev Biochem Mol Biol. 2001;36:39–106. doi: 10.1080/20014091074174. [DOI] [PubMed] [Google Scholar]
- 20.Somero GN. Adaptations to high hydrostatic pressure. Annu Rev Physiol. 1992;54:557–577. doi: 10.1146/annurev.ph.54.030192.003013. [DOI] [PubMed] [Google Scholar]
- 21.Ladenstein R, Antranikian G. Proteins from hyperthermophiles: Stability and enzymatic catalysis close to the boiling point of water. Adv Biochem Eng Biotechnol. 1998;61:37–85. doi: 10.1007/BFb0102289. [DOI] [PubMed] [Google Scholar]
- 22.Hay S, et al. Are the catalytic properties of enzymes from piezophilic organisms pressure adapted? ChembioChem. 2009;10:2348–2353. doi: 10.1002/cbic.200900367. [DOI] [PubMed] [Google Scholar]
- 23.Murakami C, et al. Cloning and characterization of dihydrofolate reductases from deep-sea bacteria. J Biochem. 2010;147:591–599. doi: 10.1093/jb/mvp206. [DOI] [PubMed] [Google Scholar]
- 24.Mozhaev VV, Lange R, Kudryashova EV, Balny C. Application of high hydrostatic pressure for increasing activity and stability of enzymes. Biotechnol Bioeng. 1996;52:320–331. doi: 10.1002/(SICI)1097-0290(19961020)52:2<320::AID-BIT12>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Kawano H, et al. Differential pressure resistance in the activity of RNA polymerase isolated from Shewanella violacea and Escherichia coli. Extremophiles. 2004;8:367–375. doi: 10.1007/s00792-004-0397-0. [DOI] [PubMed] [Google Scholar]
- 26.Luke KA, Higgins CL, Wittung-Stafshede P. Thermodynamic stability and folding of proteins from hyperthermophilic organisms. FEBS J. 2007;274:2023–2033. doi: 10.1111/j.1742-4658.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 27.Lackowicz JR. 2nd ed. New York: Plenum; 1999. Principles of fluorescence spectroscopy; pp. 12pp. 13–295. [Google Scholar]
- 28.Chilukuri LN, Bartlett DH, Fortes PA. Comparison of high pressure-induced dissociation of single-stranded DNA binding protein (SSB) from high pressure-sensitive and high pressure-adapted marine Shewanella species. Extremophiles. 2002;6:377–383. doi: 10.1007/s00792-002-0267-6. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Ziv R, Libchaber AJ. Effects of DNA sequence and structure on binding of RecA to single-stranded DNA. Proc Natl Acad Sci USA. 2001;98:9068–9073. doi: 10.1073/pnas.151242898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrin J. Polarization de la luniere de fluorescence. Vie moyenne de molecules dans l’etat excite. J Phys Radium. 1926;7:390–401. [Google Scholar]
- 31.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Privalov PL. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 33.Hummer G, Garde S, Garcia AE, Paulatis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95:1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawley SA. Reversible pressure-temperature phase diagram of chymotrypsinogen. Biochemistry. 1971;10:2436–2442. doi: 10.1021/bi00789a002. [DOI] [PubMed] [Google Scholar]
- 35.Marteinsson VT, Birrien JL, Raguns G, da Costa MS, Prieur D. Isolation and characterization of Thermus thermophilus Gy1211 from a deep-sea hydrothermal vent. Extremophiles. 1999;3:247–251. doi: 10.1007/s007920050123. [DOI] [PubMed] [Google Scholar]
- 36.Zipp A, Kauzmann W. Pressure denaturation of metmyoglobin. Biochemistry. 1973;12:4217–4228. doi: 10.1021/bi00745a028. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki K, Miyosawa Y, Suzuki C. Protein denaturation by high pressure. Measurements of turbidity of isoelectric ovalbumin and horse serum albumin under high pressure. Arch Biochem Biophys. 1963;101:225–228. doi: 10.1016/s0003-9861(63)80006-5. [DOI] [PubMed] [Google Scholar]
- 38.Herendeen SL, VanBogelen RA, Neidhardt FC. Levels of major proteins of Escherichia coli during growth at different temperatures. J Bacteriol. 1979;139:185–194. doi: 10.1128/jb.139.1.185-194.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesch H, Stadlbauer H, Friedrich J, Vanderkooi JV. Stability diagram and unfolding of a modified cytochrome c: What happens in the transformation regime? Biophys J. 2002;82:1644–1653. doi: 10.1016/S0006-3495(02)75515-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiedersich J, Kohler S, Skerra A, Friedrich J. Temperature and pressure dependence of protein stability: The engineered fluorescein-binding Liopcalin FluA shows an elliptic phase diagram. Proc Natl Acad Sci USA. 2008;105:5756–5761. doi: 10.1073/pnas.0710409105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW. An SOS response induced by high pressure in Escherichia coli. J Bacteriol. 2004;186:6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singleton SF, Simonette RA, Sharma NC, Roca AI. Intein-mediated affinity-fusion purification of the Escherichia coli RecA protein. Protein Expr Purif. 2002;26:476–488. doi: 10.1016/s1046-5928(02)00571-5. [DOI] [PubMed] [Google Scholar]