Abstract
The canonical function of the human telomerase protein (hTERT) is to synthesize telomeric DNA, but it has other biological activities, including enhancing cell proliferation, decreasing apoptosis, regulating DNA damage responses, and increasing cellular proliferative lifespan. The mechanistic relationships among these activities are not understood. We previously demonstrated that ectopic hTERT expression in primary human mammary epithelial cells diminishes their requirement for exogenous mitogens, thus giving them a proliferative advantage in a mitogen-depleted environment. Here, we show that this phenotype is caused by a combination of increased cell division and decreased apoptosis. In addition, we use a panel of hTERT mutants to demonstrate that this enhanced cell proliferation can be uncoupled not only from telomere elongation, but also from other telomerase activities, including cellular lifespan extension and regulation of DNA damage responses. We also find that the proliferative function of hTERT, which requires hTERT catalytic activity, is not caused by increased Wnt signaling, but is accompanied by alterations in key cell cycle regulators and is linked to an hTERT-catalyzed decrease in the levels of the RNA component of mitochondrial RNA processing endoribonuclease. Thus, enhanced cell proliferation is an independent function of hTERT that could provide a new target for the development of anti-telomerase cancer therapeutic agents.
Keywords: carcinogenesis, breast, senescence, shelterin
The human telomerase protein (hTERT) is a reverse transcriptase that uses the telomerase RNA (hTR) as a template to add DNA repeats onto chromosome ends (1). This elongation of telomeres by hTERT stops loss of chromosome ends during each cell division, prevents senescence, and endows cancer cells with limitless replicative potential (1, 2). Indeed, hTERT is expressed at very low levels in normal somatic cells, and is up-regulated and reactivated in a majority of human tumors (3).
In addition to its canonical role in stabilizing telomeres, hTERT may promote tumorigenicity through other functions (4, 5). Novel functions of hTERT were first suggested by in vivo studies in mice (6, 7). Murine TERT (mTERT) overexpression in transgenic mice causes spontaneous tumorigenesis in the mammary gland, with intraepithelial neoplasia progressing to invasive carcinomas (7). This phenotype is unlikely to be an effect of the overexpressed mTERT on telomeric DNA for two reasons. First, in contrast to human chromosomes, murine chromosomes have sufficiently long telomeres (8) that phenotypes caused by loss of telomeric DNA are not evident for at least four mouse generations after genetic inactivation of telomerase (9). Second, telomerase is normally active in most somatic murine tissues (10), including the breast epithelium (11), thereby providing constitutive telomere stability (12). Thereafter, studies in primary human mammary epithelial cells (HMECs) identified an apparently telomere-independent function of hTERT that enables HMECs to proliferate in mitogen-deficient conditions, a hallmark of cancer (2, 13). Thus, in addition to the telomere elongation function of hTERT, its unique functions could promote carcinogenesis in a subset of tissues, especially in the mammary gland.
There are additional studies demonstrating alternative biological activities of telomerase. mTERT overexpression in the skin epithelium of mice causes proliferation of quiescent hair follicle stem cells (14, 15). This stimulation of proliferation is caused by mTERT acting as a cofactor in the β-catenin–containing transcriptional complex and thereby directly enhancing Wnt signaling (16, 17). Transgenic mice overexpressing mTERT in keratinocytes have an increased incidence of carcinogen-induced epidermal tumors and improved wound healing (6). Ectopic telomerase expression can also confer resistance to antiproliferative and proapoptotic stimuli (18–21) and enhance proliferation of diverse cell types such as mouse embryonic fibroblasts (18), cardiac myocytes (22), and human fibroblasts (23). Furthermore, hTERT can function as a RNA-dependent RNA polymerase that can bind to non-hTR RNAs, such as the RNA component of mitochondrial RNA processing endoribonuclease (RMRP), and use them as templates to generate double-stranded RNAs that are processed into siRNAs (24). hTERT can also alter chromatin structure and thereby affect DNA damage responses (25), as well as localize to the mitochondria and impact mitochondrial DNA damage and apoptosis (21, 26, 27).
The discovery of these varied novel hTERT functions (4, 5) may have critical implications not only for tumorigenesis and telomerase-targeted anticancer therapeutics (28) but also for tissue regeneration (29), genetic disorders associated with hTERT defects (30), as well as normal biological processes such as tissue homeostasis and organismal aging (31). A key to realizing this potential lies in determining whether these hTERT functions are mediated by the same or different biochemical or molecular activities, as this would not only help us understand how hTERT performs its diverse roles, but also determine whether these different pathways could provide multiple independent therapeutic targets. We addressed this question by using a panel of hTERT mutants (32–35) and demonstrated that the ability of hTERT to enhance proliferation, which was a result of increased cell division and decreased apoptosis, could be genetically uncoupled from its functions in telomere elongation, lifespan extension, and DNA damage responses. We also identified telomere elongation-deficient mutants that were still able to extend cellular lifespan. This ability was compromised when we knocked down the levels of a telomere-capping protein, hPOT1 (human Protection of Telomeres 1), suggesting that the lifespan extension by the telomere elongation-defective mutants was likely because they retain the ability to recruit telomere-capping proteins. Finally, we found that enhanced cell proliferation by hTERT, which requires hTERT catalytic activity, was not because of altered Wnt signaling but was linked to its ability to modulate RMRP levels. Our results therefore provide critical genetic support for the idea that many hTERT functions may involve separate mechanisms of action.
Results
hTERT Enhances Proliferation by Increasing Cell Division and Survival in Mitogen-Deficient Conditions.
We have previously shown that expression of hTERT in HMECs increases cell proliferation in complete growth medium and more dramatically in mitogen-limiting conditions (13) (Fig. 1A). The relative contributions of cell division and cell death to increased cell proliferation were determined by staining for BrdU incorporation and annexin V expression, respectively. In mitogen-limiting conditions, control cells showed a small percentage of cells in S phase (i.e., BrdU-positive) and an almost equal percentage of apoptotic (i.e., annexin V-positive) cells, which together account for their very slow overall rate of proliferation (Fig. 1 B and C). In contrast, cells expressing hTERT displayed a significantly larger percentage of S-phase cells and a relatively low percentage of apoptotic cells, thus explaining their greater overall proliferation rate (Fig. 1 B and C).
Fig. 1.
Ectopic hTERT expression enhances cell proliferation. HMECs were transduced with the indicated retroviral vectors, and grown in complete medium (A) or mitogen-limited (i.e., minimal) medium (A–C). (A) Cell proliferation in complete medium (CM) or minimal medium (MM) was measured by using crystal violet staining (SI Appendix, SI Materials and Methods). The graph represents averages of triplicates ± SD from one representative of three independent experiments. (B) Cells were labeled with BrdU. Representative flow cytometry profiles and percentages of BrdU-positive cells are shown. (C) Cells were stained for annexin V. Representative flow cytometry profiles and percentages of annexin V-positive cells are shown. Graphs in B and C represent averages ± SEM of three experiments.
Enhanced Cell Proliferation and Telomere Elongation Are Separable Functions of hTERT.
Our earlier work suggested that hTERT's ability to enhance cell proliferation might be independent of telomere elongation, because this phenotype was observed in early passage HMECs that do not have critically short telomeres (13). We proposed that hTERT has at least two independent cellular functions, one necessary for telomere elongation and another involved in stimulating cell proliferation. To test this hypothesis, we evaluated a panel of 15 hTERT mutants (SI Appendix, Table S1) for their effects on these two functions. We asked if we could find separation of function mutations that would prevent hTERT from elongating telomeric DNA yet not alter its ability to enhance cell proliferation. The identification of such a mutant would constitute strong genetic evidence that telomere elongation and enhancement of cell proliferation are independent functions of hTERT.
The panel included hTERTDN (34) that harbors inactivating amino acid substitutions in the catalytic domain, a carboxyl-terminal HA epitope-tagged protein (hTERT-HA) that renders it unable to maintain telomeres (23, 35), and a set of mutants that contain six amino acid substitutions. These substitutions were in “functional” domains that are essential for hTERT catalytic activity in vitro and/or telomere elongation and cellular lifespan extension in vivo, or in “linker” regions that are dispensable for hTERT activity, telomere elongation, and lifespan extension (SI Appendix, Fig. S2 A and B) (32, 33). These mutants were transduced into HMECs, and the cells assayed for hTERT catalytic activity, telomere elongation, and enhancement of cell proliferation in mitogen-limited medium.
We identified a domain in the amino-terminal region of hTERT that was essential for telomere elongation but dispensable for enhancing cell proliferation. This 6-amino acid substitution mutation starts at +32 in domain IA of the N-terminus of hTERT (32). Although the IA mutant (hTERTIA−) was catalytically active in vitro (Fig. 2A), it was unable to elongate telomeres in vivo (Fig. 2B), possibly because of an inability to interact with an accessory protein(s) necessary for localization to the telomere, as has been observed with similar N-terminal mutants (36). In mitogen-limited conditions, HMECs expressing hTERTIA− proliferated at an increased rate like that seen with cells expressing WT hTERT (Fig. 2C). We observed a comparable trend in BrdU incorporation (Fig. 2D). To further explore why an increased number of HMECs expressing hTERTIA− are in S phase, we examined the levels of key cell cycle modulators in these cells. We observed that cells expressing WT hTERT or hTERTIA− show increased cyclin D1, cyclin A2, and E2F1 protein levels (Fig. 2E). We also observed increased hyperphosphorylation of pRB (Fig. 2E), suggesting increased cyclin-dependent kinase activity in these cells. Thus, hTERTIA− expression caused alterations in cell cycle proteins and was sufficient to confer a proliferative advantage to HMECs despite being unable to elongate telomeres.
Fig. 2.
Enhanced cell proliferation and telomere elongation are independent functions of hTERT. HMECs were stably transduced with the indicated retroviral constructs (Vec, vector; WT, WT hTERT; IA-, hTERTIA−). (A) hTERT catalytic activity. The graph represents the averages ± SEM of three TRAP assays. (B) Southern blot of telomeric terminal restriction fragments from late passage HMECs with empty vector (lane 2), WT hTERT (lane 3), or hTERTIA− (lane 4). The blot is representative of five experiments performed by using cells derived from four independent transductions. Lane 1 is the marker (kb). (C) Cell proliferation in minimal medium. Results are averages ± SD from one representative of three growth assays. (D) Cells were labeled with BrdU. Representative flow cytometry profiles and percentages of BrdU-positive cells are shown. The graph shows the averages ± SEM of three experiments. (E) Cell lysates were analyzed by Western blotting for levels of the indicated proteins, and blots shown are representative of three experiments. Actin and GRB2 serve as loading controls. ppRB represents the hyperphosphorylated form of pRB.
Cells expressing hTERT-HA had a similar separation of function phenotype. The hTERT-HA mutant is catalytically active in vitro (23, 35) (SI Appendix, Fig. S1A), yet unable to maintain telomeres in vivo (23, 35). We found that hTERT-HA conferred a cell proliferation advantage in a mitogen-deficient medium, and promoted increased BrdU incorporation, compared with vector-transduced cells (SI Appendix, Fig. S1 B and C). The results with hTERT-HA were more variable than seen with other mutants, and its efficacy in mediating a proliferation advantage appeared intermediate compared with WT hTERT. Nevertheless, these results with hTERT-HA were interesting in light of previous results in fibroblasts showing that hTERT-HA can substitute for WT hTERT and collaborate with H-ras to promote tumorigenesis in vivo in a telomere length-independent manner (23). Together, the data from the hTERTIA− and hTERT-HA mutants showed that the cell proliferation and telomere elongation functions of hTERT were separable.
Telomerase Catalytic Activity Is Necessary for Enhancing cell Proliferation.
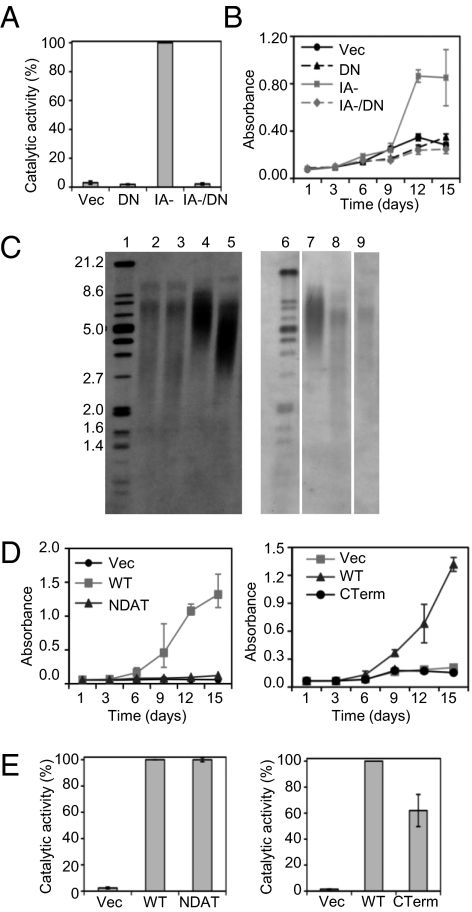
Analysis of the remaining mutants showed that there was substantial overlap in hTERT functional domains required for these two biological activities. This included the domains necessary for catalytic activity: the RNA binding domains (SI Appendix, Fig. S2C) and the enzyme's active site. In one extensively studied mutant, hTERTDN, the essential aspartic acid and valine residues at positions 710 and 711 in the active site are substituted with alanine and isoleucine, respectively (34). This mutant was catalytically inactive (Fig. 3A), failed to maintain telomeres (Fig. 3C, lanes 3 and 9) or extend lifespan (Fig. 6B), and was unable to enhance cell proliferation (Fig. 3B) (13). We introduced these two substitution mutations into hTERTIA−, thus eliminating its enzymatic activity (Fig. 3A). We found that the hTERTIA−/DN double mutant did not enhance cell proliferation (Fig. 3B), which suggested that hTERTIA− was not a gain-of-function mutant and confirmed the essential contribution of catalytic activity to the proliferation phenotype.
Fig. 3.
hTERT catalytic activity is necessary but not sufficient to enhance cell proliferation. HMECs were transduced with the indicated retroviral vectors (Vec, vector; DN, catalytically inactive hTERT; IA-, hTERTIA−, IA-/DN, hTERTIA−/DN double mutant; WT, WT hTERT; NDAT, hTERTN-DAT 116, CTerm, hTERTCTerm). (A) hTERT catalytic activity. The graph represents the averages ± SEM of two TRAP assays. (B) Cell proliferation in minimal medium by crystal violet staining. Results are averages ± SD from one assay, and were confirmed in an independent experiment. (C) Telomere length determination by Southern blotting of telomeric terminal restriction fragments in late passage HMECs with hTERTN-DAT 116 (lane 5, left blot) or hTERTCTerm (lane 8, right blot). Late passage HMECs with vector (lane 2), hTERTDN (lanes 3 and 9) or WT hTERT (lanes 4 and 7) serve as controls. Lanes 1 and 6 are molecular weight markers (kb). The blots are representative of three independent experiments. (D) Proliferation of HMECs in minimal medium. Data are averages ± SD from one representative of three growth assays. (E) hTERT catalytic activity. The graphs show the averages ± SEM of three TRAP assays.
Fig. 6.
The DNA damage response and proliferation functions of hTERT are separable. HMECs were stably transduced with the indicated retroviral vectors (WT, WT hTERT; DN, catalytically inactive hTERT; NDAT92, hTERTN-DAT 92; CDAT, hTERTC-DAT 1127). (A) hTERT catalytic activity was measured by the TRAP assay (±SEM). (B) Late passage HMECs were stained to detect SA-β-gal activity (blue). Images are representative of cells derived in two independent transductions. (C) Cell proliferation of early passage cells in minimal medium was measured by crystal violet staining. Results are averages ± SD from one assay, and were confirmed in an independent experiment. (D) Cells were labeled with BrdU. Representative flow cytometry profiles and percentage of BrdU-positive cells are shown.
Two other mutants, hTERTN-DAT 116 and hTERTC-Term, which are defective in telomere elongation (Fig. 3C), likely because they are unable to interact with accessory proteins necessary for telomere recruitment (36), also abrogated proliferation enhancement (Fig. 3D). Of significance was that hTERTN-DAT 116 and hTERTC-Term were catalytically active (Fig. 3E), indicating that hTERT catalytic activity, although necessary, was not sufficient for proliferation enhancement.
None of the mutations in hTERT linker domains, which are not required for catalytic activity (SI Appendix, Fig. S2A) or telomere maintenance (32, 33), inactivated its ability to enhance cell proliferation (SI Appendix, Fig. S2C). Therefore, we were unable to identify in this mutant set a domain of hTERT uniquely required for enhancing cell proliferation.
Finally, mutation of the hTERT 14–3-3 binding domain (37), which results in a predominantly cytoplasmic localization (33), had no effect on catalytic activity (SI Appendix, Fig. S2A). However, this mutant was unable to enhance proliferation (SI Appendix, Fig. S2C). Thus, hTERT-enhanced proliferation required nuclear localization and was probably unrelated to potential functions in the cytoplasm or mitochondria (21, 26, 27).
Cellular Lifespan Extension by hTERT Is Independent of Telomere Elongation.
Normal somatic human cells undergo a limited number of cell divisions before senescence. The stepwise erosion of telomeres during successive cell divisions is thought to underlie the counting mechanism that triggers irreversible stress responses, cellular senescence, and a permanent proliferative arrest. Expression of hTERT is therefore thought to promote extension of proliferative lifespan by preventing telomere loss (38).
The discovery of hTERT mutants that cannot elongate telomeres but nevertheless greatly prolong the proliferative lifespan of primary human fibroblasts is not entirely consistent with this hypothesis (39, 40). Our observations with HMECs confirmed these previous experiments in fibroblasts. Three mutants, hTERTIA−, hTERTN-DAT 116, and hTERTC-Term, were catalytically active but defective in telomere elongation (Fig. 2 A and B and Fig. 3 C and E). Despite being unable to elongate telomeres, late passage HMECs expressing hTERTIA− or hTERTN-DAT 116 did not show large, flat, vacuolated cells indicative of senescence, were negative for senescence-associated β-gal activity (SA-β-gal; Fig. 4A), and could be passaged for at least 5 mo (Fig. 4B). Vector-transduced HMECs always underwent irreversible senescence in approximately 6 wk (Fig. 4B). Cells expressing hTERTC-Term were heterogeneous. Late passage hTERTC-Term populations always showed some areas of large, flat, vacuolated cells with positive staining for SA-β-gal (SI Appendix, Fig. S3A). At later passages, cells expressing hTERTC-Term exhibited a brief plateau in proliferation, but eventually they demonstrated an extended lifespan (SI Appendix, Fig. S3B). When assayed at approximately 4 mo after senescence of vector control cells, HMECs with hTERTIA−, hTERTN-DAT 116, or hTERTC-Term all continued to express hTERT catalytic activity at levels similar to cells with WT hTERT (Fig. 4C and SI Appendix, Fig. S3C). Nevertheless, telomeres in cells expressing each of these mutants continuously shortened (Fig. 4D and SI Appendix, Fig. S3D). This showed that extended lifespan was not caused by activation of the alternative lengthening of telomeres mechanism of telomere elongation, and ruled out the possibilities that it was caused by reversion of the mutations or endogenous hTERT activation.
Fig. 4.
Cellular lifespan extension by hTERT can be uncoupled from telomere elongation. HMECs were stably transduced with the indicated retroviral vectors (Vec, vector; WT, WT hTERT; IA-, hTERTIA−; NDAT, hTERTN-DAT 116). (A) HMECs at passage 16 were stained for SA-β-gal (blue). (B–D) HMECs were passaged by counting cells and replating 0.2 million cells every 7 d (or all cells if less than 0.2 million were present) for 5 mo. (B) Live cell counts were determined at the specified passages. (C) hTERT catalytic activity at late passage. The graph shows the averages ± SEM of two TRAP assays. (D) HMECs with WT hTERT (lanes 2, 5, and 8), hTERTIA− (lanes 3, 6, and 9), or hTERTN-DAT 116 (lanes 4, 7, and 10) were harvested at the indicated passages and their telomere lengths compared. Lane 1 is the marker (kb). Results shown are representative of three experiments performed with cells derived from two independent transductions.
Telomere Elongation-Independent Lifespan Extension by hTERT Occurs Without Chromosomal Instability or DNA Damage Activation and Is Dependent on Telomere Capping.
All of the aforementioned mutants that conferred extended lifespan retained catalytic activity (Figs. 2A and 3E), and it remained possible that they could stably maintain very short telomeres that were sufficient to recruit telomere-binding proteins of the shelterin complex, such as the hPOT1 protein (41, 42). These proteins, together with hTERT, localize to chromosome ends creating a telomere “cap.” This cap is thought to distinguish natural chromosome ends from double strand breaks, thereby preventing the DNA rearrangements typically associated with broken chromosome ends and the triggering of a senescence-associated DNA damage response (41, 42). We therefore first examined HMECs transduced with WT hTERT, hTERTIA−, hTERTN-DAT 116, or hTERTC-Term for chromosome instability. We analyzed 20 metaphases from these HMECs at 4 mo after senescence of control cells. None showed chromosome abnormalities indicative of telomere dysfunction. In contrast, telomere-associated events were observed in many metaphases of the late passage (i.e., senescing) control HMECs (SI Appendix, Fig. S4A). This indicated that the chromosomes in HMECs with hTERTIA−, hTERTN-DAT 116, or hTERTC-Term were protected from telomere-associated DNA rearrangements despite the lack of telomere elongation.
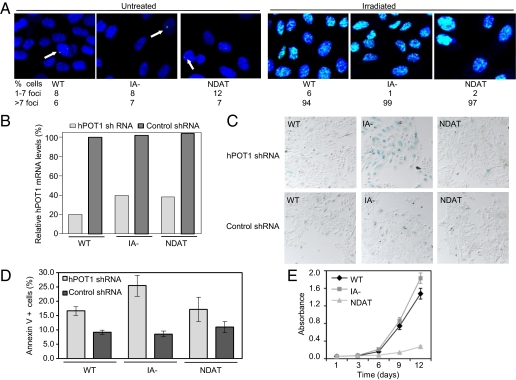
We further assessed the cells for activation of a DNA damage response resulting from telomere dysfunction by measuring formation of phospho-H2AX foci (43, 44). At 4 mo after senescence of control cells, cells expressing hTERT mutants that extended lifespan did not show more DNA damage foci than cells expressing WT hTERT (Fig. 5A and SI Appendix, Fig. S4B). As a control, we showed that all these cells remained able to assemble DNA damage foci after γ-irradiation (Fig. 5A and SI Appendix, Fig. S4B), demonstrating that the cellular responses to damaged DNA were intact. Together, these results showed that hTERT mutants that were unable to carry out telomere elongation could extend lifespan of HMECs without causing chromosome instability and activation of a DNA damage response, consistent with chromosome capping.
Fig. 5.
Telomere elongation-independent lifespan extension by hTERT occurs without DNA damage activation or enhanced proliferation and is dependent on telomere capping. HMECs were stably transduced with the indicated retroviral vectors (WT, WT hTERT; IA-, hTERTIA−; NDAT, hTERTN-DAT 116). (A) HMECs passaged for 4 mo (as described in Fig. 4) were left untreated or irradiated and stained for phospho-H2AX. Phospho-H2AX foci in at least 300 cells for each of the cell populations were scored and the average percentage of cells containing foci are shown. The images are representative of one of three independent experiments. (B–D) After 5 mo in culture, HMECs expressing indicated hTERT constructs were stably transduced with retroviral vectors expressing hPOT1 or control shRNAs. (B) hPOT1 mRNA levels were measured by qPCR and were normalized to HPRT1 levels in each sample. The results show the averages of duplicates and represent the percent hPOT1 knockdown relative to HMECs with WT hTERT plus control shRNA. (C) HMECs were stained to detect SA-β-gal activity (blue) at 4 d after drug selection. Images are representative of cells from two independent transductions. (D) Cells were stained for annexin V and analyzed by flow cytometry. Percentages of annexin V-positive cells are shown. The graph represent averages ± SEM of three experiments. (E) HMECs passaged for 5 mo (as described in Fig. 4) were analyzed for proliferation rates in minimal medium. Results are the averages ± SD from one assay, and were confirmed in another growth assay with independently transduced cells.
Finally, we directly tested whether inhibiting expression of hPOT1 could affect the ability of these hTERT mutants to extend lifespan. We stably transduced HMECs expressing the aforementioned hTERT mutants at 4 mo after senescence of control cells with an hPOT1 or control shRNA vector (45). By using quantitative real-time RT-PCR (qPCR), we observed 60% to 80% knockdown of endogenous hPOT1 mRNA expression in HMECs expressing WT hTERT, hTERTIA−, or hTERTN-DAT 116 (Fig. 5B). For HMECs expressing hTERTC-Term, we obtained cell populations expressing the control shRNA vector but were unable to isolate cells expressing the hPOT1 shRNA vector in two independent transduction attempts, suggesting that they may have a critical requirement for hPOT1. Furthermore, although we were able to isolate HMEC populations expressing hTERTIA− plus hPOT1 shRNA, these cells proliferated very slowly. We observed many large, flat, vacuolated cells, within 1 to 2 d after drug selection, and staining for SA-β-gal confirmed that a majority of these cells were senescent (Fig. 5C). In contrast, knockdown of hPOT1 in HMECs expressing WT hTERT or hTERTN-DAT 116 did not cause the rapid onset of senescence (Fig. 5C). However, these cells still proliferated more slowly than those expressing control shRNAs (SI Appendix, Fig. S5A). We therefore assessed them for apoptosis by measuring the percentage of annexin V-positive cells by flow cytometry, and observed an increased cell death in all cells expressing hPOT1 shRNAs (Fig. 5D and SI Appendix, Fig. S5B). Thus, hPOT1 knockdown in cells expressing these hTERT mutants results in decreased cell numbers associated with senescence and/or apoptosis. These results demonstrate that extended-lifespan cells with short telomeres still require shelterin proteins, such as hPOT1, and link the lifespan extension conferred by telomere elongation-defective hTERT mutants to chromosome capping.
Cellular Lifespan Extension by hTERT Can Be Uncoupled from Enhanced Proliferation.
We examined the abilities of these mutants (telomere elongation-negative and lifespan extension-positive) to cause enhanced proliferation in mitogen-limiting conditions at late passage, 4 mo after the controls had become senescent. As we had observed in young cells, hTERTIA− increased proliferation similar to WT hTERT, whereas hTERTN-DAT 116 and hTERTC-Term did not (Fig. 5E and SI Appendix, Fig. S4C). Thus, lifespan extension was observed with one mutant, hTERTN-DAT 116, and to a lesser extent with a second, hTERTC-Term, which were both defective in enhancing cell proliferation. Hence, lifespan extension and proliferative advantage functions of hTERT were independent.
An important conclusion, therefore, is that chromosome capping was insufficient to explain the proliferation advantage imparted by hTERT. At least two mutants that retained this capping function, hTERTN-DAT 116 and hTERTC-Term (as evidenced by lifespan extension without chromosome dysfunction), did not enhance cell proliferation. Conversely, hTERT-HA largely retains the proliferation function of hTERT (SI Appendix, Fig. S1 B and C), yet is unable to maintain telomeres (23, 35) or cause lifespan extension (SI Appendix, Fig. S1D). Thus, chromosome capping by hTERT was also not necessary for enhancing proliferation.
Enhanced Proliferation and Regulation of DNA Damage Signaling Are Separable Functions of hTERT.
It has been proposed that hTERT's ability to stimulate proliferation might be linked to suppression of telomere-associated DNA damage signals, as these signals might sensitize cells to the antiproliferative effect of diminished mitogens (44). Our results did not support this interpretation, because both hTERTN-DAT 116 and hTERTC-Term suppressed DNA damage responses associated with telomere erosion (Fig. 5A and SI Appendix, Fig. S4B), yet were not sufficient to enhance cell proliferation (Fig. 5E and SI Appendix, Fig. S4C).
We further tested whether these two functions were related by assessing formation of DNA damage foci in cells that had telomeres of different lengths; in early passage (i.e., passage 8) uninfected HMECs and HMECs transduced with vector control or WT hTERT at presenescent (i.e., passage 12) and late passages (i.e., passage 16). In contrast to previous studies (44), we observed that uninfected and presenescent HMECs did not show high DNA damage signaling as indicated by the similar low number of phospho-H2AX foci observed in these cell types, irrespective of whether they were transduced with hTERT (SI Appendix, Fig. S6 A and B). It was only at late passages that the difference in focus number between HMECs transduced with control vector or hTERT became apparent (SI Appendix, Fig. S6 A and B). Therefore, hTERT was able to enhance cell proliferation (Figs. 1–3) at a presenescent passage (i.e., passage 11) during which DNA damage signaling was not evident. Together, these results showed that the proliferation advantage function of hTERT was not related to its role in telomere-dependent DNA damage signaling.
It has also been reported that hTERT has a telomere-independent role in DNA damage signaling (25). In these experiments, hTERT was necessary for cells to respond normally to intrachromosomal DNA double-stranded breaks. Separation of function mutants, hTERTN-DAT 92 and hTERTC-DAT 1127, retained the ability to mediate DNA damage signaling despite being defective in telomere maintenance (25). We therefore asked whether hTERT-enhanced cell proliferation might be related to its telomere-independent effects on DNA damage signaling. We generated HMECs transduced with hTERTN-DAT 92 and hTERTC-DAT 1127. Both mutants were catalytically active (Fig. 6A), a prerequisite for enhancing proliferation (Fig. 3), but defective in conferring lifespan extension (Fig. 6B). Importantly, in mitogen-limiting conditions, HMECs expressing hTERTN-DAT 92 and hTERTC-DAT 1127 were unable to enhance proliferation (Fig. 6C). We observed a similar trend in BrdU incorporation (Fig. 6D). Together, these data showed that the telomere-independent DNA damage signaling and proliferation functions of hTERT were separable.
Enhanced Cell Proliferation Occurs Independently of Increases in Wnt Signaling But Is Linked to Decrease in RMRP Levels.
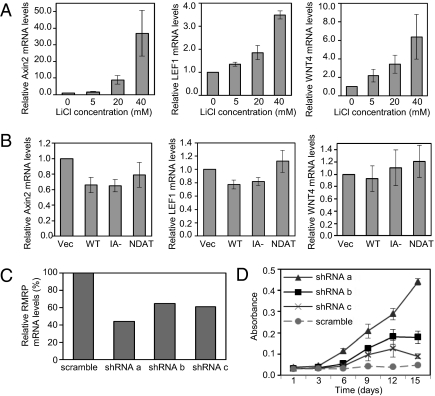
Recent studies have shown that mTERT interacts with Brg-1, which binds β-catenin, and is thereby recruited to Wnt target genes, activating their transcription (16, 17). We addressed whether this might contribute to the enhanced cell proliferation of hTERT-transduced HMECs. We first tested whether specific Wnt pathway genes, which have been reported to be targets of mTERT or important in breast cancer (16, 17, 46), were responsive to Wnt signaling in HMECs. Axin2, LEF1, WNT4, WNT11, and SMAD7 mRNA levels were increased in HMECs treated with the Wnt pathway activator LiCl (Fig. 7A and SI Appendix, Fig. S7A), whereas WNT5A and WNT1 were not. Brg-1 was expressed at equal levels in control and hTERT-transduced HMECs (SI Appendix, Fig. S7B). However, none of the Wnt-responsive genes was induced by hTERT (Fig. 7B and SI Appendix, Fig. S7C). Thus, TERT-mediated Wnt transcriptional induction likely does not contribute to enhanced cell proliferation in HMECs.
Fig. 7.
Enhanced cell proliferation occurs independently of increases in Wnt signaling but is linked to a decrease in RMRP levels. (A and B) The mRNA levels of the specified genes were measured by qPCR and were normalized to HPRT1 in each sample. (A) HMECs were treated with the indicated concentrations of LiCl for 18 h before harvesting and RNA isolation. The results are the averages ± SEM of four experiments and show the fold change in mRNA levels relative to untreated HMECs. (B) HMECs were stably transduced with the indicated retroviral vectors (Vec, vector; WT, WT hTERT; IA-, hTERTIA−; NDAT, hTERTN-DAT 116). The results are the averages ± SEM of five experiments and show the fold change in mRNA levels relative to HMECs with the vector control. (C and D) HMECs with empty vector were stably transduced with the indicated RMRP shRNAs. (C) RMRP mRNA levels were measured by qPCR and normalized to β-actin in each sample. The graph shows the averages of duplicates and represents the percent RMRP knockdown relative to HMECs with RMRP scramble shRNA control. (D) Cell proliferation in minimal medium was measured by crystal violet staining. Results are averages ± SEM and were confirmed in another experiment with independently transduced cells.
Recent studies also indicate that hTERT has terminal transferase (47) and RNA-dependent RNA polymerase (24) activities that are dependent on its catalytic domain but hTR-independent. We asked whether hTERT-mediated proliferation enhancement requires hTR by stably expressing hTR shRNAs. However, we were unable to isolate such HMEC populations, possibly because hTR suppression triggers rapid telomerase-independent growth arrest and apoptosis through ATM and ATR-mediated DNA damage checkpoint responses (48, 49). As an alternative, we used the VA13 fibroblast cell line, which lacks both hTR and hTERT expression and maintains telomeres via the alternative lengthening of telomeres pathway. We confirmed lack of endogenous hTERT expression and expression of ectopic hTERT (SI Appendix, Fig. S8A). Cells were grown in primary fibroblast media, specifically omitting FGF and EGF to obtain mitogen-limited conditions that inhibited proliferation (SI Appendix, Fig. S8B). We observed a modest but very reproducible and statistically significant enhancement of proliferation by hTERT in three experiments (SI Appendix, Fig. S8C), suggesting that the noncanonical activity of hTERT that enhances proliferation is hTR-independent.
hTERT can also use the noncoding RNA, RMRP, as a template to generate double-stranded RNAs that are processed into siRNAs; one consequence of this is feedback suppression of RMRP itself (24). The biological effects of the hTERT–RMRP pathway are unknown, and we therefore tested its role in hTERT-enhanced cell proliferation. As a baseline, we found that hTERT reduced RMRP expression by 60% in HMECs (SI Appendix, Fig. S9A), and that this amount of RMRP suppression could be mimicked by transduction of HMECs with RMRP shRNAs (Fig. 7C). Remarkably, knockdown of RMRP by shRNAs alone was sufficient to enhance HMEC proliferation (Fig. 7D). Transduction of RMRP shRNAs into HMECs expressing hTERT caused only a small further decrease in RMRP levels and a marginal additional enhancement of proliferation (SI Appendix, Fig. S9B). These data link the ability of hTERT to modulate RMRP levels to its functions in enhancing HMEC proliferation.
Discussion
We have used an extensive panel of hTERT mutants (SI Appendix, Table S1) to explore whether the diversity of biological functions attributed to hTERT are mediated by the same or different mechanisms. Our major conclusion is that the ability of hTERT to enhance proliferation, which was caused by increased cell division and decreased apoptosis, could be genetically separated from its functions in telomere elongation, lifespan extension, and DNA damage responses. Further, we found that enhanced cell proliferation in mitogen-limited conditions was not caused by altered Wnt signaling, but was accompanied by changes in key cell cycle regulators and was linked to an hTERT-catalyzed decrease in the expression of the noncoding RNA, RMRP.
We were also able to uncouple telomere elongation from lifespan extension (Fig. 4 and SI Appendix, Fig. S3) as previously seen in primary human fibroblasts ectopically expressing hTERT mutants (39, 40). All telomere elongation-defective mutants that conferred extended lifespan retained catalytic activity. It therefore remained possible that they could stably maintain very short telomeres that were sufficient to recruit telomere-binding proteins, such as those of the shelterin complex. This would support the idea that hTERT can greatly extend cellular proliferative lifespan in the absence of telomere elongation, possibly via chromosome capping (41, 42). Indeed, we found that, in these extended-lifespan cells, which have a stable karyotype, suppressing expression of the hPOT1 capping protein results in senescence and/or apoptosis (Fig. 5 B–D and SI Appendix, Fig. S5 A and B), consistent with previous hPOT1 knockdown studies that have shown that it causes apoptosis in immortalized cancer cell lines (45, 50) and senescence in primary cells (45, 50, 51). We further observed that the phenotypes resulting from hPOT1 knockdown were rapidly lost. The loss of the senescence phenotype occurred within 2 wk (compare Fig. 5C vs. SI Appendix, Fig. S5C) after knockdown, and proliferation defects were completely lost within an additional 2-wk period. We confirmed that hPOT1 levels remained very low at that time. The cause of this recovery remains to be investigated, and it is possible that a subpopulation of cells that was tolerant to low levels of hPOT1 was selected. Importantly, we also demonstrated that the lifespan extension and proliferative functions of hTERT are separable, and therefore mechanistically distinct. We found N- and C-terminal hTERT mutants that promoted lifespan extension, but were unable to confer a proliferative advantage in mitogen-limiting conditions (Figs. 4 and 5 and SI Appendix, Figs. S3 and S4).
Previous studies on alternative biological functions of hTERT showed that it could also modulate DNA damage responses. Ectopic hTERT expression altered the posttranslational modifications on histone tails and thus promoted a chromatin structure that facilitated DNA damage responses (25). hTERT mutants that were unable to maintain telomeres remained active in this DNA damage response pathway, provided they retained catalytic activity (25). We found that the same mutants (i.e., DNA damage response-positive and telomere maintenance-negative) were unable to enhance proliferation (Fig. 6). Thus, the role of hTERT in proliferation was also separable from its role in this DNA damage response, and therefore likely involved a different mechanism. In sum, our analyses showed that at least four biological actions of hTERT—enhancement of cell proliferation, telomere elongation, cellular lifespan extension, and regulation of DNA damage responses—were genetically separable and therefore, at least in part, functionally independent.
The most surprising conclusion that can be drawn from this collection of studies may be that the diverse biological roles of hTERT all require hTERT catalytic activity, even though all but one are functionally independent of telomere elongation. Further, an hTERT mutation that resulted in a predominantly cytoplasmic localization (33), but had no effect on catalytic activity (SI Appendix, Fig. S2A), was unable to enhance proliferation (SI Appendix, Fig. S2C). Thus, hTERT-enhanced proliferation required nuclear localization and was probably unrelated to potential functions in the cytoplasm or mitochondria (21, 26, 27). In addition to our experiments in HMECs, experiments in other tissue types such as cardiac myocytes (22), mouse embryonic fibroblasts (18), and human fibroblasts (23) show that catalytic activity is necessary for hTERT to increase cell proliferation. In contrast, the ability of mTERT to stimulate proliferation of skin and hair follicle stem cells (14, 15) by modulating the Wnt signaling pathway is independent of its catalytic activity (16, 17). Consistent with our findings that hTERT catalytic activity and RNA binding are required for enhanced proliferation of HMECs (Fig. 3B and SI Appendix, Fig. S2C) (13), we found that Wnt-responsive genes were not induced by hTERT in HMECs (Fig. 7 A and B and SI Appendix, Fig. S7) and that, therefore, their enhanced proliferation phenotype was not related to Wnt signaling. Instead we found that the hTERT–RMRP pathway, which results in generation of siRNAs and feedback suppression of RMRP, is linked to the enhanced cell proliferation phenotype in HMECs (Fig. 7 and SI Appendix, Fig. S9). Thus, there may be more than one mechanism by which hTERT can stimulate cell proliferation. RMRP has varied cellular functions, including processing RNAs required to generate primers for mitochondrial DNA replication, pre-rRNA processing during rRNA maturation, mRNA cleavage of cell cycle genes, and potential regulation of gene expression via complexing with hTERT to generate siRNAs (24, 52). The pathway(s) through which RMRP might impact the proproliferative effect of hTERT remains to be elucidated. Further investigation of these pathways, as well as of alternative hTERT activities and its other binding partners, would not only improve our understanding of how hTERT mediates its diverse roles but could also uncover new targets for the development of anti-telomerase cancer therapeutic agents.
Materials and Methods
Cells and Cell Culture.
HMECs were obtained and cultured as described previously (13). Retroviral vectors (SI Appendix, SI Materials and Methods and Table S1) used for expressing WT/mutant hTERT [gift from C. M. Counter (Duke University Medical Center, Durham, NC) (32, 33) and R. A. Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA) (23, 34)], hPOT1 shRNAs [gift from C. M. Counter (45)], or RMRP shRNAs were transduced into HMECs, followed by drug selection to obtain stably transduced cells. Long-term passaging was carried out for 5 mo and the live cell counts determined at each passage. VA13 cells (CCL-75.1; American Type Culture Collection) were cultured in DMEM with 10% bovine growth serum and thereafter in complete primary fibroblast medium with all supplements (SI Appendix, SI Materials and Methods). When studying effects of hTERT in mitogen-limiting conditions (i.e., minimal medium), human EGF, and FGF were omitted from the medium.
Proliferation and Cell Death Assays.
For the cell growth assay (SI Appendix, SI Materials and Methods), HMECs or VA13 cells were cultured in minimal/complete medium, fixed at indicated time points, and stained with crystal violet, which was eluted with acetic acid and absorbance measured at 595 nm. BrdU incorporation was assayed as described previously (53). Annexin V staining and analysis was carried out per manufacturer instructions by using the APOAF annexin V FITC apoptosis detection kit (Sigma).
Western Blotting.
Cells were lysed in 1% Nonidet P-40 lysis buffer, and 20 to 40 μg protein resolved by SDS-PAGE, transferred to PVDF membranes, and probed with primary antibodies (SI Appendix, SI Materials and Methods) for cyclin D1, cyclin A2, pRB, E2F1, Brg1, Actin, or Grb2. After incubation with horseradish peroxidase-conjugated secondary antibodies, bands were visualized by using the ECLPlus Western blotting detection system.
Quantitative Real-Time RT-PCR.
Cells were harvested using Qiazol lysis reagent, total RNA isolated, and cDNA synthesized by using Taqman reverse transcription reagents. Real-time PCR was carried out by using Taqman Universal PCR Master Mix and primer probe sets (SI Appendix, SI Materials and Methods) for LEF1, Axin2, WNT4, SMAD7, WNT11, and POT1 (Applied Biosystems) or SYBR Green qPCR SuperMix (Invitrogen) and RMRP primers as described previously (24). mRNA levels of each sample were normalized to expression of HPRT1 or β-actin controls in that sample.
Telomerase Repeat Amplification Protocol Assay (TRAP) and Telomere Length Determination by Terminal Restriction Fragment Analysis.
Telomerase activity and telomere length were measured per instructions in the Telomerase TeloTAGGG PCR ELISA and Telomere Length Assay Kits, respectively (no. 11854666910 and 12209136001; Roche; SI Appendix, SI Materials and Methods). TRAP PCRs were each done in duplicate.
DNA Damage Analysis.
Untreated or irradiated (10 Gy) cells were stained with an anti–phospho-histone H2AX serine 139 antibody (no. 05–636; Upstate Biotechnology; SI Appendix, SI Materials and Methods), followed by Alexa Fluor 488-conjugated goat anti-mouse antibody and TO-PRO 3 (A11029 and T-3605; Invitrogen/Molecular Probes) to visualize DNA damage foci.
Karyotyping.
Metaphase spreads were prepared from HMECs treated with colcemid (0.2 μg/mL) followed by 0.075 M KCl hypotonic treatment. Standard G-banding karyotypic analysis was performed on 20 metaphase spreads for each cell type.
Senescence-Associated β-Gal Staining.
HMECs were stained with SA-β-gal as described previously (54). Cells were imaged by using a Nikon SMZ1500 microscope.
Experimental procedures are described in further detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the J.M.R. laboratory and Drs. Laura Smith, Hemant Varma, Keith Loeb, Dan Gottschling, and Toshiyashu Taniguchi for helpful discussions. We also thank Dr. Min Fang and the Seattle Cancer Care Alliance Cytogenetics Laboratory for the karyotype analysis. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health/National Cancer Institute Grant R01 CA120294 (to J.M.R.), the Thomsen Family, and Susan G. Komen for the Cure Foundation (S.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 19847.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112414108/-/DCSupplemental.
References
- 1.Stewart SA, Weinberg RA. Telomeres: Cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 4.Bollmann FM. The many faces of telomerase: Emerging extratelomeric effects. Bioessays. 2008;30:728–732. doi: 10.1002/bies.20793. [DOI] [PubMed] [Google Scholar]
- 5.Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res. 2008;18:725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 6.González-Suárez E, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artandi SE, et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci USA. 2002;99:8191–8196. doi: 10.1073/pnas.112515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 9.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 11.Chadeneau C, Siegel P, Harley CB, Muller WJ, Bacchetti S. Telomerase activity in normal and malignant murine tissues. Oncogene. 1995;11:893–898. [PubMed] [Google Scholar]
- 12.Broccoli D, Godley LA, Donehower LA, Varmus HE, de Lange T. Telomerase activation in mouse mammary tumors: Lack of detectable telomere shortening and evidence for regulation of telomerase RNA with cell proliferation. Mol Cell Biol. 1996;16:3765–3772. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 14.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 16.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geserick C, Tejera A, González-Suárez E, Klatt P, Blasco MA. Expression of mTert in primary murine cells links the growth-promoting effects of telomerase to transforming growth factor-beta signaling. Oncogene. 2006;25:4310–4319. doi: 10.1038/sj.onc.1209465. [DOI] [PubMed] [Google Scholar]
- 19.Stampfer MR, et al. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor beta growth inhibition in p16INK4A(-) human mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudognon C, et al. Death receptor signaling regulatory function for telomerase: hTERT abolishes TRAIL-induced apoptosis, independently of telomere maintenance. Oncogene. 2004;23:7469–7474. doi: 10.1038/sj.onc.1208029. [DOI] [PubMed] [Google Scholar]
- 21.Del Bufalo D, et al. Involvement of hTERT in apoptosis induced by interference with Bcl-2 expression and function. Cell Death Differ. 2005;12:1429–1438. doi: 10.1038/sj.cdd.4401670. [DOI] [PubMed] [Google Scholar]
- 22.Oh H, et al. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc Natl Acad Sci USA. 2001;98:10308–10313. doi: 10.1073/pnas.191169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart SA, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masutomi K, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haendeler J, Hoffmann J, Brandes RP, Zeiher AM, Dimmeler S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol Cell Biol. 2003;23:4598–4610. doi: 10.1128/MCB.23.13.4598-4610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell. 2004;3:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- 28.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 29.Shay JW, Wright WE. Use of telomerase to create bioengineered tissues. Ann N Y Acad Sci. 2005;1057:479–491. doi: 10.1196/annals.1356.037. [DOI] [PubMed] [Google Scholar]
- 30.Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banik SS, et al. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol Cell Biol. 2002;22:6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn WC, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 35.Counter CM, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armbruster BN, Etheridge KT, Broccoli D, Counter CM. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol Cell Biol. 2003;23:3237–3246. doi: 10.1128/MCB.23.9.3237-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seimiya H, et al. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Wang H, Bishop JM, Blackburn EH. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M, Xu L, Blackburn EH. Catalytically active human telomerase mutants with allele-specific biological properties. Exp Cell Res. 2003;288:277–287. doi: 10.1016/s0014-4827(03)00217-9. [DOI] [PubMed] [Google Scholar]
- 41.Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 42.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 43.Paull TT, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 44.Beliveau A, et al. p53-dependent integration of telomere and growth factor deprivation signals. Proc Natl Acad Sci USA. 2007;104:4431–4436. doi: 10.1073/pnas.0700260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Ayyanan A, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lue NF, et al. Telomerase can act as a template- and RNA-independent terminal transferase. Proc Natl Acad Sci USA. 2005;102:9778–9783. doi: 10.1073/pnas.0502252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kedde M, et al. Telomerase-independent regulation of ATR by human telomerase RNA. J Biol Chem. 2006;281:40503–40514. doi: 10.1074/jbc.M607676200. [DOI] [PubMed] [Google Scholar]
- 49.Stohr BA, Blackburn EH. ATM mediates cytotoxicity of a mutant telomerase RNA in human cancer cells. Cancer Res. 2008;68:5309–5317. doi: 10.1158/0008-5472.CAN-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esakova O, Krasilnikov AS. Of proteins and RNA: The RNase P/MRP family. RNA. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]