Abstract
Mitochondrial proteins are almost exclusively imported into mitochondria from the cytosol in an unfolded or partially folded conformation. Regardless of whether they are destined for the outer or inner membrane, the intermembrane space, or the matrix, proteins begin the importation process by crossing the mitochondrial outer membrane via a specialized protein import machinery whose main component is the Tom40 channel. High-resolution ion conductance measurements through the Tom40 channel in the presence of the mitochondrial presequence peptide pF1β revealed the kinetics of peptide binding. Here we show that the rates for association kon and dissociation koff strongly depend on the applied transmembrane voltage. Both kinetic constants increase with an increase in the applied voltage. The increase of koff with voltage provides strong evidence of peptide translocation. This allows us to distinguish quantitatively between substrate blocking and permeation.
Introduction
Understanding the mechanism of protein transport across lipid membranes has been one of the most challenging topics over the past three decades (1, 2, 3, 4, 5, 6, 7). It is an essential process in all living cells whereby proteins are sorted to different cellular compartments or exported out of the cell. Most proteins are translocated through protein-conducting channels in an unfolded state, presumably driven by motor proteins attached to the channels, by a power stroke, by Brownian ratchet motion, or by entropic pulling.
Until now, most studies that reported on the mechanism of protein and peptide translocation across lipid membranes used the ion channels α-hemolysin and aerolysin reconstituted into planar lipid bilayers or nanofabricated solid-state pores as model translocation systems (8, 9, 10, 11, 12, 13). When reconstituted in planar lipid membranes, α-hemolysin and aerolysin are known to exhibit silent open pores, thus allowing electrical detection of substrate-induced channel blockages (9, 11). Analysis of the frequency and duration of single substrate-induced blockage events revealed detailed insights into the kinetics of substrate binding and substrate release, and thus the energetics of peptide transport through these pores. The increase of koff with voltage demonstrated translocation of peptides through α-hemolysin. However, to date, this has not been shown for protein translocases of mitochondria and other organelles.
Here, we study the interaction of a mitochondrial presequence peptide with the mitochondrial protein-conducting channel Tom40. Tom40 is the central component of TOM, the preprotein translocase of the outer mitochondrial membrane. It is an integral membrane protein with a β-barrel structure and an estimated inner pore diameter of 22–25 Å (14, 15, 16). Electron microscopy, mass spectrometry, and biochemical studies suggested that two or three channels are assembled within one TOM complex (15, 16, 17). Together with the single-pass membrane receptor Tom22 and the small Tom proteins Tom5, Tom6, and Tom7, it forms the general import pore (GIP) of the TOM machinery. Associated with the GIP are the single transmembrane anchored receptors Tom70 and Tom20, which form the initial recognition site for preproteins with multiple internal and amino-terminal targeting signals, respectively (18).
To elucidate the relevance of the individual partners of the TOM machinery, we performed high-resolution ion-conductance measurements to monitor individual presequence peptide translocation events through single Tom40 molecules purified from Neurospora crassa mitochondria at submillisecond resolution. Previous single-channel recordings of the mitochondrial protein-conducting channel Tom40 from Saccharomyces cerevisiae and N. crassa revealed a voltage-dependent increase in the frequency of channel closures in the presence of mitochondrial targeting peptides and provided a first glimpse into peptide interaction with TOM (14, 15, 19, 20, 21, 22, 23, 24, 25, 26, 27). However, these experiments did not allow the rate of dissociation koff to be estimated as a function of the voltage required to demonstrate translocation. Although it seemed like an obvious conclusion to interpret the channel closures as individual peptide translocation events, the analysis of such experiments was complicated by the fact that substrate-induced channel blockage had to be distinguished from spontaneous channel gating. In addition, peptide blockage does not imply translocation, because the peptide just binds to the channel surface.
To address this issue, we improved the time resolution of the ion-current recordings to resolve the peptide-binding kinetics at the single-molecule level and determined koff as a function of voltage. Our data provide a quantitative description of the relevant kinetic and electric parameters for peptide partitioning and translocation through the Tom40 channel.
Materials and Methods
Purification of Tom40
Tom40 protein was purified from the N. crassa strain GR-107 (28), which contains a hexahistidinyl-tagged form of Tom22, as previously described (15, 29) with minor modifications. Isolated mitochondria of the N. crassa strain GR-107 were solubilized at a protein concentration of 10 mg/ml in 1% (w/v) n-dodecyl β-D-maltoside (DDM; Glycon Biochemicals, Luckenwalde, Germany), 20% (v/v) glycerol, 300 mM NaCl, 20 mM imidazole, 20 mM Tris (pH 8.5), and 1 mM phenyl methyl sulfonyl fluoride (PMSF) for 30 min at 4°C. After centrifugation at 130,000 × g, the clarified extract was filtered and loaded onto an Ni-NTA column (GE Healthcare). The column was rinsed with 10 column volumes of 0.1% DDM, 10% glycerol, 300 mM NaCl, and 20 mM Tris (pH 8.5). Tom40 was directly eluted with 3% (w/v) n-octyl β-D-glucopyranoside (OG; Glycon Biochemicals, Luckenwalde, Germany), 2% (v/v) dimethyl sulfoxide (DMSO), and 20 mM Tris (pH 8.5). Tom22 and other bound proteins were eluted with buffer containing 0.1% DDM, 2% DMSO, 1 M imidazole, and 20 mM Tris (pH 8.5). Alternatively, Tom40 was isolated from purified TOM core complex. To that end, purified TOM core complex (25) was loaded onto an Ni-NTA affinity column equilibrated with 0.1% DDM, 2% DMSO, and 20 mM Tris (pH 8.5). The column was washed with 5–10 column volumes of equilibration buffer, and Tom40 was eluted with 3% OG, 2% DMSO, and 20 mM Tris (pH 8.5). The purity of the isolated protein (∼0.4 mg/ml) was assessed by means of sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and Coomassie Brilliant Blue staining.
Synthesis of blocking peptide
A polypeptide corresponding to the first 31 residues of the precursor of the S. cerevisiae F1-ATPase α-subunit (Ac-MVLPRLYTATSRAAFKAAKQS-APLLSTSWKR-NH2, pF1β, molecular mass = 3451.0 Da (15, 26)) was used as the substrate for Tom40. The polypeptide was custom-synthesized, confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and had a high-performance liquid chromatography purity of at least 95% (Biosyntan, Berlin, Germany). Fresh stock solutions of polypeptides were prepared for each experiment with peptide concentrations of 0.1 mg/ml in water and kept at 4°C for a maximum of 1 day. For channel blockage experiments, the peptide concentrations were in the micromolar range.
Single-channel conductance measurements
Purified Tom40 was reconstituted into virtually solvent free planar lipid bilayers according to standard protocols (30, 31, 32). Classical planar lipid bilayers were formed with the monolayer opposition technique across an 80–100 μm diameter circular aperture in a 25 μm thick polytetrafluoro-ethylene film (Goodfellow, Cambridge, UK) that was tightly glued between two Delrin chambers. Each chamber contained 2 ml of an aqueous bathing solution (1 M KCl, 20 mM MES, pH 6) and a bilayer was formed by layering 1 μl of a 5 mg/ml solution of 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (DPhPC; Avanti Polar Lipids, Alabaster, AL) dissolved in pentane on the buffer surface on each side. Purified Tom40 was added to the cis side of the membrane at a final concentration of 5 × 10−4 mg/ml, and protein insertion was facilitated by mixing the contents of the chamber and applying a transmembrane potential of 300 mV. Electrical recordings were made through a pair of Ag/AgCl electrodes (World Precision Instruments, Sarasota, FL). One electrode was used as the ground (cis) and the other (trans) was connected to an Axon Instruments 200B amplifier with a capacitive headstage, digitized by an Axon Digidata 1440A digitizer and computer-controlled by Clampex 10.0 software (all from Axon Instruments, Foster City, CA). The data were filtered by an analog low-pass, four-pole Bessel filter at 10 kHz, and digitally sampled at 50 kHz. Data from several 2- to 10-min current recordings were analyzed with the use of Clampfit 10.0 software (Axon Instruments, Foster City, CA). Peptide was added asymmetrically to either the cis or trans side of the same channel or a different one. Ion current blockage rates were obtained by using the standard routines available in Clampfit 10.0, giving the number of blockage events and the average residence time.
Results
Single-channel properties of the Tom40 channel
A single Tom40 channel (Fig. 1 A) was reconstituted into a planar lipid bilayer, and ion currents through the channel were recorded (Fig. 1, B and C, and Fig. S1 in the Supporting Material). Tom40 reconstituted in stable DPhPC lipid bilayers (n = 15) showed a characteristic single-channel conductance of ∼3.0 nS in 1 M KCl, 20 mM MES, pH 6, and an asymmetric channel closure with respect to the polarity of the applied voltage, in line with previous measurements (15). Fig. 1, B and C, show representative single-channel recordings of a Tom40 channel reconstituted in a planar lipid bilayer at applied voltages of −50 and +50 mV, respectively. The channel shows typical switching between open and closed states, with a prevalence of closed-state conductance at −50 mV. The gating frequency increased with the increase in the voltage (data not shown). In the case of +50 mV, the channel primarily existed in one open conductance state. The ion currents through the channel were stable without any flickering. The Tom40 channel reconstituted in the lipid membrane shows a clear asymmetry in the channel conductance with respect to the polarity of the applied voltage, suggesting an asymmetric conformation of the channel. Although we could not detect the exact orientation of Tom40 with this type of measurement, we were able to use the asymmetry in channel closure and conductance as a control for the orientation of the channel insertion. In some cases, we also observed that Tom40 channels in the lipid bilayers fluctuated between different subconductance states with very high noise (data not shown).
Figure 1.
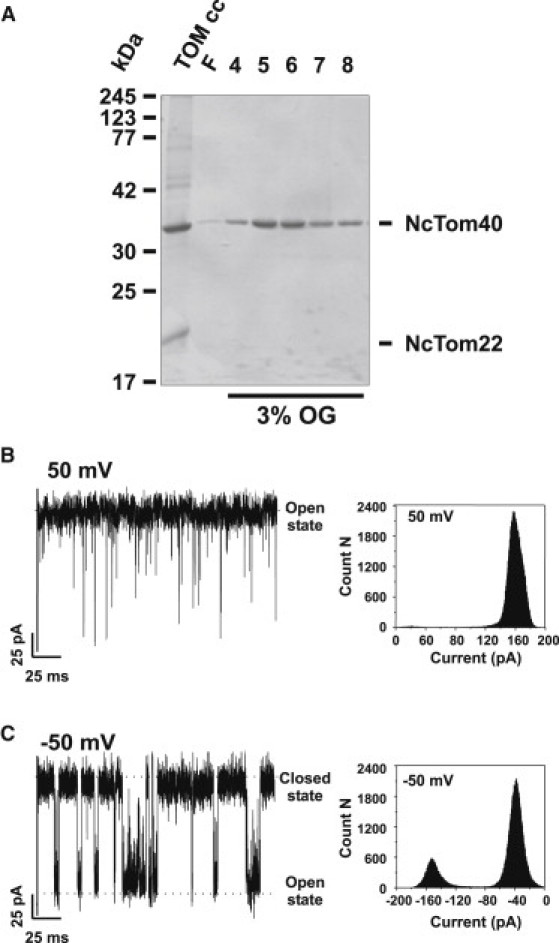
Purification and single-channel recordings of N. crassa Tom40. (A) Coomassie Blue-stained SDS-polyacrylamide gel of purified Tom40. Tom40 was purified from the TOM core complex of N. crassa mitochondria as previously described (15, 29). (B and C) Ionic currents through a single Tom40 channel at +50 and −50 mV. At 50 mV, the channel exits in one open conductance state, whereas at −50 mV the channel fluctuates between an open and a closed conductance state. The corresponding amplitude histogram is shown on the right side. Experimental conditions are 1 M KCl, 20 mM MES, pH 6, at room temperature.
Interaction of peptide with the Tom40 channel
To quantify the mitochondrial presequence peptide-channel interaction, we used high-resolution ion-conductance measurements through a single Tom40 channel. Because the Tom40 channels showed spontaneous voltage gating with characteristics similar to what would be expected for polypeptide interaction, we decided to test the influence of the substrate peptide on a channel where the intrinsic channel gating was virtually absent. We first analyzed current fluctuations of channels in the absence of peptides at voltages between −75 and +75 mV, and then tested the effect of the polypeptide interaction.
As shown in Fig. 2 and Fig. S1, polypeptides corresponding to the first 31 residues of the mitochondrial precursor of the S. cerevisiae F1-ATPase α-subunit (pF1β) block the ion current through a single Tom40 channel in a peptide-concentration-dependent manner. In the absence of the peptide, ion currents through selected single Tom40 channels were stable, without any significant ion current fluctuations (Fig. 2 A and Fig. S1). Addition of peptide to the trans side of the membrane induced ion current fluctuations, and these fluctuations increased with an increase in the concentration of the peptide (Fig. 2, B and C). It should be noted, however, that addition of the cationic pF1β peptide to the trans side of the membrane required positive potentials to promote the interaction between the peptide and Tom40, whereas no ion current blockages were observed when the orientation of the electric field was reversed (data not shown). We recently showed that nonmitochondrial peptides that were similar in length and overall charge to mitochondrial presequence peptides (i.e., pAK5) induced significantly less-frequent blocking of TOM core complex channels compared with mitochondrial presequence peptides (26). An interaction of nonmitochondrial peptides with the main constituent of the core complex, Tom40, cannot be expected.
Figure 2.

Typical ion current recordings through a single Tom40 channel in the presence of (A) 0 μM, (B) 5 μM, and (C) 10 μM pF1β peptide added to the trans side of the lipid membrane with applied voltage of +50 mV. Experimental conditions are 1 M KCl, 20 mM MES, pH 6, at room temperature.
Single-channel analysis was used to calculate the number (Fig. 3) and the average residence time (Fig. 4) of pF1β peptide-induced ion current blockage of Tom40. The frequency of ion current blockages increased with an increase in peptide concentration (Fig. 3 A) and applied voltage (Fig. 3 B). The latter indicates that the electric field pulled the peptides from the bath solution into an affinity site inside the Tom40 channel. The average residence times of peptide blockages were measured in 1 M KCl and calculated by single exponential fitting of dwell-time histograms (Fig. 4 A). They decreased from ∼230 to ∼120 μs (Fig. 4 B, n = 5) with an increase in voltage from 20 to 75 mV, respectively. Although the average residence time of the peptide inside the channel was strongly voltage-dependent, it did not depend on the concentration of the peptide used (Fig. 4 C, n = 5).
Figure 3.
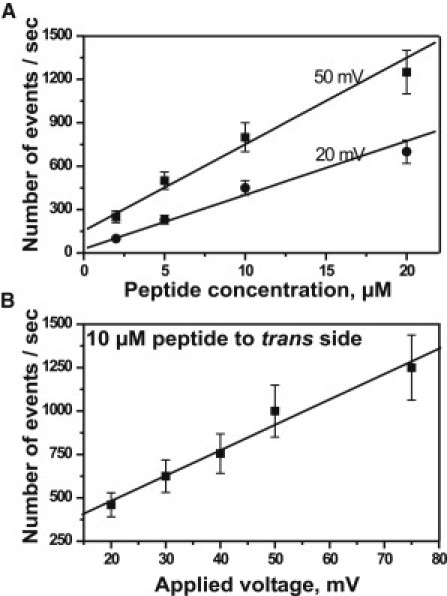
(A) Number of binding events increases with an increase in the concentration of the peptide from 2 μM to 20 μM measured at 20 mV and 50 mV. (B) The number of binding events increases with an increase in the applied transmembrane voltage from 20 mV to 75 mV at 10 μM peptide. Experimental conditions are 1 M KCl, 20 mM MES, pH 6, and peptide added to the trans side at room temperature (n = 5, mean ± SE).
Figure 4.
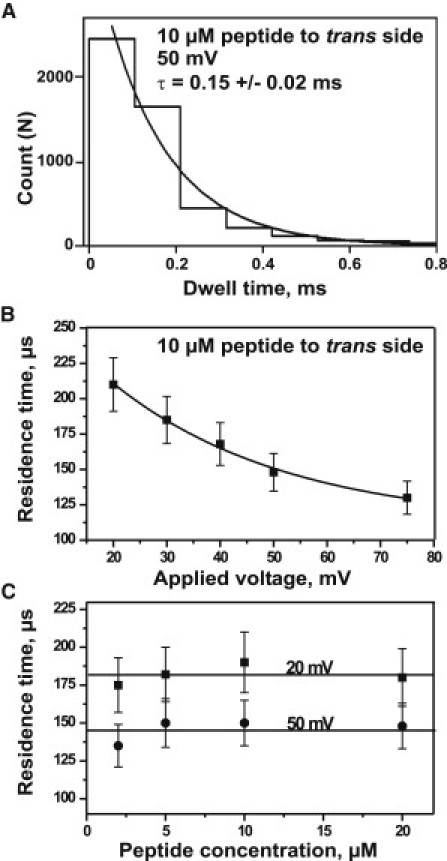
(A) Single exponential fitting of a dwell-time histogram for peptide blockage. (B) The residence time decreases with an increase in the applied transmembrane voltage from 20 mV to 75 mV at 10 μM peptide. (C) The residence time does not depend on the concentration of peptide and remains the same irrespective of an increase in peptide concentration from 2 μM to 20 μM measured at 20 mV and 50 mV. Experimental conditions are 1 M KCl, 20 mM MES, pH 6. Peptide was added to the trans side of the lipid bilayer at room temperature (n = 5, mean ± SE).
To elucidate a possible electrostatic contribution in the peptide-Tom40 interaction, we repeated the experiments at lower ionic strength. Decreasing the ionic strength of the salt solution reduced the single-channel conductance of Tom40 to ∼0.5 nS in 0.15 M KCl, 20 mM MES, pH 6, and led to a decreased signal/noise ratio. Typically, the frequency of channel closure (gating) in the absence of the peptide drastically increased, which indicates that low salt concentration facilitates the closure of the channel (data not shown). To distinguish between spontaneous and peptide-induced gating, we again selected for channels that showed only little intrinsic gating (Fig. 5 A). In the presence of pF1β peptide, the number of channel closures was clearly increased. However, separation of the number of spontaneous and peptide-induced closure events was difficult to attribute. Nevertheless, based on control measurements with the Tom40 channel in the absence of the peptide, we were able to separate endogenous channel gating with varying closure times from peptide-induced closures with constant closure times at a given voltage. A blockage time histogram in the absence and presence of the peptide revealed the channel closure time (Fig. S2). The peptide-induced closure time determined at a voltage of 50 mV was ∼400 μs (Fig. 5 B, n = 3). Closures of this length were virtually absent in the absence of the peptide. The peptide-induced closure time at low salt concentration was thus approximately three times larger than that at high salt concentration. This suggests that charge screening influences the release of presequences bound to Tom40.
Figure 5.
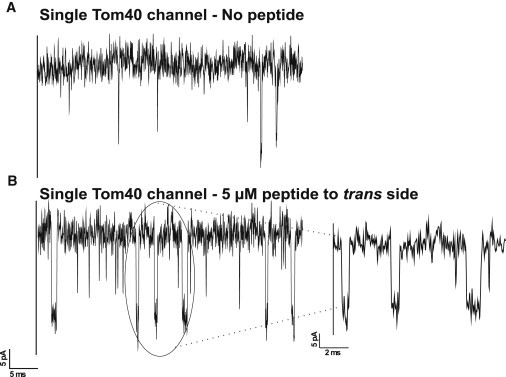
Typical ion current recordings through a single Tom40 channel in the presence of (A) 0 μM and (B) 5 μM pF1β peptide added to the trans side of the lipid membrane with applied voltage of +50 mV. Experimental conditions are 150 mM KCl, 10 mM MES, pH 6, at room temperature.
A common analysis method to study ion current fluctuation is to measure the power density spectrum, which indicates how the variance of the current is distributed with frequency. To highlight the interaction of peptide with the Tom40 channel, we performed a power spectral density analysis. Power density spectra of ion currents through single Tom40 channels obtained in the presence of various concentrations of peptide revealed excess noise compared with those obtained in the absence of peptides (Fig. 6 A). indicating a strong interaction of the peptide with the channel surface. Noise analysis of ion currents through single Tom40 also produced measurable excess noise with an increase in the applied voltage in the presence of the same concentration of peptide (Fig. 6 B). Noise spectra obtained at 50 mV revealed significant excess noise compared with those obtained at 20 mV. Noise spectral analyses of the ion current blockages by pF1β peptide showed a Lorentzian behavior and indicated that the peptide translocation process could be described by a two-state Markovian model (33). According to the chemical relaxation theory, the variation of the power density function with substrate concentration provides the on- and off-rates. This approach is identical to statistical analysis of single-channel fluctuation. Both approaches provide only the binding kinetics (33).
Figure 6.
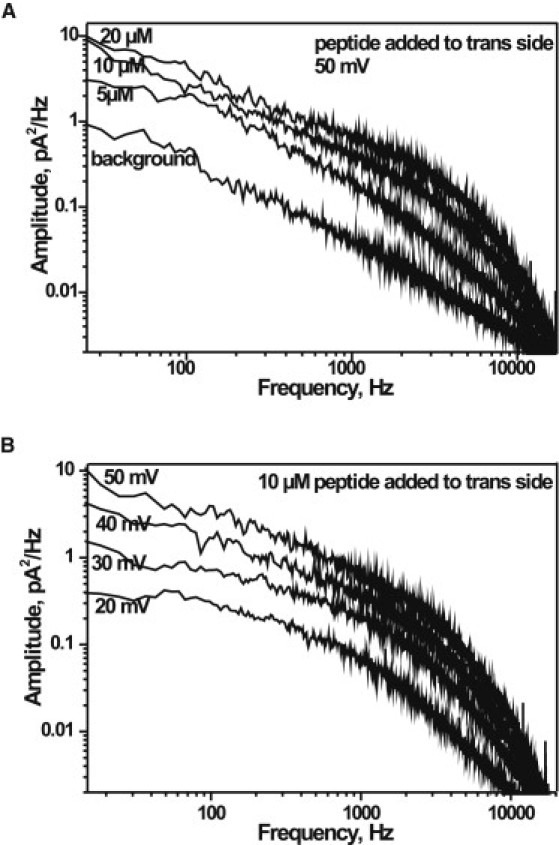
(A) Power spectral densities of the fluctuations in the ion current caused by peptide blocking a single Tom40 channel at different peptide concentrations ranging from 5 μM to 20 μM at 50 mV. (B) Power spectral densities of the fluctuations in the ion current caused by peptide blocking a single Tom40 channel at different applied voltages ranging from 20 to 50 mV at 10 μM pF1β peptide. Experimental conditions are 1 M KCl, 20 mM MES, pH 6. Peptides were added to the trans side of the channels at room temperature.
To test whether pF1β peptide blocks the Tom40 channel from both sides of the lipid membrane, we added peptide also to the cis side of the channel. Virtual orientation of the channel was concluded from the asymmetric channel closure measured at a high applied voltage of ±100 mV. The frequency of channel closure increased at negative voltage in the absence of the peptide and we selected for channels that showed only little intrinsic gating. We then set the voltage to ±50 mV, where almost no channel gating was observed (Fig. 7 A), and measured the peptide-binding kinetics from the cis and trans sides of the same Tom40 channel reconstituted into the bilayer. As noted above, the interaction of peptides on protein pores depends on the side of addition and the polarity of the applied voltage. The application of a negative potential induced fluctuations in the ion current through the Tom40 channel when peptide was added on the cis side of the chamber (Fig. 7 B). It is important to note that dilution of the peptide concentration in the bilayer chamber completely reduced the number of blockage events (data not shown). Peptide was then added to the trans side of the chamber (same channel), and application of a positive potential induced fluctuations in the ion current (Fig. 7 C). The numbers of ion current blockages caused by addition of peptides to the cis side of the bilayer depended strongly on the applied voltage (Fig. 8 A). The average residence time of the peptide (n = 5) depended on the applied voltage (Fig. 8 B) and did not depend on the concentration of the peptide used (data not shown). The average residence time of the peptide blockage inside the channel surface was the same irrespective of the side of peptide addition. It should be noted that the Tom40 channel starts to fluctuate between open and longer closed states in the absence of peptides at −75 mV. In this case, in the presence of peptide, we reduce the statistical analysis to the open time interval only (Fig. S1). A histogram analysis of kon (open time histogram) for 25 mV and 50 mV is shown in Fig. S3. Table 1 summarizes the association rate constants kon, dissociation constants koff, and equilibrium binding constants K for peptide addition to the cis and trans sides of the lipid membrane, respectively.
Figure 7.
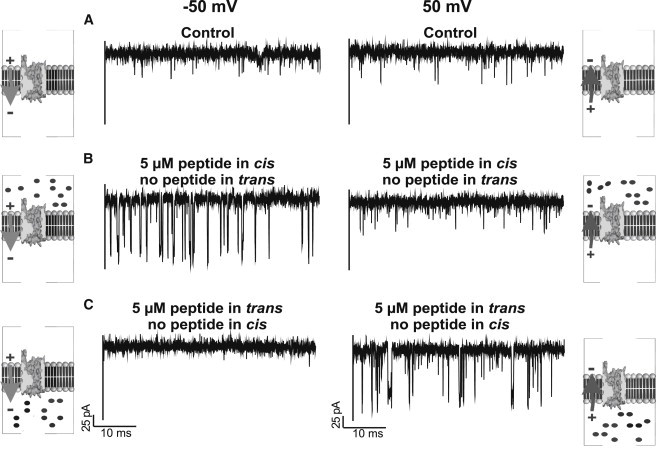
Typical ion current recordings through single Tom40 channels (A) in the absence of peptide at −50 mV and 50 mV, (B) with 5 μM peptide added to the cis side of lipid membrane with applied voltage of −50 mV and + 50 mV, and (C) with 5 μM peptide added to the trans side of lipid membrane with applied voltage of −50 mV and + 50 mV. Experimental conditions are 1 M KCl, 20 mM MES, pH 6, at room temperature. The schematic representations showing peptide translocation through N. crassa Tom40 driven by applied transmembrane potential are based on the work of Gessmann et al. (44).
Figure 8.
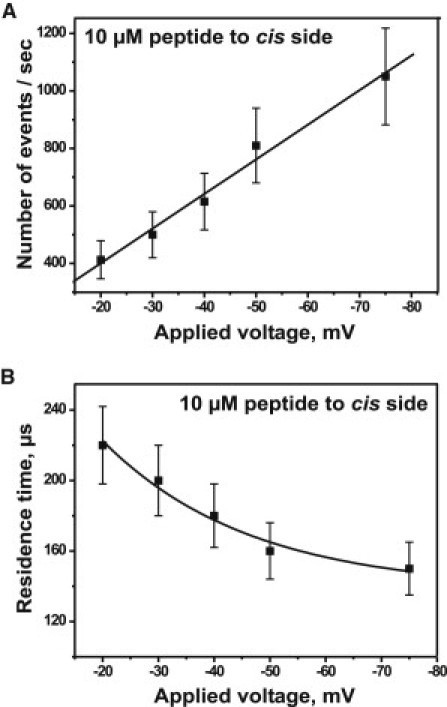
(A) Number of binding events increases with a decrease in the applied transmembrane voltage from −20 mV to −75 mV at 10 μM peptide. (B) The residence time decreases with a decrease in the applied transmembrane voltage from −20 mV to −75 mV at 10 μM peptide. Experimental conditions are 1 M KCl, 20 mM MES, pH 6, and peptide was added to the cis side of the channels at room temperature (n = 5, mean ± SE).
Table 1.
Rate constants of pF1β association kon, dissociation koff, and equilibrium binding constant K of the interaction between pF1β and Tom40
| Voltage (mV) | kontrans (M−1s−1) · 106 | kofftrans (s−1) · 103 | Ktrans (M−1) · 103 | Voltage (mV) | koncis (M−1s−1) · 106 | koffcis (s−1) · 103 | Kcis (M−1) · 103 |
|---|---|---|---|---|---|---|---|
| 20 | 46 ± 7 | 4.7 ± 0.5 | 9.5 ± 0.5 | −20 | 42 ± 6 | 4.5 ± 0.5 | 9.3 ± 0.6 |
| 50 | 100 ± 15 | 6.5 ± 0.7 | 15.0 ± 0.5 | −50 | 85 ± 13 | 6.2 ± 0.8 | 13.7 ± 0.5 |
| 75 | 125 ± 19 | 7.7 ± 0.7 | 16.0 ± 0.6 | −75 | 110 ± 17 | 7.4 ± 0.9 | 15.0 ± 0.5 |
Rate constants kontrans, koncis, kofftrans, and koffcis (mean ± SE, n = 5) were determined for pF1β peptide added to the trans and cis sides of the lipid bilayer (Figure 3, Figure 4, Figure 8), respectively, where kon = number of events s−1/[peptide], koff = 1/average residence time, and K = kon/koff.
Discussion
Most studies that addressed the mechanism of protein transport through TOM were based on in vitro import studies (18, 28, 34, 35, 36, 37) in which S35-radiolabeled mitochondrial preproteins were incubated with mitochondria or mitochondrial outer membrane vesicles purified from wild-type and mutant cells. These studies allowed the identification of numerous intermediate states along the import pathway. The mitochondrial protein import machinery includes motor complexes and chaperones that are essential components in the translocation process to drive polypeptides through passive protein-conducting channels in outer and inner mitochondrial membranes (18). According to the current view, the incoming polypeptides are pulled into the organelle by motor proteins once they have emerged at the trans side of the inner membrane of mitochondria. On the other hand, it has been also shown that chaperones bind incoming polypeptides and hinder polypeptide diffusion back into the cytosol. The global picture involves the TOM complex, with Tom40 serving as the channel that allows peptide translocation. However, quantitative information about the import kinetics and the mechanism of substrate interaction with TOM is limited by the unavailability of suitable protein import systems that allow substrate/Tom40 interactions to be monitored at the single-molecule level and with sufficiently high temporal resolution.
Translocation of proteins is a fundamental process in biology; however, most studies of peptide translocation have been limited by the lack of structural details about the TOM machinery. In previous studies, α-hemolysin was used as a model system to elucidate the kinetics of polypeptide translocation (8, 9, 38, 39). These studies revealed that peptide binding strongly depends on the charge, the length of the peptide, and the applied transmembrane voltage. In contrast, the Tom40 channel alone contains intrinsic flickering that renders a straightforward analysis more complex. A frequency analysis revealed an additional fast gating in the presence of peptide, in contrast to the slower kinetics of intrinsic fluctuation, which allows one to separate intrinsic from peptide-induced gating.
Our study provides insights into the first part of this process, i.e., the entry of presequence peptides into the TOM machinery. In particular, we focused on the role of Tom40 alone. We elucidated the kinetics of peptide translocation through Tom40, the central subunit of the TOM core complex. An ion current fluctuation analysis of single Tom40 channels in the presence of a mitochondrial presequence peptide revealed the kinetic parameters of peptide binding. We found a strong electrostatic component of the interaction between the peptide and the Tom40 channel, in agreement with previous biochemical studies that addressed the interaction of mitochondrial preproteins with the TOM machinery (34, 40). A strong attractive interaction between mitochondrial presequence peptides and Tom40 thus plays a major role in partitioning the peptide into the Tom40 channel.
In our study, we observed ion current blockage in channels when peptide was added asymmetrically to either the cis or trans side of the Tom40 channel. Addition of peptide to the cis side of the channel required trans-negative potentials to facilitate peptide-induced ion current blockages, whereas addition of peptide to the trans side reversed the voltage dependence, i.e., positive voltages enhanced ion current blockages. This indicates that the presequence peptide used in this study was able to enter the channel from both its cytosolic side and its intermembrane-space side (Fig. 7). The apparent dissociation constants for peptide binding at the trans and cis sides of the complex were virtually identical, suggesting a simple kinetic model for peptide-pore interaction with a single peptide affinity site inside the Tom40 channel. An optimal strength of interaction between the peptide and the affinity site in the channel may reduce the entropic barrier, resulting in higher translocation efficiency. In addition, this facilitated translocation was strongly dependent on the applied transmembrane voltage. We hypothesize that the applied transmembrane potential (electric field) and peptide charge can be a determining factor for the interaction that facilitates peptide transport across the channel. Our experimental results show that the applied transmembrane voltage can serve as a driving force for peptide translocation across Tom40 channels reconstituted in artificial lipid bilayers. Although the existence of a significant electrochemical potential across the outer membrane of mitochondria (41, 42, 43) is still a matter of debate, our data on peptide-Tom40 interactions can provide new insights into the energetic details of protein translocation in vivo.
We previously showed the interaction of pF1β peptide with a mitochondrial TOM core complex consisting of five subunits (Tom40, Tom22, Tom7, Tom6, and Tom5) (26, 40). The pF1β peptide-binding kinetics obtained for the TOM core complex and our similar results for the Tom40 channel alone indicate that Tom40 constitutes the central peptide-binding subunit. We conclude that the central subunit Tom40, which organizes the translocation pore that serves as an affinity site for the peptide interaction, and Tom22, Tom7, Tom6, and Tom5 are not necessarily needed for interaction of TOM channels with substrate peptides. The molecular details and the kinetic role of other TOM subunits (e.g., Tom20 and Tom70) in peptide translocation remain to be determined.
Conclusion
Binding of mitochondrial presequence peptides to the Tom40 channel does not imply translocation. The kinetic data obtained from our single-channel measurements can be used to distinguish peptide binding from peptide translocation. We have shown that the average residence time of the mitochondrial presequence peptide pF1β within the Tom40 channel decreases with the increase in applied voltage. The equivalent increase of the peptide dissociation rates kofftrans and koffcis with applied voltage demonstrates translocation of the peptide. The results presented in this work should now make it possible to characterize the protein translocation pathway through the Tom40 channel in more detail. Analysis of substrates of different charges and lengths can provide new insights into the energetic details of protein translocation into mitochondria.
Acknowledgments
We thank Beate Nitschke for expert technical assistance, Dr. Frank Nargang for providing the Tom40 N. crassa expression strain GR-107, and Dr. Roland Benz for helpful discussion.
This work was funded in part by grants from the Competence Network on Functional Nanostructures of the Baden Württemberg Stiftung (to S.N.) and the Deutsche Forschungsgemeinschaft (DFG WI 2278/18-1 to M.W.). M.R.R. received a fellowship from the International Center for Transdisciplinary Studies at Jacobs University Bremen.
Editor: William Wimley.
Footnotes
Three supplementary figures plus figure legends are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(11)05354-9.
Supporting Material
References
- 1.Pfanner N., Geissler A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 2.Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 3.Mokranjac D., Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim. Biophys. Acta. 2009;1793:33–41. doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Schleiff E., Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 5.Endo T., Yamano K., Kawano S. Structural insight into the mitochondrial protein import system. Biochim. Biophys. Acta. 2011;1808:955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 6.du Plessis D.J., Nouwen N., Driessen A.J. The Sec translocase. Biochim. Biophys. Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Robinson C., Matos C.F., et al. Mendel S. Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta. 2011;1808:876–884. doi: 10.1016/j.bbamem.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Mohammad M.M., Movileanu L. Excursion of a single polypeptide into a protein pore: simple physics, but complicated biology. Eur. Biophys. J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 9.Movileanu L., Schmittschmitt J.P., et al. Bayley H. Interactions of peptides with a protein pore. Biophys. J. 2005;89:1030–1045. doi: 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe A.J., Mohammad M.M., et al. Movileanu L. Catalyzing the translocation of polypeptides through attractive interactions. J. Am. Chem. Soc. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 11.Oukhaled G., Mathé J., et al. Auvray L. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 2007;98:158101–158104. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 12.Pastoriza-Gallego M., Rabah L., et al. Pelta J. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 2011;133:2923–2931. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 13.Hall A.R., Scott A., et al. Dekker C. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat. Nanotechnol. 2010;5:874–877. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill K., Model K., et al. Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment] Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- 15.Ahting U., Thieffry M., et al. Nussberger S. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 2001;153:1151–1160. doi: 10.1083/jcb.153.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Model K., Meisinger C., Kühlbrandt W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 2008;383:1049–1057. doi: 10.1016/j.jmb.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 17.Mager F., Sokolova L., Lintzel J., Brutschy B., Nussberger S. LILBID-mass spectrometry of the mitochondrial preprotein translocase TOM. J. Phys. Condens. Matter. 2010;22:454132. doi: 10.1088/0953-8984/22/45/454132. [DOI] [PubMed] [Google Scholar]
- 18.Chacinska A., Koehler C.M., et al. Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thieffry M., Chich J.F., et al. Henry J.P. Incorporation in lipid bilayers of a large conductance cationic channel from mitochondrial membranes. EMBO J. 1988;7:1449–1454. doi: 10.1002/j.1460-2075.1988.tb02962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallette F.M., Juin P., Pelleschi M., Henry J.P. Basic peptides can be imported into yeast mitochondria by two distinct targeting pathways. Involvement of the peptide-sensitive channel of the outer membrane. J. Biol. Chem. 1994;269:13367–13374. [PubMed] [Google Scholar]
- 21.Muro C., Grigoriev S.M., et al. Campo M.L. Comparison of the TIM and TOM channel activities of the mitochondrial protein import complexes. Biophys. J. 2003;84:2981–2989. doi: 10.1016/S0006-3495(03)70024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fèvre F., Henry J.P., Thieffry M. Reversible and irreversible effects of basic peptides on the mitochondrial cationic channel. Biophys. J. 1994;66:1887–1894. doi: 10.1016/S0006-3495(94)80982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Künkele K.P., Juin P., et al. Thieffry M. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 1998;273:31032–31039. doi: 10.1074/jbc.273.47.31032. [DOI] [PubMed] [Google Scholar]
- 24.Becker L., Bannwarth M., et al. Wagner R. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J. Mol. Biol. 2005;353:1011–1020. doi: 10.1016/j.jmb.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Ahting U., Thun C., et al. Nussberger S. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 1999;147:959–968. doi: 10.1083/jcb.147.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Ruiz M., Mahendran K.R., et al. Nussberger S. Interactions of mitochondrial presequence peptides with the mitochondrial outer membrane preprotein translocase TOM. Biophys. J. 2010;99:774–781. doi: 10.1016/j.bpj.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harsmanm A., Krüger V., et al. Wagner R. Protein conducting nanopores. J. Phys. Condens. Matter. 2010;22:454102. doi: 10.1088/0953-8984/22/45/454102. [DOI] [PubMed] [Google Scholar]
- 28.Künkele K.P., Heins S., et al. Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- 29.Poynor M., Eckert R., Nussberger S. Dynamics of the preprotein translocation channel of the outer membrane of mitochondria. Biophys. J. 2008;95:1511–1522. doi: 10.1529/biophysj.108.131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahendran K.R., Chimerel C., et al. Winterhalter M. Antibiotic translocation through membrane channels: temperature-dependent ion current fluctuation for catching the fast events. Eur. Biophys. J. 2009;38:1141–1145. doi: 10.1007/s00249-009-0495-0. [DOI] [PubMed] [Google Scholar]
- 31.Engelhardt H., Meins T., et al. Zeth K. High-level expression, refolding and probing the natural fold of the human voltage-dependent anion channel isoforms I and II. J. Membr. Biol. 2007;216:93–105. doi: 10.1007/s00232-007-9038-8. [DOI] [PubMed] [Google Scholar]
- 32.Arnold T., Poynor M., et al. Linke D. Gene duplication of the eight-stranded β-barrel OmpX produces a functional pore: a scenario for the evolution of transmembrane β-barrels. J. Mol. Biol. 2007;366:1174–1184. doi: 10.1016/j.jmb.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 33.Nekolla S., Andersen C., Benz R. Noise analysis of ion current through the open and the sugar-induced closed state of the LamB channel of Escherichia coli outer membrane: evaluation of the sugar binding kinetics to the channel interior. Biophys. J. 1994;66:1388–1397. doi: 10.1016/S0006-3495(94)80929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer A., Neupert W., Lill R. Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell. 1995;80:127–137. doi: 10.1016/0092-8674(95)90457-3. [DOI] [PubMed] [Google Scholar]
- 35.Vasiljev A., Ahting U., et al. Rapaport D. Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol. Biol. Cell. 2004;15:1445–1458. doi: 10.1091/mbc.E03-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt S., Ahting U., et al. Nussberger S. Role of Tom5 in maintaining the structural stability of the TOM complex of mitochondria. J. Biol. Chem. 2005;280:14499–14506. doi: 10.1074/jbc.M413667200. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt O., Harbauer A., et al. Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Bikwemu R., Wolfe A.J., Xing X., Movileanu L. Facilitated translocation of polypeptides through a single nanopore. J. Phys. Condens. Matter. 2010;22:454117. doi: 10.1088/0953-8984/22/45/454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng H., Detillieux D., et al. Lee J.S. Nanopore analysis of tethered peptides. J. Pept. Sci. 2010;16:701–708. doi: 10.1002/psc.1289. [DOI] [PubMed] [Google Scholar]
- 40.Stan T., Ahting U., et al. Rapaport D. Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 2000;19:4895–4902. doi: 10.1093/emboj/19.18.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemeshko V.V. Model of the outer membrane potential generation by the inner membrane of mitochondria. Biophys. J. 2002;82:684–692. doi: 10.1016/S0006-3495(02)75431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemeshko V.V. Theoretical evaluation of a possible nature of the outer membrane potential of mitochondria. Eur. Biophys. J. 2006;36:57–66. doi: 10.1007/s00249-006-0101-7. [DOI] [PubMed] [Google Scholar]
- 43.Benz R., Kottke M., Brdiczka D. The cationically selective state of the mitochondrial outer membrane pore: a study with intact mitochondria and reconstituted mitochondrial porin. Biochim. Biophys. Acta. 1990;1022:311–318. doi: 10.1016/0005-2736(90)90279-w. [DOI] [PubMed] [Google Scholar]
- 44.Gessmann D., Flinner N., et al. Mirus O. Structural elements of the mitochondrial preprotein-conducting channel Tom40 dissolved by bioinformatics and mass spectrometry. Biochim. Biophys. Acta. 2011;1807:1647–1657. doi: 10.1016/j.bbabio.2011.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


