Abstract
Pyrabactin resistance (PYR) 1 and its relatives belong to a family of soluble abscisic acid (ABA) receptors that inhibit type 2C protein phosphatases (PP2C) when in their agonist-stabilized conformation. Given their switch-like properties, we envisioned that mutations that stabilize their agonist-bound conformation could be used to activate signaling in vivo. To identify such mutations, we subjected PYR1 to site-saturation mutagenesis at 39 highly conserved residues that participate in ABA or PP2C contacts. All 741 possible single amino acid substitutions at these sites were tested to identify variants that increase basal PYR1-PP2C interactions, which uncovered activating mutations in 10 residues that preferentially cluster in PYR1's gate loop and C-terminal helix. The mutations cause measurable but incomplete receptor activation in vitro; however, specific triple and quadruple mutant combinations were constructed that promote an agonist-bound conformation, as measured by heteronuclear single quantum coherence NMR, and lead to full receptor activation. Moreover, these mutations retain functionality when introduced into divergent family members, and can therefore be used to dissect individual receptor function in vivo, which has been problematic because of redundancy and family size. Expression of activated PYL2 in Arabidopsis seeds activates ABA signaling by a number of measures: modulation of ABA-regulated gene expression, induction of hyperdormancy, and suppression of ABA deficiency phenotypes in the aba2-1 mutant. Our results set the stage for systematic gain-of-function studies of PYR1 and related ABA receptors and reveal that, despite the large number of receptors, activation of a single receptor is sufficient to activate signaling in planta.
Keywords: constitutively active receptor, pyrabactin resistance 1-like, regulatory component of abscisic acid receptor, stAR-related lipid transfer domain, seed dormancy
It has recently been discovered that the plant stress hormone abscisic acid (ABA) elicits many of its cellular responses by binding to a soluble family of receptors called pyrabactin resistance (PYR) 1/PYR1-like (PYL)/regulatory component of ABA receptor (RCAR) proteins (1–3), which belong to the large stAR-related lipid transfer domain (START) superfamily of ligand-binding proteins (4, 5). When agonists bind to these receptors, contacts between a mobile gate loop and agonist stabilize gate closure, which allows the receptors to dock into and inhibit the active site of clade A type 2C protein phosphatases (PP2Cs) (1, 6–12), which are negative regulators of ABA signaling. The resulting inhibition, in turn, allows activation of downstream SNF1-related kinase 2 (SnRK2) family members (13, 14), which regulate transcription factors, ion channels, and other proteins involved in ABA responses (recently reviewed in 15–17). Thus, PYR1 and related ABA receptors are molecular switches that function at the apex of a signaling cascade that regulates diverse ABA responses.
In addition to the important role that gate closure plays in receptor activation, other structural rearrangements are critical. For example, PYR1, PYL1, and PYL2 are homodimers in the absence of agonist (6, 7, 18) but bind to PP2Cs as monomers after ABA perception (8, 15, 17). The homodimer interface of these receptors overlaps with the PP2C interaction interface; as a consequence, the apo-receptor is blocked from binding PP2Cs and an ABA-promoted dimer-disruption step is necessary for activation of dimeric receptors (6, 18). Additionally, a tryptophan lock residue is located on a conserved loop in clade A PP2Cs; this residue inserts into a small pore on agonist-bound receptors and stabilizes the receptor–PP2C complex (6, 8, 9, 11, 12, 19).
The receptor family can be classified into different subtypes on the basis of sequence similarity, ABA sensitivity, oligomeric state, and basal activation level. PYR1, PYL1, and PYL2 are dimeric in solution, display low basal activity and require higher concentrations of ABA to elicit PP2C inhibition in comparison to monomeric PYLs because part of the energy released by ABA binding is diverted towards dimer disruption (20, 21). Ten of the 14 Arabidopsis family members (all except PYL7 and PYL11–PYL13) have been characterized in vitro; of these, PYL6 and PYL10 display substantially higher basal activity in comparison to other receptors (11, 21). The high basal activity of PYL10 is attributable, in part, to sequence differences in its gate that allow it to dock into a PP2C in its apo-state (21); however, the full sequence determinants of its high basal activity remain unresolved and incorporating PYL10's unique gate sequence into the low basal-activity receptor PYL2 only partially increases its basal activity (21). More importantly, however, the in vivo significance of the biochemical differences among receptors remains unclear, and, to date, a clear single-gene loss-of-function phenotype has only been observed in pyr1 mutants, which are insensitive to the selective ABA agonist pyrabactin. Moreover, a quadruple receptor mutant (pyr1/pyl1/pyl2/pyl4), which is deficient in the three dimeric receptors, is compromised in many aspects of ABA signaling (1), providing additional evidence that dimeric receptors are important to signaling despite their lower ABA sensitivity relative to monomeric receptors.
Both functional redundancy and the large size of the receptor family present challenges to developing a complete understanding of receptor function. One strategy to dealing with these issues is to activate individual receptor members selectively by chemical or genetic means and assess the functional consequences of these perturbations. Here, we describe mutations that can be used to create fully activated monomeric and dimeric class receptors and show that selective activation of PYL2 is sufficient to activate ABA signaling in planta to a degree that suppresses phenotypes observed in the strong ABA auxotroph aba2-1; thus, activation of a single family member is sufficient to activate signaling in vivo. Our results set the stage for systematic gain-of-function dissection of ABA receptor function.
Results
Site-Saturation Mutagenesis of PYR1.
PYR1 and related dimeric receptors do not substantially inhibit PP2C activity in the absence of ABA when tested at equimolar PP2C/receptor ratios, which is a consequence of their requirement for ABA to adopt a PP2C-binding conformation. In principle, receptor mutations that allow high-affinity binding of PP2Cs in the absence of agonist should activate the ABA signaling pathway in an ABA-independent fashion and create constitutively active (CA) receptors.
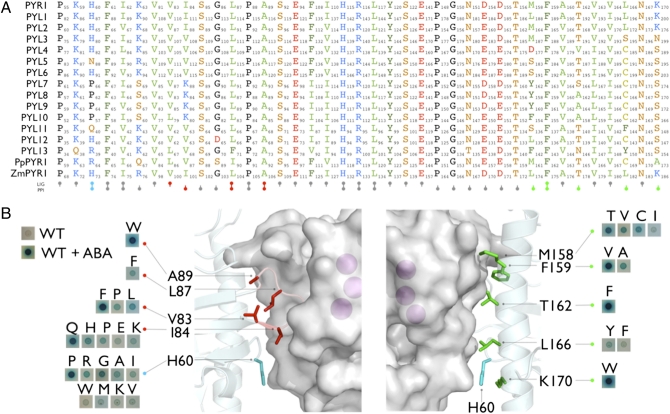
Given the value of CA receptors for gain-of-function studies, we set out to define sites systematically that can be mutated to improve interactions between receptors and PP2Cs. We conducted site-saturation mutagenesis on residues in PYR1 that normally contact ABA or PP2Cs, reasoning that specific mutations in these regions could mimic the effects of agonist occupancy. Thirty-nine residues within 5 Å of ABA, four water-molecules that contact ABA, or the PP2C HAB1 (Fig. 1A) were identified using PYR1 structure coordinates (7, 18, 19), and all possible 741 single amino acid substitutions at these sites were constructed by site-directed mutagenesis. We next examined the effect of each mutation on the PYR1-HAB1 interaction using a yeast two-hybrid assay; this revealed that 29 mutations, located in 10 different residues, increased PYR1-HAB1 interactions in the absence of ABA (Fig. 1B and Table S1). These activating mutations preferentially cluster in PYR1's mobile gate loop (V83, I84, L87, A89) and its C-terminal helix (M158, F159, T162, L166, K170), which participates in PP2C contacts and receptor homodimerization. Additionally, the residue H60, which resides on a loop adjacent to the C-terminal helix and contacts PP2Cs, is a hotspot for activating mutations, because 9 of the 19 H60 mutations increase PYR1's basal PP2C interaction level (Fig. 1B).
Fig. 1.
Site-saturation mutagenesis of PYR1 identifies partial activation mutants. (A) Sites selected for saturation mutagenesis. Thirty-nine residues involved in agonist (LIG) or PP2C (PPI) contacts were selected based on structure coordinates and subjected to site-saturation mutagenesis creating 741 PYR1 mutants; the alignment shows the identity and amino acid numbering of homologous residues in all Arabidopsis PYLs, as well as maize and physcomitrella PYR1 homologs. (B) Activating mutations identified by site-saturation mutagenesis. Of the 741 mutants constructed, 29 promoted interactions with HAB1 in the absence of ABA as measured using an established yeast two-hybrid assay; the locations of activating mutations are mapped onto PYR1-ABA-HAB1 coordinates (19). The front view to the left shows the gate residues (red), and the opposite side to the right shows the C-terminal helix residues (green); H60 is shown in cyan. Inset are images of X-Gal–stained yeast colonies for the subset of PYR1 mutants that bind HAB1 in the absence of ABA. For reference, the wild-type PYR1-HAB1 interaction in the yeast two-hybrid assay is shown in the presence and absence of 10 μM ABA. The purple balls represent active site magnesium ions.
To ascertain if the yeast assay successfully identified bona fide activating mutations, representative mutants selected from the eight strongest activating sites were expressed as 6× His fusion proteins in Escherichia coli and then used in in vitro PP2C assays. All eight mutant receptors tested increased PYR1's basal activity in comparison to wild-type PYR1 (Fig. S1 and Table S2), demonstrating that our mutagenesis strategy successfully defined true activating mutations. By definition, a fully CA PYR1 receptor should inhibit PP2C activity to the same level as that observed for ABA-saturated wild-type receptor. None of our single mutants mimic the activation levels observed for WT at ABA saturation, demonstrating that the single mutants only partially activate PYR1. We note that, to date, full CA mutants have not been described for any ABA receptor and that PYL6 and PYL10, which have high basal activities (11, 21), are not full CA receptors either (Fig. S2 and Table S2).
Combining Activating Mutations Leads to Full CA Receptors.
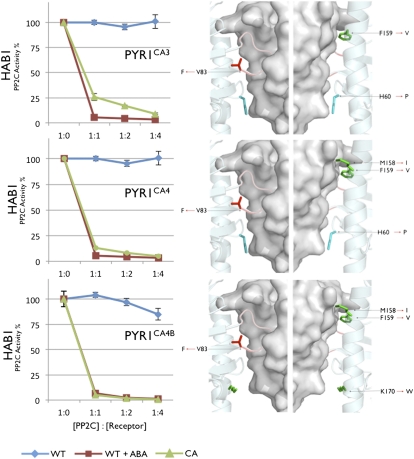
Activation of PYR1 is accompanied by gate closure and homodimer disruption, both of which are necessary for stable PP2C binding. We reasoned that combinations of mutations in the gate, C-terminal helix, and H60 might enhance basal activation further than that observed in the single mutants. We therefore constructed a series of double mutants, but these failed to enhance receptor activation substantially above that seen with single mutants (Table S2). However, when triple and quadruple combinations of gate and helix mutations were constructed, we obtained four different PYR1 variants with activation levels nearly indistinguishable from ABA-saturated wild-type PYR1 (Fig. 2, Fig. S3, and Table S2). We subsequently focused efforts on characterizing two mutants, CA3 and CA4, in further detail. Importantly, the PYR1CA3 and PYR1CA4 receptors inhibit multiple clade A PP2Cs (Fig. 3 and Table S2) in vitro. To determine if the CA3 and CA4 mutations work in the context of other receptor backbones, we introduced homologous substitutions into PYL2 and PYL9, generating PYL2CA3, PYL2CA4, and PYL9CA4 variants; these receptors are highly activated in the absence of ABA and inhibit multiple clade A PP2Cs (Fig. 3). Thus, combining a small number of specific partially activating mutations enables full activation of diverse receptors, despite only 55% and 49% amino acid sequence identity between PYL2-PYR1 and PYL9-PYR1, respectively.
Fig. 2.
Combining partial activation mutants leads to CA PYR1. Triple and quadruple combinations of partial activation mutants were made as described in the text (a complete list of all mutants characterized is provided in Table S2). Recombinant 6× His-PYR1, PYR1CA3, PYR1CA4, and PYR1CA4B were expressed in E. coli, purified, and used in PP2C assays with GST-HAB1. Reactions contain 600 nM GST-HAB1 and varying concentrations of receptor (0, 600-, 1,200-, 2,400-, or 4,800-nM receptor). PP2C activity is expressed as percentage of control (i.e., activity of PP2C in the absence of receptor and ABA but otherwise identical reaction conditions). (Left) Graphs plot average values from three technical replicates, and error bars show 1 SD. To enable comparison between a mutant protein's activation level and the degree of activation elicited by ABA on wild-type receptor, each graph shows wild-type PYR1 reactions run with either 0 μM (blue) or 10 μM (red) ABA; mutant proteins are shown in green and were assayed in the absence of ABA. The wild-type controls shown for CA3 and CA4 are the same data separated for image clarity. (Right) Locations of the specific mutations in each CA receptor mapped onto the crystal structure of PYR1-ABA-HAB1 (19). The side chains for residues mutated are shown in stick form; red corresponds to gate-, green to C-terminal helix-, and cyan to H60-located mutations.
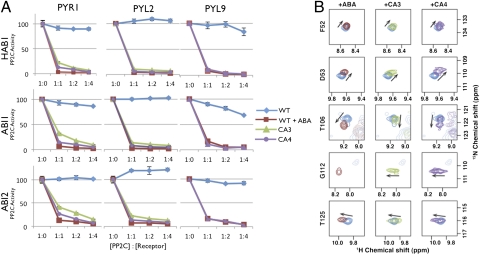
Fig. 3.
CA3 and CA4 mutations function in the context of other receptor backbones and stabilize the ABA-bound conformation of PYR1. (A) CA3 and CA4 mutations activate diverse receptors. The CA3 and CA4 mutations were introduced into homologous positions in PYL2 and PYL9, and recombinant receptors were assayed for activity on GST-HAB1, 6× His-Sumo-ABI1, and 6× His-Sumo-ABI2. Reactions contain 600 nM PP2C and varying concentrations of receptor (0, 600-, 1,200-, 2,400-, or 4,800-nM receptor). PP2C activity is expressed as percentage of control (i.e., activity of PP2C in the absence of receptor and ABA); each graph shows wild-type receptors in reactions run with either 0 μM (blue) or 10 μM (red) ABA. The graphs show average values from three technical replicates, and error bars show 1 SD. (B) HSQC NMR shows that PYR1CA3 and PYR1CA4 adopt an ABA-bound receptor conformation. 1H-15N HSQC spectra of apo-PYR1 (cyan), ABA-bound PYR1 (red), PYR1CA3 (green), and PYR1CA4 (purple) show the progressive shift of PYR1CA3 and PYR1CA4 toward a conformation resembling the ABA-bound state. Each box shows the peak of a single residue excised from a complete HSQC spectrum.
Using 2D 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectroscopy, we have previously shown that PYR1 exists in a two-state conformational equilibrium that is shifted to one conformation on agonist binding (1, 9, 10). HSQC NMR therefore provides a powerful method with which to probe ABA-mediated conformational dynamics. Given the locations of the CA mutations in the gate and helix, we envisioned that they might stabilize receptors in a conformation resembling the agonist-bound state. To examine this, we prepared 15N-labeled PYR1, PYR1CA3, and PYR1CA4 receptors and compared the HSQC signal perturbations induced by ABA binding to wild-type PYR1 with those induced by the CA3 and CA4 receptor mutations, focusing on the responses of residues distant from mutation sites. A subset of clearly resolved residues outside the ABA binding pocket and distal from the mutation sites was perturbed in a similar manner when the HSQCs of ABA-bound PYR1, PYR1CA3, and PYR1CA4 are compared with that of apo-PYR1 (Fig. 3B). Further, the PYR1CA3 and PYR1CA4 HSQCs reveal that the equilibrium of CA4 is shifted toward the ABA-like conformation more so than that of CA3, which is consistent with the enhanced activity of PYR1CA4. We note that HSQC measurements were made in the absence of a PP2C, which indicates that a PP2C is not necessary to promote the ABA-induced conformation of CA3 and CA4 receptors. Thus, the CA3 and CA4 mutations partially mimic the effects of ABA occupancy observed for wild-type PYR1.
In Planta Activation of ABA Signaling by CA Receptors.
To examine if activating mutations can increase ABA signaling above basal levels in vivo, we attempted to make transgenic plants expressing 35S-driven GFP-tagged constructs for PYL2, PYL2CA3, or PYL2CA4 in the wild-type Columbia or aba2-1 mutant background, reasoning that if CA receptors function as expected in vivo, they should activate ABA responses in WT and suppress the effects of ABA depletion in the aba2 mutant. PYL2 and its CA mutants were chosen as a first test case for in planta experiments because the CA mutations in this backbone are slightly stronger relative to the homologous mutations in PYR1 (Fig. 3 and Table S2). We note that when tested at 1:1 stoichiometry in vitro, PYL2CA3 and PYL2CA4 reduce HAB1's PP2C activity to 10.5 ± 0.5% and 6.3 ± 0.5% that of control levels, respectively, and that at saturating ABA concentrations, wild-type PYL2 reduces HAB1's PP2C activity to 4.1 ± 0.1% (Table S2). We were unable to obtain 35S::GFP-PYL2CA4 transgenic lines; however, several single-insert homozygous 35S::GFP-PYL2 and 35S::GFP-PYL2CA3 lines were obtained. We speculate that our inability to obtain 35S::GFP-PYL2CA4 transgenic lines may be a consequence of the greater constitutive activity of the CA4 receptor relative to CA3 (Fig. 3 and Table S2); however, we cannot rule out other more trivial explanations. Interestingly, all 35S::GFP-PYL2CA3 transgenics display strong GFP expression in developing, mature, and imbibed seeds but undetectable GFP in shoot tissue postgermination, despite expression from the strong 35S promoter; this restricted expression pattern was not observed with wild-type 35S::GFP-PYL2 transgenics (Fig. S4). Examination of GFP-PYL2CA3 transcript and protein levels in seeds and mature leaf tissues of transgenic plants shows that despite comparable mRNA expression levels, GFP-PYL2CA3 protein is only detectable in seeds (Fig. S4). Thus, a posttranscriptional mechanism limits expression of GFP-PYL2CA3 protein but not GFP-PYL2 protein in vegetative tissues. This observation hints at a mechanism that may reduce levels of activated receptor proteins, which is intriguing in light of the rapid transcriptional down-regulation of multiple ABA receptor mRNAs after ABA treatment of seedlings (22, 23).
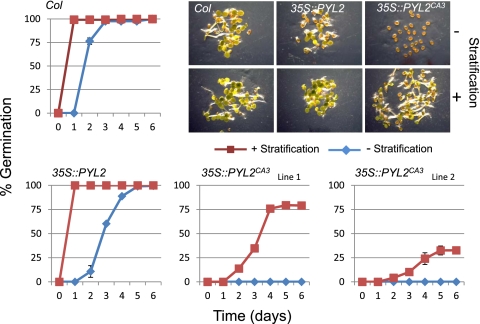
Given the restricted expression pattern of 35S::GFP-PYL2CA3, we investigated the effects of the 35S::GFP-PYL2CA3 transgene on seed ABA responses. ABA plays a major role in seed biology and controls seed dormancy, which is induced during embryogenesis, and abiotic stress responses during germination (24). It is well established that mutants with increased ABA sensitivity have higher seed dormancy; for example, the enhanced response to aba1 (era1) mutation shows a stratification requirement for germination (25). We therefore compared the seed dormancy of wild-type Columbia, 35S::GFP-PYL2, and 35S::GFP-PYL2CA3 genotypes. As shown in Fig. 4, in the wild-type Columbia background, overexpression of PYL2CA3 induces a state of hyperdormancy, as indicated by the strong stratification-dependent germination of two independent CA3 lines. Thus, overexpression of GFP-PYL2CA3 activates the ABA-regulated seed dormancy pathway in wild-type plants. We note that overexpression of wild-type GFP-PYL2 also increased seed dormancy but to a much lesser extent than the CA3 receptor (Fig. 4).
Fig. 4.
Overexpression of PYL2CA3 induces seed hyperdormancy. Seeds of the wild-type Columbia (Col), 35S::GFP-PYL2, or two independent 35S::GFP-PYL2CA3 lines were either stratified for 6 d at 4 °C or unistratified; their germination, indicated by radical emergence, was then monitored at 24-h intervals postimbibition. Graphs plot the averages of values from three biological replicates, and error bars show 1 SD. (Upper Right) Representative images at 48 h postimbibition for Columbia, 35S::GFP-PYL2, or two independent 35S::GFP-PYL2CA3 (line 1) lines.
To investigate seed ABA responses further, we examined ABA-regulated gene expression for the ABA markers LEA, RD29b, and Em6 using quantitative RT-PCR analyses of the same genotypes characterized above. As shown in Fig. S5, imbibed seeds of two independent CA transgenic lines display elevated levels of ABA-regulated genes in the absence of ABA treatment. The expression levels observed for these genes exceed those observed in wild-type seeds imbibed in the presence of 5 μM ABA. Thus, the 35S::GFP-PYL2CA3 transgene is sufficient to cause high levels of several ABA-regulated mRNAs, consistent with the conclusion that it activates ABA signaling in vivo.
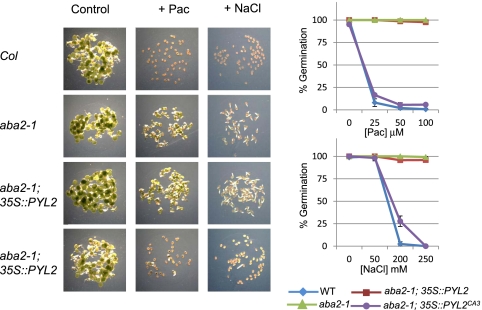
Interpreting the effects of receptor overexpression can be complicated by the presence of an endogenous agonist, which can activate wild-type receptors. We therefore examined the ability of expression of PYL2CA3 to revert phenotypes caused by ABA deficiency in the aba2-1 mutant, which has greatly reduced ABA levels because of a mutation in the enzyme xanthoxin dehydrogenase (26, 27). The aba2-1 background therefore provides a stringent test for constitutive activation of signaling in vivo, which, by definition, should be ABA-independent. We therefore examined the germination of various genotypes on paclobutrazol, an inhibitor of GA biosynthesis, and NaCl. Both of these treatments inhibit seed germination in an ABA- and ABA2-dependent manner (27, 28). Consistent with our observations made with transgenes expressed in wild-type plants, the 35S::GFP-PYL2CA3 transgene suppresses the paclobutrazol and NaCl insensitivity of aba2-1 mutants, whereas the wild-type 35S::GFP-PYL2 transgene does not (Fig. 5). Collectively, our in vivo data demonstrate that the PYL2CA3 receptor is a potent activator of multiple ABA responses and demonstrate that our CA receptors can be used to modulate ABA signaling in vivo.
Fig. 5.
PYL2CA3 suppresses phenotypes caused by ABA deficiency. Seeds of the wild-type Columbia (Col), aba2-1, aba2-1;35S::GFP-PYL2, or aba2-1;35S::GFP-PYL2CA3 genotypes were stratified for 4 d at 4 °C on agar media containing different concentrations of paclobutrazol (Pac) or sodium chloride (NaCl), and germination was scored 72 h postimbibition. The experiment was conducted in triplicate, and the SD is shown on graph points. (Left) Representative images at 72 h postimbibition for control, 50 μM paclobutrazol, or 250 mM NaCl. Values plotted in graphs are the average of three independent measurements, and error bars show 1 SD.
Discussion
We have used saturation mutagenesis to identify a series of mutations in PYR1 that increase its basal activity. Combinations of these mutations led to the rapid construction of near-fully activated PYR1 variants. The activating combinations can be incorporated into diverse PYL receptors to elicit full activation; PYL2CA4, PYL2CA4B, and PYL9CA4 are nearly indistinguishable from wild-type receptors examined under saturating ABA levels. When PYL2CA3 is expressed in vivo, it activates ABA signaling and enables near-complete suppression of two separate ABA-mediated seed responses that are deficient in the aba2-1 mutant; this stringent functional test shows that activation of PYL2 is sufficient to activate ABA signaling in vivo.
With the CA combinations we have described, we now have tools with which to activate individual family members selectively and explore phenotypic consequences. In this report, we explore the consequences of activating PYL2 in seeds. ABA can normally activate a multiplicity of receptors in the wild-type context, and it is not yet clear if different receptors have different subfunctions. To date, selective ABA receptor activation has only been achieved using pyrabactin (1, 29), which has strong agonist activity on PYR1 and PYL1 and activates a full complement of ABA-responsive gene transcription in imbibed seeds. The genetic removal of PYR1, but not PYL1, causes pyrabactin insensitivity during germination, which argues that the major cellular target of pyrabactin in seeds is PYR1 and that activating PYR1 is sufficient to activate seed ABA signaling. Pyrabactin's effects, however, are complicated by its weak partial agonist/antagonist activity on other receptors like PYL2 (10, 11) and PYL5 (21). CA receptors avoid the complication of pharmacological treatments, although they do necessitate construction of transgenic plants for full characterization, which is less convenient than pharmacological perturbations. Activation of PYL2 by the CA3 mutations mimics the effects of ABA treatment on seeds for three ABA marker genes, suppresses the salt and paclobutrazol sensitivity of aba2-1, and induces hyperdormancy. However, this conclusion is derived from the use of 35S-driven constructs; more specific information on the precise role of PYL2 may be better obtained in future experiments using native promoter-driven constructs in a pyl2-null mutant background. Nonetheless, our data demonstrate that expression of PYL2CA3 is sufficient to activate seed ABA responses. Together with pyrabactin's previously characterized effects, our data are consistent with the hypothesis that activation of a single receptor (PYR1 or PYL2) is sufficient to activate signaling in seeds and that multiple receptors need not be simultaneously activated to elicit an ABA response. The underlying mechanistic basis for this remains unknown but could be explained if receptor levels in seeds are present in excess relative to PP2Cs; future quantitative proteomics experiments may help to illuminate this hypothesis. It is also probable that the picture of receptor function will evolve as the effects of selective receptor activation are examined in vegetative tissues and/or as the activation of other receptor family members is examined. Our tools provide one powerful experimental means with which to sharpen the emerging picture of receptor function in vivo using gain-of-function approaches.
Methods
Site-Saturation Mutagenesis.
Mutants were created using one of two methods. About half of the mutants were made using the QuikChange site-directed mutagenesis kit (Stratagene) utilizing primers that contain random nucleotides (i.e., NNN) at the target position (a list of all mutagenesis primer sequences is provided in Table S3). Twenty-microliter mutagenesis reactions were conducted using pBD GAL4-PYR1 template (1) per the manufacturer's instructions (Stratagene), containing 10 pmol of NNN primer and 0.5 pmol each of M- and W-encoding primers, which were added to enrich the frequency of rare codons. Plasmid DNA for 96 colonies per site was isolated using a Bioneer AccuPrep Plasmid Mini Extraction Kit and sequenced to identify mutants, which identified ∼12 of the 19 desired mutations per target site per 96 clones sequenced. In the second mutagenesis method, we made mutations using the QuikChange lightning multisite-directed mutagenesis kit (Agilent Technologies) utilizing a phosphorylated primer that, instead of NNN at the mutagenesis target site, contained the sequence NNK, which reduces degeneracy (30). Plasmid DNA for 96 colonies per site was isolated using a Beckman Multimek 96 robot and Perfectprep Vac kit (5 Prime, Inc.) and sequenced, which identified 14 of the 19 desired mutations per 96 clones sequenced on average. Mutations not identified by sequencing of random clones were constructed with specifically designed mutagenic primers using the QuikChange lightning multisite-directed mutagenesis kit. This process was conducted for all 39 target sites to yield, ultimately, a set of 741 sequence-validated mutant PYR1 clones.
NMR Spectroscopy.
15N-labeled PYR1, PYR1CA3, and PYR1CA4 proteins for NMR were expressed in E. coli and purified as described previously (1), with the exception that the variants used for NMR were mutated to lack their last 12 unstructured amino acids, which we have found improves the suitability of recombinant proteins for structural studies without affecting PP2C inhibition. PYR1 peak assignments have been previously published and were used to assign specific residues to HSQC peaks in the current experiments (9). PYR1–ABA complexes were obtained by addition of a twofold excess of ABA to the purified protein (500 μM). NMR experiments were performed at 35 °C on a Bruker Avance 600-MHz spectrometer equipped with a 5-mm TCI CryoProbe.
Transgenic Plants.
To create the desired transgenic plants, the coding sequences of PYL2, PYL2CA3, and PYL2CA4 were cloned into a modified version of pEGAD (31) to create 35S-driven GFP-receptor fusion proteins. Prior work has demonstrated that an N-terminal GFP fusion tag does not interfere with PYR1 function in vivo (1). The constructs were sequence-validated and then introduced into Columbia or the aba2-1 mutant using Agrobacterium-mediated transformation via the floral dip method (32). For each genotype constructed, ∼40 primary transgenic plants were identified by virtue of glufosinate resistance or GFP expression in T1 seeds and/or seedlings, and single-insertion homozygous lines were then isolated from the progeny of 10 T1 lines.
Seed Assays.
To assay dormancy of seeds for Columbia, 35S::GFP-PYL2 and 35S::GFP-PYL2CA3 were divided in two aliquots and dry surface-sterilized for 2 h using chlorine gas (prepared in situ by mixing commercial bleach and 12 N of HCl). One portion of sterilized seed was stratified on one-third Murashige and Skoog medium (MS) agar plates for 6 d at 4 °C in darkness, and the second portion, which was maintained dry at room temperature, was seeded 6 d later on the 1/3 MS agar plates; both samples were transferred to a 23 °C light-tight growth chamber, and germination was scored at 24-h intervals. The homozygous 35S::GFP-PYL2 and 35S::GFP-PYL2CA3 seeds used in these experiments were ∼5 and ∼6 mo postharvest, respectively, at the time of the experiment shown.
Germination tests using paclobutrazol (Wako Chemicals) and NaCl were conducted as follows. Columbia, aba2-1, aba2-1;35S::GFP-PYL2, and aba2-1;35S::GFP-PYL2CA3 seeds were surface-sterilized and plated onto 1/3 MS agar media containing 25, 50, or 100 μM paclobutrazol or 0, 50, 200, or 250 mM NaCl. Control wells contained 1/3 MS agar (NaCl controls) or 1/3 MS agar supplemented with 0.1% DMSO, the carrier solvent for paclobutrazol. The seeds were stratified for 4 d in darkness and then transferred to continuous illumination at room temperature (23 °C). Germination was measured after 72 h; seeds showing emergent radicals at least 1/2 seed length or greater were scored as positive. Each experiment was performed in triplicate; experiments were performed on seeds that were ∼6 mo postharvest. Methods for heterologous and in vitro receptor activation assays are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Natasha Raikhel (University of California, Riverside) and Masanori Okamoto (University of California, Riverside) for valuable comments on the manuscript, and Rizaldy Garcia, Glenn Hicks, David Carter, and Clay Clark (University of California, Riverside) for valuable technical assistance. This work was supported in part by the National Science Foundation (IOS 0820508), Syngenta Corporation (S.R.C.), and United States–Israel Binational Agricultural Research and Development Postdoctoral Fellowship F1-440-2010 (to A.M.).
Footnotes
Conflict of interest statement: S.R.C. receives research funding from Syngenta Corporation for his laboratory's work on abscisic acid receptors, and A.M. and S.R.C. have filed a provisional patent application of the receptor variants described in this report.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112838108/-/DCSupplemental.
References
- 1.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 3.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 4.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Aravind L. START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- 6.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 9.Melcher K, et al. A gate-latch-lock mechanism for signal transduction by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson FC, et al. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010;17:1109–1113. doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melcher K, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010;17:1102–1108. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao Q, et al. Functional mechanism of the abscisic acid agonist pyrabactin. J Biol Chem. 2010;285:28946–28952. doi: 10.1074/jbc.M110.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner JJ, Peterson FC, Volkman BF, Cutler SR. Structural and functional insights into core ABA signaling. Curr Opin Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melcher K, Zhou XE, Xu HE. Thirsty plants and beyond: Structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol. 2010;20:722–729. doi: 10.1016/j.sbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 19.Dupeux F, et al. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 2011;156:106–116. doi: 10.1104/pp.110.170894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupeux F, et al. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 2011;30:4171–4184. doi: 10.1038/emboj.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Q, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell. 2011;42:662–672. doi: 10.1016/j.molcel.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Goda H, et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 23.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 25.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 26.Cheng W-H, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Guzmán M, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Léon-Kloosterziel KM, et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, et al. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol. 2007;3:716–721. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
- 30.Kretz KA, et al. Gene site saturation mutagenesis: A comprehensive mutagenesis approach. Methods Enzymol. 2004;388:3–11. doi: 10.1016/S0076-6879(04)88001-7. [DOI] [PubMed] [Google Scholar]
- 31.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.