Abstract
Seed development in flowering plants is initiated after a double fertilization event leading to the formation of zygotic embryo and endosperm tissues surrounded by the maternally derived seed coat. Although the seed coat does not take part in the fertilization process it develops immediately after fertilization, implicating a signaling mechanism from zygotic tissues to the surrounding maternal tissues. We addressed the question of the underlying mechanisms repressing seed coat development before fertilization and initiating seed coat development after fertilization by analyzing combinations of mutants that initiate seed development in the absence of fertilization. We discovered that seed coat development is actively repressed before fertilization by dosage-sensitive Polycomb group proteins acting in maternal tissues surrounding the female gametophyte. This repression is relieved after fertilization by a signal that is formed by the sexual endosperm. Fertilization is required for signal formation, as asexually formed endosperm fails to effectively initiate seed coat development in mutants with uncompromised maternal Polycomb group function. Mutants for the MADS-box transcription factor AGL62 initiate embryo and endosperm formation but fail to develop a seed coat, implicating AGL62 expression in the endosperm as a requirement for signal initiation. Together, our results provide evidence that fertilization of the central cell generates a signal that relieves Polycomb group-mediated repression in the surrounding maternal tissues to initiate seed coat formation.
Seed development in flowering plants is initiated by double fertilization of the female gametophyte. The female gametophyte harbors two distinct gametic cells that will have distinct fates after fertilization; the haploid egg cell will give rise to the diploid embryo and the homodiploid central cell will form the triploid endosperm (1). Embryo growth is supported by the nourishing endosperm tissue that delivers nutrients acquired from the mother plant (2). Embryo and endosperm coordinate their development with the surrounding seed coat that constitutes the nonfertilized part of the seed (2). Seed coat development initiates from ovule integuments after fertilization, implicating signaling between the fertilized products and the ovule integuments (3). Proliferation of central cell and egg cell is actively suppressed in the absence of fertilization. Similarly, the switch from integument to seed coat development does not occur in the absence of fertilization and unfertilized mature ovules degenerate after a few days (4, 5). The fertilization independent seed (fis) mutants can bypass the fertilization requirement and initiate seed development in the absence of fertilization (4, 5). There are four FIS-class genes known: MEDEA (MEA), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), FERTILIZATION INDEPENDENT SEED2 (FIS2), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) (6–11). The FIS-class genes encode for evolutionary conserved Polycomb group (PcG) proteins that assemble together into the FIS multisubunit Polycomb repressive complex 2 (PRC2) (10). PRC2 complexes have histone methyltransferase activity and repress target genes by applying histone methylation at lysine 27 on histone H3 (12). Mutations in FIS genes cause parent-of-origin–dependent seed abortion, with all seeds inheriting a mutant fis allele from the mother abort, regardless of the presence of a wild-type paternal allele (13). Development of fertilized fis mutant seeds is delayed and seeds abort with embryos arrested at late heart stage containing noncellularized endosperm with strongly overproliferated chalazal endosperm domains (6, 7, 10, 11). Whereas MEA and FIS2 are specifically expressed in the central cell and the endosperm (14, 15), expression of FIE and MSI1 extends to sporophytic tissues as well (9, 16). In sporophytic tissues, FIE and MSI1 interact with CURLY LEAF (CLF), SWINGER (SWN), and EMBRYONIC FLOWER2 (EMF2) forming the EMF complex, or, alternatively, they act together with CLF, SWN, and VERNALIZATION2 (VRN2) in the VRN complex (12). Concomitant loss of homologous genes CLF and SWN as well as EMF2 and VRN2 causes strongly enhanced phenotypes compared with the single mutants (17, 18), implicating partial functional redundancy of PcG genes. Functional redundancy has also been suggested for MEA and SWN in suppressing central cell proliferation, as penetrance of autonomous seed formation in the mea mutant can be strongly increased by concomitant loss of the MEA homolog SWN (15).
We addressed the question of whether EMF2 and VRN2, two homologs of FIS2, act redundantly with FIS2 to suppress autonomous seed formation. Our data reveal that FIS2 does not act redundantly with EMF2 and VRN2, but that instead sporophytically active PcG proteins suppress seed coat development in the absence of fertilization. Our data suggest that in response to fertilization a mobile signal is formed by the sexual endosperm to relieve PcG repression in the integuments and to initiate seed coat formation. Finally, we succeeded in identifying the type I MADS-box transcription factor AGL62 as a component required to form the mobile signal, generating evidence for the central requirement of this quickly evolving family of transcription factors in coordinating seed development.
Results
EMF2 and VRN2 Are Not Acting Redundantly with FIS2 in the Female Gametophyte.
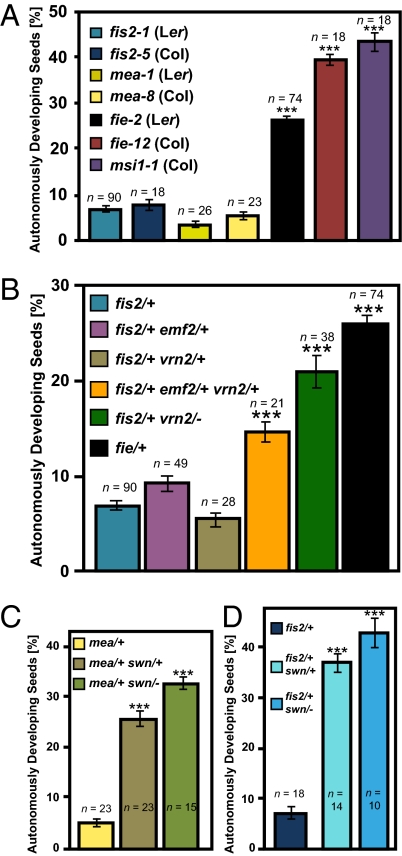
We determined the penetrance of autonomous seed development in different mutants lacking components of the FIS PcG complex (Fig. 1A). The presence of four or more nuclei in the central cell at 7 d after emasculation (DAE) was considered as criterion for autonomous endosperm and seed development. Alternatively, strongly enlarged autonomous seeds containing less than four endosperm nuclei were also counted. Whereas fis2 and mea mutants generated only very few autonomous seeds (about 5–8%; penetrance 10–16%), almost every ovule inheriting a fie or msi1 mutation initiated autonomous seed development (up to 42 or 45% autonomous seeds, penetrance 84 or 90%, respectively). Therefore, fis mutants have a strikingly different penetrance in autonomous seed formation, which is contrasted by a completely penetrant phenotype of each mutant after fertilization. Lack of any FIS complex subunit gives rise to 50% seed abortion (4–6, 10, 11), implicating no functional redundancy of FIS genes after fertilization. This suggests the presence of genes acting redundantly with MEA and FIS2 before fertilization or, alternatively, an increased capacity of fie and msi1 to undergo autonomous seed development due to unknown defects caused by loss of FIE and MSI1 function. To test whether differences in the penetrance of autonomous seed development are caused by functional redundancy of FIS2 with VRN2 or EMF2 in the female gametophyte, we analyzed penetrance of autonomous seed development in double heterozygous mutants fis2/+ vrn2/+ and fis2/+ emf2/+ (Fig. 1B). In this genetic combination we expect 25% of female gametophytes lacking FIS2 and VRN2 or FIS2 and EMF2 function, respectively. However, penetrance of autonomous seed development was increased neither in fis2/+ vrn2/+ nor fis2/+ emf2/+ double mutants. We generated a triple fis2/+ vrn2/+ emf2/+ mutant containing 12.5% ovules lacking all three homologous genes. In this triple mutant, 15% of the ovules initiated autonomous seed development (Fig. 1B), whereas autonomous seed development was never observed in vrn2/+ emf2/+ and vrn2/− emf2/+ double mutants. One explanation for this finding is that all three genes act redundantly in the female gametophyte. We investigated this hypothesis further by testing whether expression of EMF2 and VRN2 under control of the FIS2 promoter could rescue the fis2 mutant. We generated transgenic lines in the fis2 mutant background expressing the VRN2 and EMF2 genes under control of the FIS2 promoter. However, among 19 FIS2pro:VRN2 and 12 FIS2pro:EMF2 lines, none of them complemented the fis2 seed abortion phenotype, whereas expression of FIS2 under control of the same promoter provided complete complementation (of 15 lines analyzed, all lines showed complementation revealed by reduction of seed abortion rates to 25% in lines hemizygous for the construct). These findings do not support the hypothesis that VRN2 and EMF2 act redundantly with FIS2 in the female gametophyte and rather suggest that double heterozygosity for EMF2 and VRN2 causes an effect on sporophytic tissues, promoting the capacity of fis2 mutants to form autonomous seeds. We tested the hypothesis that decreased dosage of PcG proteins in sporophytic tissues enhances autonomous seed formation of fis2 by analyzing autonomous seed development in a fis2/+ vrn2/− double mutant. In contrast to a double heterozygous fis2/+ vrn2/+ mutant that did not increase penetrance of autonomous seed formation (Fig. 1B), a strongly increased penetrance was observed in the fis2/+ vrn2/− double mutant (Fig. 1B), strongly supporting the idea that reduction of PcG function in sporophytic tissues promotes autonomous seed development. Together, we conclude that FIS2 does not act redundantly with EMF2 and VRN2 in the female gametophyte, but that reduced dosage of EMF2 and VRN2 in sporophytic tissues promotes autonomous seed development of the fis2 mutant. This suggests that PcG proteins act in maternal integument tissues surrounding the female gametophyte to restrict the development of autonomous seeds.
Fig. 1.
Percentage of autonomously developing seeds in PcG mutants and mutant combinations. (A) Percentage of autonomously developing seeds in different fis mutants determined at 7 DAE. (B) Percentage of autonomously developing seed in the fis2-1/+ single mutant and in double and triple mutant combinations of fis2-1/+ with emf2-5 and vrn2-1. The fie-2 mutant served as a positive control. (C) Percentage of autonomously developing seeds in the mea-8/+ single mutant and double mutant of mea-8/+ with swn-3. (D) Percentage of autonomously developing seeds in the fis2-5/+ single mutant and double mutant of fis2-5/+ with swn-3. Asterisks indicate significant deviations from fis2/+ (A, B, and D) or mea/+ (C) determined by ANOVA testing (P < 0.001). Numbers on top of each bar indicate analyzed siliques. Error bars indicate SEM. Each silique contained on average 60 ovules or seeds.
SWN Acts in Sporophytic Tissues to Suppress Autonomous Seed Formation.
Previous studies proposed that low penetrance of autonomous seed development in the mea mutant is caused by functional redundancy of MEA and the MEA homolog SWN in the central cell (15). SWN is only weakly expressed in the central cell of the female gametophyte but strongly expressed in sporophytic tissues (15, 19), where it was previously shown to act redundantly with CLF (17). In agreement with previous findings (15), the mea/+ swn/+ double heterozygous mutant had strongly increased penetrance of autonomous seed development in comparison with the mea single mutant (Fig. 1C). If MEA and SWN act redundantly in the female gametophyte to suppress autonomous seed development, we expected that the mea/+ swn/− double mutant would have approximately twice as many autonomously developing seeds compared with the mea/+ swn/+ double heterozygous mutant. However, only a minor increase of autonomously developing seeds was observed upon complete loss of SWN function, implicating that SWN, like VRN2 and EMF2, has a role in sporophytic tissues to suppress autonomous seed development. To further explore this idea we generated and analyzed fis2/+ swn/+ and fis2/+ swn/− double mutants. These double mutant combinations caused a dramatic increase in autonomous seed formation to 37% and 43% compared with 7% in the fis2 single mutant, respectively (Fig. 1D), strongly supporting a sporophytic role of SWN to suppress autonomous seed development. Consistently, SWN is strongly expressed in the integuments (15). Similarly, we detected strong VRN2 expression in the integuments and in the seed coat after fertilization (Fig. S1), indicating that relieve of PcG suppression is not caused by reduced PcG gene expression.
FIS Genes Have a Nonredundant Role After Fertilization.
PHERES1 (PHE1) is a direct target gene of the FIS PcG complex and exclusively expressed in the endosperm (20, 21). Loss of FIS2 function causes increased expression of PHE1 in the endosperm (Fig. S2A). Consistent with the idea that FIS2 does not act redundantly with VRN2 and EMF2, expression levels of PHE1 were similarly increased in seeds of single fis2 mutants as in double and triple mutants of fis2 with vrn2 and emf2. Similarly, there was no relevant difference in the expression of PHE1 or of a PHE1pro:PHE1-EGFP reporter line in the fie mutant compared with the fis2 mutant (Fig. S2 A and B), supporting the idea that FIS2 homologs do not act redundantly in controlling expression of FIS target genes. Together with the fact that fis mutants have a completely penetrant seed abortion phenotype, we conclude that FIS genes play a nonredundant role in the endosperm after fertilization and propose that differences among fis mutants before fertilization are caused by a haploinsufficient requirement of FIE and MSI1 in suppressing seed coat development.
fis Mutants Form Two Classes of Autonomously Developing Seeds.
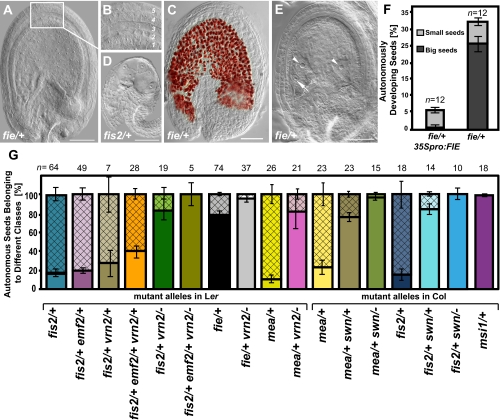
All four fis mutants generate autonomous seeds. However, we noticed that aside from the different penetrance of this phenotype, there were two distinct classes of autonomously developing seeds. Whereas most of the autonomously developing seeds in fis2 and mea remained small like unfertilized wild-type ovules (Fig. 2 D and G), the majority of autonomously formed seeds in fie and msi1 were much bigger (Fig. 2 A and G). The size of the autonomous seeds was not directly correlated with endosperm growth, because we found small seeds with up to 64 nuclei and big seeds with as few as one, two, or four endosperm nuclei (Fig. 2 A, D, and E). The second striking difference concerns the development of the integuments. Similar to unfertilized wild-type ovules, integuments of fis2 and mea autonomous seeds did not progress in their development and degenerated about 5 DAE, although they contained developing endosperm (Fig. 2D). In stark contrast, integuments of big autonomous seeds in fie and msi1 mutants differentiated into a seed coat with five clearly distinguishable cell layers (Fig. 2B) that accumulated proanthocyanidins in the endothelium layer (Fig. 2C) as a hallmark for seed coat development (22). These findings indicate that compromised sporophytic PcG function in fie and msi1 mutants promotes autonomous seed coat development, suggesting that sporophytically acting PcG complexes prevent development of the seed coat in the absence of fertilization. In agreement with this hypothesis, the majority of autonomously developing seeds in fis2/+ vrn2/− double mutants were big and had developed seed coats (Figs. 2G and 3C). Conversely, autonomous seeds of fis2/+ vrn2/+ double mutants remained small and rarely developed a seed coat (Fig. 2G). Triple heterozygous plants fis2/+ vrn2/+ emf2/+ differed from fis2 mostly in the number of autonomous seeds with developed seed coat, whereas the number of small and degenerated autonomous seeds was comparable between both genotypes (around 7%, Figs. 1B and 2G). Together, we conclude that differences in penetrance and developmental potential of autonomous seeds are controlled by sporophytically acting PcG complexes.
Fig. 2.
fis mutants form two classes of autonomously developing seeds. (A) Big autonomous fie-2 seed with developed seed coat and endosperm. Inset window enlarged in B shows five seed coat layers. (C) Proanthocyanidin accumulation in fie-2 seed. (D) Small autonomous fis2-1 seed with developing endosperm but degenerated integuments. (E) Autonomous fie-2 seed with developed seed coat and only two endosperm nuclei (arrowheads). Arrow indicates egg cell nucleus. (Scale bar, 50 μm in A–E.) (F) Percentage of autonomously developed seeds in fie-2 and fie-2 mutant expressing a wild-type copy of the FIE gene under control of the 35S promoter. Numbers on top of each bar indicate analyzed siliques. Error bars indicate SEM. (G) Percentage of each class of autonomous seeds in respective genotypes (mutant alleles in Ler: fis2-1, fie-2, emf2-5, and vrn2-1; mutant alleles in Col: fis2-5, mea-8, swn-3, and msi1-1) determined at 7 DAE. Open and hatched bars represent small and big autonomous seeds, respectively. Numbers on top of each bar indicate analyzed siliques. Error bars indicate SEM.
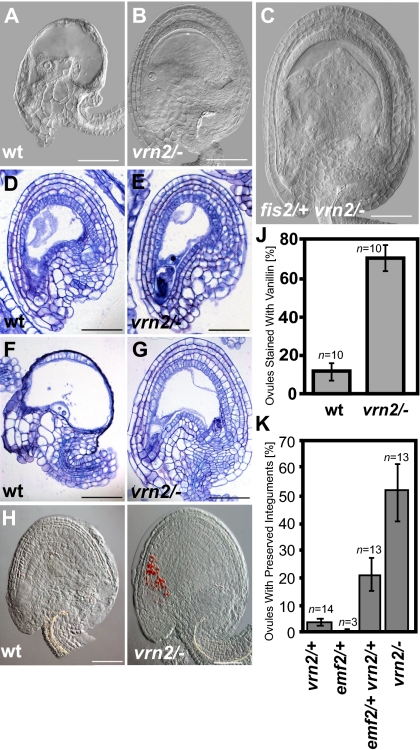
Fig. 3.
Loss of VRN2 initiates endothelium development in the absence of endosperm development. Microscopy images of cleared (A–C), sectioned (D–G), and vanillin stained (H and I) ovules and autonomous seeds at 5 DAE (D, E, H, and I) and 6 DAE (A–C, F, and G). (A, D, F, and H) Wild type (WT). (B, E, G, and I) vrn2-1/−. (C) fis2-1/+vrn2-1/−. (Scale bar for A–I, 50 μm.) (J) Percentage of wild-type and vrn2-1/− ovules stained with vanillin at 5 DAE; error bars indicate SEM. (K) Percentage of ovules with nondegenerated five integument layers at 6 DAE; error bars indicate SEM.
FIE Is Haploinsufficient and Acts in Sporophytic Tissues to Suppress Seed Coat Development.
The strong penetrance of autonomous seed formation of the heterozygous fie mutant suggests that FIE is a dosage-sensitive suppressor of integument development. We tested this hypothesis by analyzing whether we could increase the penetrance of autonomous seed formation of the fie/+ mutant by additionally depleting VRN2 function. Therefore, we generated and analyzed double mutants of fie with vrn2. Whereas about 20% of autonomously developing seeds in the fie/+ mutant had early degenerating integuments, almost all developing autonomous seeds in fie/+ vrn2/− had developed seed coats (Fig. 2G). Thus, we conclude that incomplete penetrance of the fie mutant phenotype is caused by remaining PcG function in sporophytic tissues. To further test the hypothesis that FIE acts in sporophytic tissues to suppress autonomous seed development, we expressed the FIE gene under control of the 35S promoter of the Cauliflower mosaic virus (35S). The 35S promoter is active in sporophytic tissues but is generally considered not to be active during the gametophytic stage (23) (Fig. S3). Consistently, expression of 35Spro:FIE in the fie/+ mutant did not rescue the fie seed abortion phenotype. Among 13 35Spro:FIE; fie/+ lines analyzed, we did not identify a line with reduced numbers of aborted seeds. However, we observed a complete suppression of the formation of big autonomous seeds with developed seed coats in fie/+ mutants being hemizygous for the 35Spro:FIE construct (Fig. 2F), strongly supporting the hypothesis that FIE is haploinsufficient and acts in integument tissues suppressing seed coat development.
Loss of VRN2 Initiates Endothelium Development in the Absence of Endosperm Development.
Development of wild-type ovules is accompanied by cell divisions in all layers of the integuments. This process decelerates in mature ovules at the time of anthesis. Ovules at this stage await fertilization and if fertilization does not occur, they quickly degenerate at about 5 DAE (corresponding to about 3–4 d postanthesis) (24) (Fig. 3A). We observed that vrn2/− ovules did not degenerate at the same time as wild-type ovules. Clearings and sections of unfertilized ovules revealed that even at 6 DAE, vrn2 ovules were not degenerated and five layers of integuments were clearly distinguishable (Fig. 3 B, E, G, and K).
We investigated whether unfertilized vrn2/− ovules initiate seed coat development by analyzing the formation of proanthocyanidins using vanillin staining. Whereas vanillin staining in unfertilized wild-type ovules was rarely observed, more than half of all tested vrn2/− ovules showed a strong staining in the micropylar region of the ovule (Fig. 3 H–J), which was, however, not accompanied by a size increase of vrn2/− ovules (Fig. 3 D and E and Fig. S4). Together, loss of VRN2 function initiates seed coat differentiation in unfertilized ovules, supporting the view that PcG proteins suppress seed coat development in the absence of fertilization. We tested whether seed coat development would also be initiated in heterozygous vrn2/+, emf2/+, and double heterozygous vrn2/+ emf2/+ mutants. Consistent with the finding that neither heterozygous vrn2/+ nor emf2/+ could enhance the fis2 phenotype, we did not observe initiation of autonomous seed coat development in either mutant (Fig. 3K). Similar to homozygous vrn2 ovules, about 22% of the ovules of double heterozygous vrn2/+ emf2/+ mutants remained intact and did not collapse 6 DAE (Fig. 3K), supporting the view that the observed increased penetrance of autonomous seed formation in fis2/+ vrn2/+ emf2/+ triple mutants (Fig. 1B) is caused by depletion of PcG function in the integuments, promoting seed coat formation.
The Signal for Seed Coat Development Is Generated by the Sexual Endosperm Depending on AGL62.
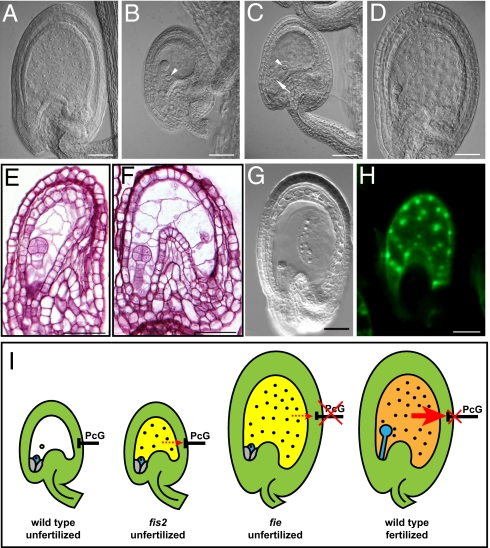
Although data from several studies support a role of the endosperm in regulating seed coat growth (3, 25–27), formal evidence for the sexual endosperm being sufficient for generating a signal initiating seed coat development is missing. To define the origin of the signal initiating seed coat development we investigated seeds of the kokopelli (kpl) mutant. The kpl mutation affects male gametogenesis, leading to the formation of single sperm cell male gametophytes and random single fertilization events (28). Consequently, seeds derived after fertilization with kpl pollen frequently contain only embryos or only endosperm, resulting from fertilization of egg or central cell, respectively (Fig. 4 A and B). Seed coat development of both seed classes was strikingly different; whereas all seeds containing only endosperm had normally developed seed coats (n = 26; Fig. 4 A and D), all seeds containing only an embryo did not initiate seed coat development and integuments appeared similar to unfertilized wild-type ovules (n = 21; Fig. 4 B and C). In this seed class, embryo development arrested at the globular stage and integuments degenerated at the same time as in unfertilized wild-type ovules (Fig. S5). Given that the autonomously formed endosperm in fis2 and mea mutants is rarely sufficient to initiate seed coat formation, we conclude that a signal initiating seed coat development in Arabidopsis is derived from the sexual endosperm.
Fig. 4.
The sexual endosperm generates a signal for seed coat development dependent on AGL62. (A) Seed developing after fertilization with kpl pollen at 2 DAP. Seed contains only endosperm and no embryo. (B) Seed developing after fertilization with kpl pollen at 2 DAP. Seed contains only an embryo and no endosperm. (C) Unfertilized ovule shortly before degeneration. (D) Seed developing after fertilization with wild-type pollen at 2 DAP. (E and F) Sections of agl62-2/− seeds at 3 DAP. (G) Cleared ttn2 seed at 3 DAP. (H) AGL62:AGL62-GFP expression in autonomous fie-12 seeds. (Scale bars, 50 μm in A–H.) Arrow and arrowheads mark egg cell and central cell, respectively. (I) Model depicting events in autonomous and sexual seed development. In wild-type ovules, sporophytically active PcG proteins repress seed coat development. In fis2 mutants, this repression remains active and autonomous endosperm division is not accompanied by seed coat development. In fie mutants, this repressive block is released and autonomous endosperm formation is associated with seed coat development. In wild type, a signal formed by the sexual endosperm (red arrow) relieves PcG repression in the seed coat, causing the initiation of seed coat development. In fis2 and fie mutants, no signal or only a weak signal is formed by the autonomous endosperm.
Disruption of the signaling pathway from the endosperm to the seed coat is expected to result in the formation of early arresting seeds containing embryo and endosperm, but without a developed seed coat. Lack of the type I MADS-box transcription factor AGL62 causes precocious endosperm cellularization after about three to four nuclei divisions and early embryo arrest (29). Although agl62 seeds initiate embryo and endosperm development, we found that this was generally not accompanied by seed coat formation. We noted a minor fraction of agl62/− seeds arresting development later with about 50 endosperm cells and a developed seed coat (2.5% of n = 1,080 analyzed seeds; Fig. S6), suggesting incomplete penetrance of the agl62-2 mutant phenotype. The majority of agl62/− seeds did not initiate seed coat formation and had collapsed integuments after about 3 days after pollination (DAP) (19.5% of n = 1,080 analyzed seeds; Fig. 4 E and F), similar to the phenotype observed in autonomously developing fis2 seeds (Fig. 2D). It is unlikely that failure of seed coat initiation is a consequence of endosperm proliferation failure, as seed coat development is clearly initiated in the titan 2 (ttn2) mutant that has severe endosperm proliferation defects and arrests development containing a comparable number of endosperm nuclei as the agl62 mutant (n = 38; Fig. 4G) (30). This strongly indicates that AGL62 is required in the endosperm to initiate seed coat formation. We analyzed AGL62 expression in wild-type seeds after fertilization and autonomously developing fie seeds using a translational fusion of the AGL62 gene under control of the native promoter fused to GFP. This construct completely complemented the agl62 mutant phenotype, indicating that the AGL62pro:AGL62-GFP expression reflects the native AGL62 expression pattern. In wild-type as in autonomously developing fie seeds, AGL62 remained exclusively expressed in endosperm nuclei (Fig. 4H), indicating that AGL62 is required to form the mobile signal rather than being the mobile signal itself.
Discussion
The data presented in this manuscript reveal that initiation of seed development is negatively controlled by PcG proteins at multiple levels; the FIS PcG complex represses autonomous replication of the central cell, whereas autonomous seed coat development is suppressed by sporophytically active PcG proteins. Therefore, specific impairment of FIS function alone in fis2 and mea mutants elicits autonomous replication of the central cell, but fails to efficiently initiate seed coat development. In contrast, concomitant loss of FIS function as well as compromised function of sporophytic PcG complexes in the integuments initiates formation of autonomous seeds containing endosperm as well as seed coat. Our analysis of heterozygous fie as well as double heterozygous emf2 and vrn2 mutants reveal that sporophytic PcG complexes are haploinsufficient to suppress autonomous seed coat formation, revealing a dosage-sensitive function of plant PcG proteins similar to the fine-tuned dosage-sensitive requirement of PcG proteins in animals (31). Our data strongly suggest that upon fertilization a mobile signal is formed in the endosperm that migrates to the integuments and relieves PcG repression at specific target loci (Fig. 4I). The importance of the endosperm for seed coat development has been implicated before (3, 25–27, 32); however, our study makes a significant advance by revealing that the sexual endosperm is required for seed coat formation, whereas asexually formed endosperm rarely suffices to initiate seed coat formation. This implicates that signal formation is initiated after fertilization and that formation of the signal is not or not sufficiently initiated in the asexual endosperm of fis mutants (Fig. 4I). We demonstrate that seed coat formation is initiated in sporophytic PcG mutants, implicating that upon loss of sporophytic PcG function signal formation is initiated in the integuments independently of a sexual endosperm or, alternatively, processes downstream of the signal are initiated upon loss of PcG function. Importantly, however, loss of sporophytic PcG function only initiates proanthocyanidin formation as one of the first processes of seed coat differentiation, whereas complete differentiation of seed coat layers was not observed. In contrast, concomitant loss of FIS function as well as impaired sporophytic PcG function caused a strong increase in seed size and the formation of five distinguishable seed coat layers, implicating that initiation of endosperm formation is required to activate all processes leading to seed coat formation. This is unlikely to be a consequence of an increased mechanical pressure through endosperm growth, as we observed enlarged autonomous seeds containing only two endosperm nuclei. Therefore, it is more likely that complete activation of seed coat formation requires a signal threshold and that formation of the signal strongly increases after central cell division. To identify the nature of the signal remains the subject of future investigations. Finally, we succeeded in identifying the type I MADS-box transcription factor AGL62 as one central component required for formation of the mobile signal. Seeds lacking AGL62 fail to initiate seed coat formation, similar to autonomously developing fis2 seeds as well as seeds containing only a developing embryo but no endosperm. It is unlikely that this is a consequence of agl62 endosperm proliferation failure and early endosperm cellularization, as mutants defective in nuclear proliferation still develop a seed coat (ref. 30 and data shown in this paper). Conversely, defects in seed coat growth negatively impact on endosperm growth (26, 32), indicating that impaired seed coat development is a cause rather than a consequence of early agl62 endosperm arrest. We could furthermore show that AGL62 is unlikely to be the signal itself, as expression of AGL62-GFP remained confined to the endosperm and was not detected in the seed coat. AGL62 has been shown to interact with type I MADS-box proteins including the specifically paternally expressed PHERES1 (33), suggesting that an AGL62-containing protein complex is required to activate downstream target genes to form the seed coat initiation signal.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana mutants fis2-1, fie-2 (5), vrn2-1 (34), emf2-5 (35), mea-1 (6), and kpl-2 (28) are in the Ler accession. Mutants fis2-5 (21), fie-12 (36), msi1-1 (10), mea-8 (37), swn-3 (17), and agl62-2 (29) are in the Col-0 accession. The ttn2-1 mutant (30) is in the Wassilewskija accession. Plants were grown in a growth cabinet under long-day photoperiods (16 h light and 8 h dark) at 23 °C. After 10 d, seedlings were transferred to soil and plants were grown in a growth chamber at 60% humidity and daily cycles of 16 h light and 8 h darkness at 21 °C.
Generation of Transgenic Lines.
Binary destination vector pB7WG2 was used to generate FIS2pro:FIS2, FIS2pro:VRN2, and FIS2pro:EMF2 plasmids. The 1,850 bp of the FIS2 promoter were amplified with primers containing SacI and XbaI sites and cloned into pB7WG2 replacing the 35S promoter. The FIS2 coding sequence was amplified from Ler seed cDNA and inserted into pENTR-D TOPO vector (Invitrogen). Entry clones for VRN2 and EMF2 (kindly provided by Lars Hennig, SLU, Uppsala, Sweden) were generated by cDNA amplification and insertion into pENTR-D TOPO vector. Expression clones were generated following the Gateway cloning protocol (Invitrogen). The coding sequence of FIE was amplified by PCR and inserted into vector pBI101 containing a double myc tag to generate 35Spro:FIE. Transgenic lines were generated by Agrobacterium tumefaciens-mediated transformation into fis2-1. Independent single transgene locus insertion T2 lines were analyzed. The AGL62:AGL62-GFP–containing vector was generated by inserting 2,500 bp of amplified AGL62 promoter and coding region fused to eGFP into pCAMBIA 1300. The construct was transformed into the agl62-2 mutant and independent single transgene locus insertion T2 lines were analyzed.
Histological Analysis.
Samples were fixed in 4% formaldehyde, 50% ethanol, and 10% acetic acid overnight at 4 °C. Samples were dehydrated in a series of ethanol dilutions (50, 70, and 100%) for 1 h each and embedded in Technovit 7100 (Kulzer) according to the manufacturer's instructions. Four-micrometer sections were prepared using a Leica RM2145 microtome. Sections were stained with 0.05% Toluidine blue or Schiff's reagent.
Microscopy.
Samples were fixed in ethanol/acetic acid in a ratio of 9:1, washed with 70% ethanol, and mounted in chloral hydrate solution (glycerol/chloral hydrate/water in a ratio of 1:8:3). For vanillin staining, ovules were manually dissected from ovaries and mounted on slides in 1% (wt/vol) vanillin (4-hydroxy-3-methoxybenzaldehyde; Sigma) in 6 N HCl solution. Slides were analyzed after 20 min of incubation. Samples were analyzed with a Leica DM2500 microscope using differential interference contrast optics. Images were recorded with a Leica DFC 300 FX digital camera. Samples from GFP lines were mounted in water and analyzed with epifluorescence optics.
Supplementary Material
Acknowledgments
We thank Sabrina Huber for excellent technical support, André Imboden for excellent plant growth support, Wilhelm Gruissem for sharing laboratory facilities, Abed Chaudhury for providing fis2-1 and fie-2 seeds, Rita Gross-Hardt and Ronny Völz for contributing the 35Spro:NLS-3xGFP line, and Nicole Schatlowski and Jordi Moreno Romero for critical comments on this manuscript. This research was supported by Swiss National Science Foundation Grant PP00P3_123362 (to C.K.) and by a Heinz Imhof Fellowship (to P.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117111108/-/DCSupplemental.
References
- 1.Drews GN, Yadegari R. Development and function of the angiosperm female gametophyte. Annu Rev Genet. 2002;36:99–124. doi: 10.1146/annurev.genet.36.040102.131941. [DOI] [PubMed] [Google Scholar]
- 2.Ingram GC. Family life at close quarters: Communication and constraint in angiosperm seed development. Protoplasma. 2010;247:195–214. doi: 10.1007/s00709-010-0184-y. [DOI] [PubMed] [Google Scholar]
- 3.Ingouff M, Jullien PE, Berger F. The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell. 2006;18:3491–3501. doi: 10.1105/tpc.106.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohad N, et al. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhury AM, et al. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 7.Kiyosue T, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M, et al. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohad N, et al. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köhler C, et al. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guitton AE, et al. Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development. 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- 12.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Köhler C, Makarevich G. Epigenetic mechanisms governing seed development in plants. EMBO Rep. 2006;7:1223–1227. doi: 10.1038/sj.embor.7400854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:13244–13249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- 17.Chanvivattana Y, et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 18.Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Spillane C, et al. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;448:349–352. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- 20.Köhler C, et al. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003;17:1540–1553. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010;6:e1001152. doi: 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Bechtold N, et al. The maternal chromosome set is the target of the T-DNA in the in planta transformation of Arabidopsis thaliana. Genetics. 2000;155:1875–1887. doi: 10.1093/genetics/155.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner DJ, Hill TA, Gasser CS. Regulation of ovule development. Plant Cell. 2004;16(Suppl):S32–S45. doi: 10.1105/tpc.015933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 2003;133:1882–1892. doi: 10.1104/pp.103.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell. 2008;20:635–647. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 1998;16:21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- 31.Mollaaghababa R, et al. Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc Natl Acad Sci USA. 2001;98:3958–3963. doi: 10.1073/pnas.061497798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia D, et al. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003;131:1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler J, Wilson A, Dean C. Arabidopsis mutants showing an altered response to vernalization. Plant J. 1996;10:637–644. doi: 10.1046/j.1365-313x.1996.10040637.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang CH, Chen LJ, Sung ZR. Genetic regulation of shoot development in Arabidopsis: Role of the EMF genes. Dev Biol. 1995;169:421–435. doi: 10.1006/dbio.1995.1158. [DOI] [PubMed] [Google Scholar]
- 36.Wolff P, et al. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 2011;7:e1002126. doi: 10.1371/journal.pgen.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngo QA, Moore JM, Baskar R, Grossniklaus U, Sundaresan V. Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development. 2007;134:4107–4117. doi: 10.1242/dev.007310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.