Abstract
The specificity of most aminoacyl-tRNA synthetases for an amino acid and cognate tRNA pair evolved before the divergence of the three domains of life. Glutaminyl-tRNA synthetase (GlnRS) evolved later and is derived from the archaeal-type nondiscriminating glutamyl-tRNA synthetase (GluRS), an enzyme with relaxed tRNA specificity capable of forming both Glu-tRNAGlu and Glu-tRNAGln. The archaea lack GlnRS and use a specialized amidotransferase to convert Glu-tRNAGln to Gln-tRNAGln needed for protein synthesis. We show that the Methanothermobacter thermautotrophicus GluRS is active toward tRNAGlu and the two tRNAGln isoacceptors the organism encodes, but with a significant catalytic preference for 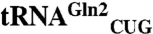 . The less active
. The less active 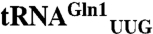 responds to the less common CAA codon for Gln. From a biochemical characterization of M. thermautotrophicus GluRS variants, we found that the evolution of tRNA specificity in GlnRS could be recapitulated by converting the M. thermautotrophicus GluRS to a tRNAGln specific enzyme, solely through the addition of an acceptor stem loop present in bacterial GlnRS. One designed GluRS variant is also highly specific for the
responds to the less common CAA codon for Gln. From a biochemical characterization of M. thermautotrophicus GluRS variants, we found that the evolution of tRNA specificity in GlnRS could be recapitulated by converting the M. thermautotrophicus GluRS to a tRNAGln specific enzyme, solely through the addition of an acceptor stem loop present in bacterial GlnRS. One designed GluRS variant is also highly specific for the 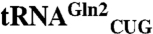 isoacceptor, which responds to the CAG codon, and shows no activity toward
isoacceptor, which responds to the CAG codon, and shows no activity toward 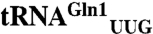 . Because it is now possible to eliminate particular codons from the genome of Escherichia coli, additional codons will become available for genetic code engineering. Isoacceptor-specific aminoacyl-tRNA synthetases will enable the reassignment of more open codons while preserving accurate encoding of the 20 canonical amino acids.
. Because it is now possible to eliminate particular codons from the genome of Escherichia coli, additional codons will become available for genetic code engineering. Isoacceptor-specific aminoacyl-tRNA synthetases will enable the reassignment of more open codons while preserving accurate encoding of the 20 canonical amino acids.
Keywords: enzyme kinetics, glutamylation, phylogeny, translation
The genetic code depends on the catalytic action of the aminoacyl-tRNA synthetases (aaRSs) that are responsible for accurately ligating amino acids to their cognate tRNAs. The high fidelity of the genetic code is derived principally from the exclusive interaction between an aaRS and its cognate tRNA, and this property of “orthogonality” has been exploited to expand the genetic codes of bacterial and eukaryotic cells to encode noncanonical amino acids (reviewed in refs. 1 and 2).
In nature, some aaRSs evolved and are selectively maintained to promiscuously recognize more than one tRNA species. Selenocysteine (Sec) is biosynthesized on its tRNA from a Ser-tRNASec precursor, which is generated by a regular seryl-tRNA synthetase (SerRS) that ligates serine to tRNASec and to tRNASer. Glutamine and asparagine are also biosynthesized on their tRNAs in many organisms (3). In these organisms, Gln-tRNAGln (4) and Asn-tRNAAsn (5) are synthesized by the action of specialized amidotransferase enzymes (6) from the respective precursors Glu-tRNAGln and Asp-tRNAAsn. In both cases, the seemingly misacylated precursor aminoacyl-tRNA is formed by a nondiscriminating aaRS—i.e., a glutamyl-tRNA synthetase (GluRS) (7) that glutamylates both tRNAGlu and tRNAGln or an aspartyl-tRNA synthetase (AspRS) (8) that forms both Asp-tRNAAsp and Asp-tRNAAsn.
There are two types of nondiscriminating GluRS (ND-GluRS). All GluRSs share the class I Rossman fold catalytic domain, but the bacterial GluRS has an α-helical bundle anticodon binding domain that is unrelated to the dual ß-barrel anticodon binding domain found in all glutaminyl-tRNA synthetases (GlnRSs) and archaeal and eukaryotic GluRSs. Although the bacterial ND-GluRS is well characterized (9, 10), the distinct architecture of the archaeal ND-GluRS indicates that its mechanism of dual tRNA recognition is also distinct and yet uncharacterized.
We recently described the crystal structure of an archaeal ND-GluRS (11). Structural comparison of the Methanothermobacter thermautotrophicus ND-GluRS with the Escherichia coli GlnRS tRNAGln complex (12) revealed two significant loops in the protein that could contribute to the high specificity of GlnRS for tRNAGln, whereas their absence in the archaeal ND-GluRS may allow the enzyme to recognize both tRNAGlu and tRNAGln (11). To investigate tRNA recognition by the archaeal ND-GluRS, we determined the enzyme kinetics of Glu-tRNAGlu and Glu-tRNAGln formation for the wild-type M. thermautotrophicus ND-GluRS (WT-GluRS) and rationally designed GluRS variants that were engineered to selectively aminoacylate tRNAGln. GlnRS evolved from an ancestor closely similar to the archaeal ND-GluRS, so the designed GluRSs represent plausible intermediate forms preceding GlnRS evolution.
Results
Biochemical Characterization of the M. thermautotrophicus Nondiscriminating GluRS.
Because the catalytic preference of the archaeal-type nondiscriminating GluRS has not been documented, we measured the kinetic constants of the ND-GluRS toward its two tRNAGln isoacceptors (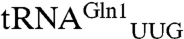 ,
, 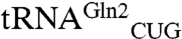 ) and tRNAGlu (Fig. 1).
) and tRNAGlu (Fig. 1).
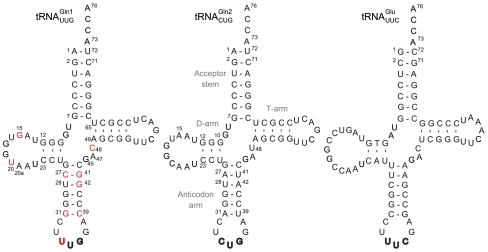
Fig. 1.
Cloverleaf structures of the M. thermautotrophicus tRNAGln and tRNAGlu molecules. Sequence differences between the tRNAGln isoacceptors (tRNAGln1 has the UUG anticodon and tRNAGln2 has the CUG anticodon) are highlighted in red on tRNAGln1. The structures are annotated with canonical tRNA numbering. The anticodon sequences are shown in bold.
The WT-GluRS showed greatest activity toward the tRNAGln2 isoacceptor with a kcat = 0.41 ± 0.04 s-1 and KM = 1.33 ± 0.10 μM (Table 1). All tRNA substrates used in this study were in vitro produced transcripts (see Methods). Although in some cases lacking base modifications can lead to inactive tRNAs (10), the kinetic values measured for WT-GluRS are within the typical range for aaRSs and indicate the role of modified bases is not critical for WT-GluRS activity. The kinetic constants of WT-GluRS for in vitro transcribed tRNAGln2 were recently measured (13), showing the enzyme to be marginally (8.5 ± 3.6-fold) less efficient in our experiments.
Table 1.
Aminoacylation kinetics of WT-GluRS
|
kcat , s-1 |
KM, μM |
kcat/KM, s-1 μM-1 |
Loss of efficiency* |
|
| WT-GluRS | ||||
| tRNAGln2 | 0.41 ± 0.04 | 1.33 ± 0.40 | 0.31 ± 0.10 | 1.0 |
| tRNAGlu | 0.04 ± < 0.01 | 3.61 ± 0.88 | 0.01 ± < 0.01 | 24 |
| tRNAGln1 | 0.006 ± < 0.001 | 5.39 ± 1.44 | 0.001 ± < 0.001 | 257 |
*Loss of catalytic efficiency (x fold) is the relative fold decrease in kcat/KM that is calculated as the ratio of kcat/KM for tRNAGln2 over the kcat/KM for the tRNA species listed in the far left column. Standard deviations are reported. Reaction conditions are given in SI Methods.
Although still a nondiscriminating enzyme, WT-GluRS shows a 24-fold catalytic preference for tRNAGln2 over tRNAGlu (Table 1). This preference did not manifest in the plateau charging reaction (Fig. 2A). Compared to tRNAGln2, WT-GluRS has a 2.7-fold greater KM for tRNAGlu and an even greater reduction (10-fold) in enzyme turnover rate. Surprisingly, tRNAGln1 is a far less catalytically competent substrate for WT-GluRS (Fig. 2A and Table 1). Whereas the KM is only 1.5-fold greater for tRNAGln1 than tRNAGlu, the larger effect is again on kcat (62-fold lower for tRNAGln1 than tRNAGln2). WT-GluRS displays a marked catalytic preference (257-fold) for tRNAGln2 over tRNAGln1.
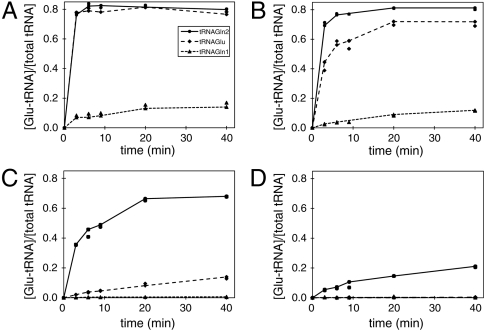
Fig. 2.
Plateau tRNA charging curve. The plot shows the fraction of Glu-tRNA formation over the total amount of tRNA during the time course of the reaction. Plateau charging levels were measured for the enzymes WT-GluRS (A), AL3-GluRS (B), ASL1-GluRS (C), and ASL2-GluRS (D) with the tRNA substrates (see Fig. 1): tRNAGln2 (●), tRNAGlu (⧫), and tRNAGln1 (▴). In the reactions, 1 μM enzyme and 10.4 μM tRNA were used.
Rational Design of a WT-GluRS Variant Specific for tRNAGln.
GlnRS evolved from an ancestor of the eukaryotic/archaeal-type nondiscriminating GluRS (11, 14–17). A series of GluRS mutant enzymes were designed in order to define the molecular basis by which WT-GluRS achieves relaxed tRNA specificity and to demonstrate how a tRNA-specific enzyme can evolve from a nondiscriminating ancestor.
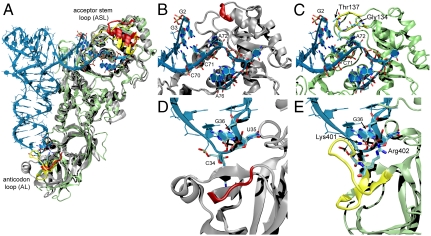
A comparison of the WT-GluRS structure (11) with the E. coli GlnRS tRNAGln complex (12) revealed two loops in GlnRS that contact tRNAGln and are absent from the ND-GluRS structure (Fig. 3). One loop (E. coli GlnRS 134–140) found in the acceptor binding domain (CP1-domain in other aaRSs) apparently acts to disrupt the first base pair of the acceptor stem (Figs. 3C and 4A), allowing the protein to recognize nucleotide identity elements in the second and third base pairs of the acceptor stem (12). The second loop is found at the opposite end of the protein (E. coli GlnRS 392–408) and includes Arg402 that makes direct contact with base 36 of the tRNA, a distinguishing feature between tRNAGln ( anticodon) and tRNAGlu(
anticodon) and tRNAGlu(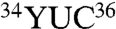 anticodon) (Figs. 3D and 4B). Arg402 is almost completely conserved among GlnRS sequences (Fig. S1B).
anticodon) (Figs. 3D and 4B). Arg402 is almost completely conserved among GlnRS sequences (Fig. S1B).
Fig. 3.
A structural comparison (A) of the M. thermautotrophicus GluRS (silver) (11) and the E. coli GlnRS (green) tRNAGln (blue) complex (12). Enlarged views of the ASL region are shown for (B) GluRS and (C) GlnRS. Larger view of the AL region are also shown for (D) GluRS and (E) GlnRS. Regions of GlnRS (yellow) swapped for regions in GluRS (red) in the ASL and AL-GluRS variants are highlighted. Sequences for each mutant are in Fig. 4. In panels showing the GluRS (B, D), the E. coli tRNAGln is included for reference.
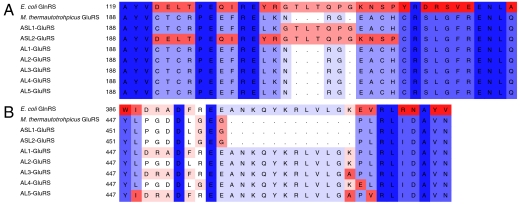
Fig. 4.
Alignment of E. coli GluRS, M. thermautotrophicus GluRS (WT-GluRS), and GluRS variants. Regions surrounding the (A) acceptor stem loop and the (B) anticodon loop are shown. GluRS variants only differ from WT-GluRS at positions indicated. The enzymes are otherwise sequence identical. Sequences are color coded according to amino acid identity (descending from blue to red).
We attempted to design a variant GluRS specific for tRNAGln by inserting one or both of the anticodon (AL) or acceptor stem (ASL) loops into the WT-GluRS (Fig. 3). We constructed a number of variants including a larger or smaller insertion of the relevant GlnRS peptide (Fig. 4). Although some (AL1, AL2, AL4) displayed low levels of protein production, a number of variants were highly produced and active in Glu-tRNA formation.
Standard aminoacylation plateau charging curves were used to screen the WT-GluRS variants for enhanced activity toward tRNAGln2 and/or decreased activity toward tRNAGlu (Fig. 2 B–D). The AL3 (Fig. 2B) and AL5 variants showed 80% charging level for Glu-tRNAGln2 production, but only AL3 showed potential discrimination against tRNAGlu with a reduction to 70% plateau level for tRNAGlu charging. The acceptor stem loop GluRS variants (ASL1 and ASL2) displayed significant preference for tRNAGln2, and ASL2-GluRS showed no detectable activity for tRNAGlu (Fig. 2 C and D). Both ASL1 and ASL2-GluRS showed no activity toward tRNAGln1 under the assay conditions (Fig. 2 C and D).
Because the AL3 mutant enzyme showed a modest increase in tRNAGln over tRNAGlu discrimination, we constructed additional mutants that contained the AL3 loop with either the ASL1 (Fig. S2A) or ASL2 (Fig. S2B) loops. Neither mutant appeared to increase specificity for tRNAGln2, so they were not further characterized.
Kinetic Characterization of GluRS Variants.
A kinetic characterization was conducted to establish the magnitude of tRNA discrimination in the AL3, ASL1, and ASL2-GluRS variants (Table 2). The AL3-GluRS is of similar catalytic efficiency (within error) toward the tRNAGlu compared to the WT-GluRS and shows slightly less tRNA discrimination than WT-GluRS.
Table 2.
Aminoacylation kinetics of AL3, ASL1, and ASL2-GluRS variants
| kcat, s-1 | KM, μM | kcat/KM, s-1 μM-1 | tRNAGln specificity* | Loss of efficiency† | |
| AL3-GluRS | |||||
| tRNAGln2 | 0.38 ± 0.09 | 2.22 ± 1.30 | 0.17 ± 0.11 | 1.0 | 1.8 |
| tRNAGlu | 0.04 ± < 0.01 | 2.89 ± 0.92 | 0.01 ± < 0.01 | 14 | 26 |
| ASL1-GluRS | |||||
| tRNAGln2 | 0.12 ± < 0.01 | 2.36 ± 0.28 | 0.05 ± 0.01 | 1.0 | 6.1 |
| tRNAGlu | 0.01 ± < 0.01 | 16.4 ± 3.6 | (8.3 ± 2.0) × 10-4 | 61 | 374 |
| ASL2-GluRS | |||||
| tRNAGln2 | 0.004 ± < 0.001 | 6.49 ± 0.90 | (5.8 ± 0.8) × 10-4 | 1.0 | 535 |
| tRNAGlu | no activity | no activity | no activity | ND | ND |
*tRNAGln specificity is calculated as the ratio of kcat/KM of the GluRS variant listed at left for tRNAGln2 over the kcat/KM of the same GluRS variant for tRNAGlu.
†Loss of catalytic efficiency (x fold) calculated as the ratio of kcat/KM of WT-GluRS toward tRNAGln2 (data in Table 1) over the kcat/KM for the GluRS variant and tRNA indicated in the first column. No activity, no aminoacylation detectable with conditions in SI Methods; ND, not determinable.
Insertion of the acceptor stem loop from E. coli GlnRS into WT-GluRS does result in GluRS variants with enhanced specificity for tRNAGln2 over tRNAGlu. The ASL1 mutant replaces residues 203–205 of WT-GluRS with 134–140 from E. coli GlnRS, whereas the insertion in the ASL2-GluRS is a larger segment of the GlnRS sequence that replaces residues 191–209 of WT-GluRS with 122–144 of E. coli GlnRS (Fig. 4A). These insertions lead to less active enzymes than WT-GluRS, but they show a greater degree of tRNAGln specificity. With only sixfold less efficiency for tRNAGln2 compared to WT-GluRS, the ASL1-GluRS catalytically favors tRNAGln2 by 61-fold, which is a 2.5-fold enhancement in specificity for tRNAGln2 as compared to the WT-GluRS. The specificity was achieved by kinetic discrimination against tRNAGlu by ASL1-GluRS resulting from an approximately fourfold reduced kcat and a fivefold increase in KM for tRNAGlu compared to WT-GluRS (Table 2).
The ASL2-GluRS, although far less catalytically efficient (by about 500-fold) than WT-GluRS toward tRNAGln2, was highly specific for tRNAGln2 and showed no activity toward tRNAGlu. The ASL2-GluRS exhibited a 100-fold reduction in kcat as compared to wild-type GluRS, but the KM for tRNAGln2 was only fivefold greater than for WT-GluRS. The data indicate that the large ASL2 insertion may have perturbed the active site structure and this could explain the larger reduction on kcat versus KM.
Behavior of GluRS Variants Toward Acceptor Stem tRNA Mutants.
Because the acceptor stem loop appears to endow the ASL2-GluRS with tRNAGln specificity, we created tRNA mutants to define which nucleotides in tRNAGln help ensure the engineered specify. The acceptor stem loop in E. coli GlnRS interacts (nonspecifically via hydrophobic contacts, see Fig. 3C) with the second base pair of the tRNA acceptor stem. In the complex structure, the first base pair (U1∶A72) is open and the U1 residue is disordered (12).
Given that the first base pair could be a critical element in tRNAGln versus tRNAGlu discrimination as it is with the archaeal-specific amidotransferase Glu-tRNAGln amidotransferase (GatDE) (18), we constructed tRNAGln2 mutants in which the wild-type A1∶U72 base pair is mutated to G1∶C72 (as in M. thermautotrophicus tRNAGlu) and to U1∶A72 as in E. coli tRNAGln. Additional details are given in SI Results (Table S1 and Fig. S3), but the data show that WT and ASL GluRSs both recognize the first base pair, but respond differently to its mutation. The ASL GluRSs display significantly decreased affinity for the G1∶C72 mutant, whereas the same mutation affects both tRNA binding and catalysis for the WT-GluRS.
tRNA Phylogeny.
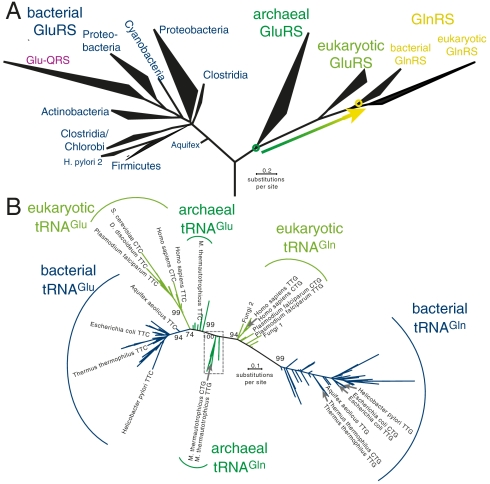
The GluRS and GlnRS phylogenetic tree is well characterized (11, 14–17), but a detailed phylogeny of their cognate tRNAs has not been presented. We calculated a large scale phylogenetic tree of tRNAGln and tRNAGlu sequences to determine if the evolution of GlnRS from the archaeal/eukaryotic GluRS is imprinted in the evolutionary history of the tRNAs.
A phylogenetic tree including all the tRNAGlu and tRNAGln sequences available from the tRNA database (19) is presented in Fig. 5B and in complete detail in Dataset S1. Although some branches are not statistically supported and collapsed in the tree, most of the major clades in the tree (i.e., the archaeal tRNAGln, eukaryotic tRNAGln, bacterial tRNAGln, archaeal tRNAGlu, eukaryotic tRNAGlu, and bacterial tRNAGlu) form well-separated and statistically significant groups, and the archaeal and eukaryotic tRNAs are more similar to each other than their bacterial counterparts as in the ribosomal RNA tree. These observations indicate that the evolutionary history of organisms is to some extent retained in the tRNA molecules. Furthermore, the tRNAGlu and tRNAGln sequences are separated (supported by 100% bootstrap confidence). Although GlnRS evolved from the archaeal/eukaryotic GluRS (Fig. 5A), a similar event (i.e., a distinct tRNAGln evolving from an archaeal/eukaryotic tRNAGlu) is not present in the tRNA phylogeny (Fig. 5B). Further details regarding the tRNA phylogeny are given in SI Results.
Fig. 5.
Phylogenetic trees showing the evolution of (A) GluRS and GlnRS, and (B) tRNAGlu and tRNAGln. The schematic GluRS and GlnRS phylogeny (A) shows only the major phylogenetic groups from a previously calculated tree (11). The arrow indicates evolution of GlnRS from an archaeal-like nondiscriminating GluRS ancestor. A maximum likelihood phylogeny is shown (B) that was calculated from an alignment of 753 tRNAGlu and tRNAGln sequences. Major clades and a few representative taxa are labeled. Bootstrap values are given for the major clades only (for a tree showing all taxa and bootstrap values see Dataset S1). Branches completely lacking support are collapsed. The boxed region is shown in complete detail in Fig. S4.
In order to understand how the two tRNAGln isoacceptors evolved in M. thermautotrophicus, we constructed a detailed phylogeny of the archaeal tRNAGln sequences (Fig. S4). The tree is well resolved with a distinct separation between crenarchaeal and euryarchaeal tRNAs and other standard taxonomic divisions are also visible. The pattern of evolutionary divergence experienced by the archaeal species is evident and, therefore, retained in the tRNA sequences.
Despite being catalytically distinct substrates, tRNAGln1 and tRNAGln2 from M. thermautotrophicus are evolutionarily more closely related to each other than to any other tRNAGln isoacceptor from a different archaeal genus. The tRNAGln2 occupies a long branch in its clade in the phylogeny (Fig. S4), so tRNAGln2 underwent a greater rate of evolutionary change than tRNAGln1. The data support a scenario in which the increase in aminoacylation efficiency of WT-GluRS for tRNAGln2 compared to tRNAGln1 is the principally the result of positive selection on tRNAGln2.
Discussion
Biochemical Properties of ND-aaRSs.
Although the existence of tRNA-dependent amino acid biosynthesis was established more than 40 y ago with the demonstration that in Bacillus species Gln-tRNAGln was synthesized from Glu-tRNAGln and not from free Gln (4), only in the last decade has attention been devoted to understanding how relaxed tRNA specificity is achieved by the aaRS enzymes in these pathways.
The Thermus thermophilus nondiscriminating AspRS (ND-AspRS2) has 11-fold higher catalytic efficiency toward tRNAAsp compared to tRNAAsn (20). A mutant of the discriminating AspRS (D-AspRS) from Pyrococcus kodakaraensis could be converted to a nondiscriminating enzyme (still showing 10-fold catalytic preference for tRNAAsp) by swapping the larger L1 loop in the anticodon binding domain of the D-AspRS with that found in the ND-AspRS (20, 21). A closely related Deinococcus radiodurans ND-AspRS2 mutant in the L1 loop (P77K) increased enzymatic discrimination threefold (22). The work proved that a loop in the anticodon binding domain of the aaRS affects tRNAAsp discrimination by differentiating the base at position 36 in the Asp versus the Asn anticodon.
The bacterial-type nondiscriminating GluRS is reminiscent of the ND-AspRSs. A crystal structure of the T. thermophilus discriminating GluRS (D-GluRS) indicated that Arg358 is the critical element that distinguishes the Glu (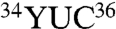 ) and Gln (
) and Gln ( ) anticodons (9) based upon the identity of the nucleotide at position 36. The Arg358Gln mutation led to an enzyme with relaxed anticodon specificity (9). A structural and biochemical characterization of the Thermosynechococcus elongatus ND-GluRS found a Gly in place of Arg358 from T. thermophilus and other bacterial-type D-GluRSs (10). Enzyme kinetics revealed that this ND-GluRS is 13-fold more catalytically efficient toward tRNAGlu compared to tRNAGln.
) anticodons (9) based upon the identity of the nucleotide at position 36. The Arg358Gln mutation led to an enzyme with relaxed anticodon specificity (9). A structural and biochemical characterization of the Thermosynechococcus elongatus ND-GluRS found a Gly in place of Arg358 from T. thermophilus and other bacterial-type D-GluRSs (10). Enzyme kinetics revealed that this ND-GluRS is 13-fold more catalytically efficient toward tRNAGlu compared to tRNAGln.
Certain bacterial D-GluRS enzymes are able to discriminate tRNAGlu from tRNAGln at the acceptor stem. The catalytic domain of the E. coli D-GluRS alone is able to discriminate tRNAGlu from tRNAGln (21). Discrimination of tRNAGln from tRNAGlu isoacceptors by the Helicobacter pylori GluRS2 is also achieved by recognizing the acceptor stem, in particular the U1∶A72 base pair in tRNAGln (23).
As the example system for archaeal-type ND-GluRSs, we found the M. thermautotrophicus GluRS to be biochemically distinct from the bacterial ND-GluRS and other ND-aaRSs characterized previously. In sharp contrast to the bacterial-type T. elongatus ND-GluRS, the M. thermautotrophicus GluRS was shown to prefer the major tRNAGln isoacceptor (tRNAGln2) by 24-fold over tRNAGlu, a twofold greater tRNA specificity than typically observed for ND-aaRSs. In addition, the D-AspRSs and bacterial type D-GluRSs typically recognize base 36 in the anticodon as a major element of discrimination between the tRNA substrates, whereas nondiscriminating relatives of the enzymes fail to recognize base 36 and are therefore able to aminoacylate both tRNA species. Our engineered GluRS variants indicate that the evolution of tRNAGln specificity in GlnRS resulted from differentiating the tRNAGlu and tRNAGln at the acceptor stem.
tRNAGln Isoacceptors: Aminoacylation Efficiency and Codon Usage.
The M. thermautotrophicus GluRS displays a striking catalytic preference (250-fold difference in kcat/KM) for tRNAGln2 compared to tRNAGln1. Interestingly, 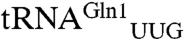 responds to the CAA codon, which appears 700 times in the 1,869 protein coding ORFs in the M. thermautotrophicus genome (24, 25). The other isoacceptor,
responds to the CAA codon, which appears 700 times in the 1,869 protein coding ORFs in the M. thermautotrophicus genome (24, 25). The other isoacceptor, 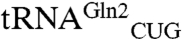 responds to the far more common glutamine codon CAG (9,286 occurrences). The catalytic competence of the tRNA as a substrate for aminoacylation is, therefore, correlated with the tRNA’s cognate codon usage. It is well known that tRNA expression levels are correlated with codon usage bias (26, 27), but there are few examples of a correlation with aminoacylation efficiency. A detailed kinetic analysis of ArgRSs and their specificity for different tRNAArg isoacceptors did not show such a correlation (28).
responds to the far more common glutamine codon CAG (9,286 occurrences). The catalytic competence of the tRNA as a substrate for aminoacylation is, therefore, correlated with the tRNA’s cognate codon usage. It is well known that tRNA expression levels are correlated with codon usage bias (26, 27), but there are few examples of a correlation with aminoacylation efficiency. A detailed kinetic analysis of ArgRSs and their specificity for different tRNAArg isoacceptors did not show such a correlation (28).
There appear to be two potential evolutionary forces responsible for the great difference in catalytic competence for tRNAGln1 versus tRNAGln2. Perhaps, as suggested by the phylogeny (Fig. S4), the enhanced aminoacylation efficiency of tRNAGln2 resulted from positive selection. The tRNAGln1 responds to fewer codons, so it is likely under a lower selective pressure than tRNAGln2 and could have acquired deleterious mutations from genetic drift. There is evidence supporting both scenarios in the nine nucleotide positions that differ between the tRNAGln isoacceptors (Fig. 1). The energetically unfavorable U28∶G42 base pair found in tRNAGln1 is the likely the most disruptive element for its catalytic performance. Because this base pair is found in no other archaeal tRNA, genetic drift may apply. At two of these positions, tRNAGln1 encodes bases (G15, C48) conserved in all other archaea, whereas tRNAGln2 has a unique mutation (A15, T48). Further experimentation could show if these changes enhanced the activity of tRNAGln2.
The Emergence of tRNAGln Specificity.
We found that insertion of the acceptor stem loop from GlnRS was sufficient to convert the archaeal-type nondiscriminating GluRS into a tRNAGln specific enzyme. Because GlnRS evolved from an ancestor similar to the archaeal-type ND-GluRS, our transplantation of tRNAGln specificity from GlnRS into the ND-GluRS recapitulates part of this evolutionary pathway.
Although Gln-tRNAGln formation predates the emergence of GlnRS (11, 14–17), GlnRS was the first (known) enzyme that evolved to specifically recognize tRNAGln and discriminate against tRNAGlu. Besides noting an increased evolutionary rate among bacterial and eukaryotic tRNAs (SI Results), we were unable to find a compelling imprint of GlnRS evolution in the tRNAGlu and tRNAGln phylogeny (Fig. 5). The finding indicates that GlnRS evolved to specifically recognize the extant tRNAGln and did not require coevolution of an entirely new type of tRNA.
A second part of the evolutionary transition from ND-GluRS ancestor to GlnRS involved active site mutations that converted amino acid specificity from Glu to Gln. In a recent study, a GlnRS mutant capable of forming Glu-tRNAGln was engineered from an impressive total of 22 amino acid replacements and one deletion (13, 29). It is, therefore, more likely that tRNAGln specificity (requiring fewer mutations) evolved first, and direct Gln-tRNAGln formation activity evolved subsequently. An analogous evolutionary intermediate exists in one of the two bacterial type GluRSs from H. pylori. One is specific for tRNAGln and the other for tRNAGlu (30, 31). If amino acid specificity had evolved first, an undesirable evolutionary intermediate enzyme would result, which could form Gln-tRNAGlu, potentially disrupting translation fidelity, whereas any Glu-tRNAGln formed by a GluRS specific for tRNAGln could be converted to Gln by the action of the GatDE amidotransferase.
Because the acceptor stem loop is critical for converting WT-GluRS to a tRNAGln specific enzyme, sequence alignment of this region sheds light on the evolution of tRNAGln specificity in the archaeal/eukaryotic type GluRS and GlnRS. The acceptor stem loop of E. coli GlnRS is found in nearly all GlnRS sequences (GTLTXXG consensus, see Fig. S1A). The loop is notably deleted in the T. thermophilus GlnRS and indicates that this enzyme may show some activity toward both tRNAGln and tRNAGlu. Eukaryotic GlnRSs likely evolved a distinct mechanism for discriminating tRNAGln from tRNAGlu because this loop is usually absent in their GlnRS sequences. Exceptions include the GlnRS from yeast and other fungi that have a similar but smaller loop (Fig. S1A).
In keeping with the prediction that all archaeal GluRSs are nondiscriminating (32), the acceptor stem loop is indeed absent from all archaea, with only one exception. The Sulfolobus solfataricus GluRS has a similar loop (Fig. S1A) that is longer by one residue and of distinct sequence from the bacterial GlnRS loop. Because the organism lacks GlnRS, the S. solfataricus GluRS must be a nondiscriminating enzyme, yet we predict the enzyme will show a significantly higher catalytic preference for tRNAGln.
Conclusion
Biochemical measurements of the M. thermautotrophicus GluRS enzyme with its three homologous tRNA substrates (tRNAGln1, tRNAGln2, tRNAGlu) served as the basis for comparison to our rationally designed enzymes. Although we expected tRNAGln specificity could be controlled by the anticodon loop in GlnRS, activity of the engineered GluRS variants indicates that the acceptor stem loop is the principle discrimination element because insertion of this loop alone enhanced the specificity of archaeal GluRS toward tRNAGln2, significantly in the case of ASL2-GluRS.
There is now an increasing need for designed aminoacyl-tRNA synthetases. Recent efforts (reviewed in refs. 1 and 2) relied on unusual and engineered aaRSs as the principal vehicle for expanding the genetic code. Although over 100 different noncanonical amino acids have been genetically encoded already, for sufficient incorporation to support efficient recombinant protein synthesis only one amino acid at a time can be added to the genetic code. Genetic code expansion strategies are limited to tRNAs that in many cases inefficiently read amber, opal, or even four-base codons. Because tRNAs that read amber and opal codons must compete with the release factor (RF), an engineered E. coli strain lacking RF1 improved the read through efficiency of amber codons (33). Whereas four-base codons might allow 200 new open codons, experiments using orthogonal ribosomes selected to enhance read-through of four-base codons still lead to mostly truncated protein with inefficient synthesis of full-length product (34). These techniques were further manipulated to include two new amino acids simultaneously (35), but unless additional open codons are created the genetic code will be limited to about 22 amino acids for efficient production of protein containing multiple noncanonical amino acids.
Although in the past an unthinkable task, the ability to “write” (36) or recode (37) an entire genome may become routine before long. By, for example, reassigning all the glutamine codons to CAA, the CAG codon would then become open. The Acidithiobacillus ferrooxidans GluRS2, which is specific for 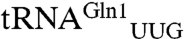 that only reads the CAA codon in E. coli (38), could be employed with the amidotransferase GatDE to ensure that CAA remains a Gln codon. In such an organism, our designed GluRS and cognate
that only reads the CAA codon in E. coli (38), could be employed with the amidotransferase GatDE to ensure that CAA remains a Gln codon. In such an organism, our designed GluRS and cognate 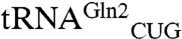 would become a vehicle for genetic code expansion. The suggestion is supported by earlier work that showed E. coli tRNA isoacceptors are poor substrates for archaeal GluRS and that established an archaeal GluRS and an engineered
would become a vehicle for genetic code expansion. The suggestion is supported by earlier work that showed E. coli tRNA isoacceptors are poor substrates for archaeal GluRS and that established an archaeal GluRS and an engineered 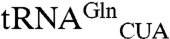 as an orthogonal amber decoding pair in E. coli (39).
as an orthogonal amber decoding pair in E. coli (39).
Additional codons could be “opened,” providing a platform for a vastly expanded genetic code. Suitable model organisms with such a dramatically expanded code would lead to breakthroughs in various fields including protein engineering, the encoded synthesis of biomaterials, and would provide a means to experimentally probe the optimality and structure of the genetic code itself. In order for an organism to accurately translate a genetic code containing many more amino acids, aminoacyl-tRNA synthetases that are specific for one (and only one) tRNA isoacceptor (itself specific for only one codon) will be needed to accurately synthesize all the necessary aminoacyl-tRNAs required to translate a maximally expanded code. Our data show that it is possible to develop an aaRS that is specific for a particular tRNA isoacceptor, suggesting that the engineered GluRS variant would be functional as an orthogonal pair in the context of a genome lacking the CAG codon. In such a background, further engineering of the GluRS active site could allow for incorporation of a selected noncanonical amino acid, and so represents an approach toward further expansion of the code.
Methods
Plasmids and Bacterial Strains.
The WT-GluRS was previously cloned (40) into the pTYB1 vector (New England Biolabs) and then transformed into an E. coli BL21/DE3 strain. Additional details are in SI Methods.
Protein and tRNA Purification and Preparation.
Pure WT-GluRS, GluRS variants, and tRNA transcripts were produced as before (40), but with slight modification (see SI Methods).
Aminoacylation Assay.
Formation of Glu-tRNA was monitored by measuring the amount of aminoacylated [32P] labeled tRNA during the reaction time course. The reaction products were separated by thin layer chromatograph (see SI Methods), and during development radioactive spots for AMP and Glu-AMP (representing free tRNA and Glu-tRNA, respectively) were separated and then visualized and quantified by phosphorimaging.
Determination of Enzyme Kinetics.
Enzyme kinetics were determined from experiments performed in duplicate, conducted independently at least twice. As the number of active molecules in an enzyme preparation of the WT-GluRS cannot be determined (13), we also assume the enzyme preparations to be fully active. Precise experimental details for all aminoacylation reactions reported are given in SI Methods.
Phylogeny and Bioinformatics.
The tRNA gene sequences and alignments were downloaded from the transfer RNA database (19). Additional details and phylogenetic calculation parameters are given in SI Methods.
Supplementary Material
Acknowledgments.
We are grateful to Ilka Heinemann for scientific discussions and a critical reading of the manuscript, and to Dr. Michael J. O’Donoghue for encouragement. This work was supported by grants to D.S. from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02-98ER20311), and the National Institute of General Medical Sciences (GM22854).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117294108/-/DCSupplemental.
References
- 1.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 2.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 3.Sheppard K, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc Natl Acad Sci USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 6.Curnow AW, et al. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker HD, Reinbolt J, Kreutzer R, Giege R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 9.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat Struct Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 10.Schulze JO, et al. Crystal structure of a non-discriminating glutamyl-tRNA synthetase. J Mol Biol. 2006;361:888–897. doi: 10.1016/j.jmb.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 11.Nureki O, et al. Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: A missing link in the evolution of Gln-tRNAGln formation. Nucleic Acids Res. 2010;38:7286–7297. doi: 10.1093/nar/gkq605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rould MA, Perona JJ, Söll D, Steitz TA. Structure of E.coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez-Hernández A, Bhaskaran H, Hadd A, Perona JJ. Synthesis of Glu-tRNAGln by engineered and natural aminoacyl-tRNA synthetases. Biochemistry. 2010;49:6727–6736. doi: 10.1021/bi100886z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JR, Doolittle WF. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 15.Lamour V, et al. Evolution of the Glx-tRNA synthetase family: The glutaminyl enzyme as a case of horizontal gene transfer. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family—rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur J Biochem. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 17.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshikane H, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 19.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charron C, Roy H, Blaise M, Giege R, Kern D. Non-discriminating and discriminating aspartyl-tRNA synthetases differ in the anticodon-binding domain. EMBO J. 2003;22:1632–1643. doi: 10.1093/emboj/cdg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta S, et al. The role of the catalytic domain of E. coli GluRS in tRNAGln discrimination. FEBS Lett. 2009;583:2114–2120. doi: 10.1016/j.febslet.2009.05.041. [DOI] [PubMed] [Google Scholar]
- 22.Feng L, Yuan J, Toogood H, Tumbula-Hansen D, Söll D. Aspartyl-tRNA synthetase requires a conserved proline in the anticodon-binding loop for tRNAAsn recognition in vivo. J Biol Chem. 2005;280:20638–20641. doi: 10.1074/jbc.M500874200. [DOI] [PubMed] [Google Scholar]
- 23.Chang KM, Hendrickson TL. Recognition of tRNAGln by Helicobacter pylori GluRS2—a tRNAGln-specific glutamyl-tRNA synthetase. Nucleic Acids Res. 2009;37:6942–6949. doi: 10.1093/nar/gkp754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DR, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 27.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 28.Fender A, Sissler M, Florentz C, Giegé R. Functional idiosyncrasies of tRNA isoacceptors in cognate and noncognate aminoacylation systems. Biochimie. 2004;86:21–29. doi: 10.1016/j.biochi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Bullock TL, Rodriguez-Hernandez A, Corigliano EM, Perona JJ. A rationally engineered misacylating aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2008;105:7428–7433. doi: 10.1073/pnas.0711812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar JC, et al. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc Natl Acad Sci USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J Mol Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 35.Neumann H, Slusarczyk AL, Chin JW. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J Am Chem Soc. 2010;132:2142–2144. doi: 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]
- 36.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 37.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Núñez H, Lefimil C, Min B, Söll D, Orellana O. In vivo formation of glutamyl-tRNAGln in Escherichia coli by heterologous glutamyl-tRNA synthetases. FEBS Lett. 2004;557:133–135. doi: 10.1016/s0014-5793(03)01460-1. [DOI] [PubMed] [Google Scholar]
- 39.Santoro SW, Anderson JC, Lakshman V, Schultz PG. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003;31:6700–6709. doi: 10.1093/nar/gkg903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J Biol Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.