Abstract
The Gag proteins of HIV-1 are central players in virus particle assembly, release, and maturation, and also function in the establishment of a productive infection. Despite their importance throughout the replication cycle, there are currently no approved antiretroviral therapies that target the Gag precursor protein or any of the mature Gag proteins. Recent progress in understanding the structural and cell biology of HIV-1 Gag function has revealed a number of potential Gag-related targets for possible therapeutic intervention. In this review, we summarize our current understanding of HIV-1 Gag and suggest some approaches for the development of novel antiretroviral agents that target Gag.
Introduction
The HIV-1 replication cycle
HIV-1, the causative agent of acquired immunodeficiency syndrome (AIDS), is a retrovirus in the genus Lentiviridae. It is an enveloped virus that encodes two envelope (Env) glycoproteins - the surface (SU) glycoprotein gp120 and the transmembrane (TM) glycoprotein gp41 (Fig. 1), four major Gag proteins—matrix (MA), capsid (CA), nucleocapsid (NC), and p6—and the pol-encoded enzymes protease (PR), reverse transcriptase (RT), and integrase (IN). HIV-1 also encodes two regulatory proteins—Tat and Rev—and several accessory proteins—Vpu, Vif, Nef, and Vpr (Fig. 1). The genome is pseudodiploid, composed of two single strands of RNA linked in a dimer.
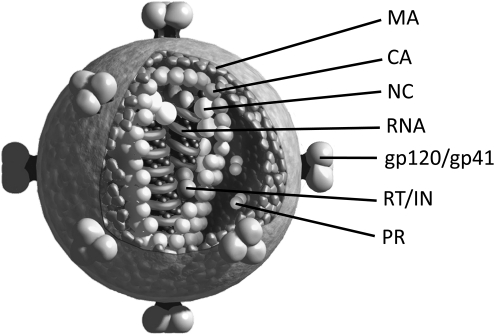
Fig. 1.
Representation of an HIV-1 virion. The locations of the Gag proteins matrix (MA), capsid (CA), and nucleocapsid (NC), the viral enzymes reverse transcriptase (RT), protease (PR), and integrase (IN), and the Env glycoproteins are indicated. The figure was generously provided by L. Henderson, National Cancer Institute, Frederick, MD.
The HIV-1 infection process begins with the attachment of gp120 to the target cell plasma membrane (Fig. 2).1–4 The principal attachment receptor for HIV-1 and other primate lentiviruses is CD4. Productive infection also requires the presence of a coreceptor, typically CXCR4 or CCR5. The binding of gp120 to CD4 and coreceptor triggers conformational changes in gp41, which in turn lead to the fusion of the viral envelope and the target cell membrane and entry of the viral core into the host cell cytoplasm. Recent evidence suggests that HIV-1 entry can also occur in a low-pH endosomal compartment after receptor-mediated endocytosis.5 Upon entry of the virion into the cytosol, the Env glycoproteins and the lipid-associated MA protein dissociate from the incoming particle at the membrane, and the poorly understood process of uncoating is initiated.
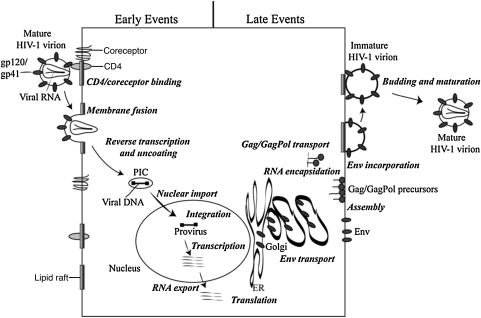
Fig. 2.
Schematic representation of the HIV-1 replication cycle. The details of the replication cycle are described in the text. Reprinted with permission from Freed (2004).19 Copyright 2004 Elsevier Inc.
The enzymes RT and IN, together with the NC protein, remain in close association with the viral RNA as it is converted to double-stranded DNA by RT-catalyzed reverse transcription.6 NC acts as a nucleic acid chaperone at several steps during reverse transcription to facilitate the conversion of RNA to DNA.7 Vpr is also a component of the reverse transcription complex (RTC). The extent to which CA remains associated with the incoming RTC has been a topic of debate. However, reverse transcription and uncoating appear to be temporally linked,8 and it is clear that some host restriction factors that block early postentry steps in the viral replication cycle target CA.9,10 The newly reverse transcribed viral DNA is translocated to the nucleus in a structure known as the preintegration complex (PIC). The nuclear import process remains incompletely understood; however, a role for CA in this process11,12 implies that some CA protein may remain associated with the viral nucleoprotein complex as it traffics to the nuclear pore. Once inside the nucleus, the double-stranded viral DNA integrates into the target cell genome through the action of the IN enzyme.13 The integrated viral DNA serves as the template for transcription from the viral promoter in the 5′ long terminal repeat (LTR) to generate the spliced viral mRNAs and full-length genomic RNAs; these are transported out of the nucleus via the action of the Rev protein.3
The Gag proteins are translated from full-length message as a polyprotein precursor containing MA, CA, NC, and p6 domains as well as two spacer peptides, SP1 and SP2.14,15 During translation of the Gag precursor, known as Pr55Gag, an occasional 1 ribosomal frameshift leads to the production of a GagPol precursor protein (Pr160GagPol), the abundance of which is approximately 5% that of Pr55Gag. The Gag and GagPol precursor polyproteins are transported to the plasma membrane, where they assemble and incorporate the viral Env glycoproteins. The membrane targeting of Gag and GagPol is regulated by the MA domain, which also plays an important role in the incorporation of the viral Env glycoproteins. Assembly takes place in cholesterol-rich membrane microdomains (lipid rafts) through direct interactions between MA and the phospholipid phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2].16,17 Interactions within the CA domain of Gag initiate the Gag assembly process.
The two copies of viral RNA are encapsidated during the assembly process via direct interactions between the genomic RNA and the NC domain of Gag.7 NC–RNA interactions also play an important role in promoting Gag–Gag interactions. The nascent virus particle buds and pinches off from the plasma membrane by recruiting components of the cellular endosomal sorting machinery, specifically, the endosomal sorting complexes required for transport (ESCRTs) and associated factors.18–20 Upon release from the infected cell, the viral PR cleaves the Gag and GagPol polyprotein precursors in a highly ordered, step-wise processing cascade. Gag and GagPol cleavage triggers virion maturation.21 In the mature virion, MA is associated with the inner leaflet of the lipid bilayer and CA reassembles to form a conical shell around the viral RNA in complex with RT, IN, and NC. With maturation, the viral replication cycle is complete and a new round of infection can take place.
Current status of antiretroviral therapy
Over the past three decades, HIV-1 infection has led to the deaths of more than 35 million people worldwide, with over 33 million currently infected (http://data.unaids.org). Close to 70% of current AIDS cases are in sub-Saharan Africa. Despite these bleak statistics, antiretroviral therapies that have been in widespread use in the developed nations since the late 1990s have had a major impact on survival rates of HIV-1-infected individuals and on limiting new infections, particularly in the context of mother–child transmission.
There are currently ∼25 FDA-approved antiretroviral drugs available to treat HIV-1-infected patients.22 The majority of these target two viral enzymes—RT and PR. RT inhibitors fall into two mechanistic classes, the nucleoside-analog RT inhibitors (NRTIs) and the nonnucleoside-analog RT inhibitors (NNRTIs). An inhibitor is also available that binds gp41 to inhibit fusion (e.g., enfuvirtide), and maraviroc, the first antiretroviral drug that targets a cellular protein, acts by binding CCR5 to prevent viral fusion and entry. In 2007, the first IN inhibitor, raltegravir, was approved. Because treatment with any single drug rapidly leads to the emergence of drug-resistant viral variants, patients are treated simultaneously with a cocktail of inhibitors, often referred to as highly active antiretroviral therapy (HAART).
Although HAART has been very effective in patients with access to therapy, drug resistance continues to be a serious problem that in many cases limits the effectiveness of available antiretroviral agents.23 Drug accessibility, toxicity, and compliance issues have also blunted the positive impact of HAART. For these reasons, it is our opinion that the HIV-1 research community should remain actively enGaged in identifying additional drugs, particularly those that target novel steps in the replication cycle. Although PR inhibitors block Gag and GagPol processing by interfering with the enzymatic activity of PR, it is noteworthy that there are currently no drugs in clinical use that act on any aspect of Gag function. This review addresses the possibility that events in the HIV-1 replication cycle promoted by Gag—virus assembly, Env incorporation, genomic RNA encapsidation, particle budding and maturation, uncoating, and possibly nuclear import—constitute viable antiretroviral targets. Much of what we propose is highly speculative, but we hope it will help guide future efforts in this aspect of HIV-1 drug discovery and development.
Gag as a Target for Antiviral Therapy
The HIV-1 particle production pathway follows a series of discrete steps that include (1) targeting of Gag to the site of virus assembly, (2) binding of Gag to cellular membranes, (3) interactions between Gag monomers leading to multimerization, and (4) budding and release of nascent virions from the infected cell. As discussed above, HIV-1 particle production is driven by the Gag polyprotein precursor, Pr55Gag (which is often referred to as “Gag”), and each domain of Gag is involved in the assembly and release process. The MA domain directs Pr55Gag to the plasma membrane of the host cell where it anchors Gag to the inner leaflet of the lipid bilayer. The CA domain, together with SP1 and NC, homooligomerizes to drive the multimerization of Gag and promote particle assembly. The NC domain also binds to the viral genomic RNA, leading to the packaging of two copies of the viral genomic RNA into each virus particle. The C-terminal p6 contains motifs known as “late domains” that interact with cellular ESCRT machinery to promote the fission and release of virions from the cell surface. As mentioned above, cleavage of Pr55Gag by the PR leads to virion maturation. During the assembly process a number of cellular factors are incorporated into virions. It has been reported that ∼250 cellular proteins are present in HIV-1 virions produced from monocyte-derived macrophages.24 In most cases, the functional significance of these virion-associated host proteins remains to be established.
The immature virus particle is composed of ∼5000 Gag molecules that are tightly packed to form a roughly spherical protein shell.25,26 High-resolution cryoelectron microscopy (EM) analyses have demonstrated that rod-shaped Gag molecules are packed within immature particles side-by-side in a radial fashion, with the N-terminal MA domain associated with the membrane and the C-terminal portion of Gag oriented toward the center of the particle.27–29 In immature virus particles, Gag molecules form a continuous but incomplete hexameric lattice.30,31
MA
The MA domain plays an important role in the trafficking of Gag to the assembly site, which in most cases is the plasma membrane. Structural analyses revealed that MA is composed primarily of α-helices, which form a globular core that is connected to the CA domain by a C-terminal extended helix.32–34 The MA domain is cotranslationally modified at its N-terminus with myristate; as is the case with many other myristylated proteins, this covalent modification is essential for the association of HIV-1 Gag with the inner leaflet of the plasma membrane. Mutations that prevent Gag myristylation have a profound effect on virus assembly and release by blocking the association of Gag with the cell membrane.35–38 Resh and colleagues proposed that Gag–membrane binding is regulated by a “myristyl switch” mechanism, whereby the myristate can shift between exposed and sequestered conformations.39 Summers and co-workers used nuclear magnetic resonance (NMR) methods to confirm this hypothesis and to demonstrate that myristate exposure can be promoted by factors that increase Gag self-association.40 Mutations near the N-terminus of MA that disrupt Gag–membrane binding41 apparently do so by preventing myristate exposure.42
HIV-1 assembly takes place in cholesterol- and sphingolipid-enriched plasma membrane microdomains known as lipid rafts.16,17,43 HIV-1 virions are thus highly enriched in cholesterol and saturated, raft-derived lipids.44–46 Gag displays behavior characteristic of a raft-associated protein in biochemical assays,47–51 cholesterol depletion of virus-producing cells inhibits virus particle production and virion infectivity,49 and treating Gag-expressing cells with unsaturated fatty acids reduces virion assembly by directing Gag to nonraft domains on the plasma membrane.52
In addition to the N-terminal myristate, Gag association with membrane is also regulated by a highly basic patch of amino acid residues located near the N-terminus of the MA domain (amino acid residues ∼17–31).53,54 This basic cluster of amino acids forms a positively charged surface at the “top” of the MA domain.32,33 Mutations in these basic residues mistarget Gag to late endosomes or multivesicular bodies (MVBs).55 A number of cellular proteins whose membrane association is regulated by fatty acylation and a basic patch of amino acid residues interact with a specific class of negatively charged lipids known as phosphoinositides. The phosphoinositides differ from one another in the position and number of phosphates on the inositol head group.56,57
Specific members of the phophoinositide family are enriched at particular membrane sites within the cell and thus their binding to proteins plays important roles in membrane and protein trafficking and signaling.58 For example, phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] is enriched in the inner leaflet of the plasma membrane57; phosphatidylinositol-(3,5)-bisphosphate [PI(3,5)P2] is concentrated on late endosomes58 and phosphatidylinositol-3-phosphate [PI(3)P] is most abundant on early endosomal membranes and on late endosomal vesicles.58 PI(4,5)P2 plays an important role in the association of HIV-1 Gag with the plasma membrane.59 Overexpression of 5-phosphatase IV, an enzyme that converts PI(4,5)P2 to PI(4)P, results in the depletion of PI(4,5)P2 from the plasma membrane and induces the retargeting of Gag from the plasma membrane to late endosomes/MVBs. Overexpressing a constitutively active mutant of ADP-ribosylation factor 6 (Arf6) leads to the formation of PI(4,5)P2-enriched vesicular structures that serve as sites for Gag assembly.59 These observations suggest that PI(4,5)P2 in the plasma membrane is required for efficient HIV-1 release, and that basic residues in MA are involved in direct interactions with PI(4,5)P2.
A direct interaction between MA and PI(4,5)P2 has been demonstrated by several groups using different approaches. Summers and co-workers used NMR spectroscopy to resolve the structure of a complex between a water-soluble, truncated form of PI(4,5)P2 and myristylated MA.60 In this study, it was shown that MA binding to PI(4,5)P2 triggered the myristyl switch, resulting in increased myristate exposure. The NMR data revealed that in addition to electrostatic interactions between MA and PI(4,5)P2, the 2′-unsaturated acyl chain of PI(4,5)P2 packs into a hydrophobic cleft within the globular core of MA (Fig. 3).
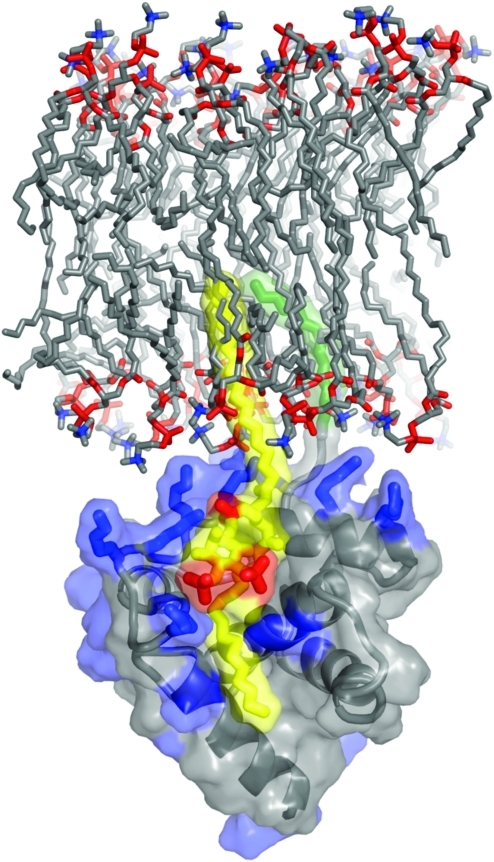
FIG. 3.
Model of HIV-1 MA binding to phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] in the membrane. The basic residues of MA (blue) interact electrostatically with phosphate groups (red) in PI(4,5)P2 (yellow and red). PI(4,5)P2 binding by MA induces exposure of the myristyl group (green), allowing it to insert into the membrane. The 2′-unsaturated acyl chain (yellow) of PI(4,5)P2 is predicted to bind to a hydrophobic cleft in MA. Unpublished image provided by M. Summers, based on the data of Saad et al. (2006).60 Color images available online at www.liebertonline.com/aid
The extrusion of the unsaturated 2′-acyl chain of PI(4,5)P2 from the lipid bilayer upon MA binding could in theory promote Gag partitioning into the saturated lipid raft microenvironment, as the saturated 1′-acyl chain would be the sole PI(4,5)P2 acyl chain left in the membrane. Whether such a conformation would be energetically favorable in the context of a lipid bilayer remains to be determined. Shkriabai et al. used a mass spectrometric protein footprinting approach with lysine-modifying agents to show that Lys-29 and Lys-31 of MA are important for the MA–PI(4,5)P2 interaction.61
These two basic residues are among those previously shown to be important for Gag localization and virus assembly.55,62,63 Ono and co-workers used liposome-binding assays to show that full-length myristylated Gag binds to PI(4,5)P2-enriched vesicles, and that mutation of residues 29 and 31 reduced binding.64 As one would predict if Gag–membrane association and HIV-1 assembly require PI(4,5)P2, this lipid is enriched in HIV-1 virions compared to the plasma membrane of the virus-producing cell. Furthermore, deletion of MA significantly reduced the incorporation of PI(4,5)P2 into virions.46 Direct interactions between MA and PI(4,5)P2 have also been reported for other retroviruses, e.g., HIV-2,65 MLV,46,66 and EIAV.67 These studies demonstrate that a broad array of retroviruses exploits an MA–PI(4,5)P2 interaction at the membrane binding step of virus assembly. In the case of EIAV, binding between MA and PI(3,5)P2 may regulate Gag association with internal membranes.68
A number of host cell proteins have been implicated in regulating the trafficking of Gag to the plasma membrane. These include the clathrin adaptor protein complexes AP-1, AP-2, and AP-3,69–71 the Arf proteins,72,73 the Golgi-localized, γ-ear-containing, Arf-binding (GGA) proteins,72,73 tail-interacting protein 47 (TIP47),74–76 calmodulin,77,78 and the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) machinery.79 The mechanism by which these factors promote Gag–membrane association remains to be defined.
In addition to playing an important role in directing HIV-1 Gag to the plasma membrane, the MA domain also functions to direct the incorporation of the Env glycoprotein complex into virus particles.80 While solid and reproducible biochemical evidence for a direct MA–gp41 interaction is lacking, abundant genetic evidence supports the existence of such an interaction.62,81–84 Host factors, including calmodulin and TIP47, have been implicated as cofactors in the Env incorporation process,76,85 although a role for these proteins awaits independent confirmation and mechanistic characterization.
In theory, a variety of strategies could be pursued for therapeutically targeting the MA domain of HIV-1 Gag. As mentioned above, in its sequestered state, the myristate moiety attached to the N-terminus of MA packs into a hydrophobic groove in the globular core of MA.40,60 This hydrophobic cleft could potentially be targeted by small molecules whose binding would dysregulate the myristyl switch. This could result in enhanced Gag–membrane association, which intuitively would seem likely to increase the efficiency of virus assembly; however, mutations that enhance membrane association have been reported to disrupt replication by inducing a postentry defect in the next round of infection.86 The PI(4,5)P2-binding cleft of MA could also be targeted by small molecules that would compete for an MA–PI(4,5)P2 association. Interestingly, in vitro studies have found that soluble PI(4,5)P2 derivatives, rather than competing with MA for PI(4,5)P2 binding and thus disrupting membrane association, actually enhance the efficiency of MA binding to liposomes.87
This raises the possibility that small molecules that dock in the PI(4,5)P2-binding groove might increase Gag–membrane association by triggering the myristyl switch. Such dysregulation of membrane binding activity could disrupt proper Gag targeting, or, as suggested above, could interfere with particle infectivity. If the putative interaction site between MA and the gp41 Env glycoprotein can be better defined, the MA–gp41 interaction could possibly be targeted to disrupt Env incorporation into virions. The association of Gag with lipid rafts could also be developed as a therapeutic target, as reviewed in detail recently,16,17 perhaps most effectively in the context of chemoprevention. Finally, the interaction between MA and host proteins could be disrupted pharmacologically.
Several issues need to be addressed before significant progress can be made in developing antiretrovirals that target the MA domain: (1) robust, high-throughput assays, preferably cell-based, need to be developed to screen for molecules that block the association of Gag with membrane and/or disrupt the proper localization of Gag at the plasma membrane, and (2) structural information needs to be obtained on putative interaction interfaces between MA and gp41 and between MA and host-cell factors. With this additional information and related screening tools, the MA domain could offer a variety of possibilities for antiretroviral development.
CA
As is the case with MA, the CA domain and the mature CA protein play multiple roles in the virus replication cycle. During assembly, the CA domain of Pr55Gag functions to promote Gag–Gag interactions that drive Gag multimerization; after release and cleavage of Pr55Gag by the viral PR, the mature CA protein plays a central role in particle maturation by reassembling into the conical core that houses the viral RNA genome and the viral enzymes RT and IN.88,89 After entry into the target cell, the CA core complex serves as the target for host restriction factors (e.g., TRIM5α). CA is composed of two independently folded domains, the N-terminal and C-terminal domains (NTD and CTD, respectively). The CA NTD and CTD are connected by a short flexible linker.89,90 The NTD (CA residues 1–145) is composed of seven α-helices (CA helices 1–7) packed in the shape of an arrowhead, with an extended, Pro-rich loop connecting helices 4 and 5. This loop binds the peptidyl-prolyl cis-trans isomerase, cyclophilin A.91–93 Deletion of the entire NTD does not disrupt particle production; however, point mutations in helices 4–6 of the NTD impair particle production, suggesting that this region of the NTD forms weak interactions during assembly or that these NTD mutations disrupt overall CA folding.94–96
The CTD (residues 151–231) is composed of a short 310-helix followed by an extended strand and four α-helices (CA helices 8–11). The CA CTD has a propensity to dimerize97,98 and plays a central role in Gag multimerization during assembly.94–99 CA dimerization takes place by mutual interactions of α-helix 9 from each monomer, with the aromatic rings of Trp-184 buried in the dimer interface.97,98 Mutation of residues 184 and 185 significantly reduces particle production in cells and CA dimerization in vitro.96,97,100,101 Near the N-terminus of the CTD, in helix 8, is a sequence of 20 amino acids that is highly conserved among retroviral CA proteins.97,102,103 This region, aptly named the major homology region (MHR), plays an important role in Gag assembly.96,102,104,105 The crystal structure of the CTD shows that the MHR is distinct from the dimer interface and forms an array of hydrogen bonds.97 The precise role of the MHR in CA function remains to be fully elucidated.
Residues at the C-terminus of CA and the N-terminus of SP1 are also important for immature particle formation, suggesting that the CA/SP1 boundary region forms a critical assembly domain.94,96,106–112 Although this region is highly flexible and disordered in crystal structures, it displays a tendency to form an α-helix.31,94,97,98,113,114 It has been suggested from cryoelectron tomography analysis that the putative CA/SP1 boundary region forms a six-helix bundle.31
In the immature particle, the CA domain assembles into a hexameric lattice, with a unit-to-unit spacing of ∼8 nm.25,115 The immature hexameric lattice is continuous but incomplete, with a large gap that allows the lattice to curve.30 During PR-mediated maturation, ∼1500 copies of CA again assemble into a hexagonal lattice, but with a unit cell spacing of ∼10 nm.116 In its mature form, the hexagonal lattice organization resembles that of a fullerene cone, with the hexameric tubes closed off at both ends by the presence of a specific number (12) of pentamers116,117 (Fig. 4). The hexameric lattice of the mature conical core is composed of rings formed by six NTDs connected to each other by CTDs (Fig. 4). Several stabilizing interfaces create the hexameric rings and link them together to form the lattice: (1) NTD–NTD intermolecular interactions create the NTD hexameric rings, (2) NTD–CTD intermolecular interactions stabilize the hexamer, and (3) the CTD–CTD dimer contacts link the rings together to form the lattice.118–120 Recently, Gronenborn and colleagues described an additional CTD–CTD interface formed by CTD dimers from three adjacent hexamers.121,122 Only relatively subtle differences are observed between the subunit interfaces of pentamers and hexamers.123,124
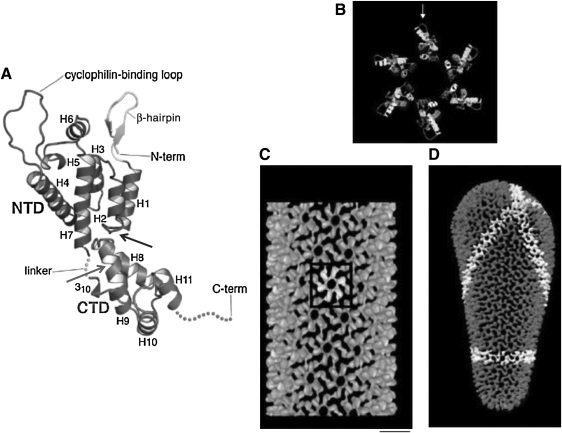
Fig. 4.
Structure of mature HIV-1 CA. (A) Tertiary structure of the mature monomeric CA with the NTD and CTD connected by the linker region. Helices 1–11 (H1–11), the N-terminal β-hairpin, and the cyclophilin A binding loop are shown. The C-terminal 11 residues of CA that are typically disordered in the crystal structure are indicated by a dashed line. The binding sites for CAI/NYAD-1/NYAD-13 and CAP1 are indicated by gray and black arrows, respectively. Reprinted with permission from Ganser-Pornillos et al. (2008).90 Copyright 2008 Elsevier Inc. (B) Molecular model of the hexameric ring formed by the NTD of CA, with the cyclophilin A-binding loop indicated with an arrow. (C) Outside view of the assembled tube structure, showing one hexamer (white) and the hexagonal CA lattice (gray). Scale bar=10 nm. (D) Model of an HIV-1 conical core. A continuous line of hexamers is highlighted in white. Reprinted with permission from Li et al. (2000).116 Copyright 2000 Macmillan Publishers Ltd: [Nature].
The detailed structural information now available on HIV-1 CA, obtained over the past dozen years by a combination of cryoelectron tomography, NMR, and x-ray crystallography methods, backed up by extensive mutations analyses, offers a diversity of targets for inhibitor development. In principle, inhibitors that bind to CA could potentially disrupt the assembly of both immature viral particles and mature cores. In initial studies, isolated CTDs were used as competitive inhibitors for CA assembly in vitro.125,126 Small peptides or organic molecules that bind at or near the CTD dimerization interface hold the potential to weaken CA dimerization. An early attempt was made by Hilpert et al. using sequence-derived peptides that inhibited the dimerization of CA in solution.127 The peptide IPVGEIYKRW, corresponding to α-helix 7 of the NTD, was reported to suppress CA dimerization in solution. However, the mechanism responsible for this inhibition remains to be clarified, and whether this peptide displays antiviral activity in cell-based systems has not, to our knowledge, been reported. Neira and co-workers designed a synthetic peptide termed CAC1 that is based on the sequence of CA helix 9, and showed that it binds to the CTD in solution.128
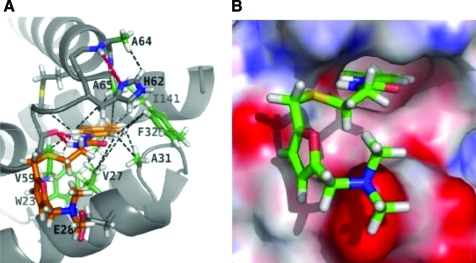
An in silico screening approach was used by Summers and colleagues to identify a small organic molecule, N-(3-chloro-4-methylphenyl)-N′-[2-[([5-[(dimethylamino)- methyl]-2-furyl]-methyl)-sulfanyl]ethyl]urea (CAP-1) that binds to the CA NTD.129 As a consequence of this interaction, virus assembly and maturation are disrupted in cell-based assays. NMR and x-ray crystallographic studies mapped the binding site of CAP-1 to a pocket formed at the point at which helices 1, 2, 4, and 7 of the NTD interact130 (Fig. 5). Binding of CAP-1 to the NTD induces the displacement of the deeply buried aromatic ring of Phe-32, thereby creating the CAP-1 binding site.130 Thus, the CAP-1 interaction with the CTD is largely hydrophobic and appears to disrupt the docking of the NTD and CTD. Although CAP-1 displays relatively weak binding affinity for CA and significant cytotoxicity, the Phe-32 pocket will likely serve as a useful target for future CA-based drug development.
FIG. 5.
Structure of CAP-1 bound to the NTD of CA calculated by restrained molecular dynamics based on hybrid x-ray/NMR data. (A) View of CAP-1 bound to the NTD. The aromatic side-chain of Phe-32 displaced from the core upon CAP-1 binding is labeled and shown in green. (B) Representation of the electrostatic surface of CAP-1 bound to the NTD of CA. Upon CAP-1 binding, the aromatic ring of CAP-1 is inserted into the pocket formerly occupied by Phe-32. Adapted from Kelly et al. (2007).130 Copyright 2007 Elsevier, with permission. Color images available online at www.liebertonline.com/aid
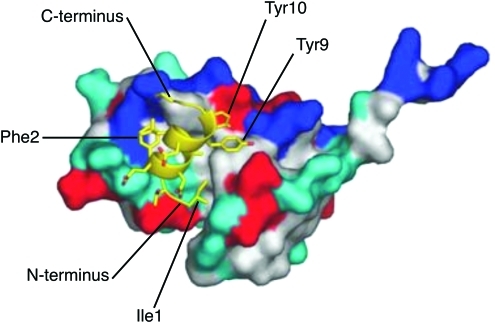
Krausslich and co-workers used a phage display screen with both full-length CA and the CTD-SP1-NC region of Gag as bait to identify a CTD-binding peptide termed CAI (CA assembly inhibitor).131 The sequence of CAI, ITFEDLLDYYGP, is not homologous to the dimerizing domain of the CTD but inhibits the in vitro assembly of both spherical particles that are analogous to immature VLPs and tubular structures that possess a mature-like CA organization.131 A high-resolution x-ray structure of the CAI–CTD complex reveals that the peptide binds in an α-helical conformation to a hydrophobic groove between helices 1, 2, and 4132 (Fig. 6). The binding of CAI allosterically disrupts the CA dimer interface and interferes with contacts between the CTD and the NTD. Barklis et al. reported that CAI induces the disassembly of CA tubes by modifying the CTD interface and also replaces NTD α-helix 4 in its binding to a CTD groove that helps to align NTDs and CTDs around the CA hexamers.133 Thus, the binding of CAI to the CTD weakens the hexamer, thereby impairing CA assembly and potentially destabilizing the viral core.
FIG. 6.
Structure of the CAI peptide bound to a hydrophobic cavity of the CA CTD. The surface of the CTD is colored based on polarity: red, positively charged; blue, negatively charged; cyan, uncharged; white, nonpolar. The residues of CAI (yellow) in direct contact with the CTD, and N- and C-termini of CAI, are labeled. Reprinted from Ternois et al. (2005)132 with permission. Copyright 2005 Macmillan Publishers Ltd. [Nature Structural and Molecular Biology]. Color images available online at www.liebertonline.com/aid
CAI itself does not display antiviral activity because it is not cell permeable; this limitation, however, was addressed by using a technique known as hydrocarbon stapling to generate cyclical, cell-penetrating derivatives with increased α-helical character.134,135 The resulting peptides, NYAD-1 and its water-soluble derivative NYAD-13, contain a covalent bridge between two amino acids separated by four residues. The sequence of NYAD-1 is ITFXDLLXYYGKKK, where X is the nonstandard amino acid (S)-2-(2′-pentenyl) alanine, which helps to stabilize the peptide. These peptides penetrate into cells and disrupt the assembly of immature VLPs, inhibit virus maturation, and exhibit an antiviral effect.135 NMR chemical shift perturbation studies indicate that CAI and NYAD-1 bind to the same hydrophobic pocket in the CTD.134,135 This hydrophobic pocket was subjected to an in silico docking-based screen for small molecules that could potentially interfere with CA CTD function. Several molecules that disrupted in vitro CA assembly and virus infectivity resulted from this effort.136
Debnath and co-workers also designed a dimer interface peptide, NYAD-201, containing the sequence AQEVKXWMTXTLLVA [based on the sequence of CA amino acids 178–192, where X represents the (S)-2-(2′-pentenyl) alanines that are connected by hydrocarbon stapling137]. This stapled peptide penetrates into cells and inhibits HIV-1 production by 3-fold. This inhibition in virus production is specific to NYAD-201, as a mutant peptide derivative in which the key dimer interface residues “WM” were changed to “AA” does not inhibit virus release. In vitro CA assembly is disrupted by NYAD-201 but not the mutant control peptide. In addition to inhibiting virus production, NYAD-201 also disrupts HIV-1 infectivity in an Env-dependent manner.137 NYAD-201 inhibits replication of a large panel of laboratory-adapted and primary HIV-1 isolates.137
As mentioned above, a Pro-rich loop in the CA NTD binds the peptidyl-prolyl isomerase cyclophilin A. Early studies demonstrated that disruption of the CA–cyclophilin A interaction with cyclosporine inhibited HIV-1 replication.138,139 More recently, a nonimmunosuppressive analog of cyclosporine, Debio-025, was shown to inhibit an early step in HIV-1 replication with increased potency and reduced cytotoxicity relative to cyclosporine.140
In a high-throughput screen for compounds that inhibit HIV-1 replication, researchers at Pfizer identified a compound, PF-1385801, that targets the CA NTD.141 Blair et al. prepared several analogs of this compound, two of which—PF-3450074 and PF-3759857—were active against most HIV-1 isolates with an effective concentration <1 μM.142 Interestingly, these compounds inhibited both early and late stages of HIV-1 replication. The early block was imposed after viral entry but before reverse transcription, and the late block was at the level of virus assembly.142 A high-resolution cocrystal structure of CA and PF-3450074 revealed that the compound binds to the pocket created by helices 3, 4, 5, and 7 of the CA NTD. Selections in PF-3450074 led to substitutions in CA that conferred resistance to the compound.142 In vitro experiments using HIV-1 particles or purified viral cores showed that PF-3450074 destabilizes the viral core, and in target cells, PF-3450074 appears to inhibit HIV-1 infection by triggering premature uncoating.143
Researchers at Boehringer Ingelheim converted the CA–NC in vitro assembly assay144 into a high-throughput format and screened for inhibitors of in vitro assembly. Two distinct classes of compounds were identified, benzodiazepines and benzimidazoles.145 Both classes of compounds appear to bind the CAP-1 pocket discussed above, and disrupt particle formation and virion maturation.145 Optimization of the benzodiazepine series led to a compound with significantly improved potency.146 Together, these studies indicate that HIV-1 CA is a potential drug target.
NC
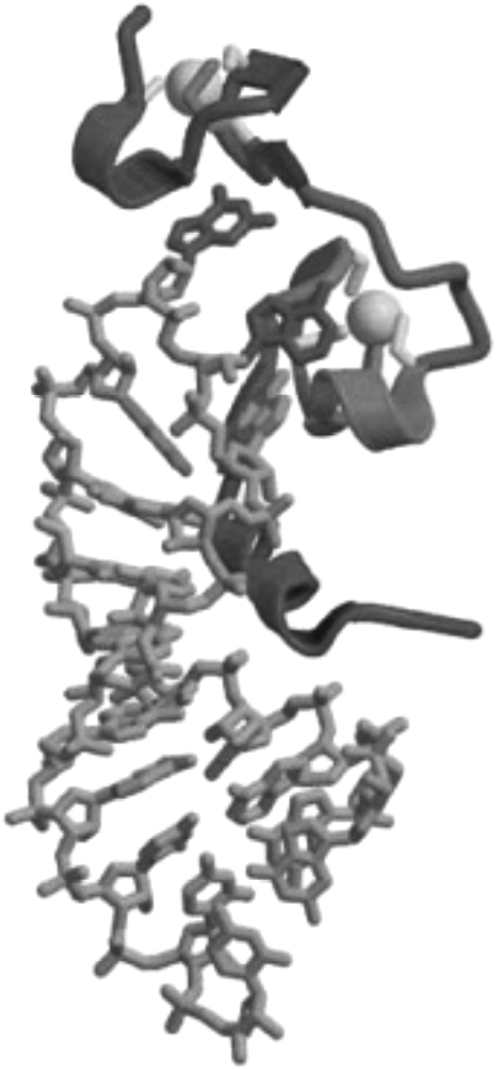
The NC domain of HIV-1 Gag is characterized by the presence of two zinc finger-like motifs flanked by highly basic sequences (Fig. 7).147,148 NC zinc fingers, typically present in one or two copies, are among the most highly conserved features of retroviral Gag proteins, and, as could be predicted from this high level of conservation, are critical for NC function. The consensus zinc-finger motif in retroviral NC domains is Cys-X2-Cys-X4-His-X4-Cys (CCHC), a sequence that is somewhat rare among cellular zinc fingers. As a domain of Pr55Gag, NC serves several primary functions during the assembly process: it enhances Gag–Gag interactions, largely as a consequence of its nucleic acid binding properties; it directs the packaging of the viral RNA genome into virus particles; and it promotes the incorporation into virions of tRNALys3, which serves as the primer for reverse transcription in the next round of infection.7 In general, the basic residues in NC are responsible for the Gag multimerization activity, again, due to their nonspecific binding to nucleic acid. Gag-bound nucleic acid serves as a multimerization platform, or scaffold, that serves to concentrate Gag molecules and facilitate their assembly.
FIG. 7.
Structure of NC bound to the SL3 stem-loop of the HIV-1 RNA packaging signal. Gray balls represent the two zinc ions that bound to the zinc-finger motifs. Reprinted from Turner and Summers (1999)278 with permission. Copyright 1999 Elsevier Inc.
In contrast, specific viral genomic RNA encapsidation involves the interaction between the NC zinc fingers and a multiple stem-loop structure in the viral RNA known as the packaging, or ψ, site.149–163 Gag selects and packages dimeric viral RNA from the cytosol.164 After liberation from Pr55Gag following PR-mediated Gag processing, the mature 7-kDa NC protein functions as a nucleic acid chaperone to increase the efficiency of reverse transcription and integration.7
The critical roles of NC in HIV-1 replication, together with the high degree of conservation and unusual sequence motif of the NC zinc fingers, suggest that NC should be a good target for drug development. Over the past nearly two decades, a number of NC-targeted HIV-1 inhibitors have been reported, most of which act by disrupting the zinc fingers. Rice and colleagues reported that cell-permeable compounds that oxidize the NC zinc-finger cysteines, thereby destroying the zinc-finger structure, inactivate virus infectivity.165,166
One such compound, 2,2-dipyridyl disulfide (also known as Aldrithiol-2 or AT-2), induces extensive cross-linking within NC and Pr55Gag and is frequently used as a research tool to chemically inactivate HIV-1.167–169 The alkylating agent N-ethylmaleimide (NEM), attacks the zinc-bound thiols in NC and consequently impairs virus infectivity.170,171 Appella and co-workers reported that pyridinioalkanoyl thioesters (PATEs) that are based on a 2-mercaptobenzamide thioester chemotype selectively target the zinc fingers of NC without affecting the zinc fingers found in other viral or cellular proteins.172–174 These PATE compounds mediate zinc ejection from NC and display low-micromolar antiviral activity with low cytotoxicity.173,174
Interestingly, one of these compounds is reportedly efficacious in an HIV-1 transgenic mouse model system to suppress the production of infectious virus particles.175,176 These initial studies led to the development of a new class of zinc-ejecting compounds, the N-substituted S-acyl-2-mercaptobenzamide thioesters (SAMTs).177,178 Unlike other NC inhibitors, the SAMTs use an S-acyl transfer mechanism rather than an oxidative mechanism to eject zinc.177,179 These compounds were reported to show antiviral activity as a vaginal microbicide in rhesus macaques in an SIV/HIV-1 (SHIV) chimeric virus infection model.180 SAMTs possess an interesting mechanism of action; they interact with NC and covalently modify the protein with an acetyl group, thereby inhibiting Gag processing and viral infectivity.181
Reportedly, the resulting mercaptobenzamide thiol can be reacetylated in cells with acetyl-CoA to regenerate the reactive thioester form of the inhibitor, which can then react with another molecule of Gag.181 This recycling mechanism could potentially increase the potency of the SAMT class of inhibitors. In another recent study, Gorelick and co-workers showed that a sulfhydryl compound [4-vinylpyridine (4-VP)] in conjunction with a membrane-permeable divalent metal ion chelator, binds to cysteine thiols in the NC zinc fingers and inactivates HIV-1 infectivity.182
The compounds described above act principally by attacking the zinc ions bound in the NC zinc fingers. The nucleic acid chaperone activity of NC could also be targeted. To this end, de Rocquigny and colleagues developed a screen for molecules that inhibit the nucleic acid destabilizing activity of NC.183 This assay is based on the ability of NC to unfold a DNA stem-loop that is labeled on one end with a fluorophore and on the other end with a fluorescence quencher. Five compounds with IC50 values in the micromolar range were identified that bind to NC zinc fingers and prevent the protein from interacting with the DNA stem-loop. These molecules block binding of NC to DNA without promoting zinc ejection.183 An alternative approach to disrupting NC function targets RNA encapsidation. Along these lines, Turner and colleagues reported that aminoglycosidic antibiotics were capable of interfering with genome recognition and encapsidation.184 The steady progress in developing ever-more-potent NC-based inhibitors suggests that this area will remain active in the future.
p6
The p6 domain, located at the C-terminus of HIV-1 Gag, functions late in the virus assembly pathway to promote the pinching off of virus particles from the cell surface. An early study observed that deletion of the p6 domain from HIV-1 Gag produced a severe block to particle production.185 Intriguingly, EM analysis showed that the p6-deleted particles were fully assembled but failed to bud off from the plasma membrane, instead remaining attached to the cell surface by a thin tether. Subsequent work identified a highly conserved Pro-Thr/Ser-Ala-Pro [P(T/S)AP] motif as being responsible for the budding activity of p6186; mutation of any of these residues recapitulated the phenotype observed upon p6 deletion.186,187 Domains with analogous functions were identified in the Gag proteins of a number of other retroviruses. These motifs were named “late domains” because of their role in virus budding.188 Three classes of late domains have been identified in retroviral Gag proteins: PT/SAP, PPXY, and YP(X)nL (where “X” is any amino acid, and n=1–4 residues).18–20,189 In many cases, retroviral Gag proteins contain multiple late domains, often located within a few residues of each other.
In the case of HIV-1, for example, the primary late domain is PT/SAP, as mentioned above, but a secondary late domain of the YP(X)nL type is found near the C-terminus of p6. It should be noted that late domains bearing these sequences are also found in other nonretroviral enveloped viruses. For example, the matrix-like VP40 protein of Ebola virus contains overlapping PTAP and PPXY motifs.190
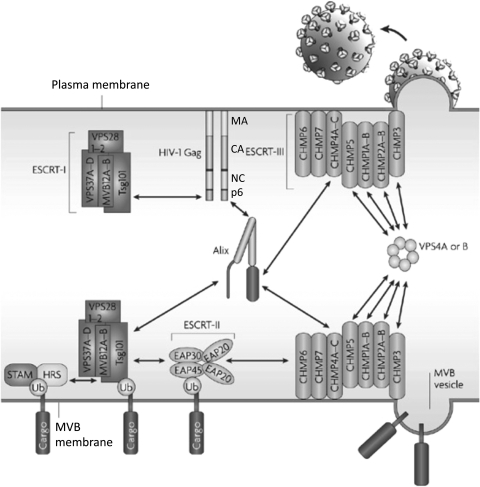
A compelling body of evidence indicates that late domains act by interacting directly with host proteins that are part of the cellular endosomal sorting pathway. At the core of this machinery are several multiprotein complexes, originally identified in yeast by the laboratory of Scott Emr, that are known as endosomal sorting complexes required for transport (ESCRT) 0, I, II, and III (Fig. 8).18,20,191,192 The ESCRT complexes play sequential roles in the sorting of cargo proteins into intralumenal vesicles that bud into late endosomes to generate MVBs.191,193 The cargo-laden MVBs ultimately fuse with the lysosome, leading to cargo degradation. Sorting of cargo proteins into the MVB pathway often requires monoubiquitination of the cytoplasmic domains of the cargo and recognition of the ubiquitin moiety by the sorting apparatus. The ESCRT machinery also functions in the membrane scission reaction that occurs during cytokinesis.194–196 Retroviruses have thus evolved to usurp a cellular machinery that directs budding and membrane fission reactions that are oriented away from the cytosol, the topological equivalent of virus budding.
FIG. 8.
Schematic representation of the ESCRT machinery and its interaction with HIV-1 Gag at the plasma membrane and ubiquitinated cargo in the MVB. The MA, CA, NC, and p6 domains of Gag are shown. Double-headed arrows denote the interaction of p6 with Tsg101 and Alix, Alix with Tsg101, and Vps4 with ESCRT-III. A ubiquitinated (Ub) cargo protein is shown interacting with the STAM/Hrs complex (often referred to as ESCRT-0). Adapted with permission from Fujii et al. (2007).279 Copyright 2007 Macmillan Publishers Ltd. (Nature Reviews Microbiology).
In addition to the four major ESCRT complexes—ESCRT-0, I, II, and III—a variety of ESCRT-accessory proteins participate in ESCRT function. These include Alix, a molecule that possesses interaction sites for both ESCRT-I and ESCRT-III, the AAA ATPase Vps4, and members of the Need4 family of ubiquitin ligases, which are involved in cargo ubiquitination. Retroviral late domains interact with various components of this sorting machinery. The PT/SAP motif binds the ubiquitin E2 variant (UEV) domain of the ESCRT-I component Tsg101 (Fig. 9),197–199 YP(X)nL motifs interact with Alix (Fig. 10),200–202 and PPPXY motifs bind Nedd4-family ubiquitin ligases.18–20
FIG. 9.
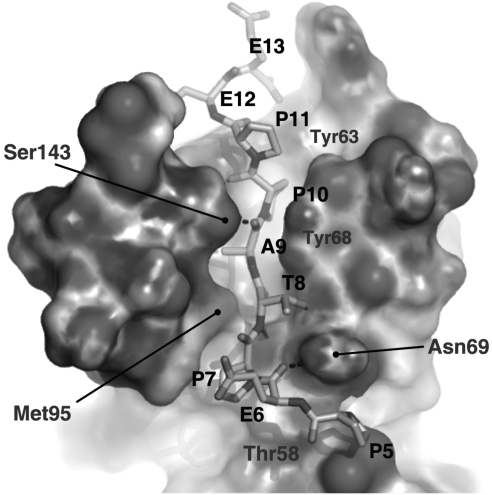
Structure of the UEV domain of Tsg101 bound to a PTAP-containing peptide derived from the sequence of HIV-1 p6. p6 residues Pro-5 (P5), Glu-6 (E6), Pro-7 (P7), Thr-8 (T8), Ala-9 (A9), Pro-10 (P10), Pro-11 (P11), Glu-12 (E12), and Glu-13 (E13) are shown, as are residues in Tsg101 that make contact with the PTAP-containing peptide. Reprinted with permission from Im et al. (2010).211 Copyright 2010 Cell Press.
FIG. 10.
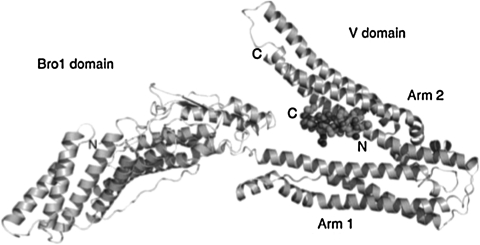
Structure of a YPXnL-containing peptide bound to the Alix V domain. Ribbon diagram of the Bro1 and V domain of ALIX in complex with the HIV-1 p6 YPXnL late-domain peptide. Adapted from Zhai et al. (2008).210 Copyright 2007 Macmillan Publishers Ltd. (Nature Structural and Molecular Biology).
The functional significance of HIV-1 late domain interactions with ESCRT or associated machinery has been demonstrated by a number of studies showing that disruption of these interactions is detrimental to HIV-1 budding. As mentioned above, early studies indicated that mutation of the primary HIV-1 late domain, PT/SAP, led to the tethering of virions at the cell surface and failure of the virions to pinch off.185,186 Subsequently, it was shown that Tsg101 depletion with siRNA or overexpression of the Gag-binding UEV domain of Tsg101 (referred to as TSG-5′) impaired virus release in much the same manner as PT/SAP mutation.187,197 Fusion of Tsg101 to Gag can also stimulate the release of particles lacking the PT/SAP late domain.203 Stable overexpression of TSG-5′ blocks the replication of feline immunodeficiency virus (FIV), which also encodes a Tsg101-binding, PT/SAP late domain.204 Disruption of the Gag–Alix interaction potently inhibits EIAV release but produces more subtle effects on virus budding in the case of HIV-1.200,201,205–207 However, overexpression of the V-domain strongly disrupts HIV-1 budding,205,206 and YP(X)nL mutations, although not producing large effects on virus release, do significantly delay HIV-1 replication in a range of cell types including primary lymphocytes and macrophages.208
While overexpression of the Tsg101 UEV domain (TSG-5′) and the Alix-V domain inhibits HIV-1 budding, these large protein fragments are not viable as therapeutics. They do, however, provide proof-of-principle that Gag interactions with cellular endosomal sorting machinery constitute viable targets for developing antiviral inhibitors. Structural information obtained from NMR and x-ray crystallographic studies has provided high-resolution detail of the PT/SAP and YP(X)nL motif interactions with Tsg101 (Fig. 9) and Alix (Fig. 10), respectively.198,209–211 Binding affinities have also been calculated. These interactions are of relatively low affinity, consistent with the other protein–protein interactions along the ESCRT pathway. In an effort to design competitive inhibitors of the PT/SAP–Tsg101 interaction that display increased affinity for Tsg101, Burke and co-workers replaced the Pro residues in the peptide PEPTAPPEE with N-substituted glycines. This was accomplished by using hydrazone and hydrazide chemistry to synthesize a large series of these peptoid derivatives.212
In a follow-up study,213 each amino acid in the PEPTAPPEE peptide was replaced by amino-oxy-containing residues. These peptides were then reacted with a series of aldehydes to obtain a library of peptoids in which each residue was modified by different functional groups. Fluorescent anisotropy was used to evaluate the binding affinity of each peptoid for the UEV domain of Tsg101. In some cases, 15- to 20-fold increases in binding affinity were observed relative to the parental PEPTAPPEE peptide.212,213 Finally, the binding conformations were stabilized and cellular uptake was increased by peptide circularization.214 Efforts are underway in our laboratory to test these peptoids for their ability to inhibit HIV-1 budding in cell-based assays. In an independent study, Tavassoli et al. designed a bacterial reverse two-hybrid system (RTHS) to identify inhibitors of the p6–Tsg101 interaction.215
They generated a large library (108) of cyclic peptides using the so-called SICLOPPS (split intein-mediated circular ligation of peptides and proteins) method, and these peptides were screened in RTHS to identify a small number of peptides that interfered with the p6–Tsg101 interaction.215 After several rounds of secondary screening, four cyclic peptides were identified that inhibited these interactions, and one of these peptides (IYWNVSGW) showed a modest ability (∼2-fold) to inhibit the release of virus-like particles in cell culture.215 The inhibitory activity of the peptide is reportedly PT/SAP-dependent, as release of an HIV-1 PTAP mutant was not inhibited. These studies represent initial steps in developing inhibitors of the interaction between p6 and Tsg101. Additional studies are required to identify small molecules capable of disrupting HIV-1 budding.
Thus far, efforts have focused on the PT/SAP–Tsg101 interface, as this is the primary late-domain interaction required for HIV-1 budding. Whether effective disruption of HIV-1 release will also require blocking the p6–Alix interaction remains to be tested. The observation that PT/SAP and YP(X)nL mutations synergize to completely block HIV-1 replication208 suggests that this might be the case. Also unresolved is whether targeting Gag interactions with the ESCRT machinery will also block vital interactions between cellular proteins and thus result in significant cytotoxicity. Efforts are currently underway in our laboratory to develop small molecule inhibitors of the PT/SAP–Tsg101 interaction.
Maturation
As discussed above, HIV-1 maturation is triggered by the cleavage of the Gag polyprotein Pr55Gag by the viral PR. The proteolytic processing of Pr55Gag takes place in a sequential cascade of events: first, Gag is cleaved into two fragments, MA–CA–SP1 and NC–SP2–p6; then cleavage occurs at the MA–CA and SP2–p6 junctions; finally, CA–SP1 and NC–SP2 are cleaved to liberate CA and NC domains.216–222 The newly generated mature CA assembles to form the conical core, a process that is essential for virus infectivity.25,223 Processing of Gag by PR is also required for activation of the fusogenic activity of the viral Env glycoprotein.83,224 Mature NC is involved in stabilization of viral dimeric RNA and condensation of the viral ribonucleoprotein complex.225,226
One of the important features of Gag processing is that it occurs in a highly ordered sequence, with the rate of processing at each step kinetically regulated in part by widely differing affinities of the PR for each of the individual cleavage sites. It is well established that interfering with the cleavage at any of the sites in Gag, or changing the order with which the sites are cleaved, results in aberrant virion morphology and loss of infectivity.94,108,219,222,227–229 These observations suggest that disrupting Gag processing event(s) could be a viable pharmacological approach for inhibiting HIV-1 replication. Although PR inhibitors (PIs) that inhibit the enzymatic activity of the enzyme and block Gag and Gag-Pol processing have been used effectively for many years, an alternative approach is to target individual Gag cleavage sites. Thus, inhibitors that block maturation by targeting Gag rather than PR are known as “maturation inhibitors” and are distinct from PIs.21,89
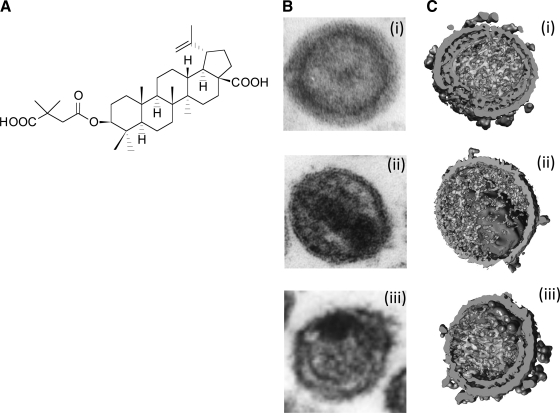
Although in theory inhibitors could be developed that block each of the five Gag cleavage sites, until now only the CA–SP1 cleavage site has been successfully targeted. The small molecule 3-O-(3′,3′-dimethylsuccinyl)betulinic acid (DSB), also known as PA-457 or bevirimat (BVM) (Fig. 11), is the first member of this class of inhibitors.229,230 BVM disrupts a late stage in PR-mediated Gag processing by blocking the cleavage of SP1 from CA, leading to the accumulation of the Gag processing intermediate CA–SP1.229,230 The accumulation of CA–SP1 interferes with virion maturation, leading to the formation of particles that lack conical cores but instead display an acentric aggregation of electron density and a crescent of Gag density near the viral membrane (Fig. 11).230,231
FIG. 11.
Bevirimat and its effect on HIV-1 maturation. (A) Structure of bevirimat. (B, C) Morphology of virions observed by thin-section EM (B) and cryoelectron tomography (C): (i) immature particles, (ii) mature particles, and (iii) particles produced from bevirimat-treated cells. Reprinted with permission from Adamson and Freed (2008)280 and Keller et al. (2011).232 Copyright 2008 Elsevier Inc. and 2011, American Society for Microbiology.
At the level of resolution provided by thin-section EM, the morphology of virions produced from BVM-treated cells is similar to that induced by mutations that block CA–SP1 processing.222 By cryoelectron tomography, however, it is clear that the crescent of Gag density present in virus particles produced from BVM-treated cells (Fig. 11) displays the hexamer–hexamer spacing typical of the immature Gag lattice, suggesting that in addition to inhibiting CA–SP1 processing, BVM may stabilize the immature Gag lattice.232
It is now clear that the CA–SP1 cleavage site in Gag is the target for BVM. Most compelling is that in vitro selection leads to the acquisition of resistance to BVM by single-amino-acid substitutions in the immediate vicinity of the CA–SP1 cleavage site (Fig. 12) rather than elsewhere in Gag or in PR.229–231,233–236 In addition, mutations engineered into the CA–SP1 boundary region can confer resistance to BVM.236 Although it is well established that BVM targets the CA–SP1 junction, the exact mechanism by which the compound inhibits CA–SP1 cleavage is not well defined.
FIG. 12.
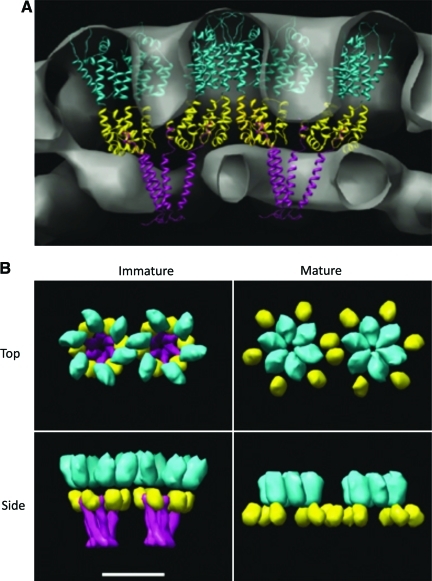
Model depicting the organization of CA in mature and immature Gag lattices. (A) Side view of the immature Gag lattice. The CA NTD (cyan), CA CTD (yellow), and SP1 (magenta) are shown. (B) Models representing the top and side views of Gag subunits in the immature (left) and mature (right) lattices. In the immature lattice, hexamers are held together by an SP1 bundle spaced by CTD interactions. SP1 is removed upon PR-mediated processing during maturation; in the mature lattice the hexamers are held together by the NTD spaced by CTD interactions. Scale bar=8 nm. Reprinted with permission from Wright et al. (2007).31 Copyright European Molecular Biology Organization. Color images available online at www.liebertonline.com/aid
The working model is that BVM binds a pocket on the assembled VLP that prevents access by PR to the CA–SP1 cleavage site. This hypothesis is supported by the observation that the ability of BVM to block CA–SP1 processing requires prior assembly of Gag.230,237 Furthermore, BVM binds to immature VLPs but not mature virions, suggesting that the binding site is destroyed by PR-mediated Gag processing238 and a number of mutations that confer BVM resistance reduce binding of labeled compound to virions.236 It is interesting to note that some BVM-resistant mutants display enhanced replication and maturation in the presence of BVM, suggesting that in these cases a simple compound exclusion mechanism is unlikely to explain resistance.231
BVM is the first Gag-targeted compound to reach clinical trials. Initial studies in a severe combined immunodeficiency (SCID)-hu Thy/Liv mouse model showed promising pharmacological results and led to the initiation of clinical trials in humans.239 After Phase I clinical trials, BVM was tested in HIV-1-infected individuals.240,241 These Phase II trials demonstrated a dose-dependent, statistically significant reduction in viral loads.241 However, McCallister et al. showed that ∼50% of BVM-treated patients did not exhibit a reduction in viral loads despite high levels of BVM in the plasma.242 The lack of response appeared to correlate with baseline polymorphisms in SP1 residues 6–8,242–244 and in vitro studies demonstrated that mutations in these residues can decrease the susceptibility of HIV-1 to BVM.233,245 These naturally occurring Gag polymorphisms are distinct from the resistance mutations selected by passaging HIV-1 in the presence of BVM in vitro.231,234 Although some mutations in SP1 residues 6–8 reduce sensitivity to BVM, mutations in PR that render the enzyme resistant to PIs can delay the acquisition of BVM resistance in vitro.234
Although BVM has demonstrated proof-of-principle that CA-SP1 cleavage can be targeted to interfere with HIV-1 replication in patients, commonly occurring polymorphisms in SP1 that reduce drug susceptibility pose a challenge for future clinical development of this compound. Research performed at Pfizer has identified a small molecule that, like BVM, blocks CA–SP1 processing. Interestingly, this compound is structurally quite distinct from BVM, even though mutations at the CA–SP1 junction can confer resistance to both compounds.246 Advances in structural biology that would allow visualization of the BVM binding site on the Gag oligomer would make possible rational, structure-based approaches to the design of more potent and broadly active inhibitors that target CA–SP1 processing.
The observation that incomplete processing at any of the Gag cleavage sites is highly detrimental to virion maturation raises the possibility that unprocessed Gag fragments can act in a dominant-negative manner to interfere with maturation and infectivity. This issue was addressed recently in several studies, which demonstrated by using low doses of PR inhibitors or a genetic approach involving cotransfection of wild-type and cleavage-site-mutant molecular clones that uncleaved Gag fragments do indeed disrupt HIV-1 infectivity.247–249 These results highlight the potential for drugs that block virus maturation by disrupting individual steps in the Gag processing cascade.
Uncoating and nuclear import
Thus far, our discussion has been focused primarily on targeting Gag during the late steps in the virus replication cycle. However, Gag domains also performs functions at early, postentry steps. As discussed previously, after the fusion of the viral and cellular membranes, the viral core is delivered into the cytoplasm. A poorly understood process known as uncoating then takes place; during this process, the viral RNA is reverse transcribed to double-stranded DNA.3 Although the details of the uncoating mechanism remain to be elucidated, it is clear that correct regulation of core uncoating is required for productive infection to take place. CA mutants that display either premature or delayed uncoating in in vitro assays display impaired reverse transcription and infectivity.250–252
The importance of core uncoating has been underscored by the discovery of host restriction factors that target CA during early steps in the retroviral replication cycle.9 The classical example of such a host restriction factor is Fv1, shown decades ago to restrict infection by certain strains of MLV253 and more recently demonstrated to encode a Gag-like product derived from an endogenous retrovirus.254 Residues in CA regulate susceptibility to Fv1,255 implicating CA as the primary target of Fv1 restriction. A somewhat analogous restriction factor was found to function in primate cells to restrict infection by a variety of lentiviruses. In 2004, Stremlau and colleagues identified this restriction factor as TRIM5α (tripartite motif 5, isoform α).256
Soon thereafter, a related gene in which TRIM5 is fused to cyclophilin A (hence named TRIMCyp) was identified as a restriction factor in owl monkey cells.257,258 TRIM proteins contain a RING finger, a B-box, and a coiled-coil domain9; the B-box and coiled-coil domains are required for TRIM multimerization and anti-HIV-1 restriction activity.259,260 The α isoform of TRIM5 contains an additional C-terminal domain referred to as PRYSPRY (or B30.2); it is this region of the protein that possesses CA-binding properties. In the case of TRIMCyp, the PRYSPRY domain is replaced by cyclophilin A, which, as discussed earlier, binds the CA NTD.256,261–267
Although the exact mechanism by which TRIM5α and TRIMCyp exert their antiviral activity is not fully understood, CA is the target for restriction. It appears that the TRIM-family restriction factors bind the incoming viral capsid complex and promote rapid and premature uncoating and, under some conditions, proteasomal degradation.265,268–272 The antiviral activity of TRIM5α is species-specific, i.e., TRIM5α from rhesus macaques blocks HIV-1 infection, whereas human TRIM5α is only weakly active against HIV-1 but strongly inhibits infection by some strains of SIV.9 Recent structural analysis indicates that TRIM5α assembles into a flexible, hexagonal lattice that uses the CA hexagonal array as a template.273
Because the assembly of hexagonal arrays of CA appears to be a highly conserved feature of retroviral core structure, such a model helps to explain how TRIM family proteins can restrict a wide array of distantly related retroviruses.
The potent antiviral activity conferred by TRIM5α raises the possibility that these host restriction factors could in some way be harnessed to interfere with HIV-1 replication. Akkina and co-workers used a gene therapy approach to deliver an HIV-1-restricting human-rhesus TRIM5α chimera in a SCID-hu mouse model system.274 Transgenic thymocytes and macrophages were refractory to HIV-1 infection in vivo in this setting, illustrating the potential for TRIM-based antiviral strategies.274 Since gene therapy approaches face many hurdles in the effective treatment of HIV-1 infection in humans, alternative strategies for exploiting TRIM5α should be considered. These include the possibility that small molecules could be designed to alter human TRIM5α conformation such that it acquires the capacity to bind and restrict incoming HIV-1 core complexes. Any efforts to modify or emulate the TRIM5α-mediated block will require more information on the structural details of the TRIM–CA interaction and a more in-depth understanding of the mechanism by which this family of proteins restricts viral infection.
The import of the PIC into the nucleus represents an additional postentry step in the replication cycle that could be targeted by antiretroviral agents. This step has been difficult to study, and the viral determinants of nuclear import have proven elusive. Mounting evidence suggests that CA plays an important role in nuclear import12,275 and a recent study developed an inhibitor of this process. KewalRamani and colleagues reported that a cytosolically localized fragment of cleavage and polyadenylation factor 6 (CPSF6) potently inhibits HIV-1 infectivity by blocking nuclear import of the PIC.276 Selection for resistance to the CPSF6 fragment revealed that a single-amino-acid change in the CA NTD provides escape from the restriction. Remarkably, the CFSF6-resistant CA mutant displays an altered specificity in terms of its requirement for nuclear import factors, suggesting that CA may interact directly with components of the nuclear pore.276,277 An increased understanding of this putative interaction could lead to novel approaches for blocking nuclear import of the HIV-1 PIC, a step in the replication cycle that is particularly critical in nondividing cells such as monocyte-derived macrophages.
Conclusions
In recent years, the HIV field has experienced an explosion of new information on the role of Gag proteins in HIV-1 assembly, release, and maturation. New insights have also been gained on the function of Gag proteins, in particular CA and NC, early in the replication cycle. Advances in structural biology have added greatly to our knowledge of Gag–Gag interactions and developments in cell biology have revealed the molecular details of host cell pathways that the virus uses to promote its replication and cellular restriction factors that impede virus infection. Despite these impressive advances, Gag proteins have not yet been successfully exploited in the development of clinically approved antiretroviral therapies. Although current anti-HIV drugs are highly effective in most patients, over time it is likely that resistance to these inhibitors will become an increasing problem. We therefore believe that attention should begin to broaden from basic studies on Gag function to the application of this information to novel inhibitor development.
Acknowledgments
We thank L. Henderson for providing the image shown in Fig. 1 and M. Summers for Fig. 3. We also thank W. Sundquist, M. Summers, E. Wright, A. Steven, M. Yeager, J. Hurley, and G. Jensen for permission to reproduce figures and members of Freed laboratory for helpful discussions and critical reading of the manuscript. Research in Freed laboratory is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the Intramural AIDS Targeted Antiviral Program.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Berger EA. Murphy PM. Farber JM. Chemokine receptors as HIV‐1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Doms RW. Beyond receptor expression: The influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276(2):229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 3.Freed EO. Martin MA. HIVs, their replication. In: Knipe DM, editor; Howley PM, editor. Fields Virology. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 2107–2185. [Google Scholar]
- 4.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: The HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyauchi K. Kim Y. Latinovic O. Morozov V. Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telesnitsky A. Goff SP. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J, editor; Varmus H, editor; Hughes S, editor. Retroviruses. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1997. pp. 121–160. [PubMed] [Google Scholar]
- 7.Levin JG. Guo J. Rouzina I. Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 8.Hulme AE. Perez O. Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci USA. 2011;108(24):9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song B. TRIM5alpha. Curr Top Microbiol Immunol. 2009;339:47–66. doi: 10.1007/978-3-642-02175-6_3. [DOI] [PubMed] [Google Scholar]
- 10.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dismuke DJ. Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80(8):3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita M. Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78(11):5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown PO. Integration. In: Coffin J, editor; Varmus H, editor; Hughes S, editor. Retroviruses. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1997. pp. 161–205. [PubMed] [Google Scholar]
- 14.Adamson CS. Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 15.Freed EO. HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology. 1998;251(1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 16.Waheed AA. Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143(2):162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waheed AA. Freed EO. The role of lipids in retrovirus replication. Viruses. 2010;2(5):1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344(1):55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Demirov DG. Freed EO. Retrovirus budding. Virus Res. 2004;106(2):87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Morita E. Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 21.Adamson CS. Salzwedel K. Freed EO. Virus maturation as a new HIV-1 therapeutic target. Expert Opin Ther Targets. 2009;13(8):895–908. doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broder S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral Res. 2010;85(1):1–18. doi: 10.1016/j.antiviral.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: Top HIV. Med. 2009;17(5):138–145. [PubMed] [Google Scholar]
- 24.Chertova E. Chertov O. Coren LV, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briggs JA. Simon MN. Gross I, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 26.Carlson LA. Briggs JA. Glass B, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4(6):592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller SD. Wilk T. Gowen BE. Krausslich HG. Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7(10):729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 28.Wilk T. Gross I. Gowen BE, et al. Organization of immature human immunodeficiency virus type 1. J Virol. 2001;75(2):759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeager M. Wilson-Kubalek EM. Weiner SG. Brown PO. Rein A. Supramolecular organisation of immature and mature murine leukemia virus revealed by electron cryo-microscopy: Implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briggs JA. Riches JD. Glass B. Bartonova V. Zanetti G. Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106(27):11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright ER. Schooler JB. Ding HJ, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26(8):2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill CP. Worthylake D. Bancroft DP. Christensen AM. Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93(7):3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massiah MA. Starich MR. Paschall C. Summers MF. Christensen AM. Sundquist WI. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244(2):198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 34.Rao Z. Belyaev AS. Fry E. Roy P. Jones IM. Stuart DI. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378(6558):743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 35.Bryant M. Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87(2):523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freed EO. Orenstein JM. Buckler-White AJ. Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68(8):5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottlinger HG. Sodroski JG. Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86(15):5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal R. Reitz MS., Jr Tschachler E. Gallo RC. Sarngadharan MG. Veronese FD. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses. 1990;6(6):721–730. doi: 10.1089/aid.1990.6.721. [DOI] [PubMed] [Google Scholar]
- 39.Zhou W. Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70(12):8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang C. Loeliger E. Luncsford P. Kinde I. Beckett D. Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101(2):517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono A. Freed EO. Binding of human immunodeficiency virus type 1 Gag to membrane: Role of the matrix amino terminus. J Virol. 1999;73(5):4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saad JS. Loeliger E. Luncsford P, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366(2):574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ono A. Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 44.Aloia RC. Tian H. Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90(11):5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brugger B. Glass B. Haberkant P. Leibrecht I. Wieland FT. Krausslich HG. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103(8):2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan R. Uchil PD. Jin J, et al. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82(22):11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindwasser OW. Resh MD. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J Virol. 2001;75(17):7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen DH. Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74(7):3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ono A. Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98(24):13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono A. Waheed AA. Joshi A. Freed EO. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: Use of a novel Gag multimerization assay. J Virol. 2005;79(22):14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickl WF. Pimentel-Muinos FX. Seed B. Lipid rafts and pseudotyping. J Virol. 2001;75(15):7175–7183. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindwasser OW. Resh MD. Myristoylation as a target for inhibiting HIV assembly: Unsaturated fatty acids block viral budding. Proc Natl Acad Sci USA. 2002;99(20):13037–13042. doi: 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X. Yu X. Lee TH. Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67(11):6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou W. Parent LJ. Wills JW. Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68(4):2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono A. Freed EO. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78(3):1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Camilli P. Emr SD. McPherson PS. Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271(5255):1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 57.McLaughlin S. Wang J. Gambhir A. Murray D. PIP(2) and proteins: Interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 58.De Matteis MA. Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6(6):487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 59.Ono A. Ablan SD. Lockett SJ. Nagashima K. Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101(41):14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saad JS. Miller J. Tai J. Kim A. Ghanam RH. Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103(30):11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shkriabai N. Datta SA. Zhao Z. Hess S. Rein A. Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45(13):4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 62.Freed EO. Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69(3):1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ono A. Orenstein JM. Freed EO. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol. 2000;74(6):2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chukkapalli V. Hogue IB. Boyko V. Hu WS. Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82(5):2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saad JS. Ablan SD. Ghanam RH, et al. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382(2):434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamard-Peron E. Juillard F. Saad JS, et al. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol. 2010;84(1):503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen K. Bachtiar I. Piszczek G. Bouamr F. Carter C. Tjandra N. Solution NMR characterizations of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry. 2008;47(7):1928–1937. doi: 10.1021/bi701984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandes F. Chen K. Ehrlich LS, et al. Phosphoinositides direct equine infectious anemia virus Gag trafficking and release. Traffic. 2011;12(4):438–451. doi: 10.1111/j.1600-0854.2010.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batonick M. Favre M. Boge M. Spearman P. Honing S. Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342(2):190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 70.Camus G. Segura-Morales C. Molle D, et al. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol Biol Cell. 2007;18(8):3193–3203. doi: 10.1091/mbc.E06-12-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong X. Li H. Derdowski A, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Joshi A. Garg H. Nagashima K. Bonifacino JS. Freed EO. GGA and Arf proteins modulate retrovirus assembly and release. Mol Cell. 2008;30(2):227–238. doi: 10.1016/j.molcel.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi A. Nagashima K. Freed EO. Defects in cellular sorting and retroviral assembly induced by GGA overexpression. BMC Cell Biol. 2009;10:72. doi: 10.1186/1471-2121-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bauby H. Lopez-Verges S. Hoeffel G, et al. TIP47 is required for the production of infectious HIV-1 particles from primary macrophages. Traffic. 2010;11(4):455–467. doi: 10.1111/j.1600-0854.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 75.Blot G. Janvier K. Le Panse S. Benarous R. Berlioz-Torrent C. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J Virol. 2003;77(12):6931–6945. doi: 10.1128/JVI.77.12.6931-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopez-Verges S. Camus G. Blot G. Beauvoir R. Benarous R. Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103(40):14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Izumi Y. Watanabe H. Watanabe N. Aoyama A. Jinbo Y. Hayashi N. Solution X-ray scattering reveals a novel structure of calmodulin complexed with a binding domain peptide from the HIV-1 matrix protein p17. Biochemistry. 2008;47(27):7158–7166. doi: 10.1021/bi702416b. [DOI] [PubMed] [Google Scholar]
- 78.Radding W. Williams JP. McKenna MA, et al. Calmodulin and HIV type 1: Interactions with Gag and Gag products. AIDS Res Hum Retroviruses. 2000;16(15):1519–1525. doi: 10.1089/088922200750006047. [DOI] [PubMed] [Google Scholar]
- 79.Joshi A. Garg H. Ablan SD. Freed EO. Evidence of a role for soluble N- ethylmaleimide-sensitive factor attachment protein receptor (SNARE) machinery in HIV-1 assembly and release. J Biol Chem. 2011;286(34):29861–29871. doi: 10.1074/jbc.M111.241521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Checkley MA. Luttge BG. Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410(4):582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dorfman T. Mammano F. Haseltine WA. Gottlinger HG. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68(3):1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freed EO. Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70(1):341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murakami T. Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74(8):3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu X. Yuan X. Matsuda Z. Lee TH. Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66(8):4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srinivas SK. Srinivas RV. Anantharamaiah GM. Compans RW. Segrest JP. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268(30):22895–22899. [PubMed] [Google Scholar]
- 86.Kiernan RE. Ono A. Englund G. Freed EO. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72(5):4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alfadhli A. Still A. Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009;83(23):12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adamson CS. Freed EO. HIV-1 assembly, release, maturation. In: Jeang K-T, editor. Advances in Pharmacology, HIV-1: Molecular Biology and Pathogenesis: Viral Mechanisms. Vol. 55. Elsevier; Amsterdam: 2007. pp. 347–387. [DOI] [PubMed] [Google Scholar]
- 89.Adamson CS. Freed EO. Novel approaches to inhibiting HIV-1 replication. Antiviral Res. 2010;85(1):119–141. doi: 10.1016/j.antiviral.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ganser-Pornillos BK. Yeager M. Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18(2):203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gamble TR. Vajdos FF. Yoo S, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87(7):1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 92.Gitti RK. Lee BM. Walker J. Summers MF. Yoo S. Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273(5272):231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 93.Momany C. Kovari LC. Prongay AJ, et al. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3(9):763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 94.Accola MA. Hoglund S. Gottlinger HG. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72(3):2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Borsetti A. Ohagen A. Gottlinger HG. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72(11):9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Schwedler UK. Stray KM. Garrus JE. Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77(9):5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gamble TR. Yoo S. Vajdos FF, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278(5339):849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 98.Worthylake DK. Wang H. Yoo S. Sundquist WI. Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 99.Rose S. Hensley P. O'Shannessy DJ. Culp J. Debouck C. Chaiken I. Characterization of HIV-1 p24 self-association using analytical affinity chromatography. Proteins. 1992;13(2):112–119. doi: 10.1002/prot.340130204. [DOI] [PubMed] [Google Scholar]
- 100.Burniston MT. Cimarelli A. Colgan J. Curtis SP. Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73(10):8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joshi A. Nagashima K. Freed EO. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, Gag–Gag interactions, gag–membrane binding, and virion maturation. J Virol. 2006;80(16):7939–7951. doi: 10.1128/JVI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Strambio-de-Castillia C. Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66(12):7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wills JW. Craven RC. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 104.Craven RC. Leure-duPree AE. Weldon RA., Jr Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69(7):4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mammano F. Ohagen A. Hoglund S. Gottlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68(8):4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abdurahman S. Hoglund S. Goobar-Larsson L. Vahlne A. Selected amino acid substitutions in the C-terminal region of human immunodeficiency virus type 1 capsid protein affect virus assembly and release. J Gen Virol. 2004;85(Pt 10):2903–2913. doi: 10.1099/vir.0.80137-0. [DOI] [PubMed] [Google Scholar]
- 107.Accola MA. Strack B. Gottlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 2000;74(12):5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krausslich HG. Facke M. Heuser AM. Konvalinka J. Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69(6):3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liang C. Hu J. Russell RS. Roldan A. Kleiman L. Wainberg MA. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J Virol. 2002;76(22):11729–11737. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liang C. Hu J. Whitney JB. Kleiman L. Wainberg MA. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1. J Virol. 2003;77(3):1772–1783. doi: 10.1128/JVI.77.3.1772-1783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Melamed D. Mark-Danieli M. Kenan-Eichler M, et al. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 gag protein is important for virion assembly and release. J Virol. 2004;78(18):9675–9688. doi: 10.1128/JVI.78.18.9675-9688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morikawa Y. Hockley DJ. Nermut MV. Jones IM. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J Virol. 2000;74(1):16–23. doi: 10.1128/jvi.74.1.16-23.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morellet N. Druillennec S. Lenoir C. Bouaziz S. Roques BP. Helical structure determined by NMR of the HIV-1 (345–392) Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging. Protein Sci. 2005;14(2):375–386. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Newman JL. Butcher EW. Patel DT. Mikhaylenko Y. Summers MF. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004;13(8):2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Briggs JA. Grunewald K. Glass B. Forster F. Krausslich HG. Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14(1):15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 116.Li S. Hill CP. Sundquist WI. Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407(6802):409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 117.Ganser BK. Li S. Klishko VY. Finch JT. Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283(5398):80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 118.Ganser-Pornillos BK. Cheng A. Yeager M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell. 2007;131(1):70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 119.Lanman J. Lam TT. Barnes S, et al. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol. 2003;325(4):759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 120.Pornillos O. Ganser-Pornillos BK. Kelly BN, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137(7):1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Byeon IJ. Meng X. Jung J, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139(4):780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wright ER. Correlative structural biology: How to investigate the fine details of viral structure. Viruses. 2010;2(1):107–110. doi: 10.3390/v2010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cardone G. Purdy JG. Cheng N. Craven RC. Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457(7230):694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pornillos O. Ganser-Pornillos BK. Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469(7330):424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.del Alamo M. Mateu MG. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J Mol Biol. 2005;345(4):893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 126.Lanman J. Sexton J. Sakalian M. Prevelige PE., Jr Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol. 2002;76(14):6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hilpert K. Behlke J. Scholz C. Misselwitz R. Schneider-Mergener J. Hohne W. Interaction of the capsid protein p24 (HIV-1) with sequence-derived peptides: Influence on p24 dimerization. Virology. 1999;254(1):6–10. doi: 10.1006/viro.1998.9526. [DOI] [PubMed] [Google Scholar]
- 128.Garzon MT. Lidon-Moya MC. Barrera FN, et al. The dimerization domain of the HIV-1 capsid protein binds a capsid protein-derived peptide: A biophysical characterization. Protein Sci. 2004;13(6):1512–1523. doi: 10.1110/ps.03555304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tang C. Loeliger E. Kinde I, et al. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327(5):1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 130.Kelly BN. Kyere S. Kinde I, et al. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373(2):355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sticht J. Humbert M. Findlow S, et al. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12(8):671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 132.Ternois F. Sticht J. Duquerroy S. Krausslich HG. Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12(8):678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 133.Barklis E. Alfadhli A. McQuaw C, et al. Characterization of the in vitro HIV-1 capsid assembly pathway. J Mol Biol. 2009;387:376–389. doi: 10.1016/j.jmb.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhattacharya S. Zhang H. Debnath AK. Cowburn D. Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid. J Biol Chem. 2008;283(24):16274–16278. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang H. Zhao Q. Bhattacharya S, et al. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J Mol Biol. 2008;378(3):565–580. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Curreli F. Zhang H. Zhang X, et al. Virtual screening based identification of novel small-molecule inhibitors targeted to the HIV-1 capsid. Bioorg Med Chem. 2011;19(1):77–90. doi: 10.1016/j.bmc.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang H. Curreli F. Zhang X, et al. Antiviral activity of alpha-helical stapled peptides designed from the HIV-1 capsid dimerization domain. Retrovirology. 2011;8:28. doi: 10.1186/1742-4690-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Franke EK. Yuan HE. Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372(6504):359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 139.Franke EK. Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology. 1996;222(1):279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- 140.Daelemans D. Dumont JM. Rosenwirth B. De Clercq E. Pannecouque C. Debio-025 inhibits HIV-1 by interfering with an early event in the replication cycle. Antiviral Res. 2010;85(2):418–421. doi: 10.1016/j.antiviral.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 141.Cao J. Isaacson J. Patick AK. Blair WS. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob Agents Chemother. 2005;49(9):3833–3841. doi: 10.1128/AAC.49.9.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blair WS. Pickford C. Irving SL, et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010;6(12):e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shi J. Zhou J. Shah VB. Aiken C. Whitby K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol. 2011;85(1):542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gross I. Hohenberg H. Krausslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249(2):592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 145.Titolo S. Mercier J-F. Wardrop E, et al. Discovery of potent HIV-1 capsid assembly inhibitors; 17th Conference on Retroviruses and Opportunistic Infections; 2010. Oral abstract 50. [Google Scholar]
- 146.Fader LD. Bethell R. Bonneau P, et al. Discovery of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of inhibitors of HIV-1 capsid assembly. Bioorg Med Chem Lett. 2011;21(1):398–404. doi: 10.1016/j.bmcl.2010.10.131. [DOI] [PubMed] [Google Scholar]
- 147.Morellet N. Jullian N. De Rocquigny H. Maigret B. Darlix JL. Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11(8):3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Summers MF. Henderson LE. Chance MR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1(5):563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aldovini A. Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64(5):1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cimarelli A. Luban J. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J Virol. 2000;74(15):6734–6740. doi: 10.1128/jvi.74.15.6734-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cimarelli A. Sandin S. Hoglund S. Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74(7):3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.D'Souza V. Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3(8):643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 153.Dannull J. Surovoy A. Jung G. Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13(7):1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dawson L. Yu XF. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251(1):141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 155.De Guzman RN. Wu ZR. Stalling CC. Pappalardo L. Borer PN. Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279(5349):384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 156.Gorelick RJ. Benveniste RE. Gagliardi TD, et al. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology. 1999;253(2):259–270. doi: 10.1006/viro.1998.9513. [DOI] [PubMed] [Google Scholar]
- 157.Gorelick RJ. Nigida SM., Jr Bess JW., Jr Arthur LO. Henderson LE. Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64(7):3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guo J. Wu T. Kane BF, et al. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 2002;76(9):4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Poon DT. Wu J. Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70(10):6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sandefur S. Smith RM. Varthakavi V. Spearman P. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag) J Virol. 2000;74(16):7238–7249. doi: 10.1128/jvi.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sandefur S. Varthakavi V. Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72(4):2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schmalzbauer E. Strack B. Dannull J. Guehmann S. Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70(2):771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tanchou V. Decimo D. Pechoux C, et al. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72(5):4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moore MD. Nikolaitchik OA. Chen J. Hammarskjold ML. Rekosh D. Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5(10):e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rice WG. Schaeffer CA. Harten B, et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361(6411):473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 166.Rice WG. Supko JG. Malspeis L, et al. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270(5239):1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- 167.Lifson JD. Rossio JL. Piatak M, Jr, et al. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res Hum Retroviruses. 2004;20(7):772–787. doi: 10.1089/0889222041524661. [DOI] [PubMed] [Google Scholar]
- 168.Lu W. Arraes LC. Ferreira WT. Andrieu JM. Therapeutic dendritic-cell vaccine for chronic HIV-1 infection. Nat Med. 2004;10(12):1359–1365. doi: 10.1038/nm1147. [DOI] [PubMed] [Google Scholar]
- 169.Rossio JL. Esser MT. Suryanarayana K, et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J Virol. 1998;72(10):7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chertova EN. Kane BP. McGrath C, et al. Probing the topography of HIV-1 nucleocapsid protein with the alkylating agent N-ethylmaleimide. Biochemistry. 1998;37(51):17890–17897. doi: 10.1021/bi980907y. [DOI] [PubMed] [Google Scholar]
- 171.Morcock DR. Thomas JA. Gagliardi TD, et al. Elimination of retroviral infectivity by N-ethylmaleimide with preservation of functional envelope glycoproteins. J Virol. 2005;79(3):1533–1542. doi: 10.1128/JVI.79.3.1533-1542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Goel A. Mazur SJ. Fattah RJ, et al. Benzamide-based thiolcarbamates: A new class of HIV-1 NCp7 inhibitors. Bioorg Med Chem Lett. 2002;12(5):767–770. doi: 10.1016/s0960-894x(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 173.Song Y. Goel A. Basrur V, et al. Synthesis and biological properties of amino acid amide ligand-based pyridinioalkanoyl thioesters as anti-HIV agents. Bioorg Med Chem. 2002;10(5):1263–1273. doi: 10.1016/s0968-0896(01)00392-3. [DOI] [PubMed] [Google Scholar]
- 174.Turpin JA. Song Y. Inman JK, et al. Synthesis and biological properties of novel pyridinioalkanoyl thiolesters (PATE) as anti-HIV-1 agents that target the viral nucleocapsid protein zinc fingers. J Med Chem. 1999;42(1):67–86. doi: 10.1021/jm9802517. [DOI] [PubMed] [Google Scholar]
- 175.Schito ML. Goel A. Song Y, et al. In vivo antiviral activity of novel human immunodeficiency virus type 1 nucleocapsid p7 zinc finger inhibitors in a transgenic murine model. AIDS Res Hum Retroviruses. 2003;19(2):91–101. doi: 10.1089/088922203762688595. [DOI] [PubMed] [Google Scholar]
- 176.Srivastava P. Schito M. Fattah RJ, et al. Optimization of unique, uncharged thioesters as inhibitors of HIV replication. Bioorg Med Chem. 2004;12(24):6437–6450. doi: 10.1016/j.bmc.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 177.Jenkins LM. Byrd JC. Hara T, et al. Studies on the mechanism of inactivation of the HIV-1 nucleocapsid protein NCp7 with 2-mercaptobenzamide thioesters. J Med Chem. 2005;48(8):2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- 178.Jenkins LM. Durell SR. Maynard AT, et al. Comparison of the specificity of interaction of cellular and viral zinc-binding domains with 2-mercaptobenzamide thioesters. J Am Chem Soc. 2006;128(36):11964–11976. doi: 10.1021/ja063329e. [DOI] [PubMed] [Google Scholar]
- 179.Loo JA. Holler TP. Sanchez J. Gogliotti R. Maloney L. Reily MD. Biophysical characterization of zinc ejection from HIV nucleocapsid protein by anti-HIV 2,2′-dithiobis[benzamides] and benzisothiazolones. J Med Chem. 1996;39(21):4313–4320. doi: 10.1021/jm960253w. [DOI] [PubMed] [Google Scholar]
- 180.Wallace GS. Cheng-Mayer C. Schito ML, et al. Human immunodeficiency virus type 1 nucleocapsid inhibitors impede trans infection in cellular and explant models and protect nonhuman primates from infection. J Virol. 2009;83(18):9175–9182. doi: 10.1128/JVI.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Miller Jenkins LM. Ott DE. Hayashi R, et al. Small-molecule inactivation of HIV-1 NCp7 by repetitive intracellular acyl transfer. Nat Chem Biol. 2010;6(12):887–889. doi: 10.1038/nchembio.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Morcock DR. Thomas JA. Sowder RC., 2nd Henderson LE. Crise BJ. Gorelick RJ. HIV-1 inactivation by 4-vinylpyridine is enhanced by dissociating Zn(2+) from nucleocapsid protein. Virology. 2008;375(1):148–158. doi: 10.1016/j.virol.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shvadchak V. Sanglier S. Rocle S, et al. Identification by high throughput screening of small compounds inhibiting the nucleic acid destabilization activity of the HIV-1 nucleocapsid protein. Biochimie. 2009;91(7):916–923. doi: 10.1016/j.biochi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 184.Turner KB. Hagan NA. Fabris D. Inhibitory effects of archetypical nucleic acid ligands on the interactions of HIV-1 nucleocapsid protein with elements of Psi-RNA. Nucleic Acids Res. 2006;34(5):1305–1316. doi: 10.1093/nar/gkl004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gottlinger HG. Dorfman T. Sodroski JG. Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88(8):3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang M. Orenstein JM. Martin MA. Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69(11):6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Demirov DG. Orenstein JM. Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76(1):105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Parent LJ. Bennett RP. Craven RC, et al. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69(9):5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Freed EO. Viral late domains. J Virol. 2002;76(10):4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Licata JM. Simpson-Holley M. Wright NT. Han Z. Paragas J. Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77(3):1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20(1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hurley JH. Emr SD. The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wollert T. Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Carlton JG. Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 195.McDonald B. Martin-Serrano J. No strings attached: The ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122(Pt 13):2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Morita E. Sandrin V. Chung HY, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26(19):4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Garrus JE. von Schwedler UK. Pornillos OW, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 198.Pornillos O. Alam SL. Rich RL. Myszka DG. Davis DR. Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21(10):2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.VerPlank L. Bouamr F. LaGrassa TJ, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98(14):7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martin-Serrano J. Yarovoy A. Perez-Caballero D. Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Strack B. Calistri A. Craig S. Popova E. Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 202.von Schwedler UK. Stuchell M. Muller B, et al. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 203.Martin-Serrano J. Zang T. Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7(12):1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 204.Luttge BG. Shehu-Xhilaga M. Demirov DG, et al. Molecular characterization of feline immunodeficiency virus budding. J Virol. 2008;82(5):2106–2119. doi: 10.1128/JVI.02337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lee S. Joshi A. Nagashima K. Freed EO. Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14(3):194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Munshi UM. Kim J. Nagashima K. Hurley JH. Freed EO. An Alix fragment potently inhibits HIV-1 budding: Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282(6):3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 207.Puffer BA. Parent LJ. Wills JW. Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71(9):6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fujii K. Munshi UM. Ablan SD, et al. Functional role of Alix in HIV-1 replication. Virology. 2009;391(2):284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pornillos O. Alam SL. Davis DR. Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9(11):812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 210.Zhai Q. Fisher RD. Chung HY. Myszka DG. Sundquist WI. Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV–1 and EIAV. Nat Struct Mol Biol. 2008;15(1):43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 211.Im YJ. Kuo L. Ren X, et al. Crystallographic and functional analysis of the ESCRT–I /HIV-1 Gag PTAP interaction. Structure. 2010;18(11):1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu F. Stephen AG. Adamson CS, et al. Hydrazone-and hydrazide-containing N-substituted glycines as peptoid surrogates for expedited library synthesis: Application to the preparation of Tsg101-directed HIV-1 budding antagonists. Org Lett. 2006;8(22):5165–5168. doi: 10.1021/ol0622211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu F. Stephen AG. Waheed AA, et al. SAR by oxime-containing peptide libraries: Application to Tsg101 ligand optimization. Chembiochem. 2008;9(12):2000–2004. doi: 10.1002/cbic.200800281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu F. Stephen AG. Waheed AA. Freed EO. Fisher RJ. Burke TR., Jr Application of ring-closing metathesis macrocyclization to the development of Tsg101-binding antagonists. Bioorg Med Chem Lett. 2010;20(1):318–321. doi: 10.1016/j.bmcl.2009.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tavassoli A. Lu Q. Gam J. Pan H. Benkovic SJ. Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem Biol. 2008;3(12):757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 216.Erickson-Viitanen S. Manfredi J. Viitanen P, et al. Cleavage of HIV-1 gag polyprotein synthesized in vitro: Sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5(6):577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 217.Krausslich HG. Schneider H. Zybarth G. Carter CA. Wimmer E. Processing of in vitro-synthesized gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J Virol. 1988;62(11):4393–4397. doi: 10.1128/jvi.62.11.4393-4397.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mervis RJ. Ahmad N. Lillehoj EP, et al. The gag gene products of human immunodeficiency virus type 1: Alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62(11):3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pettit SC. Moody MD. Wehbie RS, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68(12):8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tritch RJ. Cheng YE. Yin FH. Erickson-Viitanen S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J Virol. 1991;65(2):922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Vogt VM. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 222.Wiegers K. Rutter G. Kottler H. Tessmer U. Hohenberg H. Krausslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72(4):2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Benjamin J. Ganser-Pornillos BK. Tivol WF. Sundquist WI. Jensen GJ. Three- dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol. 2005;346(2):577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wyma DJ. Jiang J. Shi J, et al. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: A novel role of the gp41 cytoplasmic tail. J Virol. 2004;78(7):3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Paillart JC. Shehu-Xhilaga M. Marquet R. Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol. 2004;2(6):461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 226.Rein A. Henderson LE. Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem Sci. 1998;23(8):297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 227.Kaplan AH. Zack JA. Knigge M, et al. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberant virus assembly and the formation of non-infectious particles. J Virol. 1993;67:4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pettit SC. Henderson GJ. Schiffer CA. Swanstrom R. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol. 2002;76(20):10226–10233. doi: 10.1128/JVI.76.20.10226-10233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhou J. Yuan X. Dismuke D, et al. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol. 2004;78(2):922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li F. Goila-Gaur R. Salzwedel K, et al. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA. 2003;100(23):13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Adamson CS. Ablan SD. Boeras I, et al. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat) J Virol. 2006;80(22):10957–10971. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Keller PW. Adamson CS. Heymann JB. Freed EO. Steven AC. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol. 2011;85(4):1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Adamson CS. Sakalian M. Salzwedel K. Freed EO. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology. 2010;7:36. doi: 10.1186/1742-4690-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Adamson CS. Waki K. Ablan SD. Salzwedel K. Freed EO. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457) J Virol. 2009;83(10):4884–4894. doi: 10.1128/JVI.02659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li F. Zoumplis D. Matallana C, et al. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology. 2006;356(1–2):217–224. doi: 10.1016/j.virol.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 236.Zhou J. Chen CH. Aiken C. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J Virol. 2006;80(24):12095–12101. doi: 10.1128/JVI.01626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sakalian M. McMurtrey CP. Deeg FJ, et al. 3-o-(3′,3′-Dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 gag precursor assembled in vitro. J Virol. 2006;80(12):5716–5722. doi: 10.1128/JVI.02743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhou J. Huang L. Hachey DL. Chen CH. Aiken C. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J Biol Chem. 2005;280(51):42149–42155. doi: 10.1074/jbc.M508951200. [DOI] [PubMed] [Google Scholar]
- 239.Stoddart CA. Joshi P. Sloan B, et al. Potent activity of the HIV-1 maturation inhibitor bevirimat in SCID-hu Thy/Liv mice. PLoS ONE. 2007;2(11):e1251. doi: 10.1371/journal.pone.0001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Martin DE. Salzwedel K. Allaway GP. Bevirimat: A novel maturation inhibitor for the treatment of HIV-1 infection. Antivir Chem Chemother. 2008;19(3):107–113. doi: 10.1177/095632020801900301. [DOI] [PubMed] [Google Scholar]
- 241.Smith PF. Ogundele A. Forrest A, et al. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother. 2007;51(10):3574–3581. doi: 10.1128/AAC.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.McCallister S. Lalezari J. Richmond G, et al. HIV-1 Gag polymorphisms determine treatment response to bevirimat (PA-457) XVII International HIV Drug Resistance Workshop; Sitges, Spain: 2008. [Google Scholar]
- 243.Margot NA. Gibbs CS. Miller MD. Phenotyphic susceptibility to bevirimat among HIV-infected patient isolates without prior exposure to bevirimat; 16th Conference on Retroviruses and Opportunistic Infections; Montreal, Canada. 2009. [Google Scholar]
- 244.Van Baelen K. Salzwedel K. Rondelez E, et al. HIV-1 susceptibility to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag SP1. Antimicrob Agents Chemother. 2009;53:2185–2188. doi: 10.1128/AAC.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Seclen E. Gonzalez Mdel M. Corral A. de Mendoza C. Soriano V. Poveda E. High prevalence of natural polymorphisms in Gag (CA-SP1) associated with reduced response to bevirimat, an HIV-1 maturation inhibitor. AIDS. 2010;24(3):467–469. doi: 10.1097/QAD.0b013e328335ce07. [DOI] [PubMed] [Google Scholar]
- 246.Blair WS. Cao J. Fok-Seang J, et al. New small-molecule inhibitor class targeting human immunodeficiency virus type 1 virion maturation. Antimicrob Agents Chemother. 2009;53(12):5080–5087. doi: 10.1128/AAC.00759-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Checkley MA. Luttge BG. Soheilian F. Nagashima K. Freed EO. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology. 2010;400(1):137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lee SK. Harris J. Swanstrom R. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol. 2009;83(17):8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Muller B. Anders M. Akiyama H, et al. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem. 2009;284(43):29692–29703. doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Brun S. Solignat M. Gay B, et al. VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology. 2008;5:57. doi: 10.1186/1742-4690-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Fitzon T. Leschonsky B. Bieler K, et al. Proline residues in the HIV-1 NH2-terminal capsid domain: Structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000;268(2):294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- 252.Forshey BM. von Schwedler U. Sundquist WI. Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76(11):5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lilly F. Fv-2: Identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J Natl Cancer Inst. 1970;45(1):163–169. [PubMed] [Google Scholar]
- 254.Best S. Le Tissier P. Towers G. Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382(6594):826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 255.Kozak CA. Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225(2):300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 256.Stremlau M. Owens CM. Perron MJ. Kiessling M. Autissier P. Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 257.Nisole S. Lynch C. Stoye JP. Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101(36):13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sayah DM. Sokolskaja E. Berthoux L. Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 259.Berthoux L. Sebastian S. Sokolskaja E. Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci USA. 2005;102(41):14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Li X. Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82(23):11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kar AK. Diaz-Griffero F. Li Y. Li X. Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82(23):11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Langelier CR. Sandrin V. Eckert DM, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82(23):11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li X. Li Y. Stremlau M, et al. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains. J Virol. 2006;80(13):6198–6206. doi: 10.1128/JVI.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Perez-Caballero D. Hatziioannou T. Yang A. Cowan S. Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79(14):8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Stremlau M. Perron M. Lee M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Stremlau M. Perron M. Welikala S. Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Yap MW. Nisole S. Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 268.Hatziioannou T. Perez-Caballero D. Yang A. Cowan S. Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci USA. 2004;101(29):10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Black LR. Aiken C. TRIM5{alpha} disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol. 2010;84(13):6564–7569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Perron MJ. Stremlau M. Lee M. Javanbakht H. Song B. Sodroski J. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81(5):2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Shi J. Aiken C. Saturation of TRIM5 alpha-mediated restriction of HIV-1 infection depends on the stability of the incoming viral capsid. Virology. 2006;350(2):493–500. doi: 10.1016/j.virol.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 272.Anderson JL. Campbell EM. Wu X. Vandegraaff N. Engelman A. Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80(19):9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ganser-Pornillos BK. Chandrasekaran V. Pornillos O. Sodroski JG. Sundquist WI. Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108(2):534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Anderson J. Akkina R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther. 2008;19(3):217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- 275.Yamashita M. Perez O. Hope TJ. Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3(10):1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lee K. Ambrose Z. Martin TD, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7(3):221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Shah VB. Aiken C. HIV nuclear entry: Clearing the fog. Viruses. 2010;2(5):1190–1194. doi: 10.3390/v2051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Turner BG. Summers MF. Structural biology of HIV. J Mol Biol. 1999;285(1):1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 279.Fujii K. Hurley JH. Freed EO. Beyond Tsg101: The role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5(12):912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 280.Adamson CS. Freed EO. Recent progress in antiretrovirals—lessons from resistance. Drug Discov Today. 2008;13(9–10):424–432. doi: 10.1016/j.drudis.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]