RDNA AND NUCLEOLAR CHROMATIN
rDNA Structure
The nucleus is a highly organized structure. However, unlike cytoplasmic organelles, nuclear substructures are not bounded by membranes, but rather are held together by interactions between their component proteins and nucleic acids, and are thus probably best regarded as giant, extended multimolecular complexes. The nucleolus is the most prominent of these structures. It is the site of transcription by RNA polymerase I (Pol I) of the rDNA, tandemly repeated copies of the genes for three of the four ribosomal RNAs (rRNAs: s-rRNA, 5.8S-rRNA, and l-rRNA), which are called the nucleolar organizer regions (NORs) of the chromosomes (Raska et al., 2006a, 2006b). The rDNA repeats are transcribed as a single precursor RNA, which is edited into the three rRNAs by excising leading, internal, and trailing transcribed spacer sequences. The rDNA repeats are separated by untranscribed intergenic sequences, which are much shorter in plants (2–3 kb) than in mammals and many other vertebrates (10–30 kb; Hadjiolov, 1985). In many cases the size of the intergenic spacers is heterogeneous, as, for example, in Xenopus laevis (3–9 kb). The fourth rRNA, 5S, is transcribed by RNA Pol III from tandem repeats elsewhere in the nucleus (Highett et al., 1993). The nucleolus is also the location at which most of the steps of ribosome assembly and maturation occur. This means about 90 ribosomal proteins, as well as many processing and accessory factors, need to be imported from the cytoplasm to the nucleolus, and the nearly complete small and large ribosomal subunits separately exported to the cytoplasm. Since a rapidly growing cell may require millions of ribosomes to be synthesized, the nucleolus is by far the major destination and origin of nucleocytoplasmic transport in the most active cells.
Nucleolar Organization
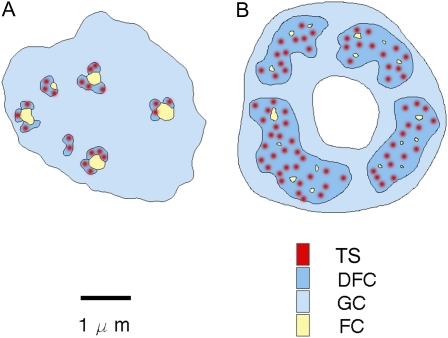
When nuclei are stained with fluorescent DNA dyes such as 4′,6-diamidino-2-phenylindole, the nucleolus is seen as a dark region within the more brightly stained nuclear chromatin. This is an indication that the active rDNA is highly dispersed within the nucleolus. On the basis of their appearance in electron microscope (EM) images, the nucleoli in many animal cells have been described in terms of a tripartite structure, with small, lightly staining regions called fibrillar centers (FCs), surrounded by densely stained material called the dense fibrillar component (DFC); the remainder of the nucleolus contains what appear to be densely packed granules (the granular component [GC]; Shaw et al., 1995; Fig. 1). Originally these granules were assumed to be entirely preribosomal particles in various stages of processing, but recent studies have indicated other types of complexes may occupy specific parts of the GC, which may be segregated into distinct regions of different compositions (Politz et al., 2005). What appear to be FCs are often seen in plants, but the regions assumed to be related to the DFC are much more extensive in plant nucleoli and often no more densely stained than the surrounding granules (Shaw and Jordan, 1995; Fig. 1). In reality the organization of the nucleolus is likely to be more complex than previously thought. A recent article has shown that within the chordates there are species that display a bipartite organization, in essence lacking FCs (Thiry et al., 2011). These authors suggest that the FCs originated with the emergence of the amniotes. A problem with this hypothesis is that some anamniote nucleoli, such as in Xenopus, do contain FCs, as indeed do plants (Raska et al., 2006b). The controversies about nucleolar organization will probably not be fully resolved until we have a better understanding of what these electron microscopic structures represent in molecular and functional terms.
Figure 1.
Comparison of the structure of animal (A) and plant (B) nucleolar organization. TS, Transcription sites. In plants (B) a central region is often present and is called the nucleolar cavity.
The molecular interpretation of these structures has been the subject of a long controversy. Immunogold EM studies showed concentrations of RNA Pol I in the FCs, which were consequently suggested to be the sites of transcription (Scheer and Rose, 1984). In situ labeling, whether at the optical or EM level, showed most rDNA in condensed heterochromatin at the nucleolar periphery, and smaller foci of rDNA labeling corresponding to FCs (Shaw et al., 1995). With more sensitive in situ techniques, fainter, diffuse rDNA labeling was subsequently seen throughout the DFC. The problem with a simple interpretation of these labeling experiments is that most Pol I, and most rDNA copies are not transcriptionally active. It was not until fluorescence detection of bromo-uridine labeling was introduced that the sites of transcription could be clearly labeled. In plant cells, this showed the transcription sites to consist of many foci within the DFC, with the smallest foci representing individual rDNA gene repeats (Thompson et al., 1997; González-Melendi et al., 2001; Koberna et al., 2002). Additional labeling with probes to transcribed spacer regions in the prerRNA and with various proteins and other small RNAs known to be involved in ribosome maturation led to a radial model, where the newly formed transcripts moved away from the dispersed genes within the DFC and then subsequently moving into the GC for later processing stages (Brown and Shaw, 1998).
Nucleolar Chromatin and Epigenetics
The number of rDNA repeats varies greatly among eukaryotes; many plants have thousands of copies, of which only a small proportion seems to be transcriptionally active at any one time. There is evidence that some repeats in human rDNA are inverted and may not be functional (Caburet et al., 2005). In rice (Oryza sativa), fiber fluorescence in situ hybridization has suggested that the repeats are regular and do not contain inversions or rearrangements (Mizuno et al., 2008). Furthermore, physical mapping studies in Arabidopsis (Arabidopsis thaliana) using pulsed field gel electrophoresis showed that restriction endonucleases that cut once per rDNA repeat gave only a single length of fragment (Copenhaver and Pikaard, 1996). Nevertheless the rDNA has not been fully sequenced in any higher eukaryote, as with current technology it is very difficult to analyze such highly repetitive sequences. We therefore do not know absolutely whether all the rDNA repeats are intact functional genes or whether other functional sequences are interspersed with them, and it is merely an assumption that all rDNA repeats are potentially transcribable. As with other genomic regions, current models suggest that rDNA chromatin can adopt three different states: inactive and condensed (heterochromatin), which corresponds to the condensed chromatin at the nucleolar periphery as well as to some intranucleolar condensed regions; active genes in an extended conformation within the DFC; and a poised or potentiated state, available for transcription but not currently being transcribed (McKeown and Shaw, 2009). This latter state may correspond to rDNA within the FCs. The balance between these states, and thus Pol I loading and transcription, is determined by DNA methylation, differences in the histone variants associated with the DNA, remodeling of the DNA, particularly the promoter regions, and the presence of histone modifications (Grummt and Pikaard, 2003). Loading of Pol I and some other factors is maintained through mitosis, and thus the chromatin state can be epigenetically inherited.
A particularly clear example of epigenetic control of rDNA is provided by nucleolar dominance in hybrids where the NORs of one parental genome are active, whereas those of the other are inactive (Tucker et al., 2010). An RNAi approach has been used with Arabidopsis suecica, a hybrid of Arabidopsis and Arabidopsis arenosa, to determine factors involved in nucleolar dominance. This has shown the involvement of the histone deacetylases Histone Deacetylase1 and Histone Deacetylase6, the de novo DNA methyltransferase Domains Rearranged Methyltransferase2 (DRM2), and the methylcytosine binding domain proteins Methyl CpG Binding Domain6 (MBD6) and MBD10 (Preuss et al., 2008). Further confirmation that nucleolar dominance in plants involves RNA-directed DNA methylation was obtained by knockdown of RNA Dependent RNA Polymerase2, Dicer-like3, and DRM2, which disrupted the silencing of the Arabidopsis-derived rRNA genes in A. suecica (Preuss et al., 2008). In mammals, rRNA genes are silenced by the nucleolar remodeling complex, NoRC, which is recruited to rRNA genes by 200- to 300-nt RNA species, termed pRNA, derived from intergenic regions of the rDNA (Guetg et al., 2010; Santoro et al., 2010).
PLURIFUNCTIONAL NUCLEOLUS
Over the last 10 to 15 years, it has become clear that the nucleolus is involved in numerous other functions than ribosome biogenesis (Pederson, 1998; Rubbi and Milner, 2003; Olson, 2004; Raska et al., 2006a; Boisvert et al., 2007). Many are RNA-related functions such as RNA processing and assembly of ribonucleoproteins (RNPs). For example, the nucleolus (and associated bodies, particularly Cajal bodies [CBs]) is involved in the maturation, assembly, and export of RNP particles such as the signal recognition particle, telomerase RNP, and processing of precursor transfer RNAs and U6 small nuclear RNA. In addition, the nucleolus has roles in cellular functions such as regulation of the cell cycle, stress responses, telomerase activity, and ageing (Pederson, 1998; Tsai and McKay, 2002; Rubbi and Milner, 2003; Olson, 2004; Raska et al., 2006a; Boisvert et al., 2007; Boulon et al., 2010). Sequestration of specific proteins in the nucleolus or their release is one mechanism by which processes such as the cell cycle or cell death are regulated. The multifunctional nature of the nucleolus is therefore reflected in the complexity of the protein and RNA composition of the nucleolus and in the dynamic composition changes in response to cellular conditions.
Protein Composition of the Nucleolus
Initial analyses of the proteome of human cells identified around 450 proteins including ribosomal proteins and proteins known to be involved in ribosome biogenesis (fibrillarin, nucleolin, B23, etc.; Andersen et al., 2002; Scherl et al., 2002). Even at this stage, unexpected proteins like splicing factors, spliceosomal proteins, and translation factors were identified. The increasing resolution of mass spectrometry techniques had led to the current characterization of the nucleolar proteome from human cells of circa 4,500 proteins (Ahmad et al., 2009). Quantitative proteomic analyses have demonstrated the dynamic behavior of nucleolar proteins such as ribosomal proteins and of protein complexes such as Pol I (Andersen et al., 2005; Lam et al., 2007). For example, quantitative changes in the relative levels of nucleolar components (reflecting accumulation or loss) were apparent upon transcriptional inhibition. The degree of change varied greatly for different proteins and showed that ribosomal proteins were highly expressed and either incorporated into ribosomal subunits or rapidly degraded (Andersen et al., 2005; Lam et al., 2007). It is also intriguing that ribosomal protein complexes associate with chromosomes and in particular transcription start sites of tRNAs, although the nature of these complexes and their function are as yet unknown (De et al., 2011).
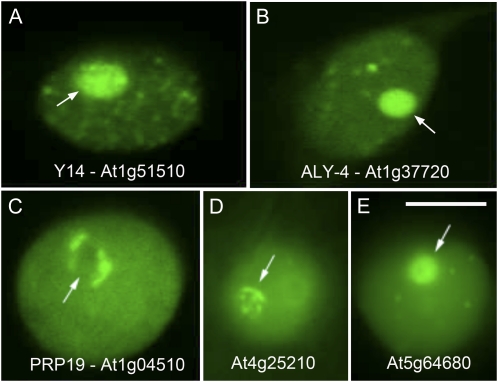
In contrast, characterization of the proteome of plant nucleoli lags significantly behind. In an initial proteomic analysis of purified Arabidopsis nucleoli 217 proteins were identified. In addition to the expected ribosomal and nucleolar proteins, a range of nonribosomal and nonnucleolar proteins including plant-specific proteins, proteins with unknown function, and splicing and translation factors was observed (Pendle et al., 2005). In particular, exon junction complex proteins (known in animal systems to associate with mRNAs after splicing) were identified and their nucleolar localization was confirmed by confocal microscopy of GFP fusions (Pendle et al., 2005; Fig. 2). One of the core exon junction complex components, eIF4A-III, was shown to redistribute from the nucleoplasm to the nucleolus and finally to splicing speckles under hypoxia stress conditions (Koroleva et al., 2009). Although the inference is that the relocalization of eIF4A-III might cause redistribution of mRNAs to the nucleolus, the exon junction complex has not yet been formally identified in plants and its association with mRNAs when in the nucleolus has not been demonstrated. A similar redistribution of an SR protein splicing factor to the nucleolus under ATP depletion has also been demonstrated (Tillemans et al., 2006). To date, this SR protein, RSZ22, is the only SR protein to show such a relocalization and again whether it relocalizes mRNAs is not known. A number of small nuclear RNP proteins were identified in the nucleolar proteome (Pendle et al., 2005) and CBs/nucleolus may be involved in production of the spliceosomal U1snRNP in plants (Lorković and Barta, 2008). Our knowledge of the nucleolar proteome in plants is still relatively limited and given the multifunctionality of this nuclear compartment, a full analysis is overdue and would be likely to provide supporting evidence for different functions or even identify new functions.
Figure 2.
Examples of transient expression of GFP fusions in suspension culture cells to proteins identified as nucleolar components. A to C, RNA binding proteins with known animal homologs. D and E, Uncharacterized, plant-specific proteins. The nucleolus is indicated in each section by an arrow. Bar = 5 μm.
RNA Complexity in the Nucleolus
The multiple functions of the nucleolus in processing of various RNAs and assembly of different RNPs are reflected in the different species of RNA. Besides rRNAs, animal cell nucleoli contain the extensive families of small nucleolar RNAs (snoRNAs) as well as snRNAs, tRNAs, 7SL RNA (signal recognition pathways), and telomerase RNA. In plants, analyses of nucleolar RNAs, excluding rRNAs, suggest that the plant nucleolus is also involved in many RNA/RNP events. Moreover, the presence of mRNAs and small-regulatory RNAs in the plant nucleolus and nucleolar-associated bodies is intriguing in terms of potentially novel functions in plants, broadening the roles of the nucleolus still further.
Cloning and sequencing of RNAs from purified Arabidopsis nucleoli identified tRNAs, snRNAs, and small cajal body-specific RNAs along with many known and novel snoRNAs (Kim et al., 2010). In particular, in addition to the expected conserved snoRNAs (U3 and MRP), orthologs of human U13 snoRNAs were discovered. The function of U13 has not been determined but it contains complementarity to the 3′ end of s-rRNA. All eukaryotes contain orphan snoRNAs, which do not have complementarity to rRNAs or snRNAs. In animals, such snoRNAs have been found to be involved in RNA editing, alternative splicing, and regulation of gene expression (Vitali et al., 2005; Kishore and Stamm, 2006; Ender et al., 2008; Saraiya and Wang, 2008; Ono et al., 2010). Recently, three mammalian snoRNAs (all encoded in the introns of a ribosomal protein gene) were shown to be involved in regulation of metabolic stress response (Michel et al., 2011). Although plants have a number of orphan snoRNAs, to date no noncanonical function in, for example, modulating gene expression has been described.
The identification of mRNA-associated proteins in the plant nucleolus suggests a function in mRNA biogenesis. In animals, only a very few mRNAs have ever been identified in the nucleolus (Pederson, 1998; Olson, 2004). In contrast, mRNAs from a wide range of genes were identified in a cDNA library from purified Arabidopsis nucleoli. Of particular interest was that aberrantly spliced mRNAs were enriched in the nucleolus and the vast majority contained premature termination codons, and therefore were likely to be turned over by the nonsense-mediated decay (NMD) pathway (Kim et al., 2009). Further, the localization of proteins involved in NMD—UPF2 and UPF3—to the nucleolus suggested a novel function for the plant nucleolus in mRNA surveillance/NMD and thereby in mRNA biogenesis (Kim et al., 2009). However, we have recently shown that mRNA transcripts with retained introns (or containing unspliced introns) are not turned over by NMD and appear to avoid the NMD pathway (Kalyna et al., 2011). While other aberrant transcripts are targeted to the nucleolus as part of the NMD pathway, intron-containing transcripts may accumulate in the nucleolus for a different function (e.g. degradation by a different pathway) or may be targeted there instead of being exported. In this regard, an association between the nucleolus and export of particular human virus RNAs is well established (Hiscox, 2007; Hiscox et al., 2010) and has been suggested to function for cellular mRNAs. In yeast (Saccharomyces cerevisiae) and animal cells the question of whether specific mRNAs/mRNPs have a nucleolar phase is still open (discussed in Jellbauer and Jansen, 2008).
Besides the novel finding of mRNAs and aberrant mRNAs in the plant nucleolus, heterochromatic small interfering RNAs, which are involved in transcriptional silencing, are produced in a region of the nucleolus or in bodies often found associated with the nucleolus called d-bodies (Pontes and Pikaard, 2008). The localization of precursor microRNAs (miRNAs) and Dicer-like1 to d-bodies also suggests a role for the nucleolus in the maturation of miRNAs (Pontes and Pikaard, 2008). Some precursor and mature miRNAs are enriched in the nucleolus of mammalian cells possibly for modification, assembly, or to regulate snoRNA activity (Politz et al., 2009; Scott et al., 2009). There is also an evolutionary relationship between miRNA precursors and snoRNAs, with some miRNAs being processed from snoRNA precursors and some miRNA precursors retaining snoRNA features (Saraiya and Wang, 2008; Politz et al., 2009; Scott et al., 2009; Ono et al., 2011). In addition, snoRNA-derived small RNAs were found to be associated with Argonaute proteins of RNA silencing pathways in both animals and Arabidopsis (Taft et al., 2009) and small RNAs from a human snoRNA reduced expression of gene targets (Ender et al., 2008). The complexity of RNA/RNP processes involving the nucleolus suggests that it is a center of RNA activity. For plants, the presence of mRNAs and small regulatory RNAs in the nucleolus allows us to speculate that the nucleolus is involved in regulation of expression, possibly in response to cellular conditions.
The Nucleolus and Virus Infection
From the integral nature of the nucleolus to many RNA processing and RNP assembly pathways, it is not surprising that many animal and plant viruses exploit the nucleolus in production and transport of viral RNPs. The involvement of the nucleolus in infection cycles of animal and human viruses is well established (Hiscox, 2007; Hiscox et al., 2010; Taliansky et al., 2011). In plants, a growing number of viruses show some interaction with the nucleolus (and other nuclear bodies such as CBs) and roles for the nucleolus and nucleolar proteins are now emerging (Taliansky et al., 2011). For example, plant viruses can recruit nucleolar proteins for assembly of viral RNP particles, virus replication and movement, and to counteract host-viral defense systems. In the best-studied system to date, the ORF3 long-distance movement protein of Groundnut rosette virus trafficks to the nucleolus via CBs, causing reorganization of CBs into multiple CB-like structures that fuse with the nucleolus. ORF3 then recruits fibrillarin (an abundant nucleolar RNA binding protein, known to be an RNA methylase) for assembly of cytoplasmic infectious viral particles (Kim et al., 2007a, 2007b; Canetta et al., 2008). The nucleolar localization of ORF3 is essential for systemic infection. Viral proteins from other plant viruses also target the nucleolus and the function of this localization and their interactions with nucleolar proteins are being established. For example, the coat protein and coat protein read-through proteins of Potato leaf roll virus (PLRV) are targeted to the nucleolus (Haupt et al., 2005) and systemic infection of PLRV is inhibited in fibrillarin-silenced plants, suggesting that fibrillarin is also involved in long-distance movement of PLRV.
Another aspect of nuclear/nucleolar targeting of viral proteins is to interfere with host defenses. Most plant viruses encode silencing suppressor proteins to counteract the silencing triggered by infection. The NIa/VPg protein of Potato virus A has suppressor activity dependent on the localization of VPg to the nucleolus and CBs, raising the question of whether this protein targets components or pathways involved in RNA silencing that are found in the nucleus or nucleolus and associated bodies (Pontes and Pikaard, 2008). The VPg domain of NIa interacts with fibrillarin in the nucleolus and CBs, and depletion of fibrillarin reduces accumulation of the virus, suggesting that fibrillarin is involved in the infection process (Rajamäki and Valkonen, 2009). The silencing suppressor protein of Cucumber mosaic virus, CMV 2b, also localizes to the nucleus and nucleolus where it interacts with Argonaute1 and Argonaute4 (González et al., 2010). However, these interactions are not sufficient for suppression of RNA silencing and hence their biological relevance remains so far unclear (González et al., 2010). Other viral proteins are also found in the nucleus/nucleolus (e.g. the P3 protein of Tobacco etch virus and the P6 protein of Cauliflower mosaic virus) but the function of the nucleolar localization is unknown (Taliansky et al., 2011). How different viral proteins target CBs and the nucleolus, their interactions with host proteins like fibrillarin, and the impact of usurping normal functions on nucleolar structure and function are important questions for the future that are likely to provide insights into nucleolar biology.
DYNAMICS OF THE NUCLEOLUS
The nucleolus, like the other parts of the nucleus, is dynamic at a number of levels. First, it breaks down and reforms during the cell cycle; second, the nucleolus structure itself is dynamic, changing shape, size, and position within the nucleus; third, the nucleolar constituents undergo exchange with pools both within the nucleolus and outside in the nucleoplasm and cytoplasm.
Cell Cycle Dynamics
The nucleolus disassembles at the end of G2 as most transcription ceases and the nuclear envelope breaks down, and then reassembles with the onset of rDNA transcription at the beginning of the following G1 (Hernandez-Verdun, 2011). During disassembly, the GC components are lost first, followed by the DFC components. Certain proteins, such as Pol I subunits and upstream binding factor, which modulates DNA conformation, remain with the rDNA arrays; the presence of upstream binding factor alone is sufficient to produce a secondary constriction in the mitotic chromosome (Prieto and McStay, 2008). Some nucleolar components diffuse throughout the mitotic cytoplasm, whereas others, such as the protein B23, associate with the periphery of the mitotic chromosomes as chromosomal passengers. When rDNA transcription is halted during mitosis, unprocessed pre-rRNA transcripts persist through the mitotic cell, demonstrating that prerRNA transcript processing is also halted. The nucleolus reforms at the end of mitosis. First, small round bodies, called prenucleolar bodies are formed (Hernandez-Verdun, 2011). When transcription of the rDNA is reinitiated, the prenucleolar bodies disappear as new nucleoli are formed. Where more than one active NOR is present in the nucleus, separate nucleoli generally initially form at each active NOR. In plants, these small nucleoli then often fuse together to a single nucleolus as interphase progresses (Shaw and Jordan, 1995).
Protein Mobility
For many years cell biologists visualized fixed cells and thus had a tendency to regard the structures seen as stationary and long lived. Live-cell imaging studies, however, have revealed a much more dynamic picture. Studies using fluorescence recovery after photobleaching of nucleolar and nuclear proteins fused to GFP have shown that virtually all nucleolar and nuclear proteins are in constant flux, exchanging between the nucleolus and cytoplasm. The mean nucleolar residence time of even well-characterized nucleolar proteins is only a few tens of seconds (Phair and Misteli, 2000; Chen and Huang, 2001; Olson and Dundr, 2005). The distinction between nuclear and nucleolar proteins is that nucleolar proteins spend a greater proportion of their time in the nucleolus. The structure and even the existence of the nucleolus as a discrete structure must depend on the rDNA nucleating a small subpopulation of proteins that then form a structure on which all the other proteins assemble and disassemble dynamically. The nucleolus (and other nuclear bodies such as CBs) thus represents a steady-state flux of proteins in rapid equilibrium with the surrounding nucleoplasm (Raska et al., 2006a).
CBs and Intranucleolar Bodies
A number of dynamic nuclear bodies are either associated with the nucleolus or contained within it or both (Mao et al., 2011). The most familiar are the CBs. These bodies were first identified more than 100 years ago by Ramon y Cajal in neuronal cells, were originally called accessory bodies, and were proposed to have a connection with the nucleolus (Gall, 2000). They were later rechristened coiled bodies because of their appearance in the EM, but have now been renamed in honor of their original discoverer. Live-cell imaging of CBs by GFP showed that CBs move, fuse, and split within the nucleus of both plant (Boudonck et al., 1999) and animal (Platani et al., 2002) cells, often migrating dramatically to the nucleolar periphery or even being contained within the nucleolus. The detailed function of CBs is still not well understood, but their role is in maturation of RNA complexes such as spliceososomal subcomplexes, small RNAs, and RNA complexes involved in silencing (Stanek and Neugebauer, 2006). CBs share many components with the nucleolus, particularly a number of small nucleolar RNAs, and RNA processing proteins such as fibrillarin, which methylates various RNA species, and dyskerin/Cbf5p, which isomerizes uridine to pseudouridine in RNAs.
A new intranucleolar body (INB) has recently been described, based on colocalization of about 20 well-characterized components (Hutten et al., 2011). The composition of INBs seems distinctly different from CBs; in particular they contain small ubiquitin-like modifier, a peptide modifier, probably conjugated to other substrate proteins. The INBs seem to have an involvement in DNA damage response, since treatments that caused DNA damage induced INBs. The INBs are completely enveloped in the nucleolus, and may be contained in or overlap with a central region of the nucleolus often called the nucleolar cavity, a structure that is particularly prominent in many plant cells and that has been shown to swell and contract dynamically.
THE NUCLEOLUS, STRESS, AND DNA DAMAGE SENSING
There are several lines of evidence suggesting that the nucleolus has a role in sensing and responding to stresses (Boulon et al., 2010). In mammals there is considerable evidence linking the P53 DNA damage-sensing pathway to the nucleolus. P53 is normally held at a low level by MDM2, an E3 ubiquitin ligase that ubiquitinates P53 and targets it for degradation. MDM2 can be inhibited by ARF, which is normally sequestered in the nucleolus. In one model, release of ARF from the nucleolus would then allow it to inhibit MDM2, with p53 levels consequently rising (Olson, 2004; Raska et al., 2006a). In an elegant series of experiments it was shown that the P53 pathway is induced by many different treatments that targeted the nucleolus (Rubbi and Milner, 2003), suggesting that the nucleolar structure itself is a direct sensor of DNA damage. In this respect the induction of INBs by DNA damage as described above is very interesting and may contribute to the sensing mechanism.
rDNA has been associated with stress response to DNA damage and with aging, the pioneering studies of this going back several decades (Johnson and Strehler, 1972). rDNA copies are multiplied by repeated recombination, and their homogeneity is maintained by gene conversion events between the tandem repeats (Kobayashi, 2008). Recombination is induced by Fob1, which causes double-strand break formation. In yeast, at least, the histone deacetylase Sir2p also has a role in rDNA copy number regulation; in a sir2 mutant copy number fluctuated wildly, whereas in a fob1 mutant rDNA repeat fluctuation was reduced or eliminated (Kobayashi, 2008). In yeast, SIR1 and FOB1 affect cellular aging, sir1 mutants having a shorter lifespan and fob1 mutants a longer lifespan (Kobayashi, 2008). It was earlier shown that budding yeast cells accumulate extrachromosomal circles from the rDNA repeats preferentially in the mother cells as they age and that this accumulation in fact is a cause of aging (Sinclair and Guarente, 1997).
It is possible that there are cell types or developmental stages in some organisms that require many more rDNA copies than are normally transcribed, but even in the yeast only about half the (approximately 150) copies are transcribed, and in a number of organisms, including yeast, viable mutants have been made with only a fraction of the normal number of rDNA copies (Takeuchi et al., 2003). There is a broad correlation between genome size and number of rDNA copies (Prokopowich et al., 2003), and this has led to a hypothesis that the rDNA may be acting as a sensor for DNA damage, protecting the rest of the genome by inducing DNA repair mechanisms or apoptosis. The extra copies present in the rDNA repeats would initially presumably buffer such damage, ensuring that sufficient undamaged copies were available for ribosome biosynthesis (Kobayashi, 2008). rDNA chromatin also seems to be able to affect the stability of heterochromatic repeats in trans, as loss of the NoRC component TIP5 leads to instability of microsatellite repeats (Guetg et al., 2010) and reduction in the number of Drosophila rDNA repeats themselves leads to a general release of heterochromatin silencing throughout the nucleus (Paredes and Maggert, 2009). Similarly, loss of chromatin assembly factor 1 activity in Arabidopsis led to loss of telomeric and rDNA repeats in successive generations, as well as enhanced sensitivity to DNA damage (Mozgová et al., 2010). As rDNA repeats are the most common gene in the genome, these effects may occur due to disrupted balance between euchromatin and heterochromatin. However, at least in yeast, there is evidence that rDNA organization can also affect the wider genome by regulating the global distribution of condensin (Wang and Strunnikov, 2008).
CONCLUSION
After many years in which the nucleolus was believed to have a well-understood and limited function in ribosome biogenesis, many novel results in recent years have pointed to a wide range of biological activities being localized to this region of the nucleus. As a great variety of species from different kingdoms has been used for these studies, we cannot as yet tell whether all these various functions are carried out in the nucleoli of all species. For the most part, what unifies these activities is the involvement of RNA at some level, usually in the biogenesis or assembly of RNA/protein machinery. Thus the nucleolus may be better regarded as an RNA processing center, rather than as purely a ribosome factory. There is clearly much to be done to clearly define all the activities of the nucleolus, and further to explain why these activities need to be partitioned together within a defined nuclear domain. The answer may lie in efficiency—increasing the local concentrations of limiting factors by sequestering them to the nucleolus. Alternatively, the answer may lie in evolutionary history; the RNA biosynthetic activities may have colocalized for reasons such as the use of common factors in different pathways. This may have left the various processes inextricably linked.
References
- Ahmad Y, Boisvert F-M, Gregor P, Cobley A, Lamond AI. (2009) NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res (Database issue) 37: D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. (2005) Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Boisvert F-M, van Koningsbruggen S, Navascués J, Lamond AI. (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. (1999) The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell 10: 2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert F-M, Lamond AI. (2010) The nucleolus under stress. Mol Cell 40: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS, Shaw PJ. (1998) Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. (2005) Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res 15: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canetta E, Kim SH, Kalinina NO, Shaw J, Adya AK, Gillespie T, Brown JWS, Taliansky M. (2008) A plant virus movement protein forms ringlike complexes with the major nucleolar protein, fibrillarin, in vitro. J Mol Biol 376: 932–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang S. (2001) Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS. (1996) Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J 9: 273–282 [DOI] [PubMed] [Google Scholar]
- De S, Varsally W, Falciani F, Brogna S. (2011) Ribosomal proteins’ association with transcription sites peaks at tRNA genes in Schizosaccharomyces pombe. RNA 17: 1713–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. (2008) A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Gall JG. (2000) Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300 [DOI] [PubMed] [Google Scholar]
- González I, Martínez L, Rakitina DV, Lewsey MG, Atencio FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol Plant Microbe Interact 23: 294–303 [DOI] [PubMed] [Google Scholar]
- González-Melendi P, Wells B, Beven AF, Shaw PJ. (2001) Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J 27: 223–233 [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. (2010) The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J 29: 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov AA. (1985) The Nucleolus and Ribosome Biogenesis, Vol 12. Springer Verlag, Wien, NY, pp 40–41 [Google Scholar]
- Haupt S, Stroganova T, Ryabov E, Kim SH, Fraser G, Duncan G, Mayo MA, Barker H, Taliansky M. (2005) Nucleolar localization of potato leafroll virus capsid proteins. J Gen Virol 86: 2891–2896 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. (2011) Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2: 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highett MI, Beven AF, Shaw PJ. (1993) Localization of 5 S genes and transcripts in Pisum sativum nuclei. J Cell Sci 105: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Hiscox JA. (2007) RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA, Whitehouse A, Matthews DA. (2010) Nucleolar proteomics and viral infection. Proteomics 10: 4077–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Prescott A, James J, Riesenberg S, Boulon S, Lam YW, Lamond AI. (2011) An intranucleolar body associated with rDNA. Chromosoma 120: 481–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellbauer S, Jansen R-P. (2008) A putative function of the nucleolus in the assembly or maturation of specialized messenger ribonucleoprotein complexes. RNA Biol 5: 225–229 [DOI] [PubMed] [Google Scholar]
- Johnson R, Strehler BL. (1972) Loss of genes coding for ribosomal RNA in ageing brain cells. Nature 240: 412–414 [DOI] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Milne L, McNicol J, et al. (2011) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JWS. (2009) Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Macfarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JWS, Taliansky M. (2007a) Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad Sci USA 104: 11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JWS, Taliansky M. (2007b) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Spensley M, Choi SK, Calixto CPG, Pendle AF, Koroleva O, Shaw PJ, Brown JWS. (2010) Plant U13 orthologues and orphan snoRNAs identified by RNomics of RNA from Arabidopsis nucleoli. Nucleic Acids Res 38: 3054–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S. (2006) The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232 [DOI] [PubMed] [Google Scholar]
- Kobayashi T. (2008) A new role of the rDNA and nucleolus in the nucleus:rDNA instability maintains genome integrity. Bioessays 30: 267–272 [DOI] [PubMed] [Google Scholar]
- Koberna K, Malínský J, Pliss A, Masata M, Vecerova J, Fialová M, Bednár J, Raska I. (2002) Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J Cell Biol 157: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JW, Shaw PJ. (2009) Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Barta A. (2008) Role of Cajal bodies and nucleolus in the maturation of the U1 snRNP in Arabidopsis. PLoS ONE 3: e3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. (2011) Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown PC, Shaw PJ. (2009) Chromatin: linking structure and function in the nucleolus. Chromosoma 118: 11–23 [DOI] [PubMed] [Google Scholar]
- Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, Behlke MA, Ory DS, Schaffer JE. (2011) Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab 14: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Sasaki T, Matsumoto T. (2008) Characterization of internal structure of the nucleolar organizing region in rice (Oryza sativa L.). Cytogenet Genome Res 121: 282–285 [DOI] [PubMed] [Google Scholar]
- Mozgová I, Mokros P, Fajkus J. (2010) Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO. (2004) Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004: pe10. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M. (2005) The moving parts of the nucleolus. Histochem Cell Biol 123: 203–216 [DOI] [PubMed] [Google Scholar]
- Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. (2011) Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res 39: 3879–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yamada K, Avolio F, Scott MS, van Koningsbruggen S, Barton GJ, Lamond AI. (2010) Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol Biol Cell 21: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S, Maggert KA. (2009) Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA 106: 17829–17834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res 26: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ. (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Lamond AI, Swedlow JR. (2002) Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol 4: 502–508 [DOI] [PubMed] [Google Scholar]
- Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T. (2005) A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell 16: 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Hogan EM, Pederson T. (2009) MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS. (2008) siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev 18: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, et al. (2008) Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. (2008) Pseudo-NORs: a novel model for studying nucleoli. Biochim Biophys Acta 1783: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Prokopowich CD, Gregory TR, Crease TJ. (2003) The correlation between rDNA copy number and genome size in eukaryotes. Genome 46: 48–50 [DOI] [PubMed] [Google Scholar]
- Rajamäki M-L, Valkonen JPT. (2009) Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21: 2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. (2006a) New insights into nucleolar architecture and activity. Int Rev Cytol 255: 177–235 [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. (2006b) Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334 [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Schmitz KM, Sandoval J, Grummt I. (2010) Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. (2008) snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog 4: e1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Rose KM. (1984) Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci USA 81: 1431–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. (2009) Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 5: e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Highett MI, Beven AF, Jordan EG. (1995) The nucleolar architecture of polymerase I transcription and processing. EMBO J 14: 2896–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. (1995) The nucleolus. Annu Rev Cell Dev Biol 11: 93–121 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. (2006) The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 115: 343–354 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. (2009) Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliansky ME, Brown JWS, Rajamaki ML, Valkonen JPT, Kalinina NO. (2011) Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv Virus Res 77: 119–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Lamaye F, Lafontaine DL. (2011) The nucleolus: when 2 became 3. Nucleus 2: 289–293 [DOI] [PubMed] [Google Scholar]
- Thompson WF, Beven AF, Wells B, Shaw PJ. (1997) Sites of rDNA transcription are widely dispersed through the nucleolus in Pisum sativum and can comprise single genes. Plant J 12: 571–581 [DOI] [PubMed] [Google Scholar]
- Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. (2006) Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18: 3218–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. (2002) A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 16: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S, Vitins A, Pikaard CS. (2010) Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol 22: 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. (2005) ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol 169: 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B-D, Strunnikov A. (2008) Transcriptional homogenization of rDNA repeats in the episome-based nucleolus induces genome-wide changes in the chromosomal distribution of condensin. Plasmid 59: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]