Abstract
Direct infusion electrospray ionization triple quadrupole precursor scanning for three oxidized fatty acyl anions revealed 86 mass spectral peaks representing polar membrane lipids in extracts from Arabidopsis (Arabidopsis thaliana) infected with Pseudomonas syringae pv tomato DC3000 expressing AvrRpt2 (PstAvr). Quadrupole time-of-flight and Fourier transform ion cyclotron resonance mass spectrometry provided evidence for the presence of membrane lipids containing one or more oxidized acyl chains. The membrane lipids included molecular species of phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, digalactosyldiacylglycerol, monogalactosyldiacylglycerol, and acylated monogalactosyldiacylglycerol. The oxidized chains were identified at the level of chemical formula and included C18H27O3 (abbreviated 18:4-O, to indicate four double bond equivalents and one oxygen beyond the carbonyl group), C18H29O3 (18:3-O), C18H31O3 (18:2-O), C18H29O4 (18:3-2O), C18H31O4 (18:2-2O), and C16H23O3 (16:4-O). Mass spectral signals from the polar oxidized lipid (ox-lipid) species were quantified in extracts of Arabidopsis leaves subjected to wounding, infection by PstAvr, infection by a virulent strain of P. syringae, and low temperature. Ox-lipids produced low amounts of mass spectral signal, 0.1% to 3.2% as much as obtained in typical direct infusion profiling of normal-chain membrane lipids of the same classes. Analysis of the oxidized membrane lipid species and normal-chain phosphatidic acids indicated that stress-induced ox-lipid composition differs from the basal ox-lipid composition. Additionally, different stresses result in the production of varied amounts, different timing, and different compositional patterns of stress-induced membrane lipids. These data form the basis for a working hypothesis that the stress-specific signatures of ox-lipids, like those of oxylipins, are indicative of their functions.
Biotic and abiotic stresses result in lipid oxidation, and there is strong evidence for the importance of oxidized free fatty acids, also known as oxylipins, in plant stress responses (Imbusch and Mueller, 2000; Vollenweider et al., 2000; Stintzi et al., 2001; Howe and Schilmiller, 2002; Stenzel et al., 2003; Thoma et al., 2003; Taki et al., 2005; Sattler et al., 2006; Thines et al., 2007; Chehab et al., 2008; Katsir et al., 2008; Mueller et al., 2008). Recent studies indicate that oxidized fatty acyl chains also occur in complex polar lipids and that plants produce complex oxidized lipids (ox-lipids) under stress conditions, including wounding (Buseman et al., 2006), bacterial infection (Andersson et al., 2006; Grun et al., 2007; Kourtchenko et al., 2007), fungal infection (Thoma et al., 2003), extended dark (Seltmann et al., 2010), aging (Xiao et al., 2010), and osmotic stress (Seltmann et al., 2010).
A number of plastid-derived, complex lipid molecular species that contain oxophytodienoic acid (OPDA) and dinor-oxophytodienoic acid (dnOPDA) have been characterized (Stelmach et al., 2001; Hisamatsu et al., 2003, 2005; Andersson et al., 2006; Buseman et al., 2006; Kourtchenko et al., 2007; Glauser et al., 2008; Maeda et al., 2008). Some of the characterized ox-lipid species have been shown to occur in thylakoid membranes (Böttcher and Weiler, 2007). In Arabidopsis (Arabidopsis thaliana), some of the OPDA- and dnOPDA-containing monogalactosyldiacylglycerols (MGDGs) contain two esterified oxidized fatty acid chains or, when the 6-position of the MGDG Gal ring is acylated, three oxidized chains. These lipid species with multiple isopentenone (OPDA or dnOPDA)-containing chains are sometimes called arabidopsides (Hisamatsu et al., 2003, 2005; Andersson et al., 2006). Characterized Gal-acylated MGDG (acMGDG) molecular species include OPDA/dnOPDA MGDG with OPDA on the Gal (arabidopside E) and a tri-OPDA MGDG species (arabidopside G; Andersson et al., 2006; Kourtchenko et al., 2007). OPDA also has been identified in phosphatidylglycerol (PG; Buseman et al., 2006). MGDG and digalactosyldiacylglycerol (DGDG) contain, in addition to OPDA and dnOPDA, 16- and 18-carbon ketols, both in combination with normal chains and with OPDA (Buseman et al., 2006). Membrane lipids also contain other oxidized acyl species, including phytoprostanes and hydroxy fatty acids (Imbusch and Mueller, 2000; Thoma et al., 2003; Grun et al., 2007).
In some studies of oxidized membrane lipids, the oxidized fatty acyl chains have been analyzed after releasing the chains from the membrane lipids (Thoma et al., 2003; Grun et al., 2007), while in other studies, the intact membrane lipid species have been measured directly (Stelmach et al., 2001; Andersson et al., 2006; Buseman et al., 2006; Böttcher and Weiler, 2007; Kourtchenko et al., 2007; Thiocone et al., 2008; Seltmann et al., 2010). Several mass spectrometry strategies have been utilized for intact lipid oxylipin-containing plant lipid profiling. Buseman et al. (2006) used precursor ion scanning by direct infusion electrospray ionization (ESI) triple quadrupole mass spectrometry (MS) to quantify multiple oxylipin-containing complex lipids. Liquid chromatography or liquid chromatography-mass spectrometry approaches have also been used (Stelmach et al., 2001; Andersson et al., 2006; Böttcher and Weiler, 007; Kourtchenko et al., 2007; Glauser et al., 2008; Thiocone et al., 2008; Seltmann et al., 2010). However, most analyses have been limited to fewer than 20 oxidized membrane lipid species.
In this work, we utilized a direct-infusion ESI triple quadrupole MS strategy to quantify a larger group of oxidized membrane lipids. By precursor scanning in negative mode, 86 peaks representing combinations of intact ion mass-to-charge ratio (m/z) and oxidized acyl fragment m/z were identified. The chemical formulas of the fatty acyl substituents of each peak were determined by accurate mass analysis. In so doing, oxidized Arabidopsis phosphatidylcholine (PC) and phosphatidylethanolamine (PE) molecular species, as well as PG, DGDG, MGDG, and acMGDG species, were characterized. We tested the hypothesis that different environmental cues trigger different changes in ox-lipid profiles of Arabidopsis by challenging wild-type Arabidopsis with mechanical wounding, infection with avirulent and virulent bacteria, and low temperature and by monitoring the oxidized membrane lipid changes. Changes in stress-associated phospholipase products were also monitored.
RESULTS
Triple Quadrupole MS Precursor Scanning for Lipids with 18-Carbon Oxidized Acyl Chains
With the goal of investigating the formation of ox-lipids in Arabidopsis leaves on a broad scale and in an expeditious manner, ionizable membrane lipids with at least one oxidized fatty acid of known chemical formula were identified using a semitargeted, direct infusion MS approach. In the polar lipid fraction of Arabidopsis, five 18-carbon oxidized fatty acids (first five entries in Table I) can be detected with three ESI triple quadrupole MS scans in negative mode. The five 18-carbon oxidized fatty acids include C18H27O3 (designated as 18:4-O; nomenclature indicates “acyl carbons:double bond equivalents beyond the acid carbonyl-number of oxygens in addition to the carbonyl group”), C18H29O3 (18:3-O), C18H31O3 (18:2-O), C18H29O4 (18:3-2O), and C18H31O4 (18:2-2O). 18:3-2O and 18:2-2O each undergo a water loss during collision-induced dissociation to produce 18:4-O and 18:3-O, respectively. The dehydration allows 18:3-2O and 18:2-2O to be detected by scans for 18:4-O and 18:3-O, respectively (Buseman et al., 2006; Maeda et al., 2008). Thus, scanning for precursors of 291.2 (Pre 291.2) detects precursors of 18:4-O and 18:3-2O, scanning for Pre 293.2 detects precursors of 18:3-O and 18:2-2O, and scanning for Pre 295.2 detects precursors of 18:2-O. Using a single scan to detect membrane lipids with two different acyl species reduces scan time. Also, importantly, using Pre 291.2 and Pre 293.2 to detect lipids containing oxidized acyl chains with m/z 309.2 (e.g. 18:3-2O) and 311.2 (e.g. 18:2-2O) provides increased specificity compared with scanning for Pre 309.2 and Pre 311.2, because anions of 18:3-2O and 18:2-2O share the same nominal m/z as normal-chain fatty acyl chains 20:1 and 20:0, respectively. 20:1 and 20:0 are not detected by scans for Pre 291.2 and Pre 293.2, because 20:1 and 20:0 do not undergo water losses.
Table I. Oxidized fatty acyl chains detected in extracts from leaves of Arabidopsis infected with PstAvr for 24 h.
Examples of possible structures and fragmentation of ox-lipids from each class are shown in Supplemental Figure S1.
| Triple Quadrupole MS Precursor Scan | Chemical Formula of Oxidized Fatty Acyl Anion | m/z of Anion | Abbreviation | Examples of Compounds Consistent with Detected Formula |
| Acyl formulas directly scanned | ||||
| Pre 291.20 | C18H27O3 | 291.1966 | 18:4-O | OPDA, keto 18:3 |
| Pre 293.21 | C18H29O3 | 293.2122 | 18:3-O | Hydroxy 18:3, keto fatty acid |
| Pre 295.23 | C18H31O3 | 295.2279 | 18:2-O | Hydroxy 18:2 |
| Pre 291.20 | C18H29O4 | 309.2071 | 18:3-2O | Ketol fatty acid, hydroperoxy 18:3, dihydroxy 18:3 |
| Pre 293.21 | C18H31O4 | 311.2228 | 18:2-2O | Hydroperoxy 18:2, dihydroxy 18:2 |
| Acyl formulas identified by scanning of the above anions or as a result of QTOF MS analysis | ||||
| – | C16H23O3 | 263.1653 | 16:4-O | dnOPDA |
| – | C16H25O3 | 265.1809 | 16:3-O | Hydroxy 16:3 |
| – | C16H25O4 | 281.1758 | 16:3-2O | Ketol fatty acid, hydroperoxy 16:3, dihydroxy 16:3 |
| – | C18H25O4 | 305.1758 | 18:5-2O | – |
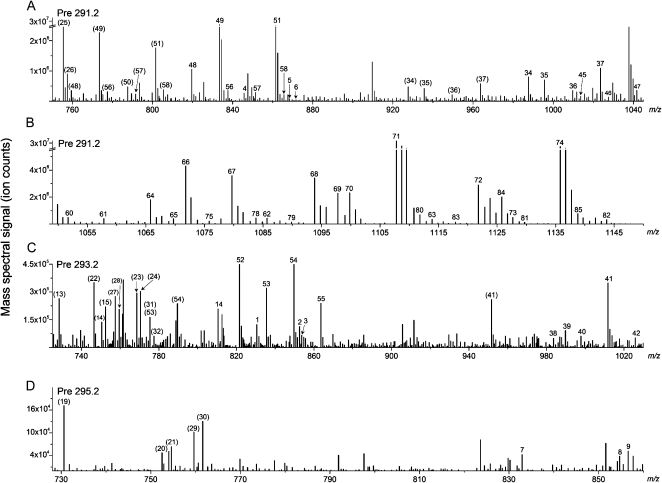
Scanning for negatively charged precursors of m/z 291.2, 293.2, and 295.2 while infusing an extract of Arabidopsis leaves infected with Pseudomonas syringae pv tomato DC3000 expressing the AvrRpt2 avirulence gene (PstAvr) reveals the spectral peaks shown in Figure 1. Scans were repeated on a series of similar samples, and peaks detected in any PstAvr-infected sample were numbered 1 through 86. With the samples dissolved in solvent containing ammonium acetate, for various lipids, peaks represent [M – H]– (indicated by peak numbers in parentheses in Fig. 1), [M + C2H3O2]–where C2H3O2 is acetate (indicated by peak numbers without parentheses), or both adducts.
Figure 1.
ESI triple quadrupole MS precursor ion spectra acquired on extracts of leaves of Arabidopsis plants infected with PstAvr for 24 h. A and B, Pre 291.2. C, Pre 293.2. D, Pre 295.2. Pre 291.2 scan detects ox-lipids containing 18:4-O and 18:3-2O, Pre 293.2 scan detects ox-lipids containing 18:3-O and 18:2-2O, and Pre 295.2 scan detects ox-lipids containing 18:2-O. Peaks with labels in parentheses are [M − H]− adducts. Peak labels with no parentheses indicate [M + C2H3O2]− adducts. Note that the intensity and m/z scales of the spectra differ. A and B have breaks in the intensity axes. Details of peaks numbered 1 to 86 are shown in Table II.
Identification of the Detected Oxidized Complex Lipids Using Quadrupole Time-of-Flight and Fourier Transform Ion Cyclotron Resonance MS
Identifying information for the observed peaks is presented in Table II and Supplemental Tables S1 and S2. Quadrupole time-of-flight (QTOF) MS aided in definition of the compounds. The extracts were batch fractionated by normal-phase chromatography. A fraction or the whole extract (as indicated in Supplemental Table S1) was directly infused into the ESI source, operating in negative mode, of a QTOF mass spectrometer. Each ox-lipid precursor ion, previously detected by precursor scanning by triple quadrupole MS (Fig. 1), was selected with the first quadrupole and subjected to collision-induced dissociation. The fragments were scanned with the time-of-flight analyzer to obtain accurate m/z ratios of the acyl anions; the m/z values were used to determine the chemical formulas of the acyl chains. Together, the precursor and fragment m/z values allowed the identification of lipid species indicated by nearly all of the peaks detected by precursor spectral scanning (Fig. 1; Table II; Supplemental Table S1). The identities of 24 of the 86 observed peaks were additionally confirmed at the level of intact ion chemical formula by the determination of accurate m/z values of precursor ions in positive mode by Fourier transform ion cyclotron resonance (FTICR) MS. The compounds are indicated in Table II, which summarizes the evidence for each identification, and the FTICR MS data are shown in Supplemental Table S2. To help the reader in visualizing the observed compounds, Supplemental Figure S1 shows examples of structures consistent with the data for some compounds. As described in the legend, the depicted structures are possibilities only.
Table II. Lipids detected by ESI MS/MS negative ion precursor ion scans, Pre 291.2, Pre 293.2, and Pre 295.2.
Supplemental Tables S3 and S4 indicate the compounds detected in a simplified form. M, Mass spectrometry abbreviation used for molecule, used to indicate the uncharged molecule.
| No.a | M Mass | M Formula | Identification | Detection Methodb | Identification; Evidencec |
| 1 | 771 | C42H78O9PN | 18:3-O/16:0 PC | Pre 293.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 2 | 793 | C44H76O9PN | 18:3-O/18:3 PC | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 3 | 795 | C44H78O9PN | 18:3-O/18:2 PC | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 4 | 787 | C42H78O10PN | 18:3-2O/16:0 PC | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 5 | 809 | C44H76O10PN | 18:3-2O/18:3 PC | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 6 | 811 | C44H78O10PN | 18:3-2O/18:2 PC | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 7 | 773 | C42H80O9PN | 18:2-O/16:0 PC | Pre 295.2, [M + C2H3O2]− | Supplemental Table S1 |
| 8 | 795 | C44H78O9PN | 18:2-O/18:3 PC | Pre 295.2, [M + C2H3O2]− | Supplemental Table S1 |
| 9 | 797 | C44H80O9PN | 18:2-O/18:2 PC | Pre 295.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 10 | 789 | C42H80O10PN | 18:2-2O/16:0 PC | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 11 | 811 | C44H78O10PN | 18:2-2O/18:3 PC | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 12 | 813 | C44H80O10PN | 18:2-2O/18:2 PC | Pre 293.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 13 | 729 | C39H72O9PN | 18:3-O/16:0 PE | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 14 | 751 | C41H70O9PN | 18:3-O/18:3 PE | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 15 | 753 | C41H72O9PN | 18:3-O/18:2 PE | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 16 | 745 | C39H72O10PN | 18:3-2O/16:0 PE | Pre 291.2, [M − H]− | Supplemental Table S1 |
| 17 | 767 | C41H70O10PN | 18:3-2O/18:3 PE | Pre 291.2, [M − H]− | Supplemental Table S1 |
| 18 | 769 | C41H72O10PN | 18:3-2O/18:2 PE | Pre 291.2, [M − H]− | Supplemental Table S1 |
| 19 | 731 | C39H74O9PN | 18:2-O/16:0 PE | Pre 295.2, [M − H]− | Supplemental Tables S1 and S2 |
| 20 | 753 | C41H72O9PN | 18:2-O/18:3 PE | Pre 295.2, [M − H]− | Supplemental Table S1 |
| 21 | 755 | C41H74O9PN | 18:2-O/18:2 PE | Pre 295.2, [M − H]− | Supplemental Table S1 |
| 22 | 747 | C39H74O10PN | 18:2-2O/16:0 PE | Pre 293.2, [M − H]− | Supplemental Tables S1 and S2 |
| 23 | 769 | C41H72O10PN | 18:2-2O/18:3 PE | Pre 293.2, [M − H]− | Tentative; Supplemental Table S1 |
| 24 | 771 | C41H74O10PN | 18:2-2O/18:2 PE | Pre 293.2, [M − H]− | Tentative; Supplemental Table S1 |
| 25 | 756 | C40H69O11P | 18:4-O/16:1 PG | Pre 291.2, [M − H]− | Buseman et al. (2006) |
| 26 | 758 | C40H71O11P | 18:4-O/16:0 PG | Pre 291.2, [M − H]− | Buseman et al. (2006) |
| 27 | 758 | C40H71O11P | 18:3-O/16:1 PG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 28 | 760 | C40H73O11P | 18:3-O/16:0 PG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 29 | 760 | C40H73O11P | 18:2-O/16:1 PG | Pre 295.2, [M − H]− | Supplemental Table S1 |
| 30 | 762 | C40H75O11P | 18:2-O/16:0 PG | Pre 295.2, [M − H]− | Supplemental Table S1 |
| 31* | 776 | C40H73O12P | 18:2-2O/16:1 PG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 31* | 776 | C43H68O12 | 18:3-O/16:4-O MGDG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 32 | 778 | C40H75O12P | 18:2-2O/16:0 PG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 33 | 922 | C49H78O16 | 18:4-O/16:3 DGDG | Pre 291.2, [M − H]− | Supplemental Table S1 |
| 34 | 928 | C49H84O16 | 18:4-O/16:0 DGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 35 | 936 | C49H76O17 | 18:4-O/16:4-O DGDG | Pre 291.2, [M + C2H3O2]− | Hisamatsu et al. (2005) |
| 36 | 950 | C51H82O16 | 18:4-O/18:3 DGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 37 | 964 | C51H80O17 | 18:4-O/18:4-O DGDG | Pre 291.2, [M + C2H3O2]− | Hisamatsu et al. (2005); Buseman et al. (2006) |
| 38 | 924 | C49H80O16 | 18:3-O/16:3 DGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 39 | 930 | C49H86O16 | 18:3-O/16:0 DGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 40 | 938 | C49H78O17 | 18:3-O/16:4-O DGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 41 | 952 | C51H84O16 | 18:3-O/18:3 DGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 42 | 966 | C51H82O17 | 18:3-O/18:4-O DGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 43 | 940 | C49H80O17 | 18:3-2O/16:3 DGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 44 | 946 | C49H86O17 | 18:3-2O/16:0 DGDG | Pre 291.2, [M − H]− | Supplemental Table S1 |
| 45′ | 954 | C49H78O18 | 18:3-2O/16:4-O DGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 45′ | 954 | C49H78O18 | 18:4-O/16:3-2O DGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 46 | 968 | C51H84O17 | 18:3-2O/18:3 DGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 47 | 982 | C51H82O18 | 18:3-2O/18:4-O DGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 48 | 760 | C43H68O11 | 18:4-O/16:3 MGDG | Pre 291.2, [M − H]− | Supplemental Table S2; Stelmach et al. (2001); Buseman et al. (2006) |
| 49 | 774 | C43H66O12 | 18:4-O/16:4-O MGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S2; Hisamatsu et al. (2003); Buseman et al. (2006) |
| 50 | 788 | C45H72O11 | 18:4-O/18:3 MGDG | Pre 291.2, [M − H]− | Buseman et al. (2006) |
| 51 | 802 | C45H70O12 | 18:4-O/18:4-O MGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S2; Hisamatsu et al. (2003); Buseman et al. (2006) |
| 52 | 762 | C43H70O11 | 18:3-O/16:3 MGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 53 | 776 | C43H68O12 | 18:3-O/16:4-O MGDG | Pre 293.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 54 | 790 | C45H74O11 | 18:3-O/18:3 MGDG | Pre 293.2, [M − H]− | Supplemental Table S1 |
| 55 | 804 | C45H72O12 | 18:3-O/18:4-O MGDG | Pre 293.2, [M + C2H3O2]− | Supplemental Table S1 |
| 56 | 778 | C43H70O12 | 18:3-2O/16:3 MGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 57 | 792 | C43H68O13 | 18:3-2O/16:4-O MGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 58 | 806 | C45H74O12 | 18:3-2O/18:3 MGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 59 | 820 | C45H72O13 | 18:3-2O/18:4-O MGDG | Pre 291.2, [M + C2H3O2]− | Buseman et al. (2006) |
| 60 | 992 | C59H92O12 | 18:4-O/16:3/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 61 | 998 | C59H98O12 | 18:4-O/16:3/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 62 | 1,026 | C61H102O12 | 18:4-O/18:3/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 63a | 1,054 | C63H106O12 | 18:4-O/18:3/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 63b | 1,054 | C63H106O12 | 18:4-O/18:2/18:1 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 64 | 1,006 | C59H90O13 | 18:4-O/16:4-O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 65 | 1,010 | C59H94O13 | 18:4-O/16:4-O/16:1 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 66 | 1,012 | C59H96O13 | 18:4-O/16:4-O/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 67a | 1,020 | C59H88O14 | 18:4-O/16:4-O/16:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 67b | 1,020 | C61H96O12 | 18:4-O/18:3/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 68a | 1,034 | C61H94O13 | 18:4-O/16:4-O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 68b | 1,034 | C61H94O13 | 18:4-O/18:4-O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 69a | 1,038 | C61H98O13 | 18:4-O/16:4-O/18:1 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 69b | 1,038 | C61H98O13 | 18:4-O/18:4-O/16:1 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 70a | 1,040 | C61H100O13 | 18:4-O/16:4-O/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 70b | 1,040 | C61H100O13 | 18:4-O/18:4-O/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 71a | 1,048 | C61H92O14 | 18:4-O/18:4-O/16:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2; Andersson et al. (2006) |
| 71b | 1,048 | C63H100O12 | 18:4-O/18:3/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 72a | 1,062 | C63H98O13 | 18:4-O/18:4-O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 72b | 1,062 | C61H90O15 | 18:4-O/18:5-2O/16:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 73 | 1,068 | C63H104O13 | 18:4-O/18:4-O/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 74 | 1,076 | C63H96O14 | 18:4-O/18:4-O/18:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2; Kourtchenko et al. (2007) |
| 75 | 1,016 | C59H100O13 | 18:3-2O/16:0/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 76a | 1,044 | C61H104O13 | 18:3-2O/18:3/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 76b | 1,044 | C61H104O13 | 18:3-2O/18:0/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 77a | 1,072 | C63H108O13 | 18:3-2O/18:3/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 77b | 1,072 | C61H100O15 | 18:3-2O/18:2/16:3-2O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 77c | 1,072 | C63H108O13 | 18:3-2O/18:2/18:1 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 78′ | 1,024 | C59H92O14 | 18:3-2O/16:4-O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 78′ | 1,024 | C59H92O14 | 18:4-O/16:3-2O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 79′ | 1,030 | C59H98O14 | 18:3-2O/16:4-O/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 79′ | 1,030 | C59H98O14 | 18:4-O/16:3-2O/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 80a′ | 1,052 | C61H96O14 | 18:3-2O/16:4-O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 80a′ | 1,052 | C61H96O14 | 18:4-O/16:3-2O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 80b | 1,052 | C61H96O14 | 18:3-2O/18:4-O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 81a | 1,070 | C61H98O15 | 18:3-2O/16:3-2O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 81b | 1,070 | C61H98O15 | 18:3-2O/18:3-2O/16:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 81c | 1,070 | C63H106O13 | 18:3-2O/18:2/18:2 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 82′ | 1,084 | C61H96O16 | 18:3-2O/16:3-2O/18:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 82′ | 1,084 | C61H96O16 | 18:3-2O/18:3-2O/16:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 83a | 1,058 | C61H102O14 | 18:3-2O/18:4-O/16:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 83b | 1,058 | C61H102O14 | 18:3-2O/16:4-O/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Tentative; Supplemental Table S1 |
| 84a | 1,066 | C61H94O15 | 18:3-2O/18:4-O/16:4-O acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 84b | 1,066 | C63H102O13 | 18:3-2O/18:3/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Tables S1 and S2 |
| 85 | 1,080 | C63H100O14 | 18:3-2O/18:4-O/18:3 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
| 86 | 1,086 | C63H106O14 | 18:3-2O/18:4-O/18:0 acMGDG | Pre 291.2, [M + C2H3O2]− | Supplemental Table S1 |
Each number (1–86) represents a peak observed in triple quadrupole MS spectra (Fig. 1). An asterisk indicates peaks resulting from two compounds of different lipid classes with the same ion mass. A prime symbol indicates peaks with at least two possible identifications, where it is unclear whether the peak represents one or both compounds. Numbers followed by a, b, or c signify that accurate m/z analysis indicates that the peak represents multiple lipid species detected by the stated precursor scan (Supplemental Tables S1 and S2).
Peaks were identified in triple quadrupole MS spectra with three negative precursor scans, Pre 291.2, Pre 293.2, and Pre 295.2; each species was observed as the [M − H]−/or [M + C2H3O2]− ion.
QTOF MS peak data are provided in Supplemental Table S1. FTICR MS peak data are provided in Supplemental Table S2. Peak identification is indicated as “tentative” if QTOF MS m/z values for one acyl group (or more) were greater than 10 ppm from the theoretical m/z and the compound was not previously identified or identified by accurate m/z of the intact compound in FTICR MS spectra. Previously identified peaks/compounds are marked with corresponding references.
Twelve oxidized PC (ox-PC; 1–12) and 12 oxidized PE (ox-PE; 13–24) species were identified by precursor scanning and confirmed by QTOF MS analysis (Table II). Each detected ox-PC and ox-PE molecular species has a normal-chain fatty acid, 16:0, 18:3, or 18:2, in combination with an oxidized chain, 18:3-O, 18:3-2O, 18:2-O, or 18:2-2O. The detected combinations are analogous to the most common Arabidopsis PC and PE species, which are 16:0/18:3, 16:0/18:2, 18:3/18:3, 18:3/18:2, and 18:2/18:2 (Devaiah et al., 2006), if 18:3 were substituted with 18:4-O (OPDA), 18:3-O, and 18:3-2O, and 18:2 were substituted with 18:2-O and 18:2-2O. Eight oxidized PG (ox-PG) species (25–32) were identified by triple quadrupole and QTOF MS (Table II). These species included a normal chain, 16:0 or 16:1, in combination with 18:4-O, 18:3-O, 18:2-O, or 18:2-2O. The 18:4-O ox-PG species were identified previously (Buseman et al., 2006). The acyl combinations found in ox-PG are again analogous to the most common PG species, 18:3/16:1, 18:2/16:1, 18:3/16:0, and 18:2/16:0 (Devaiah et al., 2006). No ox-PC, ox-PE, or ox-PG species with multiple oxidized fatty acid chains were detected.
Fifteen oxidized DGDG (ox-DGDG) species (Table II; 33–47) were identified by triple quadrupole MS precursor scanning. These include normal chains 16:3, 16:0, or 18:3, or oxidized chains 16:4-O or 18:4-O, in combination with oxidized chains 18:4-O, 18:3-O, or 18:3-2O. Five of the detected species were previously identified (Hisamatsu et al., 2005; Buseman et al., 2006). Again, the acyl combinations found in the oxidized molecular species were structurally related to the major molecular species of Arabidopsis DGDGs, 18:3/16:3, 18:3/16:0, and 18:3/18:3 (Devaiah et al., 2006). Twelve oxidized MGDG (ox-MGDG) species (48–59) were characterized (Table II). 16:3, 16:4-O, 18:3, or 18:4-O were found in combination with 18:4-O, 18:3-O, or 18:3-2O. The combinations observed also are analogous to the major molecular species of MGDG, 18:3/16:3 and 18:3/18:3 (Devaiah et al., 2006). The identities of the detected diacyl compounds (i.e. compounds 1–59) are summarized in brief form in Supplemental Table S3.
Twenty-seven oxidized acMGDG (ox-acMGDG) peaks (60–86) were identified by precursor scanning (Table II). Previous work has indicated that MGDG can be acylated at the 6-position on the Gal when plant leaves are wounded by grinding or stressed by bacterial infection (Heinz, 1967a; Heinz and Tulloch, 1969; Andersson et al., 2006). Our analysis did not determine the positions of the three chains (i.e. on the glycerol backbone or on the Gal). Besides the two previously identified acMGDGs, with combinations of 16:4-O and 18:4-O only, additional acMGDGs included various combinations of nonoxidized chains, 16:3, 16:1, 16:0, 18:3, 18:2, 18:1, 18:0, and oxidized chains, 16:4-O, 18:4-O, 16:3-2O, and 18:3-2O. The identities of the detected acMGDGs are summarized in brief form in Supplemental Table S4.
In addition to the species detected by precursor scanning, QTOF MS analysis detected two additional PCs (Supplemental Table S1; 87 and 88), two PEs (89 and 90), one PG (91), five DGDGs (92–96), two MGDGs (97 and 98), and 17 acMGDGs (99–115). These entries represent oxidized and nonoxidized species with the same nominal m/z values as peaks detected by triple quadrupole MS precursor scanning. However, 87 to 115 were not targeted by the precursor scans, because most do not contain the scanned precursor fragment. Thus, although they were noted as being present in pathogen infection, lipids 87 to 115 were not determined, or quantified, in the remaining analyses.
Most of the 86 peaks listed in Table II represented a single combination of a head group and acyl species, and their identifications were straightforward (Table II; Supplemental Table S1), but a few identifications require some explanation. The identifications of several peaks (45, 78, 79, 80a, and 82), indicated by a prime symbol in Table II, were ambiguous. The precursor m/z, in combination with the acyl chains observed, indicated that one fragment was dehydrated. Because the QTOF mass spectra did not detect the hydrated fragment, its identity is not clear. Thus, both possible acyl chain combinations are indicated for peaks with the prime symbol; the peaks detected by triple quadrupole MS scanning may represent one or both of the indicated species. Additionally, peak 31, representing an ox-PG species, occurred at the same nominal m/z as the [M – H]– ion of an ox-MGDG species. The same ox-MGDG species was detected separately in peak 53 as its [M + C2H3O2]– ion. Lastly, 13 spectral peaks (63, 67, 68, 69, 70, 71, 72, 76, 77, 80, 81, 83, and 84) observed by triple quadrupole MS scanning represented a combination of at least two members of the acMGDG class. These species are designated with the same number, but with a different letter, in Table II and Supplemental Tables S1 and S2. The multiple identifications arose from two situations: (1) ox-acMGDG species with the same chemical formula but multiple acyl combinations (e.g. 68a and 68b; 18:4-O/16:4-O/18:3 acMGDG and 18:4-O/18:4-O/16:3 acMGDG); and (2) ox-acMGDG species with different chemical formulas with the same nominal m/z (e.g. 84a and 84b; 18:3-2O/18:4-O/16:4-O acMGDG and 18:3-2O/18:3/18:3 acMGDG).
In this work, acyl components of the membrane lipids were identified at the level of chemical formula. Potential identities of the observed oxidized acyl anions are indicated in Table I, and possible structures for some detected compounds are shown in Supplemental Figure S1. Previous data indicated that in Arabidopsis, 18:4-O and 16:4-O in the complex lipids represent primarily OPDA and dnOPDA (Stelmach et al., 2001; Hisamatsu et al., 2003, 2005; Buseman et al., 2006). 18:3-O may be a keto fatty acid (Vollenweider et al., 2000) and/or a hydroxy fatty acid, as may 16:3-O. 18:2-O also may be a hydroxy fatty acid. 18:3-2O and 16:3-2O may be ketols (Hamberg, 1988; Weber et al., 1997), fatty acid hydroperoxides, and/or dihydroxy fatty acids (Hamberg et al., 2003). 18:2-2O may also represent a dihydroxy fatty acid or a fatty acid hydroperoxide. In future work, as links are established between specific oxidized membrane lipid species and physiological function via quantitative analyses of intact membrane lipid molecular species, the fatty acyl structures associated with the identified chemical formulas of the physiologically relevant membrane molecular species will be determined. The experiment described next is a start toward establishing functional links.
Experimental Design and Measurements of Oxidized Complex Leaf Membrane Lipids during Stress Responses
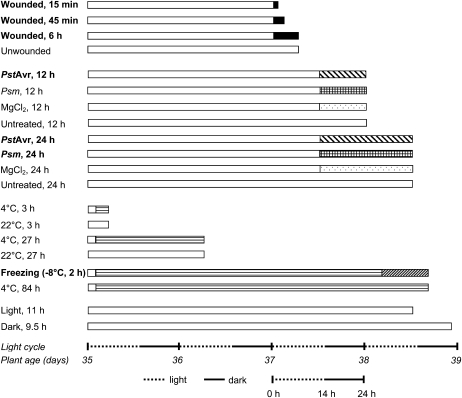
Figure 2 shows the design of stress treatments for the quantitative analysis of oxidized polar lipid molecular species in Arabidopsis accession Columbia (Col-0). In order to compare the patterns of ox-lipids produced in various stresses, plants were grown simultaneously in a growth chamber at 22°C under 14-h-light/10-h-dark cycles. The experimental sample collection occurred in one 24-h period on 5-week-old plants. The experimental design included three subexperiments: wounding with a hemostat (first four conditions); infection with PstAvr and infection with Pseudomonas syringae pv maculicola (Psm; next eight conditions); and cold acclimation and freezing (next six conditions). Each subexperiment included appropriate control conditions, including sampling at 4°C at various time points and mock-inoculated sampling at 12 and 24 h (Fig. 2; see “Materials and Methods”). Two additional controls (last two conditions) at 22°C tested the effect of light cycle changes (time of day). Three leaves from one plant made up each sample, and five biological replicate plants were sampled for each condition.
Figure 2.
Treatments of 5-week-old wild-type Arabidopsis plants. Each bar represents five plants under a single treatment, except that in the freezing treatment the plants were cold acclimated (horizontally striped bar) at 4°C before being gradually frozen (diagonally hatched bar); see “Materials and Methods.” For each treatment, the white bar at the left indicates normal growth conditions, while patterned bars to the right indicate the stress conditions. The far right end of each bar indicates the harvest point. The last two samples were controls intended to identify any effects due to time of day at harvest. Total ox-lipid signals for all samples are shown in Figure 3. Detailed lipid profiles of the samples indicated in boldface are shown in Figure 4. Detailed profiles of the other samples are shown in Supplemental Figure S3.
The experimental design was influenced by previous experiments (data not shown) that demonstrated the need to minimize variation among controls and to apply treatments consistently. Variation in plant growth conditions can affect the basal levels of some ox-lipid compounds, making comparison across multiple stress treatments difficult. Thus, an important aspect of this study was the careful limiting of variation in the control conditions; plants were grown and treated together, and care was taken to apply the stress treatments consistently. It should be noted that all treatments were sublethal to the plants. Arabidopsis accession Col-0 is resistant to PstAvr, with PstAvr causing a hypersensitive response, which occurred within the first 12 h of inoculation. Bacterial growth in plants treated with PstAvr between 12 and 24 h of infection was not significant. In contrast, Arabidopsis accession Col-0 is susceptible to Psm. Bacterial numbers increased 18-fold between 12 and 24 h after Psm infection (Supplemental Fig. S2). By 72 h post inoculation, the Psm-inoculated leaves were chlorotic and eventually died.
For all treatments, harvested leaves were immediately extracted with solvents. To quantitatively compare the pattern of ox-lipids in various stresses, the amount of each ox-lipid detected in the precursor scans was normalized to the signal of an internal standard, 18:0/16:0 MGDG, and then divided by the dry mass of the extracted tissue. A normalized mass spectral signal of 1 indicates the same amount of signal produced by 1 nmol of internal standard. A single internal standard was used because of the impracticality of obtaining internal standards similar in structure to each of the ox-lipid components. Normalizing to the signal of an internal standard corrected for any variation in response that might occur due to variable ion suppression among samples. Because the amount of mass spectral signal depends on the scan mode employed and the ability of individual compounds to undergo ionization and fragmentation, the normalized signal can be considered only a rough estimate of ox-lipid level (in nmol normalized to dry mass). Normalized mass spectral signal per dry mass is an appropriate value for comparison of ox-lipid species levels among samples and for qualitative comparison of the levels of various compounds within different samples.
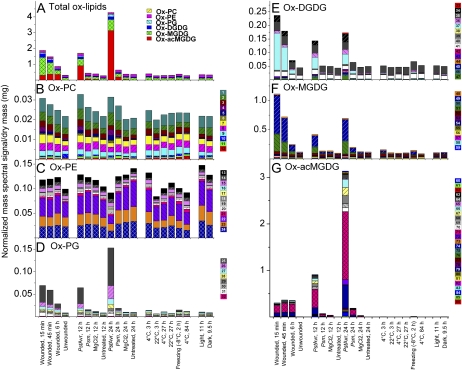
Total Ox-Lipid Accumulation as a Function of Stress
The entire ox-lipid data set is available in Supplemental Table S5, as individual sample values, and in Supplemental Table S6, as averages. Total ox-lipid levels, represented by total bar heights in Figure 3A, were similar in all control conditions. In addition, cold acclimation at 4°C and infection with virulent pathogen Psm for 12 h did not produce significant changes in overall ox-lipid amounts. Wounding produced a high level of total ox-lipids (6.1 times the average basal signal level) within 15 min, with decreased levels seen at 45 min and a further decrease at 6 h. PstAvr infection produced the highest levels of lipid oxidation (13.8 times the average basal level), but the ox-lipids were produced much more slowly, with levels increasing between 12 and 24 h of infection. Thus, as previously shown by Andersson et al. (2006), an avirulent strain of P. syringae was able to induce high levels of ox-lipids. Although Psm damaged the plants severely, infection with this pathogen for 24 h caused a relatively small increase in total ox-lipid signal (2.0 times the average basal level). Freezing also produced only a very modest accumulation of oxidized membrane lipids (1.2 times the average basal level). Per mg of dry mass, the treatment producing the highest ox-lipid mass spectral signal (PstAvr infection for 24 h) produced the same amount of mass spectral signal as 4.3 nmol of internal standard (18:0/16:0 MGDG), while control samples, on average, produced approximately the same amount of mass spectral signal as 0.5 nmol of internal standard. In comparison, on average, the normal-chain phospholipids and galactolipids in these samples produced the same amount of mass spectral signal as 259 nmol of internal standards. The entire normal-chain lipid data set is available in Supplemental Table S7, as individual sample values, and in Supplemental Table S8, as averages.
Figure 3.
Oxidized membrane lipid levels following the treatments shown in Figure 2. A, Total ox-lipids with colors indicating classes. B, ox-PC. C, ox-PE. D, ox-PG. E, ox-DGDG. F, ox-MGDG. G, ox-acMGDG. In B to G, colors indicate individual peaks detected by triple quadrupole MS precursor scanning in negative mode (Table II). The size of each color-coded block represents the quantity of the ox-lipid classes (A) or of individual peaks 1 to 86 (B–G). Vertical axes have different scales.
Patterns of Ox-Lipid Accumulation as a Function of Stress
Figures 3 and 4 display key aspects of the ox-lipid data. Figure 3 shows the ox-lipid levels during each stress condition, emphasizing differences in amount and compositional pattern. Each color within the bars in Figure 3A represents the total signal from an ox-lipid head group class. The components within each ox-lipid class are shown in Figure 3, B to G. Compositional patterns vary among stresses, with ox-MGDG and ox-DGDG predominating in wounding stress, while ox-acMGDG and ox-PG are more elevated in PstAvr stress. More details of the ox-lipid changes during each stress are described in the following sections.
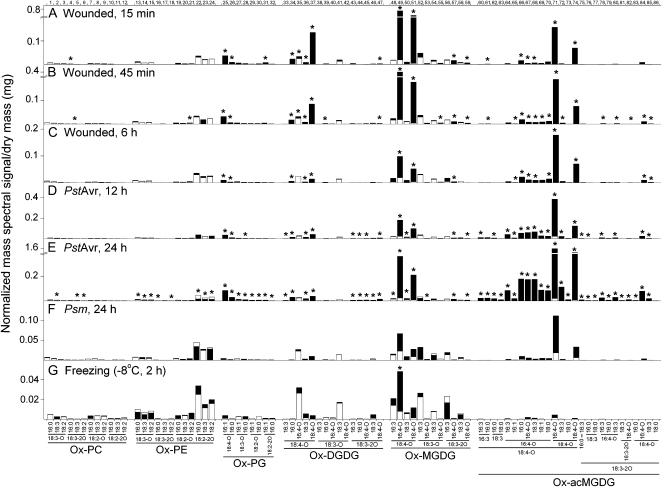
Figure 4.
Oxidized membrane lipids under stress conditions as quantified by triple quadrupole MS precursor scanning. A, Wounded, 15 min. B, Wounded, 45 min. C, Wounded, 6 h. D, PstAvr, 12 h. E, PstAvr, 24 h. F, Psm, 24 h. G, Freezing (−8°C, 2 h). Numbers along the top x axis refer to peaks/compounds in Table II. In each panel, the white bars denote the basal amount as determined under the corresponding control condition: unwounded, for A, B, and C; MgCl2, 12 h, for D; MgCl2, 24 h, for E and F; and 4°C, 84 h, for G. The black bars denote the amount of each ox-lipid measured in each stress treatment. Both white and black bars start at the x axis. The smaller of the white and black bars is “in front” of the other bar. Increments on the vertical scales of A and B (below break) and C are the same; so are those on the vertical scales of D and E (below break). P < 0.05 (n = 5).
Figure 4 shows the individual signals of each ox-lipid under each stress condition (black bar height) along with the average levels of oxidized polar lipids in the 22°C control samples (i.e. average basal levels; white bar height). Figure 4 indicates the changes that are statistically significant at P < 0.05 after correction for the false discovery rate. Data from control and other samples with only low levels of ox-lipids are in Supplemental Figure S3. Among the 22°C and 4°C control samples, there were no significant differences in levels of any ox-lipid. The mock-inoculated control samples had just slightly elevated levels of several ox-lipid species (Supplemental Fig. S3).
Patterns of Ox-Lipid Accumulation in Wounding
Wounding quickly induced a number of ox-lipids (Figs. 3 and 4, A–C). Among ox-PC and ox-PE molecular species, significant increases were observed for 4 and 5 (18:3-2O-containing PCs), which increased approximately 4-fold at 15 and 45 min, respectively, after wounding, while other ox-PC and ox-PE species were unchanged. PGs, DGDGs, and MGDGs containing 18:4-O and 18:3-2O fatty acyl chains were rapidly induced. Ox-PG, ox-DGDG, and ox-MGDG species that were induced at least 30-fold, and were highest at 15 min and lowest at 6 h, included 25 (for peak information, see Table II), 18:4-O/16:1 PG; 26, 18:4-O/16:0 PG; 34,18:4-O/16:0 DGDG; 37, 18:4-O/18:4-O DGDG; 42, 18:3-O/18:4-O DGDG; 47, 18:3-2O/18:4-O DGDG; 49, 18:4-O/16:4-O MGDG; 51, 18:4-O/18:4-O MGDG; 57, 18:3-2O/16:4-O MGDG; and 59, 18:3-2O/18:4-O MGDG. Galactolipid species with two oxidized acyl chains, 18:4-O/18:4-O DGDG (37), 18:4-O/16:4-O MGDG (49), and 18:4-O/18:4-O MGDG (51), correspond to the formulas for arabidopsides D, A, and B, respectively. The other compounds, except 34 and 42, were previously identified as OPDA- and/or dnOPDA-containing species (for references, see Table II). Among the rapidly induced group, 6 h after wounding, the level of 18:4-O/16:4-O MGDG (49) was reduced to 15%, while 18:4-O/18:4-O DGDG (37) was at 10% of its peak level at 15 min after wounding.
Additionally, large quantities of ox-acMGDGs were rapidly generated during wounding. Ox-acMGDG levels were more stable than levels of nonacylated oxidized galactolipid species containing multiple oxidized fatty acyl chains. Between 15 min and 6 h after wounding, the level of the ox-acMGDG with the highest signal (71) varied less than 20%; the 6-h level was 1.1 times the 15-min level. Meanwhile, the signal of the most abundant nonacylated ox-MGDG (49) at 6 h dropped to 15% of its 15-min signal. Thus, acMGDG formed a larger fraction of the total ox-lipid species at 6 h (41%) than at 15 min (16%) after wounding, because of its apparent stability in this time frame. The time course for the formation of ox-MGDGs and oxidized acMGDGs during wounding is shown in Supplemental Figure S4A. In wounding, ox-acMGDG species corresponding to arabidopsides E (18:4-O/18:4-O/16:4-O acMGDG; a major component of 71) and G (18:4-O/18:4-O/18:4-O acMGDG; 74) predominated among ox-acMGDGs, but 25% to 30% of the overall signal detected for ox-acMGDGs in wounding was due to previously undetected species. Those significantly induced include species with three oxidized acyl chains (82 and 84), with two oxidized acyl chains and one normal chain (65, 66, 68, 69, 70, 72, 78, 80, and 85), and with only one oxidized acyl chain (61). In wounding, the most abundant of the “minor” acylated species (behind 71 and 74 in abundance) was 18:4-O/16:4-O/16:0 acMGDG (66).
Patterns of Ox-Lipid Accumulation in Bacterial Infection
Figures 3 and 4, D and E, show ox-lipids formed upon infection with PstAvr, and Figure 4F shows the ox-lipids formed upon infection with Psm. PstAvr infection generated significant increases in 34 species of ox-lipids at 12 h and in 63 species at 24 h, including some ox-lipids in every class.
Three PC species that contain 18:3-O (2) and 18:3-2O (5 and 6) were induced significantly in PstAvr infection, while PE species containing both 18:3-O and 18:3-2O (13, 14, 15, 16, and 18) were also increased. Interestingly, 16:0/18:2-2O PE, 18:3/18:2-2O PE, and 18:2/18:2-2O PE (22, 23, and 24), which are present at low and similar concentrations in all other treatments and controls, decreased significantly at the 24-h time point of PstAvr treatment as compared with the basal level.
Every detectable ox-PG also increased in PstAvr infection at 24 h. ox-DGDG species containing 18:4-O (33–37) and 18:3-2O (43–47) species were increased, while none of the ox-DGDG species containing 18:3-O (38–42) were increased. Major nonacylated ox-MGDGs containing 18:4-O (OPDA) or 16:4-O (dnOPDA; e.g. 48–51) were significantly increased. Twenty-six of the 27 ox-acMGDG species were significantly increased. Besides 18:4-O/18:4-O/16:4-O acMGDG (arabidopside E; a major component of 71) and 18:4-O/18:4-O/18:4-O acMGDG (arabidopside G; 74), the species with the highest signals included 18:4-O/16:4-O coupled with 16:3, 16:0, 16:4-O, 18:3, 18:1, and 18:0 (64 and 66–70, respectively), 18:4-O/18:4-O/18:3 (72), and 18:3-2O/18:4-O/18:3 (85) acMGDGs. The time course for the formation of ox-MGDGs and oxidized acMGDGs during PstAvr infection is shown in Supplemental Figure S4B.
The nonacylated ox-MGDGs (49 and 51) with the highest levels were slightly over twice as high at 24 h after PstAvr infection compared with 12 h after infection. Even so, they were only 50% and 70%, respectively, as high in PstAvr infection at 24 h as at the highest level in wounding (15 min). Our observation of the prolonged presence of nonacylated ox-MGDGs in PstAvr infection was in contrast to the data of Kourtchenko et al. (2007), who found that these species dropped to basal levels by 24 h after infection with avirulent bacteria.
In comparison, the levels of 71 (includes 18:4-O/18:4-O/16:4-O; arabidopside E) and 74 (18:4-O/18:4-O/18:4-O acMGDG; arabidopside G) increased about 4-fold between 12 and 24 h after PstAvr infection. 71 was more than 7-fold higher at 24 h after PstAvr infection than at the highest level in wounding. The very high induction of arabidopside E in avirulent bacterial infection is consistent with the data of Andersson et al. (2006) and Kourtchenko et al. (2007). However, similar to their observation with 49 and 51, Kourtchenko et al. (2007) observed that arabidopsides E and G (71 and 74) peaked early and decreased by 8 h post inoculation. A potential cause of the different results may be that, in the work of Kourtchenko et al. (2007), leaf discs were punched immediately following Pst inoculation and were incubated in water until harvest, while in our study, leaves remained on the plants until harvest. It is possible that the continued production of oxidized membrane lipids requires the tissues to be attached.
Psm infection showed a tendency to generate oxidized complex polar lipids, but the amounts were much lower than in wounding or PstAvr infection and no increases for any molecular species were significant (Figs. 3 and 4F). This occurred even though the plants were seriously affected by Psm infection, and eventually, the infected leaves died. The time course for the formation of ox-MGDGs and oxidized acMGDGs during Psm infection is shown in Supplemental Figure S4C.
Patterns of Ox-Lipid Accumulation in Freezing
Ox-lipid profiles of leaves of plants subjected to freezing at −8°C after cold acclimation are shown in Figures 3 and 4G. Freezing induced the formation of a relatively low amount of ox-lipids. Only 18:4-O/16:4-O MGDG (49) was significantly increased compared with the cold-acclimated control (4°C, 84 h). This oxidized galactolipid species increased 6.1-fold. Levels of acMGDGs with oxidized acyl chains during freezing were very low.
Other Stress-Associated Lipids
For comparison with ox-lipids, levels of several lipid hydrolysis products were also determined. The time courses of PA production in wounding, PstAvr infection, and Psm infection are shown in Supplemental Figure S4, D, E, and F, respectively. Levels of lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), and phosphatidic acid (PA) for the various control and stress treatments are shown in Supplemental Figure S5. Only two treatments, PstAvr (12 h) and freezing (−8°C, 2 h), induced significant increases in total LPC and LPE. PA was induced more than LPC and LPE, with significant accumulation at each time point in wounding, PstAvr, and freezing stress. In PstAvr infection, PA accumulation occurred later than did LPE and LPC. These data clearly indicate that freezing stress was much more effective in producing PA than in producing ox-lipids (Fig. 4 compared with Supplemental Fig. S5).
DISCUSSION
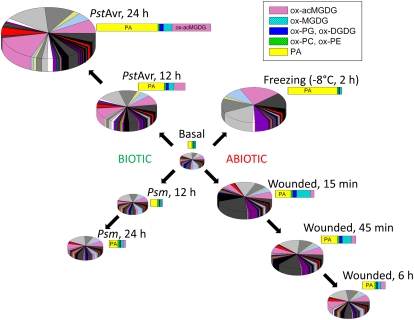
Several trends in the profiles of the lipid metabolites are apparent. First, there was a clear difference between the amount of stress induction of lipids originating in plastids (i.e. ox-PGs, ox-DGDGs, ox-MGDGs, and ox-acMGDGs) and lipids originating in the endoplasmic reticulum (i.e. ox-PCs and ox-PEs). Levels of extraplastidic ox-lipids (i.e. ox-PCs and ox-PEs) changed subtly in response to the examined stresses, while plastidic ox-lipid pools were highly responsive (Figs. 3 and 4). Second, the acyl species in different lipid classes differed. 18:4-O (consistent with being OPDA) and 16:4-O (dnOPDA) were found only in the plastidic lipid classes (PG, MGDG, DGDG, and acMGDG), while 18:2-O and 18:2-2O were found only in PC, PE, and PG (Table II; Supplemental Tables S3 and S4). Fatty acyl chains with the formulas 18:3-O and 18:3-2O were found in all the diacyl lipid classes. acMGDG molecular species were very diverse (Supplemental Table S4), although 18:4-O (OPDA) was very prominent. In some cases, such as with 18:2-O and 18:2-2O, the limitation on distribution is likely to be the limited distribution of the corresponding normal-chain lipids (i.e. 18:2 is rare in galactolipids), while in other cases, the distribution likely reflects the localization of key enzymes in the oxidization reactions; for example, OPDA is not found in PC and PE because enzymes required for its formation, such as allene oxide synthase, are plastid localized (Ferro et al., 2003; Vidi et al., 2006; Ytterberg et al., 2006). Third, particularly for the plastidic lipids, the induced pools were different compositionally from the basal lipid pools (compare Figs. 3 and 4 with Supplemental Fig. S3). This can also be seen in Figure 5, which summarizes our findings. Basal pools were rich in diacyl galactolipids with one oxidized acyl chain (Buseman et al., 2006; Supplemental Fig. S3), while induced pools were richer in ox-acMGDG and diacyl galactolipids with two oxidized acyl chains (Buseman et al., 2006; Fig. 4). Fourth, the induced pool composition varied with time. For example, plastidic diacyl ox-lipids, as exemplified by ox-MGDG (Supplemental Fig. S4), were formed very rapidly in wounding stress, while ox-acMGDGs were formed more slowly. These data are consistent with the conversion of ox-MGDG to ox-acMGDG by a transacylation reaction involving the transfer of an acyl chain from MGDG to the 6-position on the Gal of a second MGDG molecule, as demonstrated in spinach (Spinacia oleracea) for normal-chain MGDGs by Heinz (1967b). It appears that in Arabidopsis, this reaction occurs preferentially between oxidized galactolipid species rather than among the total MGDG pool, although the data suggest that other fatty acids found in galactolipids (particularly 16:0) can be transferred. Fifth, the amount and composition of the stress-induced pool depended on the stress treatment (Figs. 3–5). Freezing induced relatively few ox-lipids but strongly induced the production of PA (Figs. 3–5; Supplemental Fig. S5). The molecular species composition of PA induced in wounding was consistent with its origin largely in PC, but the presence of small amounts of 18:3/16:3 PA in freezing and PstAvr infection suggests an origin for this species in MGDG, since the 18:3/16:3 acyl combination is not found in phospholipids (Fig. 5; Supplemental Tables S7 and S8; Welti et al., 2002). This plastidic PA may be formed by the phosphorylation of diacylglycerol generated from MGDG (Moellering et al., 2010). Wounding rapidly induced high levels of plastidic ox-lipids, particularly ox-MGDGs (Figs. 3 and 4), while in bacterial infection, acMGDGs were more prominent. The fact that essentially the full complement of induced polar lipids was produced in the first 15 min after wounding may suggest that ox-lipids (and PA) are produced mainly by the short-lived activation of existing enzymes in the wounding response. This situation contrasts with that during pathogen infection, in which ox-acMGDGs were very prominent and more were formed in the second 12 h after bacterial infection than in the first 12 h (Figs. 3–5; Supplemental Fig. S4), suggesting either increased activation or induction of the enzyme(s) involved in ox-acMGDG formation.
Figure 5.
Summary of changes in ox-lipids and PA in response to stress treatments. Each pie and each bar represent all the ox-lipids and PAs analyzed. Pie graphs show the 86 individual molecular species of ox-lipids and 12 molecular species of PA. The bars show the indicated lipid classes (see key). (The colors in the bar and pie graphs are unrelated.) The area of each pie and the length of each bar for each condition represent the size of the combined ox-lipid and PA pools. The basal pool shown here is an average of the pools determined for unwounded; MgCl2, 12 h; untreated, 12 h; MgCl2, 24 h; untreated, 24 h; 4°C, 3 h; 22°C, 3 h; 4°C, 27 h; 22°C, 27 h; 4°C, 84 h; light, 11 h; and dark, 9.5 h (Fig. 2).
In mammalian systems, specific oxidized membrane lipids have been identified as regulators of many cell types. Oxidized animal membrane lipids mediate both beneficial and detrimental functions, including inflammation, apoptosis, phenotype switching in smooth muscle cells, and innate immunity (Deigner and Hermetter, 2008; Hazen, 2008). Specific ox-PCs regulate the expression of over 1,000 genes in endothelial cells (Gargalovic et al., 2006; Berliner et al., 2009), and data suggest that oxidized phospholipids act by binding specific receptor proteins (Deigner and Hermetter, 2008).
In plants, oxidized membrane lipids may represent alterations that have occurred to prevent oxidative damage elsewhere in the cell, they may function as mediators signaling stress responses, or they may be long-term modifications that might function as stress “memory” (Wang, 2004; Andersson et al., 2006; Hisamatsu et al., 2006; Wang et al., 2006; Gális et al., 2009; Mène-Saffrané et al., 2009). Recent work by Mène-Saffrané et al. (2009) suggested that ox-lipids are indeed involved in preventing damage elsewhere in the cell. These authors showed that nonenzymatic oxidation of trienoic fatty acyl species, correlating with malondialdehyde production, reduced lesion spread in the oxidatively stressed disease-lesion mimic, acd2-2. Trienoic fatty acids were implicated as reactive oxygen species (ROS) sinks that reduce ROS levels and protect against negative ROS effects in fungal infection and chronic oxidative stress responses. The notion of ox-lipids as signals is reasonable given the well-documented roles of the oxidized free fatty acids found in membrane lipids as signals via transcriptional regulation (Taki et al., 2005; Sattler et al., 2006) and the stress-specific induction of particular ox-lipids. The involvement of intact oxidized membrane lipids in signaling is also supported by their demonstrated biological activities. For example, Andersson et al. (2006) showed that 18:4/18:4-O/16:4-O MGDG (arabidopside E; a major component of 71) had greater bactericidal activity than the same molar amount of OPDA. Hisamatsu et al. (2006) showed that 18:4-O/16:4-O MGDG (arabidopside A; 49) promoted senescence in barley (Hordeum vulgare) leaves. However, recently, Seltmann et al. (2010) showed that levels of six arabidopsides (35, 37, 49, 51, 71a, and 74) were increased when plants were placed in the dark for 3 d or subjected to osmotic stress with sorbitol treatment, but arabidopside levels were not increased strongly by natural senescence; these authors suggest that oxidized fatty acids related to jasmonic acid may play only a secondary role in senescence.
Our data identify “ox-lipid signatures,” extending the notion of the “oxylipin signature” and the working hypothesis that these signatures reflect physiological status and affect stress response (Weber et al., 1997; Kramell et al., 2000). To better understand the importance and function of oxidized membrane lipids during plant stress responses, the occurrence and timing of specific ox-lipid species must be documented systematically. Our work here is a step toward that goal. This work lays the foundation for further, ongoing work aimed at associating the formation of specific oxidized membrane lipids with the action of specific gene products and with particular stress-response phenotypes.
CONCLUSION
ESI MS/MS has previously been utilized as a quick and quantitative strategy for profiling oxidized membrane lipids (Buseman et al., 2006; Maeda et al., 2008, Xiao et al., 2010). In this study, we demonstrated a direct-infusion ESI triple quadrupole MS method that detects oxidized membrane lipid species that include ox-PC, ox-PE, ox-PG, ox-DGDG, ox-MGDG, and ox-acMGDG, expanding the number of compounds previously characterized by Stelmach et al. (2001), Hisamatsu et al. (2003, 2005), Andersson et al. (2006), Buseman et al. (2006), and Kourtchenko et al. (2007). In particular, the data indicate that the number of acMGDGs (60–86 in Table II) is much larger than previously described. The precursor scanning method takes 20 to 25 min to analyze each sample. Together with the ESI triple quadrupole MS procedure for membrane lipid profiling (Welti et al., 2002; Devaiah et al., 2006), this analysis provides a useful foundation to investigate the effects of abiotic and biotic stresses on plant lipid profiles and the roles of various enzymes in response to those stresses.
MATERIALS AND METHODS
Growth Conditions and Sampling for Stress Treatments
Arabidopsis (Arabidopsis thaliana) accession Col-0 seeds were sprinkled on moist soil. The soil and seeds were placed at 4°C for 48 h before moving to a growth chamber for germination. Seedlings were covered with a plastic dome until transplantation. Seven days after germinating, two seedlings were transplanted to each 3.5-inch-square pot. Plants were kept in a growth chamber at a temperature of 22°C, humidity of 60%, and a photoperiod of 14/10 h at 100 μmol m−2 s−1. Twenty-eight days after transplanting, which corresponds to 35 d (5 weeks) postgermination, plants were ready for treatment (5 weeks old). Each treatment or control set contained three randomly mixed pots (a total of six plants with five randomly chosen plants used as biological replicate samples). Three leaves from each plant (leaf numbers 6, 7, and 8) were sampled at each time point. For all samples, the leaves were cut and immediately immersed in 3 mL of isopropanol with 0.01% butylated hydroxytoluene (BHT) at 75°C for 15 min. Totals of 1.5 mL of chloroform and 0.6 mL of water were added, and samples were then stored at −20°C until analysis.
Stress Treatments
Except for the low-temperature treatment, all treatments were applied on leaf numbers 6, 7, and 8, where leaf number 1 is the first true leaf (Weigel and Glazebrook, 2002). For wounding, a piece of paper tape was wrapped around the lower side of the hemostat grip so that wounding was reproducible and uniform. Each leaf was clamped perpendicular to the midvein in two places about 2 cm apart. Leaves were sampled at 15 min, 45 min, and 6 h after wounding. One unwounded set of plants was sampled at the same time as the 6-h time point set.
For bacterial treatment, suspensions of Pseudomonas syringae pv maculicola and Pseudomonas syringae pv tomato DC3000 expressing the AvrRpt2 avirulence gene at 107 cells mL−1 in magnesium chloride (10 mm) were infiltrated into the abaxial surface of leaves of plants with a 1-mL syringe. The control (mock) treatment was infiltration with 10 mm magnesium chloride. Leaves were sampled 12 and 24 h after injection. Bacterial counting was performed on additional simultaneously infiltrated plants (six for each condition) treated identically to the plants used for lipid extraction.
In the low-temperature treatment, plants were cold acclimated at 4°C for 72 h before treatment in a freezing chamber programmed so that the temperature dropped from 4°C to −2°C in 2 h (3°C h−1), stayed at −2°C for 2 h, dropped to −8°C in 6 h (1°C h−1), and finally stayed at −8°C for 2 h. Freezing-treated leaves were harvested after 3, 27, and 84 h of cold acclimation and after 2 h at −8°C. Plants at 22°C were also harvested at each time point.
Bacterial Counts
For bacterial counting, the pathogen-inoculated leaves (6, 7, and 8) were homogenized in a total volume of 1 mL of 10 mm magnesium chloride. Ten microliters each of 102, 103, and 104 dilutions were streaked on 1.5% King’s agar plates containing appropriate antibiotics. PstAvr (strain PV288) was streaked on plates made with rifampicin (50 μg mL−1) and kanamycin (50 μg mL−1). Psm was streaked on plates made with streptomycin (100 μg mL−1). Colonies were counted after incubation for 48 h in the dark at room temperature. The leaf homogenates were lyophilized, dried in an oven at 110°C, and their masses were determined.
Lipid Extraction
Lipid extraction was carried out using a combination of procedures described by Bligh and Dyer (1959) and Markham et al. (2006). Briefly, each lipid extract in isopropanol with BHT, chloroform, and water (see “Growth Conditions and Sampling for Stress Treatments” above) was transferred to a new glass tube using a Pasteur pipette, leaving the leaves in the original tube. Four milliliters of chloroform:methanol (2:1) was added to the leaves, the samples were shaken for 30 min, and the solvent was transferred, combining it with the first extract. The addition, shaking, and transfer steps were performed totally four times. Finally, 4 mL of solvent H (the organic phase of isopropanol:hexane:water [55:20:25, v/v]; Markham et al., 2006) was added to the leaf material, the samples were incubated at 60°C for 15 min, and the solvent was removed and combined with the previous extracts. This step was performed four times, combining all extracts. Finally, the extracted leaves were dried in an oven at 110°C, and the extracted dry leaf mass was determined. The solvent was evaporated from the extract in a CentriVap centrifugal vacuum concentrator (Labconco), and each sample was dissolved in 1 mL of chloroform. Extracts were stored at −80°C.
Tissue Mass Determination
Extracted dry leaf mass (for extracted samples) and lyophilized leaf homogenates (for bacterial counts) were weighed on a Mettler Toledo AX balance that provided mass data to the nearest 2 μg. To determine precision, a dried leaf sample was weighed seven times, indicating a mass of 4.095 mg with a sd of 0.007 mg (0.17%). To determine accuracy, six brass weights with official masses ranging from 1 to 20 g were weighed. The balance weighed, on average, 0.17% (sd of 0.08%) too high. Taken together, these data indicate that weighing dried leaf tissue was not a major source of error in quantifying the compounds in the leaves.
Extracts Used for QTOF and FTICR MS Analysis and Fractionation for QTOF Analysis
Crude extracts from all replicate samples of PstAvr (24 h) or wounding (15 min) treatment were combined for FTICR MS analysis. For lipid class fractionation and QTOF MS, Arabidopsis accession Col-0 plants were grown and infiltrated with PstAvr for 24 h as described above. Total lipid was bulk extracted by the combined extraction method (Bligh and Dyer, 1959; Markham et al., 2006). Total unfractionated lipid extract was used for QTOF analysis or fractionation. Fractionation of lipid classes was carried out as described by Buseman et al. (2006). Briefly, activated silicic acid (Unisil; Clarkson Chemical) in chloroform was packed into a 1.5-cm-diameter column (40-mL column volume). Total lipid extract from 150 mg of leaf dry mass in 15 mL of chloroform was applied to the column, and the column was batch eluted in five fractions: fraction 1, 200 mL of chloroform:acetone (1:1, v/v); fraction 2, 400 mL of acetone; fraction 3, 400 mL of chloroform:methanol (19:1, v/v); fraction 4, 400 mL of chloroform:methanol (4:1, v/v); and fraction 5,800 mL of chloroform:methanol (1:1, v/v). Each fraction was evaporated and redissolved in 15 mL of chloroform.
Triple Quadrupole MS Analysis of Ox-Lipids
Samples were prepared for mass spectral analysis by diluting a volume of each unfractionated lipid extract (in chloroform) derived from approximately 0.2 mg of leaf dry mass. An internal standard, 2.008 nmol of 18:0/16:0 MGDG (a component of hydrogenated MGDG; Matreya) was added to each sample, and the sample was diluted such that the final volume was 1.2 mL and the solvent composition was chloroform:methanol:300 mm ammonium acetate in water (300:665:35, v/v/v). Scans for precursors of 291.2, 293.2, 295.2, and 283.2 (for the 18:0 component of the internal standard) were carried out in negative mode using a triple quadrupole mass spectrometer (ABI 4000; Applied Biosystems) equipped with an ESI source. The samples were directly infused at 30 μL min−1. For precursor scans, the parameters were as follows: collision gas, 2 (arbitrary units); curtain gas, 20 (arbitrary units); ion source gases 1 and 2, 45 (arbitrary units); ion spray voltage, −4,500 V; source temperature, 100°C; declustering potential, −100 V; entrance potential, −10 V; collision energy, −45 V; collision cell exit potential, −20 V; and interface heater “on.” Spectra for precursors of 291.2, 293.2, and 295.2 were acquired from 700 to 1,150 m/z at 6 s per cycle for 65 cycles. The precursor spectrum for the internal standard (m/z 283.2) was acquired from 800 to 825 m/z at 0.5 s per cycle for 65 cycles.
A custom Analyst “add-on,” supplied by Applied Biosystems and called “MultiplePeriodProcessing,” was used to process and export data from Analyst to Excel. Precursor spectra generated by the triple quadrupole MS were smoothed by the software, with a smoothing option of 0.4 for previous and next point weight and 1 for current point weight, and baseline subtracted with a window width of 20 u. Peaks with intensity values lower than 50 counts per second were removed before spectra were exported as peak lists. Once exported, target m/z of peaks 1 to 86 were looked up in the appropriate precursor spectra with an m/z tolerance of ±0.4 from the theoretical m/z for the peak/compound listed in Table II. Peak intensity was corrected for isotopic distribution of precursor ions minus the fragment ion (i.e. for isotopic distribution within each spectrum).
Mass spectral signals (intensities) were normalized to the signal for 2.008 nmol of internal standard,18:0/16:0 MGDG, as its [M + C2H3O2]− ion. 18:0/16:0 MGDG is an unnaturally occurring lipid species that was added as an internal standard to the portion of the sample being analyzed. The mass spectral signal was normalized by the following formula: (mass spectral intensity of each lipid molecular species × amount of the internal standard in nmol) ÷ (mass spectral intensity of the internal standard). Thus, a signal of 1 is the same amount of signal produced by 1 nmol of internal standard, 18:0/16:0 MGDG. Finally, the data were corrected for isotopic overlap due to the fatty acyl portions (fragment ions), adjusted to account for the fraction of sample analyzed, and normalized to the sample dry mass to give the “normalized mass spectral signal per dry mass.” No corrections for varying mass spectral response to the various molecular species were applied. The calculated data are appropriate for direct comparison among samples. Samples were initially analyzed as soon as extraction was complete; however, extended storage at −80°C of samples containing BHT (added at the first step of the extraction), followed by reanalysis, suggests that most ox-lipids are quite stable.
Triple Quadrupole MS Analysis of Normal-Chain Lipids
Samples were prepared for mass spectral analysis by diluting a volume of each unfractionated lipid extract (in chloroform) derived from approximately 0.2 mg of leaf dry mass. Precise amounts of internal standards, obtained and quantified as described previously (Welti et al., 2002), were added as described by Xiao et al. (2010). The sample and internal standard mixture was combined with solvents, such that the ratio of chloroform:methanol:300 mm ammonium acetate in water was 300:665:35, and the final volume was 1.2 mL. Sample introduction was as for ox-lipids. Sequential precursor and neutral loss scans of the extracts were carried out as described by Xiao et al. (2010), except that only limited m/z ranges around the target m/z values of the analytes and internal standards were scanned. The scan speed was 100 mass units per second. For each spectrum, 20 to 150 continuum scans were averaged in multiple channel analyzer mode. The spectral data were smoothed and the baseline subtracted. Peaks with intensity values lower than 50 counts per second were removed, peak areas integrated, and data exported as peak lists.
The LipidomeDB Data Calculation Environment (Zhou et al., 2011) was used to locate target compound peak data, deconvolute isotopes, and quantify lipids in each class in comparison with the two internal standards of that class (Brügger et al., 1997; Welti et al., 2002). Correction for chemical and/or instrumental noise was performed as described previously (Xiao et al., 2010). Finally, the data were corrected for the fraction of the sample analyzed and normalized to the sample “dry weights” to produce data in nmol mg−1.
QTOF MS Analysis
PstAvr 24-h infection extract and fractions from its silicic acid column separation (described above) were dissolved in chloroform at a concentration of 10 mg dry tissue mass mL−1. For product ion analysis, aliquots were combined with solvents (chloroform:methanol:300 mm ammonium acetate in water, 300:665:35 [v/v/v]) so that the final concentrations were 0.33 to 2.5 mg leaf dry mass mL−1.
Spectra were acquired on an MDS SCIEX/Applied Biosystems QStar Elite hybrid QTOF MS apparatus (Applied Biosystems), with daily calibration of the instrument using a lipid standard mixture. Samples were introduced by continuous infusion into the ESI source at a rate of 30 μL min−1 using the integrated Harvard syringe pump. MS/MS product ion scans were carried out in negative ionization mode, with individual fragmentation of anions of oxidized PC, PE, PG, DGDG, MGDG, and acMGDG intact lipid species that had been detected from triple quadrupole MS precursor scans. PC species were analyzed by fragmentation of [M + C3H3O2]− ions in the unfractionated extract. PE and PG species were analyzed by fragmentation of [M – H]− ions in the unfractionated extract and silicic acid-separated fraction 5. MGDG and DGDG species were analyzed by fragmentation of [M – H]−/or [M + C3H3O2]− ions in the unfractionated extract and, additionally for MGDG species, silicic acid-separated fraction 2. acMGDG species were analyzed by fragmentation of [M + C3H3O2]− ions in the unfractionated extract and silicic acid-separated fraction 1. The resultant acyl anion fragments allowed the identification of acyl moieties in intact lipids. The ion spray voltage was set at −4.5 kV, the source temperature at 150°C, the curtain gas at 25 (arbitrary units), and the ion source gases at 20 and 30 (arbitrary units). The declustering potential was −80 V, the declustering potential 2 was −15 V, and the focusing potential was −300 V. The collision gas, nitrogen, was set at 4 (arbitrary units), and the collision energy ranged from 40 to 70 V. For each precursor ion, product ion data were collected over the range of m/z 100 to 1,150, resulting in 300 to 1,800 cumulative scans during an acquisition for 5 to 30 min. Data were collected and smoothed using Analyst QS 2.0 software. Accurate masses of the product ions were determined to ten thousandths of a mass unit.
FTICR MS Analysis
Accurate m/z data were collected using small m/z windows, which optimize sensitivity (Southam et al., 2007). Briefly, samples at a concentration of 0.014 mg dry mass mL−1 were analyzed using direct-infusion ESI on an LTQ FTICR hybrid linear quadrupole ion trap FTICR mass spectrometer (Thermo Finnigan). The ESI source was operated in positive mode with a spray voltage of 2.8 kV, a tube lens offset of 140 V, and a capillary temperature of 200°C. The instrument was calibrated using an automatic routine based on a standard calibration solution containing caffeine, peptide Met-Arg-Phe-Ala, and Ultramark 1621 (all products of Sigma-Aldrich). The 700 to 1,200 m/z range was divided into multiple 30 m/z selected ion monitoring intervals with 5 m/z overlaps at both ends. Each interval was scanned for 2 min (approximately 10 scans) using a selected ion monitoring MS target automated gain control value of 2 × 105 accumulated ions with a resolution of 500,000 at m/z 400. FTICR mass spectra from individual scans within a mass interval were averaged using Xcalibur 2.0 software, and peaks with average counts greater than 1,000 were exported to Excel 2010.
Statistical Analysis
Significance was determined at P < 0.05, after correcting for the false discovery rate, using Excel 2010. Comparisons were between stress samples and their controls.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Possible ox-lipid structures and their MS fragmentation.
Supplemental Figure S2. Bacterial counts per leaf dry mass.
Supplemental Figure S3. Ox-lipids from control and low-level treatments.
Supplemental Figure S4. Ox-MGDG, ox-acMGDG, and PA in wounding and bacterial infection.
Supplemental Figure S5. LPC, LPE, and PA under treatments.
Supplemental Table S1. QTOF MS data supporting Table 2 ox-lipid identifications.
Supplemental Table S2. FTICR MS data supporting Table 2 ox-lipid identifications.
Supplemental Table S3. Simplified designations of diacyl ox-lipids (1–59).
Supplemental Table S4. Simplified designations of acMGDGs (60–86).
Supplemental Table S5. All data: ox-lipids under treatments.
Supplemental Table S6. Averages and sds: ox-lipids under treatments.
Supplemental Table S7. All data: normal lipids under treatments.
Supplemental Table S8. Averages and sds: normal lipids under treatments.
Acknowledgments
We thank Gail Ragan for helpful suggestions. We are grateful to Mark Ungerer for generously allowing us to use his freezing chamber.
References
- Andersson MX, Hamberg M, Kourtchenko O, Brunnström A, McPhail KL, Gerwick WH, Göbel C, Feussner I, Ellerström M. (2006) Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside E. J Biol Chem 281: 31528–31537 [DOI] [PubMed] [Google Scholar]
- Berliner JA, Leitinger N, Tsimikas S. (2009) The role of oxidized phospholipids in atherosclerosis. J Lipid Res (Suppl) 50: S207–S212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Weiler EW. (2007) cyclo-Oxylipin-galactolipids in plants: occurrence and dynamics. Planta 226: 629–637 [DOI] [PubMed] [Google Scholar]
- Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. (1997) Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA 94: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, Roth MR, Esch SW, Shah J, Williams TD, et al. (2006) Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol 142: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, Dehesh K. (2008) Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS ONE 3: e1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigner H-P, Hermetter A. (2008) Oxidized phospholipids: emerging lipid mediators in pathophysiology. Curr Opin Lipidol 19: 289–294 [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugière S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Gális I, Gaquerel E, Pandey SP, Baldwin IT. (2009) Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ 32: 617–627 [DOI] [PubMed] [Google Scholar]
- Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, et al. (2006) Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA 103: 12741–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G, Grata E, Rudaz S, Wolfender J-L. (2008) High-resolution profiling of oxylipin-containing galactolipids in Arabidopsis extracts by ultra-performance liquid chromatography/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 22: 3154–3160 [DOI] [PubMed] [Google Scholar]
- Grun C, Berger S, Metther D, Mueller MJ. (2007) Early accumulation of non-enzymatically synthesised oxylipins in Arabidopsis thaliana after infection with Pseudomonas syringae. Funct Plant Biol 34: 65–71 [DOI] [PubMed] [Google Scholar]
- Hamberg M. (1988) Biosynthesis of 12-oxo-10,15(Z)-phytodienoic acid: identification of an allene oxide cyclase. Biochem Biophys Res Commun 156: 543–550 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Rodriguez MJ, Calvo AP, Castresana C. (2003) Activation of the fatty acid α-dioxygenase pathway during bacterial infection of tobacco leaves: formation of oxylipins protecting against cell death. J Biol Chem 278: 51796–51805 [DOI] [PubMed] [Google Scholar]
- Hazen SL. (2008) Oxidized phospholipids as endogenous pattern recognition ligands in innate immunity. J Biol Chem 283: 15527–15531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E. (1967a) [Acylgalactosyldiglyceride from leaf homogenates]. Biochim Biophys Acta 144: 321–332 [PubMed] [Google Scholar]
- Heinz E. (1967b) [On the enzymatic formation of acylgalactosyldiglyceride]. Biochim Biophys Acta 144: 333–343 [PubMed] [Google Scholar]
- Heinz E, Tulloch AP. (1969) Reinvestigation of the structure of acyl galactosyl diglyceride from spinach leaves. Hoppe Seylers Z Physiol Chem 350: 493–498 [DOI] [PubMed] [Google Scholar]
- Hisamatsu Y, Goto N, Hasegawa K, Shigemori H. (2003) Arabidopsides A and B, two new oxylipins from Arabidopsis thaliana. Tetrahedron Lett 44: 5553–5556 [Google Scholar]
- Hisamatsu Y, Goto N, Hasegawa K, Shigemori H. (2006) Senescence-promoting effect of arabidopside A. Z Naturforsch C 61: 363–366 [DOI] [PubMed] [Google Scholar]
- Hisamatsu Y, Goto N, Sekiguchi M, Hasegawa K, Shigemori H. (2005) Oxylipins arabidopsides C and D from Arabidopsis thaliana. J Nat Prod 68: 600–603 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL. (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230–236 [DOI] [PubMed] [Google Scholar]
- Imbusch R, Mueller MJ. (2000) Analysis of oxidative stress and wound-inducible dinor isoprostanes F(1) (phytoprostanes F(1)) in plants. Plant Physiol 124: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA. (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko O, Andersson MX, Hamberg M, Brunnström A, Göbel C, McPhail KL, Gerwick WH, Feussner I, Ellerström M. (2007) Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol 145: 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramell R, Miersch O, Atzorn R, Parthier B, Wasternack C. (2000) Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves: implications for different signaling pathways. Plant Physiol 123: 177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Sage TL, Isaac G, Welti R, Dellapenna D. (2008) Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell 20: 452–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG. (2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281: 22684–22694 [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L, Dubugnon L, Chételat A, Stolz S, Gouhier-Darimont C, Farmer EE. (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J Biol Chem 284: 1702–1708 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C. (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S. (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mène-Saffrané L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D. (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell 18: 3706–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann MA, Stingl NE, Lautenschlaeger JK, Krischke M, Mueller MJ, Berger S. (2010) Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol 152: 1940–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam AD, Payne TG, Cooper HJ, Arvanitis TN, Viant MR. (2007) Dynamic range and mass accuracy of wide-scan direct infusion nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry-based metabolomics increased by the spectral stitching method. Anal Chem 79: 4595–4602 [DOI] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Gebhardt S, Schubert-Zsilavecz M, Weiler EW. (2001) A novel class of oxylipins, sn1-O-(12-oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl diglyceride, from Arabidopsis thaliana. J Biol Chem 276: 12832–12838 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C. (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al. (2005) 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thiocone A, Farmer EE, Wolfender JL. (2008) Screening for wound-induced oxylipins in Arabidopsis thaliana by differential HPLC-APCI/MS profiling of crude leaf extracts and subsequent characterisation by capillary-scale NMR. Phytochem Anal 19: 198–205 [DOI] [PubMed] [Google Scholar]
- Thoma I, Loeffler C, Sinha AK, Gupta M, Krischke M, Steffan B, Roitsch T, Mueller MJ. (2003) Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J 34: 363–375 [DOI] [PubMed] [Google Scholar]
- Vidi P-A, Kanwischer M, Baginsky S, Austin JR, Csucs G, Dörmann P, Kessler F, Bréhélin C. (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem 281: 11225–11234 [DOI] [PubMed] [Google Scholar]
- Vollenweider S, Weber H, Stolz S, Chételat A, Farmer EE. (2000) Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J 24: 467–476 [DOI] [PubMed] [Google Scholar]
- Wang X. (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Li M, Welti R. (2006) Profiling lipid changes in plant response to low temperatures. Physiol Plant 126: 90–96 [Google Scholar]
- Weber H, Vick BA, Farmer EE. (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94: 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase D α in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen Q-F, Chan S-W, Zheng S-X, Ma J, Wang M, Welti R, Chye ML. (2010) Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier J-B, van Wijk KJ. (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts: a surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Marepally SR, Nune DS, Pallakollu P, Ragan G, Roth MR, Wang L, Lushington GH, Visvanathan M, Welti R. (2011) LipidomeDB data calculation environment: online processing of direct-infusion mass spectral data for lipid profiles. Lipids 46: 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]