Abstract
Nitrogen (N) is an essential macronutrient for plants. N levels in soil vary widely, and plants have developed strategies to cope with N deficiency. However, the regulation of these adaptive responses and the coordinating signals that underlie them are still poorly understood. The aim of this study was to characterize N starvation in adult Arabidopsis (Arabidopsis thaliana) plants in a spatiotemporal manner by an integrative, multilevel global approach analyzing growth, metabolites, enzyme activities, and transcript levels. We determined that the remobilization of N and carbon compounds to the growing roots occurred long before the internal N stores became depleted. A global metabolite analysis by gas chromatography-mass spectrometry revealed organ-specific differences in the metabolic adaptation to complete N starvation, for example, for several tricarboxylic acid cycle intermediates, but also for carbohydrates, secondary products, and phosphate. The activities of central N metabolism enzymes and the capacity for nitrate uptake adapted to N starvation by favoring N remobilization and by increasing the high-affinity nitrate uptake capacity after long-term starvation. Changes in the transcriptome confirmed earlier studies and added a new dimension by revealing specific spatiotemporal patterns and several unknown N starvation-regulated genes, including new predicted small RNA genes. No global correlation between metabolites, enzyme activities, and transcripts was evident. However, this multilevel spatiotemporal global study revealed numerous new patterns of adaptation mechanisms to N starvation. In the context of a sustainable agriculture, this work will give new insight for the production of crops with increased N use efficiency.
Nitrogen (N) is quantitatively the most important nutrient for plants. N constitutes approximately 2% of plant dry matter and is an essential component of key macromolecules, such as proteins, nucleic acids, and secondary metabolites (Crawford and Forde, 2002). Under temperate climates and in cultivated soils, N is taken up by the roots in the form of nitrate. After the reduction of nitrate by nitrate reductase (NR) and nitrite reductase, ammonium is incorporated into amino acids by the Gln synthetase/GOGAT cycle. In addition, glutamate dehydrogenase (GDH) catalyzes the reductive amination of 2-oxoglutarate (2OG) and the oxidative deamination of Glu (Melo-Oliveira et al., 1996; Turano, 1998; Restivo, 2004). Depending on the species and the environmental conditions, the energetically costly N assimilation takes place in the roots or shoots (Smirnoff and Stewart, 1985; Masclaux-Daubresse et al., 2010). N assimilation and carbon (C) metabolism are highly interconnected, as C skeletons and energy, which are necessary to reduce nitrate and to produce amino acids, are supplied by photosynthesis, photorespiration, and respiration (for review, see Krapp and Truong, 2005).
In soils, N is often a significant factor limiting plant growth, and plants frequently encounter nutrient deficiency in their natural habitats. Nitrate has a very weak affinity to form surface complexes with soil minerals (Strahm and Harrison, 2006), which leads to nitrate losses by microbial conversion to N2 gas and, in particular, by the leaching of soil water carrying dissolved nitrate. The formation of depletion areas, such as in the rhizosphere, depends on the soil type, fertilizer addition, and microbial activity, among other factors (Miller et al., 2007). For example, in a transect across four arable fields, the soil nitrate concentration was found to have varied by almost 100-fold (Lark et al., 2004). Accordingly, nutrient deprivation is a controlling factor in biomass production and yield (Marschner, 1995), and plant production has been increased during the last 50 years by the extensive use of N fertilizers (Tilman et al., 2001). However the main problem is the fact that the recovery of the N in fertilizers by crop plants is low (Peoples et al., 1995; Sylvester-Bradley and Kindred, 2009) and the remaining N is partly lost from the agroecosystem; moreover, fertilizer runoff into aquatic systems may lead to eutrophication (Johnson et al., 2007). Therefore, new strategies are needed to engineer N-efficient crops to ensure sustainable agriculture. This necessitates a detailed understanding of the complex morphological, physiological, and biochemical adaptations of plants to N starvation.
To face N deprivation, the sessile plant increases its capacity to acquire N by stimulating root growth relative to shoot growth, which leads to an increased root-shoot ratio and a strong modification of the root system architecture (Drew, 1975; Scheible et al., 1997). In addition, N starvation induces the expression of high-affinity transport systems for nitrate and ammonium (Crawford and Glass, 1998; Forde, 2000; von Wirén et al., 2000). Furthermore, the remobilization of N from source organs is stimulated. Indeed, N assimilated into biomolecules can be released back to inorganic N (ammonium) under different physiological contexts in plant cells by various physiological processes, such as photorespiration, the biosynthesis of phenylpropanoids, and the remobilization of stored reserves (Miflin and Lea, 1980). Efficient reassimilation mechanisms reincorporate liberated ammonium into metabolism and maintain the N economy in the plant. However, if these adaptation mechanisms do not provide a sufficient nutrient supply, the sessile plant is forced to respond with further adaptive metabolic strategies to safeguard survival or to complete its life cycle. These metabolic changes in response to nutrient deprivation involve alterations in the expression levels of a number of genes (Ohlrogge and Benning, 2000).
Beyond its essential role in plant metabolism, nitrate has been shown to have important signaling functions. Indeed, extensive transcriptome studies have characterized the primary nitrate signal response (Wang et al., 2000, 2003, 2004; Scheible et al., 2004; Orsel et al., 2005; Gutiérrez et al., 2007). Nitrate not only rapidly induces genes that are responsible for its transport (e.g. NRT1.1 and NRT2.1) and assimilation (e.g. NIA1, NIA2, and NII) but also triggers a change in the expression of approximately 1,000 nitrate-responsive genes in Arabidopsis (Arabidopsis thaliana). Processes such as amino acid and nucleic acid biosynthesis, transcription, RNA processing, ribosome and hormone biosynthesis, N assimilation, the generation of reductants, and trehalose metabolism respond within 20 min to 3 h of nitrate induction. Indeed, as rapidly as 5 min after nitrate resupply, changes in gene expression have been described, and the genes with modified steady-state mRNA levels only partly overlapped with the genes identified as nitrate regulated in longer (20-min) induction studies (Krouk et al., 2010; Castaings et al., 2011). Furthermore, several studies have differentiated the direct molecular responses to nitrate from the general responses to N supply using nitrate reductase null mutants (Wang et al., 2004) and mutants of the nitrate sensor NRT1.1/CHL1 (Muños et al., 2004; Hu et al., 2009; Wang et al., 2009). Taken together, these data suggest that nitrate is rapidly and specifically sensed by plant cells and that a nitrate-signaling pathway adjusts the expression of a large set of genes to adapt the cell and organ metabolism and growth of the organism to the N availability. In addition to these rapid effects of nitrate addition to N-starved plants, Scheible et al. (2004) have compared N-starved and N-replete seedlings and found that the expression of similar gene sets was modified by N starvation and nitrate induction, but in the opposite way.
In contrast to the analysis of a sudden addition of nitrate or completely N-starved seedlings, the analysis of plants grown under different N supply (Bi et al., 2007; Tschoep et al., 2009) has revealed that permanently limiting N availability caused a markedly different transcriptome response. The nitrate-inducible enzymes involved in N assimilation did not vary between plants grown in limiting and ample N, yet a different set of transcription factors were found to have changes in their expression levels. Furthermore, in 2007, Gutiérrez and coworkers first suggested that N assimilation could also be posttranscriptionally regulated by noncoding short RNAs. Indeed, many plant microRNAs (miRNAs) are involved in developmental processes and regulate the expression of transcription factors, but a subset of the target genes are involved in metabolism. For example, Gifford et al. (2008) demonstrated the link between miRNA167 and the regulation of the expression of auxin-responsive factor 8, which controls lateral root architecture in response to N treatment. Other examples, such as the posttranscriptional regulation by miRNAs of phosphate and sulfate assimilation in roots or the identification of nutrient-responsive miRNA in Arabidopsis (Chiou, 2007; Kawashima et al., 2009; Pant et al., 2009; Zhao et al., 2011), illustrate the growing importance of this type of regulation in response to nutrient deprivation.
The goal of this work was to obtain detailed insight into the integrated and multilevel plant responses to a sudden, total N starvation, as may occur under certain conditions in the soil. We set up a highly controlled hydroponic growth system that allowed us to follow the kinetics of N starvation throughout a 10-d interval. We analyzed separately the root and shoot responses, as it has been shown that 88% of the total genes that respond to a treatment responded in an organ-specific manner (Aceituno et al., 2008). Here, we present a comprehensive survey of global gene expression, metabolite levels, central enzyme activities, and N uptake with the aim to thoroughly elucidate the responses to a period of intermediate length (mid-term) and long-term N starvation. Different adaptation strategies of roots and shoots are discussed.
RESULTS AND DISCUSSION
A Spatiotemporal Approach to Monitor N Starvation
The aim of this work was to characterize the organ-specific response kinetics to sudden and complete N starvation of adult Arabidopsis plants growing in a hydroponic device under a short-day cycle. As mentioned above, previous studies have either characterized complete and long-term N starvation in seedlings (Scheible et al., 2004; Morcuende et al., 2007) or N limitation in mature plants (Bi et al., 2007; Tschoep et al., 2009), but none of them has studied the integrative responses of metabolism and genome-wide gene expression of the roots and shoots of mature plants after a mid-term or long-term complete N starvation. Thus, to study in a spatiotemporal manner the impact of complete N starvation, Arabidopsis plants were cultivated on high-nitrate supply (6 mm NO3−) for 5 weeks and then transferred to N-free medium for 10 d. After 1, 2, 4, and 10 d of starvation, root and shoot samples were collected for the analysis of growth, metabolite contents, enzyme and nitrate uptake activities, and transcriptome and metabolome analyses. Targeted analyses were performed on two independent biological experiments, with three to five individual plants analyzed at each time point. The metabolomic analysis is representative of two biological experiments using three independent replicates each. The transcriptome analysis was conducted through three independent experiments with each of them representing pools of 12 plants.
Early N Remobilization in Shoots to Support Root Growth
At the onset of starvation, the plants had reached the rosette stage with a fresh weight of 240 mg, which corresponds to developmental stage 3.7 (Boyes et al., 2001). During the 10 d of complete N starvation, the relative shoot growth rate (fresh weight basis) slowed down in a logarithmic manner (Supplemental Fig. S1). The shoot biomass doubled during these 10 d to reach 480 mg fresh weight (Fig. 1A). In comparison, plants grown for 45 d on ample N reached a shoot biomass that was 3.2 times higher (1,536 mg). The root relative growth rate was increased during the first 4 d of N starvation and then slowed. The root biomass increased 8-fold during the 10 d (Fig. 1A). The capacity of plants to grow in the absence of an external N supply demonstrates the large capacity to remobilize N from internal stores. The different growth patterns of the shoots and roots led to a strong decrease in the shoot-root ratio from 5 to 1.1 after 10 d of starvation (Fig. 1A). This adaptive behavior has been well described (Drew, 1975; Scheible et al., 1997). Under our conditions, a significantly decreased shoot-root ratio was already observed after 2 d of N starvation, showing a very rapid impact of the N starvation on root and shoot growth.
Figure 1.
Biomass and the levels of nitrate, total amino acids, total protein, soluble sugars, and starch in the roots and shoots of Arabidopsis during N starvation. A, Biomass (circles) and the shoot-root ratio (triangles). B, Nitrate. C, Total amino acids. D, Total protein. E, Glc (circles) and Fru (triangles). F, Suc (circles) and starch (triangles). White symbols, Roots; black symbols, shoots. Arabidopsis plants were grown hydroponically for 35 d in a solution containing 6 mm NO3−were then transferred to a 0 mm nitrate solution (day 0) for 10 d (under irradiation of 150 μmol photons m−2 s−1). Samples were collected at days 0, 1, 2, 4, and 10. The values are means ± se of three replicates (pooling three plants for 0, 1, and 2 d). FW, Fresh weight.
To characterize the N starvation kinetics under our conditions and to explain the rapidly changing growth rates, we first measured the levels of N assimilation-related metabolites, such as nitrate, total amino acids, total proteins, soluble sugars, and starch, in the roots and shoots. The nitrate content was three times higher in the shoots than in the roots after 35 d on 6 mm NO3− (Fig. 1B). The sudden deprivation of N in the growth medium led to a rapid decrease of nitrate in both organs (Fig. 1B). After 24 h, the shoot nitrate content declined to 70% of the initial nitrate content and was only 50% after 2 d. In the roots, the nitrate content decreased to 70% and 25% of the initial value after 1 and 2 d, respectively. After 4 d, nitrate was barely detectable in the roots (1 μmol g−1 fresh weight) but was still at approximately 10% of the initial level in the shoots (18 μmol g−1 fresh weight). After 10 d of starvation, the nitrate levels were undetectable in roots and shoots.
The total amino acid levels in the shoots did not change during the first 24 h of starvation but decreased rapidly over the next 3 d to 61% of the initial value. However, longer starvation resulted in only a slight further decrease (reaching 51% of the initial value; Fig. 1C). A minimal pool size of amino acids might be vital for the synthesis of essential proteins or other compounds with a high turnover. In the roots, the total amino acid content increased during the first day of starvation and then returned to the initial level at the end of the starvation period (Fig. 1C). The initial increase might have been due to either a remobilization of amino acids from the shoots to the roots or to the degradation of proteins in the roots. The latter hypothesis has been proposed previously for C-starved roots (Brouquisse et al., 1998). In N-starved Arabidopsis seedlings, amino acid levels have been reported to be low and to rise within 3 to 8 h after nitrate resupply and then to decrease again when protein synthesis commences (Scheible et al., 2004). However, when plants are grown under continuous N limitation, the total amino acid levels in the shoots have been found to be higher than under high-N conditions (Tschoep et al., 2009). This surprising observation has been explained by a decreased utilization of amino acids for protein synthesis and growth. In our conditions, the root growth rate increased in response to sudden N starvation, and amino acids were remobilized to sustain root growth.
The total protein content decreased by 35% and 22% after 10 d of starvation in the shoots and roots, respectively (Fig. 1D). The lower decrease in the roots could be correlated with the fact that the amino acids were exported from the shoots to the roots. The N remobilization from the shoots to the growing roots through the transport of amino acids seems to be one of the adaptive strategies to N starvation.
The intimate relationship between N and C metabolism has been well established (for review, see Stitt and Krapp, 1999); for example, starch typically accumulates under low N (Scheible et al., 1997; Stitt and Krapp, 1999). In addition, sugars, especially Glc, Fru, and Suc, are important signaling molecules (Krapp et al., 2002; Rolland and Sheen, 2005; Rook et al., 2006; Li et al., 2011), and starch is a main indicator for growth (Sulpice et al., 2009). Thus, we analyzed the sugar accumulation kinetics in response to a sudden N starvation in the roots and shoots. Soluble sugar and starch levels were measured using spectrophotometrical assays. In shoots, the starch and soluble sugar contents increased dramatically during starvation (Fig. 1, E and F). The starch level tripled in 48 h and increased by 17 times after 10 d of starvation, whereas the Suc level did not change significantly during the first 48 h but increased rapidly over the following 8 d by 4.3 times. The Glc and Fru levels increased from 24 h onward and, at 10 d of starvation, reached 14 and five times higher levels, respectively. In the roots, the levels of the soluble sugars Glc, Fru, and Suc increased from the beginning of the starvation period, increasing to 3.8, 10, and 4.7 times the initial level after 10 d of starvation, whereas the starch level was below the limit of detection. The starch level tripled in 48 h and increased by 17 times after 10 d of starvation, whereas the Suc level did not change significantly during the first 48 h but increased rapidly over the following 8 d, by 4.3 times. Glc and Fru levels increased from 24 h onward and reached at 10 d of starvation 14 and five times higher levels, respectively. In roots, the levels of the soluble sugars Glc, Fru, and Suc increased from the beginning of starvation, increasing 3.8, 10, and 4.7 times after 10 d of starvation, whereas the starch level was below the detection limit.
Carbohydrates are synthesized in the shoot, but as the root is the main growing sink under N starvation conditions, Suc is translocated to the roots. In our experiment, Suc accumulation was indeed detected from day 1 of starvation, whereas in the shoot, it occurred only from day 4; the hexose accumulation pattern was similar between the roots and shoots. It has been shown previously that invertase activities increase under abiotic stress conditions (Yamada et al., 2010), which then leads to the production of hexoses. However, we observed that Fru reached a plateau after 2 d of starvation, whereas Glc continued to increase during longer starvation. Glc is produced not only by the degradation of Suc but also by the diurnal turnover of starch in the leaves, which is then transported to the roots. However, in stress situations, such as N starvation, starch accumulates in very high amounts and is not completely turned over during the night. A negative correlation between the starch content and growth has been observed in such stress situations, and starch has been demonstrated to be a major integrator for growth (Sulpice et al., 2009).
Linking the morphological adaptation to the changes of nitrate, total amino acids, and carbohydrates, our data indicate that a full N starvation leads to morphological adaptation long before the internal N stores in the shoots are low. It was obvious that the remobilization from the shoots to roots was initiated very early to support root growth and that it involved both amino acids and Suc.
The responses in the roots and shoots concerning the principal traits, growth and nitrate, amino acid, and carbohydrate levels, were different from what might be expected due to the different growth responses, differences in metabolic regulation, and differences in remobilization strategies. Therefore, we analyzed the individual amino acid levels of roots and shoots in more detail throughout the entire kinetic profile.
Minor Amino Acid Levels Increase in Both Organs, Especially in Roots, in Response to N Starvation
Individual amino acids such as Leu and Arg have specific functions for plant metabolism, and recent results have demonstrated their potential role as regulatory molecules (Hannah et al., 2010; Mollá-Morales et al., 2011). Furthermore, the synthesis and degradation of individual amino acids occur through a variety of enzymatic reactions, with different C components as the substrate or product, respectively. Thus, we quantified individual amino acid levels by targeted HPLC.
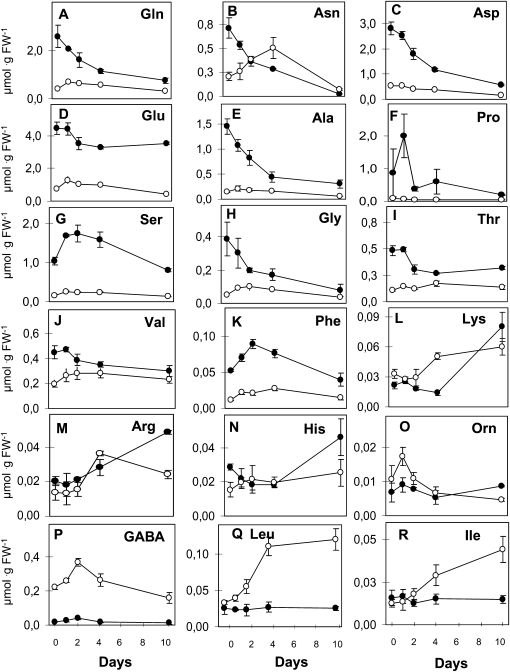
In shoots, the most pronounced decrease after 10 d of N starvation was observed for Asn level (27-fold). Asp, Ala, and Gln levels decreased 3- to 5-fold, whereas Glu level stayed rather constant (decreased by only 25%). Pro and Ser levels increased transiently but dropped after 10 d of starvation to 20% and 78% of the initial value, respectively. Gly level decreased steadily during the entire starvation period, reaching 20% of its initial level after 10 d. Val and Thr levels decreased slightly, whereas Leu, Ile, and Orn levels did not change significantly. Interestingly, the levels of other minor amino acids, such as Lys, Arg, and His, increased, especially during the long-term starvation. (Fig. 2).
Figure 2.
Individual amino acid levels during N starvation. Amino acids were measured by HPLC on the same plant samples as described in Figure 1. The values are means ± se of three replicates (pooling three plants for 0, 1, and 2 d). FW, Fresh weight.
In the roots, the kinetics of the amino acid depletion during starvation was different. Transient increases were observed for Glu, Gln, and Asn levels (Fig. 2), but after 10 d of starvation, the levels of Glu, Gln, Asp, and Asn were reduced to 50%, 75%, 40%, and 25%, respectively. However, no major change in the Gln-Glu ratio was observed. The total quantity of minor amino acids increased slightly during the first 4 d of starvation, and the quantity of some minor amino acids (i.e. Lys, Arg, and Tyr) doubled (Fig. 2). Interestingly, the minor amino acids represented 22% of the total amino acids at the end of the starvation period (compared with 10% at the start of the starvation period). This increased proportion was also observed for the shoot, increasing from 1.3% to 7%.
Fritz et al. (2006) analyzed the impact of the C and N status on amino acid profiles in tobacco (Nicotiana tabacum) leaves, where under N limitation conditions, a decrease of all the amino acids in the shoots was observed, with the biggest changes for His and Gln. In our experiment of total N starvation in Arabidopsis, the main decrease was found for Asn, and several minor amino acids were also increased in the shoots at 10 d of starvation. These different observations might reflect a different physiological response to complete N starvation and/or major differences between these two species. Amino acids also serve as precursors of secondary compounds in metabolic pathways that vary between tobacco and Arabidopsis. However, the intriguing observation that Glu levels were rather stable, independent of the N status (Fritz et al., 2006), was confirmed by our study, especially during the long-term starvation. In addition, the changes in amino acid levels in response to N stress are undoubtedly difficult to dissect: changes in amino acid levels are known to occur under other abiotic stresses (for review, see Joshi et al., 2010), but in the case of N stress, a stress response occurs and the N management of the plant is also perturbed.
Organic Acid and Carbohydrate Metabolisms Adapt to N Starvation in an Organ-Specific Manner
In addition to the different responses of sugar and N metabolism to N starvation in the roots and shoots, other metabolic pathways are known to be modified by N starvation (Scheible et al., 2004). Therefore, we undertook a global metabolomic analysis and concentrated on the time points of 2 d and 10 d of the kinetic profile. We chose these two time points with the aim of distinguishing between mid-term and long-term responses to N starvation, which could correspond physiologically to a direct response to N starvation (2 d) and to a remobilization situation (10 d), respectively. At 2 d, the roots were already short on nitrate, whereas the shoots still had 50% of their initial nitrate content; at 10 d, the shoots were also depleted of nitrate. For most of the amino acid and carbohydrate levels that we measured at 2 d of N starvation, both organs showed only slight changes, whereas dramatic changes occurred for several metabolites at 10 d of starvation.
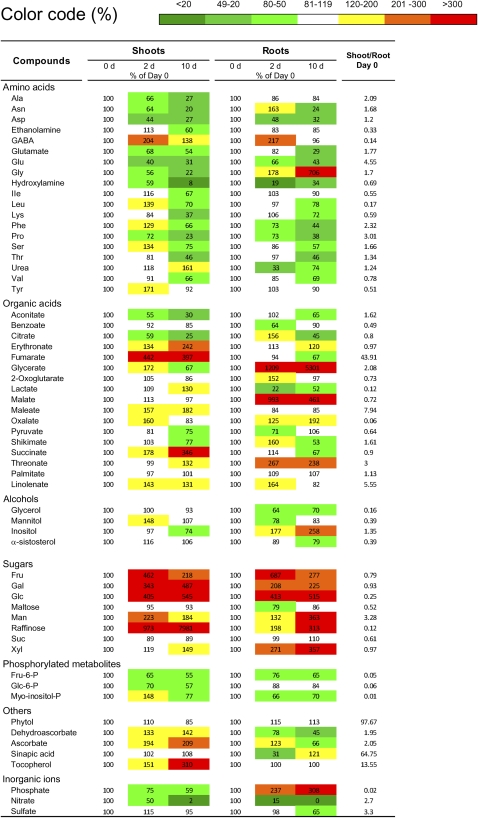
Metabolic profiling using gas chromatography (GC)-time of flight-mass spectrometry (MS) allowed us to analyze the relative amounts of 57 identified and 20 unidentified metabolites. The data obtained by the targeted analyses of amino acids and sugars (Figs. 1 and 2) were confirmed, and an additional 38 metabolites were analyzed. Table I shows the relative variations of all the identified compounds. In addition to Glc, Fru, and Suc, the levels of other carbohydrates increased upon N starvation. Man and Gal levels increased rapidly in the shoots but varied much less in the roots, whereas the opposite was observed for Xyl levels. For the shoots, the compound that varied the most was raffinose (80 times), whereas raffinose increased only three times in the roots. Raffinose has also been shown to accumulate under other abiotic stresses, such as cold and drought (Taji et al., 2002), and a role in osmoprotection and in the stabilization of cellular membranes has been proposed. In transgenic plants with increased levels of raffinose, no effect on cold acclimation has been observed (Zuther et al., 2004). However, a recent in-depth analysis of plants lacking raffinose synthase indicates that raffinose is involved in stabilizing PSII of cold-acclimated leaf cells against damage during freezing (Knaupp et al., 2011). A further hypothesis suggests that raffinose and its precursor, galactinol, may act as scavengers of hydroxyl radicals (Nishizawa et al., 2008) and, therefore, protect cells from oxidative stress, which is known to occur under several stress conditions, such as N starvation (Shin et al., 2005).
Table I. Metabolite levels in shoots and roots during N starvation.
Data are given as the percentage of day 0, and for comparison, the shoot-root ratio at day 0 is indicated. Plants were cultivated as described in Figure 1. Metabolite contents were determined by GC-MS and anion HPLC. Data are means of three independent replicates.
The levels of organic acids also showed different profiles between shoots and roots in response to N starvation (Table I). We focused on the intermediates of the tricarboxylic acid (TCA) cycle and its closely related metabolites (Supplemental Fig. S2). For roots, the main increase was observed for malate levels at 2 d of starvation, whereas in shoots, malate levels changed only slightly, but fumarate levels increased approximately four times by day 2 and stayed high until day 10 of starvation. The role of malate for N metabolism has been discussed previously (Smith and Raven, 1979). Malate production has been shown to buffer the alkalization produced by nitrate metabolism, and an increase in shoot malate content has frequently been observed after the addition of nitrate (Scheible et al., 2004). The opposite might be expected in the case of N starvation. However, in our experiment, malate levels did not change in the shoots but increased 9-fold after 2 d of N starvation in the roots. In addition to the significant increase in the malate content in the roots, at day 2, citrate and 2OG levels increased transiently by 30%; however, this was not observed in the shoots. The significant increase in malate levels might be related to its role as a substrate for the TCA cycle and, thus, increase citrate and 2OG levels. These C skeletons could then flow into γ-aminobutyric acid (GABA) pools, which were also increased in the roots at day 2.
The significant increase of fumarate in the shoots was unexpected, as fumarate levels have been found to decrease in N-limited plants (Tschoep et al., 2009). However, fumarate appears to behave as a C sink for photosynthate in a manner similar to starch. It has been reported that when starch accumulation is prevented in the phosphoglucomutase1 mutant, much more C is incorporated into fumarate (Chia et al., 2000). Under N starvation, an excess of C in the shoot might lead to an observed increase in the fumarate pool. Analyses of a cytosolic fumarase null mutant (fum2; Pracharoenwattana et al., 2010) have revealed a link between amino acids and fumarate. Interestingly, in the study of Tschoep et al. (2009) on N-limited plants, in addition to fumarate, amino acid levels also changed in an opposite direction, as was observed in our study of total N starvation (see above).
Other metabolites were also altered in an organ-specific manner after N starvation (Table I). An unexpected observation was the 5-fold increase in the phosphate content in the roots in comparison with the 2-fold decrease in the shoots. However, the interaction of nitrate and phosphate has not been well studied. Recently, Kant et al. (2011) have shown that the nitrate and phosphate supply have antagonistic interactions in their accumulation in plants, and the shoot phosphate level increases under N-limiting conditions. Conversely, the shoot phosphate content decreased in our study of total N limitation. The model for the cross talk of nitrate and phosphate by Kant and coworkers (2011) suggests that nitrate inhibits phosphate uptake by the roots; therefore, more phosphate is taken up under low external nitrate, which then accumulates in the shoots. In the case of N starvation, we propose that the increased uptake of phosphate leads to an increase in the root phosphate pool, as resources are directed to this growing sink under N stress conditions. Fine-tuning of the cross talk between the regulation of nutrient ion homeostasis might be a general feature, as has been described for phosphorus and sulfur (Rouached et al., 2011).
As for the changes in the main C and N compounds described above, we also showed that the global metabolic changes in response to N starvation were dependent on the nature of the organ. To give clearer indications of the metabolic adaptations in these two organs, we analyzed the N metabolism-related enzyme activity and N uptake capacity.
Long-Term N Starvation Increases N Remobilization Enzyme Activities in Shoots and the Capacity of High-Affinity Nitrate Uptake in Roots
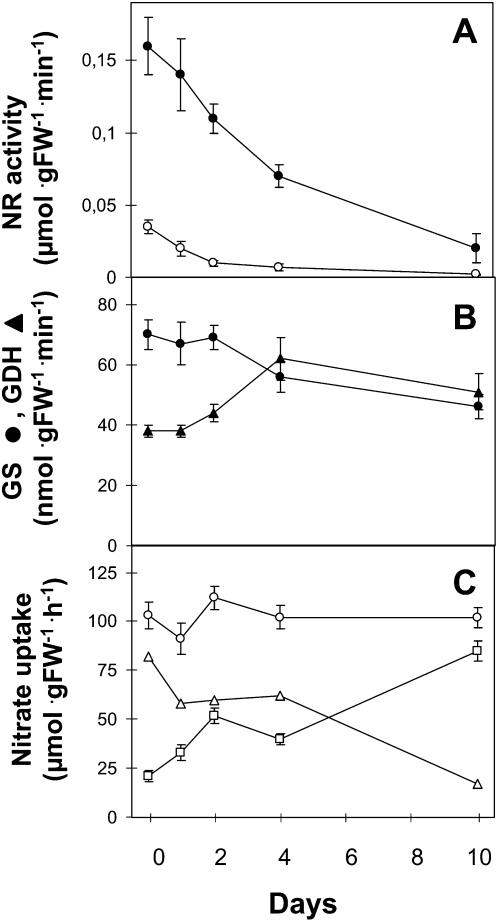
Steady-state metabolite levels are the result of fluxes in the substrates and enzyme activities. In characterizing the mid-term and long-term effects of N starvation, it is clear that changes in metabolite levels may be accompanied by adaptive modifications of protein abundance and activities. Therefore, we studied the activities of the key enzymes in N assimilation and remobilization. We measured NR activity in the shoots and roots in the absence of magnesium to detect total NR activity and in the presence of magnesium to distinguish between total activity and posttranslationally activated NR (Kaiser and Huber, 1994). Under our conditions, the total shoot NR activity was approximately four times higher than the total root NR activity, indicating that, in Arabidopsis, the majority of nitrate assimilation occurred in the shoots (Fig. 3A). The maximal extractable NR activity continuously decreased from the onset of starvation to barely undetectable levels after 4 and 10 d in the roots and shoots, respectively (Fig. 3A), while the activation state of NR was slightly increased in the roots and shoots (Supplemental Fig. S3).
Figure 3.
N assimilation enzymes and nitrate uptake activities during N starvation. A and B, Nitrate reductase (A) and GDH (triangles) and GS (circles; B) were measured in the same plants as described in Figure 1. The values are means ± se of three replicates (pooling three plants for 0, 1, and 2 d). Roots, white circles; shoots, black circles. C, Root 15NO3− influx was measured after a 5-min labeling with a complete nutrient solution containing either 0.2 mm (squares) or 6 mm (circles) 15NO3−. Plants were grown as in Figure 1. LATS (triangles) was calculated as the uptake at 6 mm minus the uptake at 0.2 mm. The values are means ± se of five replicates. FW, Fresh weight.
Glutamine synthetase (GS) and GDH (NAD-GDH and NADH-GDH) activities were measured only in shoot extracts, because NR activity was higher in the shoots than in the roots under our conditions, again indicating that most N assimilation occurred in the shoots under our conditions. The GS activity remained constant during the first 2 d of starvation and decreased by a factor of 2 after 10 d of starvation (Fig. 3B). In plants, a change in total GS is the result of both chloroplastic GS2 and cytosolic GS1 fluctuations; thus, because GS2 was the most represented GS isoenzyme in the shoots, the decrease in total GS activity can be attributed to a decrease in GS2.
In contrast to GS activity, GDH activity increased during starvation. GDH catalyzes a reversible reaction, and the in vivo role of this enzyme is still discussed. Therefore, we measured both the NADH-dependent formation of Glu and the NAD-dependent formation of ammonium and 2OG. NADH-dependent GDH activity increased after 2 d of starvation and reached 150% of the initial activity after 10 d of starvation. NAD-dependent GDH activity increased slightly faster, reaching a maximal level of 160% of the initial activity at day 4, and remained high (140%) at day 10 (Fig. 3B; Supplemental Fig. S4). Although the function of GDH in planta is still under discussion, its role in amino acid catabolism might explain the involvement of GDH in N remobilization during N starvation, as has been proposed for senescence (Diaz et al., 2006; Masclaux-Daubresse et al., 2006). The deamination catalyzed by GDH appears to be an important source of ammonia, which can be reassimilated into N transport compounds (e.g. amino acids, particularly Gln and Asn). However, evidence that GDH is a stress-responsive protein that may reflect an additional/alternative route of the GS/GOGAT pathway for ammonia assimilation has been growing (Skopelitis et al., 2006). In tobacco and grape (Vitis vinifera), there is evidence for a role of GDH in recycling ammonium in companion cells (Dubois et al., 2003; Tercé-Laforgue et al., 2004; Fontaine et al., 2006). Indeed, intracellular ammonia, due to exogenous ammonium (Restivo, 2004; Tercé-Laforgue et al., 2004), senescence-induced high proteolytic activities (Masclaux et al., 2000; Loulakakis et al., 2002), or abiotic stress (Lutts et al., 1999; Hoai et al., 2003), has resulted in increased aminating GDH activity in vitro. Under N starvation conditions, ammonia is produced by several catabolic processes, and GDH activity has also been shown to increase by 200% in plants grown under limited N (Tschoep et al., 2009).
To measure the activity of the high-affinity nitrate transport system (HATS), the root nitrate influx was measured at a concentration of 0.2 mm 15NO3− (Fig. 3C). As expected, HATS activity increased by 2.4-fold during the first 2 d of N starvation and then decreased after 4 d in N-free nutrient solution. However, after an extended starvation (10 d), the 15NO3− root high-affinity influx capacity increased again to 4.2 times higher than that at the onset of starvation. During the same experiments, the root influx was measured at 6 mm 15NO3−and the influx at 0.2 mm 15NO3− was subtracted to evaluate the activity of the low-affinity nitrate transport system (LATS). There was no significant change in the 6 mm 15NO3− influx during the N starvation experiment; therefore, the calculated LATS activity decreased as HATS activity increased. At the beginning of N starvation, HATS activity represented approximately 20% of the total 15NO3− root influx measured at 6 mm, whereas it reached 86% after 10 d in N-free nutrient solution.
This time course resembles that typically described for nitrate influx (Clarkson, 1986; Lejay et al., 1999), which has been interpreted by the action of two antagonistic regulatory mechanisms: the initial stimulation of carrier synthesis due to relief from the repression by N metabolites, and the subsequent turnover of the carrier related to the decreased induction by nitrate (Clarkson, 1986). However, HATS and LATS capacities have not been studied in Arabidopsis during a time course of long-term starvation, and the increase in HATS after 10 d of starvation is surprising and might indicate a new (undescribed) regulatory mechanism.
Global Transcriptome Responses Highlight a Spatiotemporal Adaptation toward N Starvation
The adaptation of plants to the environment includes a reprogramming of transcription and changes in the steady-state levels of transcripts. This has been shown to occur during N starvation in young seedlings (Scheible et al., 2004), but it has not been analyzed in an organ-specific manner for mature plants and after mid-term (2 d) and long-term (10 d) N starvation. Using our highly controlled hydroponic growth system, we performed a transcriptomic analysis using the CATMA array (version 2.2 or 2.3) containing 24,576 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). In addition to chloroplast and mitochondria gene-specific probes, version 2.3 of this full-genome microarray also contains 465 nonredundant 60-mer probes that are specific for known miRNA precursors and predicted small RNA precursors, which have been designed based on the identification of stable stem-loop structures throughout the Arabidopsis genome using a bioinformatic analysis in collaboration with O. Voinnet (CNRS-Institut de Biologie Moléculaire des Plantes Strasbourg/Eidgenössisch Technische Hochschule Zurich). The experimental design was arranged to compare the transcriptional status in the plant at 2 d and 10 d of N starvation with that before the onset of starvation. In all cases, the root and shoots were analyzed separately. Three independent replicates were used and analyzed during a time period of 4 years (three independent biological samples). Overall, 638 and 772 genes (of the 22,554 genes) were found to be differentially expressed in roots and shoots, respectively, at the significance threshold of Bonferroni P < 0.05 (Supplemental Table S1). This strategy allowed us to obtain a very robust profile of the transcriptome. Using real-time reverse transcription (RT)-PCR, we confirmed the observed expression changes for 25 of the 26 genes analyzed (Supplemental Fig. S5).
Mid-Term and Long-Term Responses in Shoots and Roots
For the roots, after 2 and 10 d of starvation, 232 and 608 genes, respectively, were found to be differentially expressed. For the shoots, the transcriptional response to N starvation appeared to be delayed, with only 150 genes differentially expressed at day 2 but with 697 at day 10. Indeed, the comparison between days 2 and 10 of starvation, which emphasizes late gene expression changes, confirmed this observation (224 for roots and 603 for shoots; Table II). This result underlines the kinetic difference of the response to N starvation between these organs and might reflect the different strategies developed by plants at the organ level to face N starvation. Genes whose expression was modified were compared using Venn diagrams to identify the genes that were specifically induced or repressed after mid-term starvation (2 d) or long-term starvation (10 d; Supplemental Fig. S6). We found that 18 and 22 genes were differentially expressed in a transient manner after 2 d of N starvation in the roots and shoots, respectively, whereas the number of genes whose expression was specifically changed in roots or shoots after long-term N starvation was much more significant, especially in the roots (244 genes) compared with the shoots (103 genes; Table II). Tables III and IV show the 15 most differentially expressed genes (at least a 2-fold induction or repression) for the kinetically differentially regulated genes in the roots and shoots. Among the differentially expressed genes in both organs, only a few genes displayed opposing regulation at early and late N starvation (eight and 21, respectively, in roots and three and 14, respectively, in shoots).
Table II. Mid-term and long-term gene expression changes.
Total numbers of differentially expressed genes in roots and shoots for each comparison are given. Genes were classified as “all” differentially expressed and as “specific” differentially expressed genes for the given comparison. These data are presented as a Venn diagram in Supplemental Figure S6.
| Organ | Roots |
Shoots |
||||||||||
| Comparison | 0 to 2 d |
2 to 10 d |
0 to 10 d |
0 to 2 d |
2 to 10 d |
0 to 10 d |
||||||
| Expression | All | Specific | All | Specific | All | Specific | All | Specific | All | Specific | All | Specific |
| Up-regulated genes | 62 | 7 | 162 | 16 | 275 | 103 | 92 | 17 | 274 | 22 | 314 | 40 |
| Down-regulated genes | 170 | 11 | 62 | 1 | 333 | 141 | 58 | 5 | 319 | 32 | 383 | 63 |
| Total | 232 | 18 | 224 | 17 | 608 | 244 | 150 | 22 | 603 | 54 | 697 | 103 |
Table III. The most differentially expressed genes in roots.
Of all the differentially expressed genes, only those with at least 2-fold changes, and maximal 15 genes for each different response pattern, are listed. Expression changes are given as log2. Expression changes in boldface correspond to genes differentially expressed at the significance threshold of Bonferroni P < 0.05. md, Missing data.
| Arabidopsis Genome Initiative No. | Gene Description | Root |
Shoot |
||||
| 0 to 2 d | 2 to 10 d | 0 to 10 d | 0 to 2 d | 2 to 10 d | 0 to 10d | ||
| Transiently differentially expressed at day 2 | |||||||
| AT4G23700 | ATCHX17; monovalent cation:proton antiporter | 1.20 | −0.20 | 0.03 | −0.06 | 0.03 | −0.04 |
| AT2G36690 | Oxidoreductase | −1.13 | 0.34 | −0.26 | −0.02 | −0.10 | 0.05 |
| Rapidly changed and stayed high/low | |||||||
| AT3G03910 | Oxidoreductase; Glu dehydrogenase, putative | 1.36 | 0.44 | 1.93 | 0.09 | 0.59 | 1.46 |
| AT1G70260 | Unknown protein; nodulin MtN21 family protein | 1.33 | 0.20 | 1.41 | 1.14 | −0.10 | 0.73 |
| AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE1) | 1.28 | 0.36 | 2.12 | −0.79 | 0.04 | 0.38 |
| AT3G30775 | ERD5 (PRO OXIDASE) | 1.27 | 0.20 | 0.89 | −0.11 | 0.77 | 0.75 |
| AT4G39795 | Unknown protein; senescence-associated protein-related | 1.21 | 0.52 | 2.64 | 0.06 | 0.03 | 0.19 |
| AT4G17670 | Unknown protein; senescence-associated protein-related | 1.06 | 0.19 | 1.50 | 0.21 | 0.13 | 0.87 |
| AT5G24660 | Unknown protein | −1.53 | −0.14 | −1.42 | −0.09 | 0.02 | md |
| AT5G54770 | THI1 (THIAZOLE REQUIRING); thiazole biosynthetic enzyme | −1.57 | 0.05 | −1.43 | 0.25 | 0.59 | 1.74 |
| AT1G73120 | Unknown protein | −1.58 | −0.48 | −2.67 | 0.04 | −0.06 | −0.42 |
| AT2G15620 | NIR1; ferredoxin-nitrate reductase; ferredoxin-nitrite reductase, putative | −1.60 | 0.14 | −0.82 | −0.45 | −0.55 | −1.02 |
| AT4G19690 | IRT1; cadmium ion transporter/iron ion transporter (IRT1) | −1.61 | −0.22 | −2.06 | 0.09 | −0.01 | 0.14 |
| AT1G24280 | G6PD3; Glc-6-P 1-dehydrogenase ATP-binding | −1.63 | −0.29 | −1.29 | −0.32 | −0.58 | −1.06 |
| AT5G07680 | ANAC079/ANAC080; transcription factor; no apical meristem (NAM) family protein | −1.67 | 0.22 | −0.82 | 0.09 | 0.28 | 0.50 |
| AT5G52790 | Unknown protein; CBS domain-containing protein-related | −1.77 | −0.37 | −1.74 | 0.13 | 0.02 | 0.04 |
| AT3G46900 | COPT2; copper ion transporter; copper transporter, putative | −1.77 | −0.20 | −1.39 | 0.41 | 0.25 | −0.37 |
| AT4G34950 | Unknown protein; nodulin family protein | −1.78 | −0.20 | −1.06 | −0.75 | 0.59 | 1.12 |
| AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE21) | −1.82 | −0.10 | −2.11 | −1.95 | −0.08 | −1.88 |
| AT4G05230 | Unknown protein; ubiquitin family protein | −1.82 | −0.38 | −1.95 | −0.02 | −0.25 | −0.02 |
| AT1G49500 | Unknown protein | −2.21 | −0.11 | −1.80 | −1.15 | −2.49 | −3.61 |
| AT1G77760 | NIA1 (NITRATE REDUCTASE1); nitrate reductase 1 (NR1) | −2.38 | −0.11 | −2.58 | −0.40 | −0.60 | −1.07 |
| AT4G29905 | Unknown protein | −2.58 | −0.42 | −3.45 | 0.11 | −0.74 | −1.47 |
| Differential expression increased during starvation | |||||||
| AT5G04120 | Catalytic; phosphoglycerate/bisphosphoglycerate mutase family protein | 1.41 | 2.53 | 3.40 | 0.08 | −0.03 | 0.00 |
| AT5G45380 | Solute:sodium symporter/urea transporter; sodium:solute symporter family protein | 3.52 | 2.45 | 5.20 | 0.88 | 1.52 | 2.62 |
| AT4G37220 | Unknown protein; stress-responsive protein, putative | 0.68 | 2.34 | 2.04 | −0.15 | −0.07 | −0.65 |
| AT4G32930 | Unknown protein GAMMA-VPE; Cys-type endopeptidase/legumain | 1.16 | 1.71 | 2.61 | 1.37 | 2.34 | 4.54 |
| AT1G05250 | Peroxidase | 1.12 | 1.67 | 2.38 | 0.00 | −0.09 | −0.06 |
| AT5G63840 | RSW3 (RADIAL SWELLING3); hydrolase, putative | 1.29 | 1.65 | 2.87 | 0.01 | 0.05 | 0.23 |
| AT1G73220 | Carbohydrate transporter/transporter | 1.70 | 1.59 | 3.61 | 0.07 | −0.08 | 0.27 |
| AT4G32950 | Catalytic/protein phosphatase type 2C | 3.96 | 1.58 | 3.54 | 0.12 | −0.01 | −0.13 |
| AT5E22650 | EUGENE prediction | 1.12 | 1.33 | 2.31 | 0.06 | −0.01 | −0.01 |
| AT4G33040 | Electron transporter/thiol-disulfide exchange intermediate; glutaredoxin family protein | 1.17 | 1.11 | 2.03 | 0.84 | 1.74 | 2.29 |
| AT3G48340 | Cys-type endopeptidase/Cys-type peptidase | 1.26 | 1.09 | 2.32 | −0.12 | 0.04 | 0.01 |
| AT5G42610 | Unknown protein | 1.39 | 1.02 | 2.53 | 0.16 | 0.61 | 1.33 |
| AT1G64590 | Oxidoreductase; short-chain dehydrogenase/reductase (SDR) family protein | 1.76 | 1.00 | 2.09 | −0.02 | 0.07 | 0.08 |
| AT5G60770 | ATNRT2.4; nitrate transporter; high-affinity nitrate transporter, putative | 0.97 | 0.70 | 2.22 | 0.08 | −0.11 | −0.20 |
| AT4G12360 | Lipid binding; protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.06 | 0.64 | 0.94 | −0.05 | 0.02 | 0.14 |
| AT5G09220 | AAP2 (AMINO ACID PERMEASE2); amino acid permease | 1.23 | 0.58 | 2.30 | 0.36 | 0.81 | 2.11 |
| AT5G01740 | Unknown protein | −1.86 | −0.61 | −2.66 | 0.09 | −0.17 | 0.14 |
| AT3G25190 | Unknown protein; nodulin, putative | −1.64 | −0.63 | −3.02 | 0.24 | −1.25 | −1.53 |
| AT3G07720 | Unknown protein; kelch repeat-containing protein | −1.33 | −0.63 | −2.55 | −0.74 | −0.85 | −1.66 |
| AT2G25450 | Unknown protein; 2OG-dependent dioxygenase, putative | −1.35 | −0.70 | −2.67 | 1.05 | 0.17 | 0.41 |
| AT1E59250 | EUGENE prediction | −1.49 | −0.71 | −1.64 | −0.27 | −0.10 | 0.11 |
| AT1G09560 | GLP5 (GERMIN-LIKE PROTEIN5); nutrient reservoir | −1.61 | −0.75 | −2.26 | 0.17 | −0.26 | −0.22 |
| AT1G01580 | Ferric-chelate reductase/oxidoreductase; ferric-chelate reductase, putative | −1.99 | −0.78 | −2.83 | −0.05 | −0.02 | −0.03 |
| AT5G10210 | Unknown protein | −1.22 | −0.80 | −3.25 | 0.62 | −0.18 | 0.70 |
| AT4G19750 | Glycosyl hydrolase family 18 protein | −1.33 | −0.89 | −2.82 | −0.31 | −0.19 | −0.34 |
| AT2G22122 | Unknown protein | −3.00 | −0.95 | −4.65 | −0.05 | −0.20 | −0.03 |
| AT4G04810 | Unknown protein; Met sulfoxide reductase domain-containing protein | −1.36 | −0.99 | −2.66 | 0.05 | 0.04 | 0.04 |
| AT4G04830 | Unknown protein; Met sulfoxide reductase domain-containing protein | −1.38 | −1.08 | −3.37 | 0.04 | 0.15 | 0.59 |
| AT5G23220 | Catalytic; isochorismatase hydrolase family protein | −2.53 | −1.25 | −4.25 | 0.09 | −0.02 | −0.11 |
| mirspot446a | MIR156E | −1.44 | −1.53 | −2.34 | 0.36 | −2.03 | −0.99 |
| AT2G05440 | Unknown protein; Gly-rich protein | −1.76 | −2.12 | −5.10 | 0.05 | 0.09 | −0.03 |
| Differential expression increased from day 2 | |||||||
| AT1G49320 | Unknown protein; BURP domain-containing protein | 0.43 | 2.63 | 2.17 | 0.00 | −0.17 | 0.35 |
| AT1G12940 | ATNRT2.5; nitrate transporter; high-affinity nitrate transporter, putative | md | 2.54 | 4.92 | −0.03 | 0.68 | 1.08 |
| AT3G50970 | XERO2; dehydrin xero2 (XERO2)/low-temperature- induced protein LTI30 (LTI30) | 0.21 | 1.78 | 1.89 | −0.52 | 0.62 | 0.08 |
| mirspot1123a | MIR169H | 0.16 | 1.63 | 1.72 | 0.15 | 0.31 | 0.54 |
| AT3G58550 | Lipid binding; protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 0.39 | 1.60 | 1.76 | −0.04 | 0.04 | 0.28 |
| AT5G06510 | Transcription factor; CCAAT-binding transcription factor family protein | 0.31 | 1.51 | 2.20 | 0.62 | 1.17 | 2.46 |
| AT1G72830 | HAP2C; transcription factor | 0.33 | 1.49 | 2.50 | 0.16 | 0.61 | 0.73 |
| AT1G73810 | Unknown protein | 0.05 | 1.41 | 2.35 | −0.21 | 0.34 | 0.67 |
| AT1G01380 | ETC1 (ENHANCER OF TRY AND CPC1); myb family transcription factor | −0.04 | 1.32 | 2.13 | −0.01 | −0.05 | 0.15 |
| AT1G54575 | Unknown protein | 0.65 | 1.25 | 2.01 | 0.16 | 1.10 | 2.47 |
| AT1G21890 | Unknown protein; nodulin MtN21 family protein | −0.07 | 1.21 | 2.79 | −0.01 | 0.05 | 0.32 |
| AT3G05690 | HAP2B; transcription factor | 0.15 | 0.92 | 1.82 | 0.08 | 1.08 | 1.76 |
| mirspot1002 | MIR160C | 0.47 | 0.72 | 2.00 | −0.32 | 0.50 | 1.62 |
| mirspot690a | MIR836 | 0.18 | 0.70 | 1.93 | 0.06 | −0.07 | −0.10 |
| AT3G21420 | Oxidoreductase; oxidoreductase, 2OG-Fe(II) oxygenase family protein | 0.64 | 0.69 | 1.84 | 0.04 | −0.02 | 0.31 |
| AT3G58990 | Hydrolyase; aconitase C-terminal domain- containing protein | −0.66 | −0.61 | −1.44 | −0.51 | −1.55 | −2.17 |
| AT3G18080 | Hydrolase, hydrolyzing O-glycosyl compounds; glycosyl hydrolase family 1 protein | −0.65 | −0.62 | −1.76 | −0.03 | −0.04 | 0.05 |
| AT5G10180 | AST68; sulfate transporter | −0.56 | −0.66 | −1.67 | −0.17 | −0.84 | −1.66 |
| AT2G36120 | Unknown protein; pseudogene, Gly-rich protein | −0.61 | −0.69 | −2.51 | −0.21 | −0.58 | −1.40 |
| AT2G36830 | GAMMA-TIP; water channel; major intrinsic family protein/MIP family protein | −0.55 | −0.72 | −1.87 | −0.85 | −1.12 | −2.96 |
| AT5G23020 | MAM-L (METHYLTHIOALKYMALATE SYNTHASE-LIKE); 2-isopropylmalate synthase 2 | −0.54 | −0.74 | −1.90 | −0.10 | −1.03 | −0.39 |
| AT3G61430 | PIP1A; water channel; plasma membrane intrinsic protein 1A | −0.41 | −0.78 | −2.00 | −0.05 | −0.87 | −1.46 |
| AT4G01470 | GAMMA-TIP3/TIP1;3; water channel; major intrinsic family protein/MIP family protein | −0.39 | −0.84 | −1.78 | 0.09 | 0.22 | 0.23 |
| AT3G02885 | GASA5; gibberellin-regulated protein 5 (GASA5)/ gibberellin-responsive protein 5 | −0.59 | −0.85 | −1.79 | 0.17 | −0.09 | −1.16 |
| AT2G33790 | Unknown protein; pollen Ole e 1 allergen and extensin family protein | 0.01 | −0.87 | −1.97 | −0.09 | 0.08 | 0.23 |
| AT4G17340 | DELTA-TIP2/TIP2;2; water channel; major intrinsic family protein/MIP family protein | −0.25 | −0.97 | −2.30 | 0.01 | −0.02 | 0.08 |
| AT5G47450 | AtTIP2;3; water channel; major intrinsic family protein/MIP family protein | −0.23 | −0.98 | −2.37 | 0.05 | −0.02 | −0.04 |
| AT1G48750 | Lipid-binding clathrin-binding | −0.54 | −1.24 | −2.62 | 0.13 | −0.10 | 0.42 |
| AT3G05950 | Nutrient reservoir; germin-like protein, putative | −0.10 | −2.10 | −3.77 | −0.07 | −0.07 | −0.05 |
| AT4G25100 | FSD1 (FE SUPEROXIDE DISMUTASE1) | −0.37 | −2.33 | −2.23 | 0.14 | −1.02 | 0.15 |
Identifiers corresponding to genes encoding precursors of known or predicted new small RNA are listed as shown on the Flagdb++ Web site (http://urgv.evry.inra.fr/projects/FLAGdb++/HTML/index.shtml; http://urgv.evry.inra.fr/CATdb).
Table IV. The most differentially expressed genes in shoots.
Of all the differentially expressed genes, only those with at least 2-fold changes, and maximal 15 genes for each different response pattern, are listed. Expression changes are given as log2. Expression changes in boldface correspond to genes differentially expressed at the significance threshold of Bonferroni P < 0.05
| Arabidopsis Genome Initiative No. | Gene Description | Shoot |
Root |
||||
| 0 to 2 d | 2 to 10 d | 0 to 10 d | 0 to 2 d | 2 to 10 d | 0 to 10 d | ||
| Transiently differentially expressed at day 2 | |||||||
| AT1G70260 | Unknown protein; nodulin MtN21 family protein | 1.14 | −0.10 | 0.73 | 1.33 | 0.20 | 1.41 |
| AT2G33380 | RD20 (RESPONSIVE TO DESICCATION20) | 1.00 | −0.35 | 0.22 | 0.11 | 0.03 | −0.07 |
| AT2G28000 | CPN60A; ATP-binding/protein-binding; Rubisco subunit-binding protein α-subunit | −1.04 | −0.25 | −0.72 | 0.08 | −0.36 | 0.11 |
| Rapidly changed and stayed high/low | |||||||
| AT3G19700 | Protein kinase/branched-chain-amino acid transaminase | 1.27 | −0.24 | 1.27 | −0.51 | −0.18 | −0.77 |
| AT5G23010 | MAM1; 2-isopropylmalate synthase 3 (IMS3) | 0.95 | −0.34 | 1.14 | −0.70 | −0.51 | −1.28 |
| AT1G62560 | Disulfide oxidoreductase/monooxygenase, FMO family protein | 0.95 | 0.04 | 0.90 | −0.11 | 0.02 | −0.23 |
| AT4G03060 | AOP2 (ALKENYL HYDROXALKYL-PRODUCING2) | 0.90 | −0.33 | 0.94 | 0.12 | −0.16 | −0.16 |
| AT1G67870 | Unknown protein; Gly-rich protein | 0.86 | 0.41 | 0.94 | −0.76 | 0.08 | −0.32 |
| AT4G30530 | Catalytic; defense-related protein, putative | 0.85 | −0.05 | 1.22 | −0.64 | 0.25 | −0.97 |
| AT4G28220 | NADH dehydrogenase/disulfide oxidoreductase unknown protein | 0.83 | 0.55 | 1.11 | −0.34 | −0.12 | −0.17 |
| AT1G21440 | Catalytic/isocitrate lyase; mutase family protein | 0.83 | −0.30 | 1.06 | −0.45 | −0.50 | −1.23 |
| AT2G17470 | Unknown protein | 0.79 | −0.14 | 0.89 | 0.14 | 0.09 | −0.04 |
| AT3G09390 | MT2A (METALLOTHIONEIN2A)_ metallothionein protein, putative (MT2A) | 0.79 | 0.52 | 1.26 | 0.05 | 0.46 | 0.48 |
| AT4G24010 | ATCSLG1; cellulose synthase family protein | 0.78 | 0.40 | 1.36 | −0.04 | −0.04 | −0.01 |
| AT4G12030 | Bile acid:sodium symporter; bile acid:sodium symporter family protein | 0.73 | −0.24 | 1.14 | −0.30 | −0.15 | −0.54 |
| AT1G22160 | Unknown protein; senescence-associated protein-related | 0.73 | 0.30 | 1.85 | 0.26 | 0.10 | 1.88 |
| AT5G07460 | PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE2), putative | 0.71 | −0.29 | 0.95 | 0.13 | 0.34 | −0.11 |
| AT1G54410 | Unknown protein; dehydrin family protein | 0.69 | 0.18 | 1.20 | −0.46 | 0.08 | −0.43 |
| AT1G73480 | Catalytic/hydrolase; hydrolase, α/β-fold family protein | −0.75 | −0.25 | −2.03 | 0.27 | −0.08 | 0.18 |
| AT2G31430 | Invertase/pectin methylesterase inhibitor family protein | −0.79 | 0.03 | −0.93 | −0.01 | −0.12 | −0.23 |
| AT3G58610 | Ketol-acid reductoisomerase; ketol-acid reductoisomerase | −0.80 | −0.18 | −1.00 | −0.05 | −0.52 | −0.25 |
| AT2G24050 | RNA binding; MIF4G domain-containing protein/MA3 domain-containing protein | −0.80 | 0.02 | −0.92 | 0.24 | −0.24 | 0.17 |
| AT1G72370 | P40; structural constituent of ribosome; 40S ribosomal protein SA (RPSaA) | −0.81 | −0.47 | −0.98 | 0.23 | −0.77 | 0.02 |
| AT5G22650 | HD2B (HISTONE DEACETYLASE2B) | −0.81 | −0.01 | −1.08 | 0.74 | −0.62 | 0.33 |
| AT3G48990 | AMP binding/catalytic; AMP-dependent synthetase and ligase family protein | −0.83 | 0.26 | −1.20 | −1.06 | −0.18 | −1.44 |
| AT3G44990 | XTR8; xyloglucan:xyloglucosyl transferase, putative | −0.88 | −0.19 | −0.92 | 0.02 | 0.25 | 0.69 |
| AT1G55490 | CPN60B (CHAPERONIN60 BETA); Rubisco subunit-binding protein β-subunit | −0.92 | −0.09 | −1.65 | 0.06 | −0.16 | −0.08 |
| AT5G35630 | GS2 (GLN SYNTHETASE2) | −0.95 | −0.31 | −2.77 | −0.04 | 0.40 | 0.07 |
| AT3G24420 | Catalytic/hydrolase; hydrolase, α/β-fold family protein | −0.97 | −0.21 | −1.04 | −0.17 | 0.08 | −0.25 |
| AT3G56090 | ATFER3; binding/ferric iron-binding | −1.04 | 0.00 | −1.19 | 0.29 | 0.05 | 0.70 |
| AT5G63310 | NDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE2) | −1.10 | −0.54 | −1.73 | 0.00 | −0.12 | −0.19 |
| AT2G43820 | UDP-glycosyltransferase/transferase | −1.13 | −0.34 | −1.57 | −0.34 | −0.54 | −1.19 |
| AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE21) | −1.95 | −0.08 | −1.88 | −1.82 | −0.10 | −2.11 |
| Differential expression increased during starvation | |||||||
| AT1G66390 | PAP2 (PRODUCTION OF ANTHOCYANIN PIGMENT2) | 1.30 | 3.41 | 5.11 | 0.15 | −0.04 | −0.07 |
| AT5G17220 | ATGSTF12 (GLUTATHIONE S-TRANSFERASE26); glutathione transferase | 1.56 | 2.99 | 4.92 | −0.29 | 0.13 | −0.40 |
| AT4G32930 | Unknown protein GAMMA-VPE; Cys-type endopeptidase/legumain | 1.37 | 2.34 | 4.54 | 1.16 | 1.71 | 2.61 |
| AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE) | 1.11 | 2.82 | 4.21 | 0.13 | 0.00 | 0.07 |
| AT3G16150 | Asparaginase; l-asparaginase, putative/l-Asn amidohydrolase, putative | 1.13 | 2.45 | 4.11 | 0.49 | 0.39 | 1.38 |
| AT1G56650 | PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT1) | 1.37 | 1.52 | 3.91 | 0.07 | −0.08 | −0.12 |
| AT5G07990 | TT7 (TRANSPARENT TESTA7); flavonoid 3prim-monooxygenase | 0.86 | 2.54 | 3.88 | −0.07 | 0.13 | −0.09 |
| AT3G22840 | ELIP1 (EARLY LIGHT-INDUCIBLE PROTEIN) | 0.82 | 2.18 | 3.66 | 0.65 | 0.05 | −0.07 |
| AT2G47880 | Arsenate reductase | 1.64 | 1.70 | 3.59 | 0.05 | 0.15 | 0.26 |
| AT1G34060 | C-sulfur lyase; alliinase family protein | 1.03 | 2.44 | 3.35 | 0.27 | 0.16 | 0.87 |
| AT5G15500 | Protein binding; ankyrin repeat family protein | 1.17 | 1.85 | 2.88 | 0.00 | 0.07 | −0.19 |
| AT5G54060 | Transferase, transferring glycosyl groups; glycosyltransferase family protein | 0.62 | 2.42 | 2.82 | −0.02 | 0.05 | 0.02 |
| AT5G53420 | Unknown protein | 0.72 | 1.47 | 2.75 | 0.52 | 0.58 | 1.67 |
| AT1G23130 | Unknown protein; Bet v I allergen family protein | 0.81 | 1.10 | 2.72 | −0.20 | −0.06 | −0.15 |
| AT1G62710 | BETA-VPE; Cys-type endopeptidase | 0.68 | 1.47 | 2.68 | 0.08 | 0.35 | 1.13 |
| Differential expression increased from day 2 | |||||||
| AT2G28900 | Protein translocase/Tim17/Tim22/Tim23 family protein | 0.06 | 2.10 | 2.59 | −0.31 | 0.23 | 0.33 |
| AT5G13930 | CHS (CHALCONE SYNTHASE) | 0.29 | 2.05 | 2.51 | −0.11 | 0.04 | −0.40 |
| AT4G22880 | LDOX (TANNIN-DEFICIENT SEED4); leucoanthocyanidin dioxygenase, putative | 0.44 | 1.80 | 2.65 | 0.04 | 0.19 | 0.08 |
| AT1G28330 | DRM1 (DORMANCY-ASSOCIATED PROTEIN1) | 0.25 | 1.80 | 2.29 | 0.39 | 0.75 | 1.05 |
| AT5G37600 | ATGSR1; Glu-ammonia ligase; Gln synthetase, putative | 0.39 | 1.73 | 2.55 | 0.65 | 0.41 | 0.87 |
| AT5G11670 | Malic enzyme | 0.14 | 1.60 | 2.61 | −0.40 | 0.27 | 0.17 |
| AT1G10070 | Branched-chain-amino acid transaminase/catalytic unknown protein | 0.28 | 1.45 | 2.72 | −0.07 | 0.71 | 0.35 |
| AT1G76520 | Auxin:hydrogen symporter; auxin efflux carrier family protein | 0.54 | 1.43 | 2.40 | 0.06 | 0.64 | 0.32 |
| AT2G25940 | ALPHA-VPE; Cys-type endopeptidase | 0.41 | 1.30 | 2.45 | 0.53 | 1.38 | 1.03 |
| AT2G03590 | ATUPS1; allantoin transporter | 0.28 | 1.27 | 2.35 | 0.86 | 0.28 | 1.26 |
| AT3G52180 | Phosphoprotein phosphatase (PTPKIS1) | 0.56 | 1.21 | 2.48 | 0.03 | 0.16 | 0.21 |
| AT4G19430 | Unknown protein | 0.51 | 1.15 | 2.55 | −0.02 | 0.06 | −0.07 |
| AT1G76530 | Auxin:hydrogen symporter; auxin efflux carrier family protein | 0.37 | 1.12 | 2.43 | 0.06 | 0.75 | 0.43 |
| AT1G54575 | Unknown protein | 0.16 | 1.10 | 2.47 | 0.65 | 1.25 | 2.01 |
| AT2G29670 | Unknown protein | −0.10 | 1.09 | 2.25 | 0.33 | 0.18 | 0.56 |
| AT5G25460 | Unknown protein | −0.59 | −0.69 | −2.18 | 0.29 | −0.42 | 0.41 |
| AT4G16980 | Nutrient reservoir; arabinogalactan-protein family | −0.55 | −0.83 | −2.36 | −0.16 | −0.23 | −0.16 |
| AT2G41560 | ACA4 (AUTOINHIBITED CA2+-ATPASE, ISOFORM4) | −0.26 | −1.05 | −2.37 | −1.07 | −0.51 | −0.69 |
| AT2G27420 | Cys-type endopeptidase/Cys-type peptidase | 0.15 | −1.18 | −2.23 | −0.04 | 0.01 | 0.04 |
| AT2G36120 | Unknown protein; pseudogene, Gly-rich protein | −0.17 | −1.20 | −2.22 | −0.61 | −0.69 | −2.51 |
| AT3G08740 | Translation elongation factor; elongation factor P (EF-P) family protein | −0.58 | −1.21 | −2.22 | −0.06 | −0.27 | −0.47 |
| AT1G29070 | Structural constituent of ribosome; ribosomal protein L34 family protein | −0.51 | −1.28 | −2.35 | −0.14 | −0.40 | −0.42 |
| AT3G63160 | Unknown protein | −0.47 | −1.33 | −2.19 | −0.02 | −0.01 | −0.04 |
| AT3G10360 | RNA binding; pumilio/Puf RNA-binding domain-containing protein | −0.40 | −1.34 | −2.20 | −0.36 | −0.28 | −0.63 |
| AT1G02820 | Late embryogenesis abundant 3 family protein/LEA3 family protein | 0.44 | −1.39 | −2.89 | −1.17 | 0.07 | −1.71 |
| AT1G11850 | Unknown protein | −0.46 | −1.43 | −2.45 | −0.03 | 0.00 | −0.16 |
| AT1G14880 | Unknown protein | 0.05 | −2.09 | −2.70 | −0.37 | 0.75 | −0.01 |
| AT4G27290 | S-locus protein kinase, putative | 0.21 | −2.14 | −2.55 | −0.20 | 0.14 | −0.21 |
| AT5G18600 | Arsenate reductase (glutaredoxin) | −0.58 | −2.16 | −3.04 | −0.08 | 0.03 | −0.14 |
| AT3G19030 | Unknown protein | −0.17 | −2.51 | −4.45 | −0.32 | 0.53 | −0.14 |
We then compared the expression pattern between the roots and shoots to identify genes that were specifically modified for their expression in either organ. For early and transiently modified gene expression, only one of the genes, whose expression was increased, was common between the roots and shoots (AT1G78370). Of all the differentially expressed genes at any kinetic point during the N starvation (638 and 772 genes for roots and shoots, respectively), only 142 were differentially expressed in both organs. Among these 142 genes, only 20 displayed opposing regulation in roots and shoots.
Biological Processes Affected by N Starvation
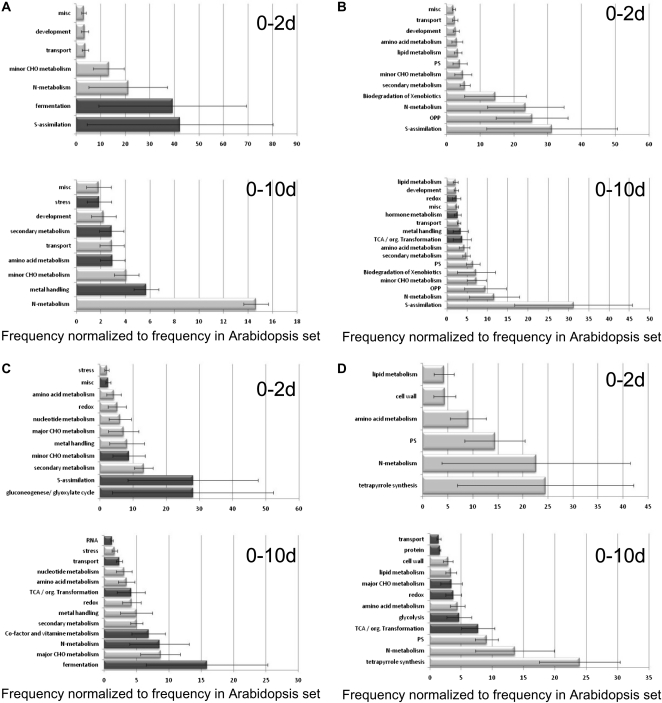
The biological processes that were most significantly and specifically affected by early and late N starvation in shoots and roots were analyzed using the classification superviewer tool from the Bio-Array Resource for Plant Biology (http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) using the MapMan classification as the source. Only those biological pathways that were significantly overrepresented for the roots and shoots at 2 and 10 d of N starvation conditions compared with day 0 were selected and are presented in Figure 4.
Figure 4.
Biological pathways with significant overrepresentation of up- and down-regulated genes (P < 0.05) in the roots or shoots under 0 to 2 d and 0 to 10 d of starvation conditions. Functional enrichment is shown for differentially expressed genes analyzed using the classification superviewer tool from the Bio-Array Resource for Plant Biology (http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) using the MapMan classification as the source. PS, Photosynthesis. Specific pathways observed only in up- or down-regulated genes are colored in dark grey. A and C, Up-regulated genes: roots (A) and shoots (C). B and D, Down-regulated genes: roots (B) and shoots (D).
In summary, for roots at days 2 and 10, the up-regulated genes that were significantly overrepresented are involved in minor carbohydrate miscellaneous whereas root-specific overrepresentation of regulated genes were in sulfur metabolism and fermentation at day 2 and metal handling, amino acid metabolism, transport, and stress at day 10. Genes whose steady-state expression decreased in the roots were overrepresented for the same functional classes, except metal handling, TCA cycle, hormone metabolism, and redox, specifically overrepresented at day 10. In contrast, the main metabolic processes affected by mid-term and long-term starvation were rather different in the shoots. For up-regulated genes, after 2 d of starvation, gluconeogenesis, sulfur assimilation, minor carbohydrate metabolism, and miscellaneous were specifically overrepresented, whereas at 10 d, the functional classes fermentation, N metabolism, cofactor and vitamin metabolism, TCA cycle, transport, and RNA processing were significantly enriched. The situation was different for the down-regulated gene set in shoots: most of the overrepresented classes were affected from day 2 onward and remained low at 10 d of starvation.
This was the case for tetrapyrrole biosynthesis, photosynthesis, N metabolism, amino acid metabolism, cell wall, and lipid metabolism. TCA cycle, glycolysis, redox, major carbohydrate metabolism, protein, and transport were overrepresented in the genes down-regulated in shoots at 10 d specifically. As early as 2 d of starvation, when nitrate levels in shoots were still high, the metabolic functional classes were already overrepresented in down- and up-regulated genes. However, other functional classes, for example, minor carbohydrate metabolism, redox, and stress, were significantly enriched but to a lesser extent. Therefore, it appeared that the shoots adjusted their gene expression to N withdrawal by rapidly modifying the expression of metabolic genes and genes of stress and protection machinery.
We wish to emphasize that we found both up- and down-regulated genes within a single functional class. Metabolic pathways are complex, and the specific regulation of subsets of one pathway or the feedback regulation inside one functional class is certainly possible. In general, these results confirm the previous analysis by Morcuende et al. (2007). However, our analysis indicates that we have been able to analyze two different levels of starvation status: a mid-term starvation, when many secondary processes, such as RNA processing and the stress response, had not yet been affected, and a long-term starvation, when stress and catabolic mechanisms had been invoked. The separate analysis of the roots and shoots reveals that the adaptation to N stress at the transcriptome level follows different patterns in each organ, similar to the contrasting adaptation of the root and shoot concerning growth and metabolism.
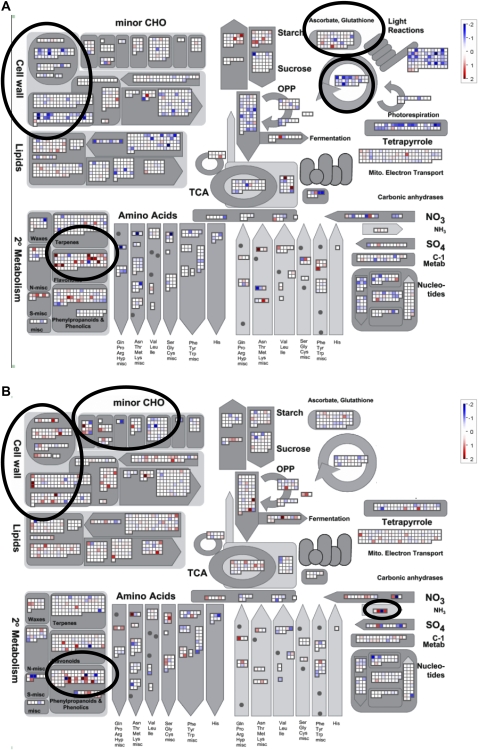
We used the MapMan tool to further analyze and compare early and late gene expression in the roots and shoots. This tool allows the visualization of modified gene expression in functional classes in more detail and also provides the possibility of directly comparing individual genes in each pathway. The overview of general metabolism indicated important differences between the differentially expressed genes for the roots and shoots after 10 d of starvation compared with the onset of starvation (Fig. 5). A significant difference was observed for genes involved in cell wall synthesis: whereas many genes were down-regulated in the shoots, they were up-regulated in roots. This difference is in agreement with the stimulation of root growth and the slowing down of shoot growth. Many genes involved in galactolipid synthesis were down-regulated after 10 d of starvation in the shoots, in contrast to the minor influence of starvation on the expression of these genes in the roots. It has been shown that N deficiency in higher plants results in a coordinated degradation of galactolipids and chlorophyll, with the deposition of specific fatty acid phytyl esters in the thylakoids and plastoglobules of chloroplasts (Gaude et al., 2007).
Figure 5.
Metabolic gene expression changes at late N starvation analyzed by the MapMan tool. (A) Shoots. (B) Roots. Ratios are given for day 10 to day 0. Plants were cultivated as described in Figure 1, and the transcriptome data were obtained as described in Figure 4.
Genes involved in secondary metabolism have been shown to respond to N starvation (Scheible et al., 2004). However, in our study, flavonoid synthesis genes were mainly differentially expressed in the shoots, whereas other phenylpropanoid synthesis genes were mostly affected in the roots. This observation was expected, as anthocyanins mainly accumulated in the shoots (Supplemental Fig. S7) after long-term N starvation.
Genes involved in starch metabolism were differentially expressed, mainly in the shoots, and different ADPG isoforms were up-regulated during starvation. Whereas APL3 (AT4G39210) expression levels were increased early, APL4 (AT2G21590) expression was modified later during starvation in the shoots. Global differences were less striking for other metabolic processes; however, specific isoforms were differentially expressed after N starvation in the root and shoots. For primary C metabolism, several gene families showed organ-specific regulation. As an example, N starvation-regulated genes involved in phosphoenolpyruvate and pyruvate metabolism were different between the roots and shoots at day 2 (Supplemental Fig. S8).
Several studies have established regulatory interactions between assimilatory sulfate and nitrate reduction (Koprivova et al., 2000). Cys synthetase, the last enzyme of the sulfate assimilation pathway, is critical because the precursor, o-acetyl-serine, is derived from the C and N assimilation pathways. An overrepresentation of sulfur assimilation transcripts was found in our study in the roots and shoots in response to N starvation. The expression of genes involved in sulfur metabolism was modified to a higher extent in the roots than in the shoots. Cys synthetase expression was decreased only in the roots, such as for APR3, whereas in the shoots, APR1 and APR2 were the most differentially expressed PAPS reductase-encoding genes. Sulfate transporters were down-regulated in the roots from day 2 onward.
Interesting differences can be deduced from our data for redox metabolism during N starvation. Whereas genes for glutathione and ascorbate metabolism were differentially expressed after N starvation mainly in the shoots, class 3 peroxidases were predominantly up-regulated in the roots (Supplemental Fig. S9).
It has been shown that several phytohormones, such as cytokinins, jasmonic acids, and salicylic acids, play important roles during the adaptation to limited N. Striking differences were observed when comparing the expression of genes involved in hormone metabolism between the roots and shoots already after 2 d of starvation (Supplemental Fig. S10).
Expression Changes for Regulatory Genes
Despite the overrepresentation of changes in the expression of metabolic genes, regulatory genes, such as transcription factors, protein kinase, and protein phosphatases, also showed modified expression during N starvation that was often in an organ-specific manner (Supplemental Fig. S10). However, of the genes most altered by N starvation (top lists in Tables III and IV), only eight regulatory genes are listed, such as two protein phosphatases, AT4G32950, one of the highest up-regulated gene in the roots, and AT3G52180, which is up-regulated by long-term starvation in the shoots. Six transcription factors were highly differentially expressed during N starvation, including PAP2, one of the major up-regulated genes in the shoots. The Myb protein PAP2 is one of the transcriptional regulators of anthocyanin biosynthesis and has been shown to be repressed by nitrate and induced during starvation (Scheible et al., 2004). In roots, up-regulation was found for three genes of the CCAAT transcription factor family (NF-YA2 [AT3G0569], NF-YA8 [AT1G72830], and NF-YA10 [AT5G06510]) and for ECR1 (AT1G01380), a Myb transcription factor encoding gene. Only one regulatory gene (ANA079/80 [AT5G07680]) was found between the main down-regulated genes in the roots. Except for NF-YA2 and NF-YA10, the differential expression of these genes during N starvation was organ specific.
The rather low number of regulatory genes that was found among the most highly differentially expressed genes might be due to the fact that our experimental systems induced many metabolic and morphologic changes, which is reflected by a high number of metabolic genes whose expression had been modified most after starvation, even as early as 2 d after the onset of starvation. In addition, the activity of many regulatory proteins is regulated not only at the transcription level but also by posttranscriptional modifications (Gutiérrez et al., 2007). However, the expression level of regulatory genes is often low, and transcripts of such genes are barely detectable with microarray approaches.
It has been shown that some miRNAs are differentially expressed after N starvation (Pant et al., 2009; Zhao et al., 2011), but no spatiotemporal regulation for miR genes has been described. The CATMA version 2.3 analysis highlighted the deregulation of seven miRNA genes (pri-miR156e, pri-miR160c, pri-miR169h, pri-miR172b, pri-miR172e, pri-miR419, and pri-miR836a) during N starvation. The array data were validated by quantitative (q)PCR for three of them, pri-miR160c, pri-miR156e, and pri-miR836a, which were differentially expressed in roots only. Pri-miR156e was repressed after both 2 and 10 d of starvation, and pri-miR160c and pri-miR836a were induced only after 10 d of starvation. None of the known targets of miR160c and miR156e were differentially expressed in our experiment. For miR836a, no targets have been described yet. However, using the psRNAtarget tool (Zhang, 2005; http://biocomp5.noble.org/psRNATarget/), a target search in WMD3 (Ossowski et al., 2008; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi?page=Home;project=stdwmd), and the prediction tool in the University of East Anglia plant small RNA toolkit (Moxon et al., 2008), we identified a putative target (AT2G40360). Expression of this gene was down-regulated at day 10 in the roots under our conditions, which would agree with the increase in pri-miR836a (array result and confirmation by qPCR; Supplemental Fig. S5). For all these miRNA precursors, further analysis should reveal whether the steady-state expression levels of mature miRNAs varied in our samples at 2 and/or 10 d of N starvation.
Several other probes corresponding to stable stem-loop precursors of eight loci predicted to encode putative new small RNAs were also differentially regulated. Six of them, named HypmiR-348339, -349340, -350341, -374365, -400391, and -1000918, were confirmed by qPCR (Supplemental Fig. S5) and were all found to be differentially expressed after long-term starvation. With the exception of HypmiR-374365 and -400391, which were repressed and induced only in the roots, respectively, the genes were induced in both the roots and shoots. Of utmost interest are the results obtained for HypmiR-374365 and -400391, as the precursors of these predicted genes were up-regulated in the dcl1-9 mutant background under N starvation conditions (data deposited in the CATdb database [http://urgv.evry.inra.fr/CATdb/], project Gnp06-01b_AgriArray/), suggesting that they might correspond to new miRNA genes.
Previously, it has been shown that the expression of miR169 precursors is regulated by N starvation (Pant et al., 2009; Zhao et al., 2011). Indeed, several genes encoding nuclear factor YA subunits, which are known to be targets of miR169s, were up-regulated in our experiment (AT5G12840/NF-YA1, AT3G05690/NF-YA2, AT1G72830/NF-YA8, and AT5G06510/NF-YA10; see discussion above). To confirm that our experiment was consistent with these results, we checked the expression pattern of three other isoforms of miR169 precursors (pri-miR169a, -b, and -d) and their predicted targets in our samples (Supplemental Fig. S5). Indeed, we confirmed our results that showed the opposite response in expression between the premiR169s and NF-YAs. It is known that miR169 can influence drought tolerance via the inhibition of NF-YA5 in Arabidopsis and that HAP2-1, the miR169 target in Medicago truncatula, is a regulator for the differentiation of nodule primordia (Combier et al., 2006; Li et al., 2008). The regulation of the miR169 family under N starvation conditions (and under abiotic stress, in general) reflects the importance of these regulators in the development of plants integrating several external signals. Furthermore, Zhao et al. (2011) have shown that plants overexpressing miR169a were more sensitive to N stress, which was the first report of a functional role of miR169a in the adaptation to N stress.
Comparison with Publicly Available Microarray Data
We first compared the transcriptome data obtained in this study with other published data on the transcriptome response to N availability. Although Scheible et al. (2004) have analyzed 3 d of starvation in 10-d-old whole seedlings, no other total starvation transcriptome data are available for Arabidopsis. However, N-limited transcriptome data are available for Arabidopsis shoots for two different levels of N deprivation (mild [1 mm NO3−] and severe [0.3 mm NO3−]; Bi et al., 2007), and we compared these transcriptome data with our data for total N starvation. A detailed comparison is provided in Supplemental Table S3, and a condensed version is provided in Table V.
Table V. Comparisons with published microarray data.
Synthesis of the number of genes in common between our microarray data and data obtained from 3 days N-starved seedlings (Scheible et al., 2004), the shoots of adults plants subjected to mild or severe N limitation (Bi et al., 2007), the roots and shoots of seedlings after 20 minutes of nitrate induction (Wang et al., 2003) and the roots of adult plants after 0.5, 1.5 and 8 h of nitrate induction (Patterson et al., 2010).
| Study | Scheible et al. (2004) |
Bi et al. (2007) |
Wang et al. (2003) |
Patterson et al. (2010) |
This Study |
Organ Specificity |
|||||||||||||||||
| Organ | Seedlings | Shoots, Mild |
Shoots, Severe |
Roots |
Shoots |
Roots 0.5 to 1.5 to 8 h |
Roots and Shoots | Roots |
Shoots |
Shoots | Roots | Roots and Shoots | |||||||||||
| Regulated Gene | Down | Down | Down | Down | Up | Down | Up | Up | Down | Up and down | 0 to 2 d down | 0 to 10 d down | 0 to 2 d up | 0 to 10 d up | 0 to 2 d down | 0 to 10 d down | 0 to 2 d up | 0 to 10 d up | |||||
| No. of genes in common/total gene in each study | 280/1418 | 0/1 | 71/190 | 4/77 | 37/261 | 0/2 | 27/77 | 50/637 | 4/307 | 306/766 | 45/306 | 103/306 | 2/306 | 4/306 | 35/306 | 217/306 | 2/306 | 5/306 | 197/306 | 80/306 | 29/306 | ||
| Percentage | 20 | 37 | 5 | 14 | 36 | 8 | 1 | 40 | 15 | 34 | 1 | 1 | 11 | 8 | 1 | 2 | 64 | 26 | 9 | ||||
| Comparison with roots and roots and shoots regulated genes (%) | 4 | 34 | 46 | 4 | 41 | 94 | 2 | 4 | |||||||||||||||
| Comparison with shoots and roots and shoots regulated genes (%) | 31 | 12 | 15 | 96 | 1 | 2 | |||||||||||||||||
| Organ | Seedlings | Shoots, Mild |
Shoots, Severe |
Roots |
Shoots |
Roots 0.5 to 1.5 to 8 h |
Roots and Shoots | Roots |
Shoots |
Shoots | Roots | Roots and Shoots | |||||||||||
| Regulated gene | Up | Up | Up | Down | Up | Down | Up | Up | Down | Up and down | 0 to 2 d Down | 0 to 10 d Down | 0 to 2 d Up | 0 to 10 d Up | 0 to 2 d Down | 0 to 10 d Down | 0 - 2 d Up | 0 to 10 d Up | |||||
| No. of genes in common/total gene in each study | 197/1142 | 11/51 | 96/270 | 36/77 | 12/261 | 2/2 | 6/77 | 9/637 | 55/307 | 257/653 | 8/257 | 9/257 | 24/257 | 136/257 | 0/257 | 3/257 | 49/257 | 144/257 | 111/257 | 105/257 | 42/257 | ||
| Percentage | 17 | 22 | 36 | 47 | 5 | 1 | 8 | 1 | 18 | 39 | 3 | 4 | 9 | 53 | 1 | 20 | 56 | 43 | 5 | 16 | |||
| Comparison with roots and roots and shoots regulated genes (%) | 24 | 8 | 6 | 37 | 5 | 6 | 16 | 93 | |||||||||||||||
| Comparison with shoots and roots and shoots regulated genes (%) | 7 | 63 | 1 | 4 | 2 | 32 | 94 | ||||||||||||||||
Approximately 40% of the differentially expressed genes in our study (both organs and both time points; 40% and 39% for down- and up-regulated genes, respectively) have been found previously. In a comparison between N-starved seedlings (Scheible et al., 2004) and the hydroponically grown adult plants in this study, 20% and 17% of the down-regulated and up-regulated genes, respectively, were found common.
When comparing complete N starvation (this study) with severe N limitation (Bi et al., 2007; Table V; Supplemental Table S3), both analyzed in adult plants, 31.4% of the down-regulated genes were observed under both conditions, only taking into account those genes differentially regulated in the shoots. Of these 71 genes, 23 were differentially expressed by 2 d of starvation, and all of them were differentially expressed by 10 d of starvation. However, the proportion of genes up-regulated in common with genes differentially expressed in the shoots was more significant (68%). Of the 96 genes that were up-regulated by severe N limitation and N starvation, 36 were differentially expressed at 2 d of starvation, whereas all were differentially expressed by 10 d of starvation. For mild N limitation, seven of the 11 common genes differentially expressed at 10 d of N starvation were already up-regulated at 2 d of starvation. Given the variations due to different culture conditions, gene expression changes resulting from either limitation of N or total absence of N might involve a common and a specific response pattern. It is evident that, in the case of limitation, nitrate was still present at the root surface; thus, signals possibly perceived by the total absence of N should be missing. However, the comparison of severe limitation and N starvation showed that, even in very different growth conditions (soil versus hydroponic), 68% of the differentially expressed genes were in common between these two treatments. In addition to this result, Bi et al. (2007) have reported very low nitrate levels (0.25 mg nitrate g−1 fresh weight) in the shoots of plants grown under severe limitation, suggesting that the physiological status of plants after 10 d of N starvation and after growth on low nitrate might be rather similar.
We also included nitrate-regulated genes in our analysis, using those that have been reported by Wang et al. (2004; seedlings separated into root and shoots) and Patterson et al. (2010; adult plants grown hydroponically with root responses exclusively). Between these and our experiments, we found 1% to 47% of the genes to be overlapping. As expected, for 89% of all the common genes, inverse steady-state transcript level changes were observed between N starvation and nitrate induction (Table V).
In summary, the comparison of our data with previously published data (Table V) clearly showed that our work is in agreement with earlier studies, but we have added a new level of resolution by separating root- and shoot-specific responses and by analyzing mid- and long-term starvation. Furthermore, our study, which was conducted under more physiologically realistic growth conditions, adds new N starvation-regulated genes (Supplemental Table S4) to the present knowledge base.
In a second step, we compared the main differentially expressed genes (Tables III and IV) with the transcriptome data from public databases to identify either similarities with other transcriptome analyses obtained in response to different stimuli or in different genetic backgrounds or a specificity of the N starvation response. A clustering analysis was performed using our top list of differentially regulated genes and all the available data sets from CATdb (Supplemental Fig. S11). A K means calculation mainly clustered our data with other N starvation-like experiments (in vitro N starvation in young seedlings on either 0.1 or 10 mm nitrate [R. Berthomé and J.-P. Renou, unpublished data]). However, for the roots, the K means calculation also clustered the genes of our top list in a series of experiments corresponding to other treatments or the analysis of specific mutants. With the exception of the experiment analyzing the impact of mutations in genes involved in starch metabolism and circadian rhythms, all the other experiments in our cluster involved biotic or abiotic stresses responses. These works were performed to study the involvement of Patatin-Like Protein2 in Botrytis cinerea resistance signaling (La Camera et al., 2009), the role of ataxia telangiectasia mutated in plants using irradiation (Ricaud et al., 2007), the Arabidopsis response to Rhodococcus fascians bacteria (Depuydt et al., 2009), and the elucidation of the role of nitric oxide or GABA in the plant response to cadmium or high-salinity treatment (Besson-Bard et al., 2009; Renault et al., 2010). GABA is a nonprotein amino acid that has been reported to accumulate in a number of plant species when subjected to high salinity and many other environmental constraints (Kinnersley and Turano, 2000). Recently, Renault et al. (2010) suggested that GABA links N and C metabolism in the roots in response to salt; we also observed an accumulation of GABA in N-depleted roots.
Similar results were obtained using the data available on www.genevestigator.com (Hutz et al., 2008). Clustering that was obtained with the genes on our shoot and root top lists and all the available shoot or root data showed a rather specific pattern for our N starvation gene set (Supplemental Fig. S12). However, several genes were also differentially expressed after other treatments or in some mutant backgrounds. A striking result was the differential expression of the genes on our top list in a series of experiments performed to identify circadian transcripts that were coregulated with cytosolic calcium concentration oscillations. To this aim, wild-type plants were treated with nicotinamide, a metabolic inhibitor of ADPR cyclase that abolishes the circadian oscillations of cytosolic calcium concentrations, and compared with untreated plants during a 3-d interval (Dodd et al., 2007). In addition, the results were compared with the expression patterns of the toc1-1 mutant, which is defective for the pseudoresponse regulator TIMING OF CAB EXPRESSION1 (TOC1 or PRR1). Calcium signaling is a possible component of the responses to N starvation. Recent studies have shown cross talk between reactive oxygen species, NO, and calcium signaling (for review, see Mazars et al., 2010). Reactive oxygen species signaling has already been proposed to play a role in the adaptive response to N availability (Shin et al., 2005). Many N metabolism-related genes are regulated by diurnal cycles, and Gutiérrez et al. (2007) have clearly demonstrated the relationship of the circadian clock with N regulation. Our results further underline the importance of diurnal regulation for N metabolism and signaling. However, this meta-analysis showed that N starvation not only interfered with the circadian clock but was also a possible connection with calcium signaling.
Integration of the Metabolite, Protein Activity, and Expression Changes
A low correlation between the mRNA expression levels, enzymatic activities, and protein levels indicates that transcriptome data are not sufficient to understand genome-wide protein dynamics and biochemical regulation (Gibon et al., 2006; Sulpice et al., 2009, 2010). Thus, we compared the expression changes on levels of RNA, enzyme activities, and nitrate transport activities (Supplemental Table S5). As in earlier studies, we observed that the correlations between transcript and activity changes are not evident. However, in some cases, for which the different genes of one family are well known, the data can be better interpreted. For example, in roots, NR activity decreased during N starvation. Although NIA2 transcript levels increased from day 0 to day 10, it should to be taken into account that, in roots, the more abundant transcript is NIA1 compared with NIA2 (Cheng et al., 1991). Changes in NR activity in roots, therefore, might correlate better to NIA1 transcript levels. In general, NR activity and transcript levels were decreasing in the roots and shoots. Regardless, NR is a highly regulated enzyme (for review, see Daniel-Vedele et al., 2010), and a correlation between transcript level and activity is thus difficult to interpret in this case.
For nitrate uptake, we classified all the genes of the NRT2 family as HATS transporters However, experimental evidence is available only for NRT2.1, NRT2.2, and NRT2.7 (Filleur et al., 2001; Chopin et al., 2007; Li et al., 2007); we did not include NRT2.7 on our list, as this protein is mainly expressed in seeds (Chopin et al., 2007). Concerning LATS, we concentrated on NRT1.1 and NRT1.2, which are responsible for root low-affinity nitrate uptake (Tsay et al., 1993; Huang et al., 1999). However, NRT1.1 has been shown to act as a dual-affinity transporter (Wang et al., 1998; Liu et al., 1999). Following phosphorylation by CIPK23, the unphosphorylated LATS form is converted into a HATS form (Ho et al., 2009). As with NR activity, a correlation between activity and transcript levels in this case might be low due to posttranslational modifications. However, concerning HATS activity, the significant increase in the expression of NRT2.4 and NRT2.5 might be correlated to the increase in HATS after 10 d of N starvation in the roots (Supplemental Table S5). Previous studies have revealed a clear correlation between HATS and steady-state NRT2.1 mRNA levels under many different conditions and genotypes (Lejay et al., 1999; Filleur et al., 2001; Monachello et al., 2009). Our results indicate that the activity of nitrate transporter(s) other than NRT2.1 might explain the increase in HATS during starvation.
Several genes are known to encode GS and GDH activities. The main GS protein in leaves is GS2, which evokes the hypothesis that the decrease in transcript levels of GS2 was responsible for the decrease in GS activity. However, in N-depleted leaves, Gln synthesis during N recycling and reassimilation has been proposed to be catalyzed by newly expressed GS1 isoforms (Guiboileau et al., 2010). Based on functional genomics and quantitative genomic approaches performed in rice (Oryza sativa) and maize (Zea mays), the importance of GS1 in N management, growth rate, crop yield, and grain filling has been emphasized (for review, see Bernard and Habash, 2009). However, Arabidopsis has five GS1 isoenzymes. Under long-term starvation in our experiment, only GLN1.1 was induced in the shoots. In addition to GS, GDH also has a controversial role in N remobilization (see above). The transcript levels of the three GDH-encoding genes either increased or decreased in our study (Supplemental Table S5). Thus, a direct correlation with GDH activity did not seem possible.
We further checked the correlations between changes in the metabolite levels and transcript levels using KaPPA-View4 tools (http://kpv.kazusa.or.jp/kpv4/). Figure 6 presents the results for nitrate assimilation, photorespiration, TCA cycle, and Asp metabolism. The choices of the metabolic pathway were guided by the availability of the metabolite data. Figure 6 also shows the fold changes of the metabolites and transcript levels in the root and shoots after 10 d of starvation. In the case of nitrate assimilation, decreased transcription levels of either NIA1 or NIA2 and NII were correlated with a decrease in ammonium in the roots and shoots. For the ensuing incorporation of ammonium into amino acids, some transcript levels encoding GS isoenzymes were reduced in the shoots (see above) only and not in the roots. However, as the levels of the ammonium and Glu substrates were low, this alone might explain the low levels of product (Gln); however, we observed a decrease in total GS activity (Fig. 3). As 2OG levels were unchanged in both organs, a decrease of GOGAT activity might be the reason for the decrease in the Glu content. Transcripts of one of the isoenzymes encoding GOGAT (AT5G53460) were decreased in the roots and shoots. In addition to its acquisition from the environment, photorespiration is also a major source of ammonium. We found that the transcript levels for two isoforms of Gly decarboxylase were decreased, but as the Gly (a product of photorespiration) level was low, no clear relationship could be drawn.
Figure 6.
Effects of N starvation on N assimilation, the TCA cycle, and Asp and Asn metabolism. The pathway maps are shown according to the transcriptome and metabolome analysis of 10 d of N depletion in plants in comparison with the onset of starvation. Substrates and products are in blue text, and enzymes are in black text. Boxes and circles correspond to the transcriptome log2 ratio and the metabolite accumulation ratio, respectively, of 0 to 10 d long-term starvation. Uncolored symbols represent missing data. [1] and [2] represent roots and shoots, respectively. Log2 ratios and metabolite accumulation magnitude are illustrated by the color key at bottom. The map was generated based on the figures generated using KaPPA-View4 (http://kpv.kazusa.or.jp/kappa-view/), with modifications.
Except for isoforms of isocitrate dehydrogenase, one isoform of aconitase, and one isoform of succinate dehydrogenase, transcript levels of enzymes involved in the TCA cycle were either not differentially expressed or were decreased at day 10 of N starvation. In contrast, the levels of some intermediates of the TCA cycle were increased, and a clear correlation was not apparent. However, as many different metabolic pathways are interconnected, it is impossible to merely examine one pathway. For example, Asp degradation results in the formation of malate via oxaloacetate, and oxaloacetate is also a substrate for Asp synthesis. Nevertheless, as fluxome studies in plants are still difficult, our integrated study adds to a detailed understanding of the metabolic adaptations in N starvation.
CONCLUSION
This thorough analysis of the adaptive responses to complete N starvation in the roots and shoots of adult Arabidopsis plants adds to the previous studies in seedlings and reveals different adaptation strategies of the photosynthetic shoot and the heterotrophic root. We showed that Arabidopsis resists N deficiency by the regulation of networks that are tuned to the environmental challenge in an organ-specific way.
Here, we illustrated the capacity of plants to continue to grow in the absence of any external N supply, which demonstrates the large potential for N remobilization from internal stores. Our data suggest that N reallocation from the shoots to the growing roots occurs mainly through the transport of amino acids. Indeed, transcriptome analysis revealed an increased expression of two amino acid transporters in shoots in long-time starvation. In addition, increased amino acid transport might arise simply from increased substrate levels or by posttranscriptional modification of transporter activities. Indeed, remobilization from the shoots to roots, either in the form of amino acids or Suc, was initiated long before the internal N stores in the shoots were low. This adaptation strategy may allow for the support of root growth before detrimental consequences of N depletion take place.
Nitrate is depleted more rapidly in the roots than in shoots: the root nitrate content relies on nitrate uptake, nitrate reduction in this organ, and nitrate translocation toward aerial parts. The decrease of nitrate reductase activity in the roots (becoming undetectable at day 4) together with the absence of nitrate in the external medium suggest that nitrate was continuously translocated toward the shoot, where it was reduced into amino acids, which, in turn, were allocated to the roots. The NRT1.5 nitrate transporter (Lin et al., 2008) might play a role in this nitrate transport within the plant. In addition to the dramatic decreases in the content of several major amino acids, the levels of several minor amino acids increased, especially during the long-term starvation. The percentage of minor amino acids in the total amino acid pools increased during the 10 d of N starvation. In the shoots, an excess of carbohydrate accumulated mainly as starch and raffinose but also as fumarate. Moreover, our data also revealed a cross talk between phosphate and nitrate management, with particulars due to the complete N starvation.
Under our conditions, nitrate assimilation, which occurred predominantly in the shoots, dropped rapidly, partly due to the reduced NR transcripts (mainly NIA2). N reassimilation during N remobilization cannot be clearly attributed to one enzyme; however, our results add to the evidence that GDH may reflect an additional/alternative route to the GS/GOGAT pathway for ammonia assimilation. During long-term N starvation, the high-affinity nitrate uptake capacity increased, which was not correlated with the steady-state transcript levels of the known HATS components, NRT2.1 and NRT2.2. This indicates that other nitrate transporters were responsible for the initial nitrate uptake after the long-term N starvation, and transcriptome analysis revealed NRT2.4 and NRT2.5 as candidates for this activity.
Our global expression study further revealed organ-specific transient changes for a low number of genes in the shoots and roots as an early response, whereas this number was much more significant after the long-term N starvation, especially in the roots. These organ-specific expression changes were also observed for genes encoding regulatory proteins, proteins involved in hormone synthesis and degradation, and the precursors of miRNAs. The results presented here also contribute to the increasing indications that miR169 plays a role in the adaptation to N starvation by targeting several transcription factors (NF-As); more interestingly, we showed that other known and unidentified putative small RNA genes displayed an altered transcriptional response to N starvation, mainly in the roots. These findings open new investigation tracks about the role of small RNAs in the adaptation to plant nutrition.
Our transcriptome data add a new level of resolution by dissecting root- and shoot-specific responses and by analyzing both mid- and long-term starvation. Several new N starvation-regulated genes were uncovered, and similarities and differences were revealed between N starvation in seedlings and adult plants and between total N starvation and mild or severe N limitation. Meta-analysis showed the rather specific transcriptome responses for N starvation, despite some overlap with other abiotic and biotic stress responses. Interestingly, GABA treatments clustered together with our results. Furthermore, exciting indications for a role of calcium signaling and of the diurnal clock need to be investigated further.
Finally, our integrative study analyzing global transcript levels and many metabolite levels together with enzyme activities, as performed during a time course, gives a multifaceted image of the changes occurring after mid- and long-term N starvation in root and shoots. This set of data sheds light on the necessity of analyzing organ-specific responses before fully appreciating the analysis of cell-specific responses in plants and other multicellular organisms. However, additional time points might be needed to decipher the network and the very early events after the withdrawal of nitrate from the growth medium. It is also obvious that our data represent only a snapshot at one time point in the day and that many of the parameters undergo substantial changes during the diurnal cycle (Matt et al., 2002; Fritz et al., 2006). Nevertheless, our integrated study adds to a detailed understanding of the adaptations in N starvation and highlights spatiotemporal responses.
MATERIALS AND METHODS
Material and Growth Conditions
Seed stocks of Arabidopsis (Arabidopsis thaliana) from the Columbia accession were used for all experiments. Plants were grown under hydroponics culture conditions in a Sanyo growth chamber with an 8-h-light/16-h-dark cycle at 21°C/17°C, respectively, 80% relative humidity, and 150 μmol m−2 s−1 irradiation. Seeds were sterilized and stratified in water at 4°C for 5 d before sowing. Each seed was sown on top of a cut Eppendorf tube filled with half-concentrated agar medium containing 0.8% agar and soaked in nutrient solution. Plants were supplied with nutrient medium containing 6 mm NO3− containing the following nutrients: 3 mm KNO3, 1.5 mm Ca(NO3)2, 2 mm MgSO4, 2 mm KH2PO4, 1 mm K2SO4, 0.7 mm CaCl2, 10 μm MnSO4, 24 μm H3BO3, 3 μm ZnSO4, 0.9 μm CuSO4, 0.04 μm (NH4)6Mo7O24, and 10 mg L−1 iron-EDTA (Sequestrene; CIBA-GEIGY). Nutrient solutions were changed every 2 d and, during the first 2 weeks, used at half the final concentration. At 35 d after sowing, plants were harvested for time point 0 and then transferred to N-free solution containing 2.5 mm K2SO4, 2.2 mm CaCl2 instead of 1 mm K2SO4, and 0.7 mm CaCl2. Pools of three or 12 plants were harvested at 1, 2, 4, and 10 d after transfer in N-free medium. All samples were taken 2 to 3 h after onset of the day. Results from a representative experiment are shown.
Root 15NO3− Influx
Influx of 15NO3− was assayed as already described by Delhon et al. (1995) The plants were transferred first to 0.1 mm CaSO4 for 1 min, then to complete nutrient solution containing either 0.2 or 6 mm 15NO3− (atom % 15N, 99%) for 5 min, and finally to 0.1 mm CaSO4 for 1 min. Roots were separated from the shoots immediately after the final transfer to CaSO4 and frozen in liquid N. After grinding, an aliquot of the powder was dried overnight at 80°C and analyzed using the ANCA-MS system (PDZ Europa). Influx of 15NO3− was calculated from the 15N content of the roots (2 mg dry weight). The values are means of four or five replicates.
Enzyme Activities
Enzymes were extracted from frozen leaf material stored at –80°C as described by Ferrario-Méry et al. (2000). Soluble protein content was determined in crude leaf extracts according to Bradford (1976). All standard assays contained optimized levels of substrates and, where necessary, activators, pH, and ionic conditions to allow maximum activity. NR maximal extractable activity and activation state were measured as described by Ferrario-Méry et al. (2000). The activation state of NR is defined as the ratio of the activity measured in the presence of 10 mm MgCl2 divided by the activity in the presence of 5 mm EDTA and is expressed as a percentage. GS was measured according to the method of O’Neal and Joy (1973). GDH aminating and deaminating activities were assayed as described by Masclaux et al. (2000).
Analysis of Individual Metabolites
Ethanol Extraction
An aliquot of the powder was weighed (50 mg fresh weight) and extracted in a four-step ethanol water procedure. The first step consisted of an extraction during 30 min at 80°C using 500 μL of 80% (v/v) ethanol. The subsequent steps completed this extraction using 250 μL of 80% (v/v) ethanol, 250 μL of 50% (v/v) ethanol, and 250 μL of water at 80°C for 30 min. Supernatants of the different extraction steps were collected and mixed thoroughly.
Nitrate, Free Amino Acid, and Carbohydrate Measurements
For determination of the nitrate content (μmol g−1 fresh weight), ethanol extracts were evaporated and diluted in water before analyzing by HPLC using a DX-120 (Dionex). The same extracts were also subjected to a Rosen evaluation of free amino acid concentration (μmol g−1 fresh weight), and soluble carbohydrate contents were determined using the Roche Suc/d-Glc/d-Fru UV method kit.
Starch content was determined on the insoluble pellets of the ethanol extraction. The pellets were dried for 1 h at 50°C and incubated in 200 μL of water at 100°C for 2 h. Starch digestion was performed overnight at 50°C adding 500 μL of acetate sodium buffer at 0.2 mm and pH 4.8, 1 mg mL−1 amyloglucosidase from Aspergillus niger (70 units mg−1; Fluka), and 25 μg mL−1 α-amylase from Bacillus amyloliquefaciens (Boehringer Mannheim). Glc equivalents were measured on the supernatant using the Roche d-Glc UV method kit. The values are means of three replicates.
Metabolome Analysis
A total of 40 mg of Arabidopsis shoots and roots (fresh material) was extracted with 1 mL of extraction buffer (Gullberg et al., 2004). The extraction buffer (chloroform:methanol:water, 1:3:1 [v/v/v] at −20°C) allowed the extraction of lipophilic and hydrophilic metabolites in one phase. All samples were vortexed for 3 min and then centrifuged for 10 min at 3,000 rpm and 4°C. A total of 300 μL of supernatant was dried under vacuum, and the dried pellets were conserved under argon and stored at –80°C prior to analysis. The dried extracts were derivatized using a two-stage process based on the method of Fiehn (2006). Twenty microliters of 40 mg mL−1 methoxyamine hydrochloride in pyridine was added to the dried extracts and held at 28°C for 90 min. This was followed by the addition of 180 μL of N-methy-N-(trimethylsilyl) trifluoroacetamide for 30 min at 37°C. The samples were analyzed by GC-MS (Waters).
Transcriptome Studies
Microarray analysis was carried out at the Research Unit in Plant Genomics in Evry, France, using the CATMA array containing 24,576 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). One biological replicate set was analyzed with CATMA array version 2.2 and the two others with version 2.3. Total RNA extractions from three independent biological replicates were performed using the Qiagen RNAeasy plant minikit according to the manufacturer’s instructions. Each biological replicate was composed of starved and unstarved plants, each sample corresponding to pooled roots or shoots from 12 plants harvested at 0, 2, and 10 d after the onset of the N starvation. For each comparison (starved versus unstarved at each time point), one technical replication with fluorochrome reversal was performed for each biological triplicate. The labeling of antisense amplified RNA with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization to the slides, and the scanning were performed as described by Lurin et al. (2004).
Statistical Analysis of Microarray Data
Experiments were designed with the Bioinformatic and Predictive Genomics group at the Unité de Recherche en Génomique Végétale in Evry, France. Specific statistics were developed to analyze CATMA hybridizations. For each array, the raw data comprise the logarithm of median feature pixel intensity (in log base 2) at wavelengths of 635 nm (red) and 532 nm (green). No background was subtracted. The normalization method used was described by Lurin et al. (2004). To determine differentially expressed genes, we performed a paired t test on the log ratios averaged on the dye swap. A trimmed variance was calculated from spots that did not display extreme variance. The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate (with a type I error equal to 5%). We also adjusted the raw P values to control a false discovery rate using Benjamini-Yetkutieli at a level of 1%. For complete statistical analysis, biological replicates 2 and 3 (both performed on CATMA array version 2.3) were analyzed and all differentially expressed genes were checked against the results of biological replicate 1 (analysis done on CATMA array version 2.2). For 0 to 2 d, 2 to 10 d, and 0 to 10 d comparisons, the number of genes differentially expressed with false discovery rate control were, respectively, 368, 1,355, and 1,865 for shoots and 376, 1,147, and 1,457 for roots. Nonetheless, in the CATMA analysis pipeline, family-wise error rate proved to be the best solution to balance the estimated number of false positives and false negatives (Ge et al., 2003). As described by Gagnot et al. (2008), when the Bonferroni P value was lower than 0.05, the gene was declared differentially expressed. The complete data set is given as Supplemental Tables S7 and S8.
Clustering
A matrix was prepared after elimination of the controls used on the microarray and the missing data provided from our samples and those corresponding to 142 publicly available CATMA projects corresponding to 1,178 hybridizations extracted from the CATdb database. The Genesis software version 1.6.0 beta 1 developed by A. Sturn (Graz University of Technology Institute for Genomics and Bioinformatics; www.tugraz.at) was used for clustering analysis. Hierarchical as well as K means clustering (10 clusters selected, 50 maximum iterations) using Pearson correlation as distance calculation were performed. Hierarchical clustering was also realized on the Genevestigator Web site (https://www.genevestigator.com/gv/) using our combined shoot and root top lists and all available shoot and root data.
cDNA Synthesis and Quantitative Real-Time PCR
Quantitative real-time RT-PCR validation was performed for 32 genes on the same organs described in the microarray section but from two independent extractions for each sample. The primers for RT-PCR were selected with Primer3 (http://fokker.wi.mit.edu/primer3/imput-030.htm; optimal temperature of 60°C; Supplemental Table S6). The primer pairs were first tested on a dilution series of genomic DNA (5, 0.5, 0.05, and 0.005 ng) to generate a standard curve and assess their PCR efficiency, which ranged between 90% and 99%. Twelve independent cDNA synthesis reactions were made for the duplicated samples (roots at 0, 2, and 10 d, shoots at 2 and 10 d). RT was performed on 1 μg of total RNA with oligo(dT) primer (18-mer) and the SuperScript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. At least three replicate PCRs for each of the cDNAs were included in every run.
For each gene investigated using qPCR, a dilution series covering 3 orders of magnitude was prepared from a cDNA stock solution (1, 1/10, 1/100). Three replicates of each of the three standards were included with every qPCR experiment together with three no-template controls. qPCR was performed in 15 μL, with 0.1 μL of RT reaction, 900 nm final concentration of each primer pair, and SYBR Green PCR master (Eurogentec). Corresponding minus-RT controls were performed with each primer pair.
All reactions were performed with the ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) as follows: 95°C for 10 min; 40× 95°C for 15 s and 60°C for 1 min; and a dissociation step to discriminate primer dimers from the PCR product. Using the SDS software provided by the manufacturer, the optimal cycle threshold (Ct) was determined from the dilution series, with the raw expression data derived.
Six housekeeping genes were assessed in this experiment, and the two best control genes, consistently expressed, were selected to calculate the average normalization factor: AT3G18780 and AT4G24820 for each sample pair. Normalized (Norm) ΔCt for each differentially expressed gene was calculated as following: Norm ΔCt = −(rawΔCt − Norm factor).
Microarray data from this article were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE8031, GSE29103, and GSE29104 and at CATdb (http://urgv.evry.inra.fr/CATdb/) under projects Gnp03 B04, Gnp06-01_AgriArray, and Gnp06-01b_AgriArray, according to the Minimum Information about a Microarray Experiment standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relative growth rate during N starvation.
Supplemental Figure S2. Metabolites related to the TCA cycle during N starvation.
Supplemental Figure S3. NR activation state during N starvation.
Supplemental Figure S4. NAD and NADH-GDH activities in shoots during N starvation.
Supplemental Figure S5. qPCR validations of transcriptome data.
Supplemental Figure S6. Venn diagram showing the total number of differentially expressed genes (P < 0.05) during N starvation.
Supplemental Figure S7. Anthocyanin levels in shoots during N starvation.
Supplemental Figure S8. Metabolic gene expression changes at early N starvation analyzed by the MapMan tool.
Supplemental Figure S9. Big multigene family expression changes, including peroxidases at late N starvation, analyzed by the MapMan tool.
Supplemental Figure S10. Regulatory gene expression changes at early N starvation analyzed by the MapMan tool.
Supplemental Figure S11. Clustering analysis within CATdb.
Supplemental Figure S12. Clustering analysis within Genevestigator public data.
Supplemental Table S1. Differentially expressed genes during N starvation.
Supplemental Table S2. Biological pathways with significant genes overrepresented (P < 0.05) in the roots or shoots under 0 to 2 d and 0 to 10 d N starvation conditions.
Supplemental Table S3. Comparisons with published microarray data.
Supplemental Table S4. Genes regulated by N starvation specific to our study.
Supplemental Table S5. Comparison of transcript levels and enzyme activity or nitrate uptake capacity for major N metabolism enzyme activities.
Supplemental Table S6. Oligonucleotides used for qRT-PCR.
Supplemental Table S7. Complete transcriptome data obtained from root samples for 0 to 2, 2 to 10, and 0 to 10 d comparisons.
Supplemental Table S8. Complete transcriptome data obtained from shoot samples for 0 to 2, 2 to 10, and 0 to 10 d comparisons.
Acknowledgments
We are grateful to Dr. Christian Meyer for inspiring discussions during the whole project and Dr. Christophe Tatou for the pleasant collaboration during the Génoplante B4 project. We thank Dr. Olivier Voinnet for early access to the CATMA version 2.3 arrays, Pascal Tillard for 15N analyses, Gilles Clement for advice on GC-MS analyses, and Dr. Marie-Laure Martin-Magniette for advice on statistical analysis.
References
- Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA. (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 182: 608–620 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Gravot A, Richaud P, Auroy P, Duc C, Gaymard F, Taconnat L, Renou JP, Pugin A, Wendehenne D. (2009) Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol 149: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brouquisse R, Gaudillere JP, Raymond P. (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol 117: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Marchive C, Meyer C, Krapp A. (2011) Nitrogen signalling in Arabidopsis: how to obtain insights into a complex signalling network. J Exp Bot 62: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Dewdney J, Goodman HM, Conkling MA. (1991) Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol 96: 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DW, Yoder TJ, Reiter WD, Gibson SI. (2000) Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 211: 743–751 [DOI] [PubMed] [Google Scholar]
- Chiou TJ. (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30: 323–332 [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel-Vedele F. (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT. (1986) Regulation of the absorption and release of nitrate by plant cells: a review of current ideas and methodology. Lambers H, Neeteson JJ, Stulen I, , Fundamental Ecological and Agricultural Aspects of Nitrogen Metabolism in Higher Plants. Martinus Nijhoff Publishers, Dordrecht, The Netherlands, pp 3–27 [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N, Glass A. (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395 [Google Scholar]
- Crawford NM, Forde BG. (2002) Molecular and developmental biology of inorganic nitrogen nutrition. The Arabidopsis Book 1: e0011, doi/10.1199/tab.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouzé P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al. (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Vedele F, Krapp A, Kaiser WM. (2010) Cellular biology of nitrogen metabolism and signaling. Hell R, Mendel RR, , Cell Biology of Metals and Nutrients. Springer-Verlag, Berlin, pp 145–172 [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L. (1995) Diurnal regulation of NO3 uptake in soybean plants: I. Changes in NO3 influx, efflux, and N utilization in the plant during the day/night cycle. J Exp Bot 46: 1585–1594 [Google Scholar]
- Depuydt S, Trenkamp S, Fernie AR, Elftieh S, Renou JP, Vuylsteke M, Holsters M, Vereecke D. (2009) An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiol 149: 1366–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Gonçalves J, et al. (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318: 1789–1792 [DOI] [PubMed] [Google Scholar]
- Drew MC. (1975) Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol 75: 479–490 [Google Scholar]
- Dubois F, Tercé-Laforgue T, Gonzalez-Moro MB, Estavillo MB, Sangwan R, Gallais A, Hirel B. (2003) Glutamate dehydrogenase in plants: is there a new story for an old enzyme? Plant Physiol Biochem 41: 565–576 [Google Scholar]
- Ferrario-Méry S, Suzuki A, Valadier M-H, Roux Y, Hirel B, Foyer CH. (2000) Modulation of amino acid metabolism in transformed tobacco plants deficient in Fd-GOGAT. Plant Soil 221: 67–79 [Google Scholar]
- Fiehn O. (2006) Metabolite profiling in Arabidopsis. Methods Mol Biol 323: 439–447 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F. (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Fontaine JX, Saladino F, Agrimonti C, Bedu M, Tercé-Laforgue T, Tétu T, Hirel B, Restivo FM, Dubois F. (2006) Control of the synthesis and subcellular targeting of the two GDH gene products in leaves and stems of Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol 47: 410–418 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Fritz C, Mueller C, Matt P, Feil R, Stitt M. (2006) Impact of the C-N status on the amino acid profile in tobacco source leaves. Plant Cell Environ 29: 2055–2076 [DOI] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Bréhélin C, Tischendorf G, Kessler F, Dörmann P. (2007) Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J 49: 729–739 [DOI] [PubMed] [Google Scholar]
- Ge Y, Dudoit S, Speed TP. (2003) Resampling-based multiple testing for microarray data hypothesis. Test 12: 1–44 [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A, Sormani R, Meyer C, Masclaux-Daubresse C. (2010) Senescence and death of plant organs: nutrient recycling and developmental regulation. C R Biol 333: 382–391 [DOI] [PubMed] [Google Scholar]
- Gullberg J, Jonsson P, Nordström A, Sjöström M, Moritz T. (2004) Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal Biochem 331: 283–295 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Caldana C, Steinhauser D, Balbo I, Fernie AR, Willmitzer L. (2010) Combined transcript and metabolite profiling of Arabidopsis grown under widely variant growth conditions facilitates the identification of novel metabolite-mediated regulation of gene expression. Plant Physiol 152: 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hoai NTT, Shim IS, Kobayashi K, Usui K. (2003) Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa) seedlings. Plant Growth Regul 41: 159–164 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PT, Chase JM, Dosch KL, Hartson RB, Gross JA, Larson DJ, Sutherland DR, Carpenter SR. (2007) Aquatic eutrophication promotes pathogenic infection in amphibians. Proc Natl Acad Sci USA 104: 15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Joung JG, Fei Z, Jander G. (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39: 933–947 [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Huber SC. (1994) Posttranslational regulation of nitrate reductase in higher plants. Plant Physiol 106: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T. (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Kinnersley AM, Turano FJ. (2000) γ-Aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19: 479–509 [Google Scholar]
- Knaupp M, Mishra KB, Nedbal L, Heyer AG. (2011) Evidence for a role of raffinose in stabilizing photosystem II during freeze-thaw cycles. Planta 234: 477–486 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S. (2000) Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol 122: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Ferrario-Méry S, Touraine B. (2002) Nitrogen and signalling. Foyer C, Noctor G, , Advances in Photosynthesis [Google Scholar]
- Krapp A, Truong HN. (2005) Regulation of C/N interaction in model plant species. Goyal S, Tischner R, Basra A, , Enhancing the Efficiency of Nitrogen Utilization in Plants. Haworth Press, New York, pp 127–173 [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. (2010) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, Roby D, Heitz T. (2009) The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant Microbe Interact 22: 469–481 [DOI] [PubMed] [Google Scholar]
- Lark RM, Milne AE, Addiscott TM, Goulding KWT, Webster CP, O’Flaherty S. (2004) Scale- and location-dependent correlation of nitrous oxide emissions with soil properties: an analysis using wavelets. Eur J Soil Sci 55: 611–627 [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A. (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S. (2011) Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci USA 108: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD. (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis KA, Primikirios NI, Nikolantonakis MA, Roubelakis-Angelakis KA. (2002) Immunocharacterization of Vitis vinifera L. ferredoxin-dependent glutamate synthase, and its spatial and temporal changes during leaf development. Planta 215: 630–638 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S, Majerus V, Kinet JM. (1999) NaCl effects on proline metabolism in rice (Oryza sativa) seedlings. Physiol Plant 105: 450–458 [Google Scholar]
- Marschner M. (1995) Mineral Nutrition in Higher Plants. Academic Press, London [Google Scholar]
- Masclaux C, Valadier M-H, Brugière N, Morot-Gaudry J-F, Hirel B. (2000) Characterization of the sink/source transition in tobacco ( Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211: 510–518 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A. (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt P, Krapp A, Haake V, Mock HP, Stitt M. (2002) Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J 30: 663–677 [DOI] [PubMed] [Google Scholar]
- Mazars C, Thuleau P, Lamotte O, Bourque S. (2010) Cross-talk between ROS and calcium in regulation of nuclear activities. Mol Plant 3: 706–718 [DOI] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi G. (1996) Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 93: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Lea PJ. (1980) Ammonia assimilation. Miflin BJ, , The Biochemistry of Plants. Vol c5. Amino Acids and Their Derivatives. Academic Press, New York, pp 169–202 [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Mollá-Morales A, Sarmiento-Mañús R, Robles P, Quesada V, Pérez-Pérez JM, González-Bayón R, Hannah MA, Willmitzer L, Ponce MR, Micol JL. (2011) Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J 65: 335–345 [DOI] [PubMed] [Google Scholar]
- Monachello D, Allot M, Oliva S, Krapp A, Daniel-Vedele F, Barbier-Brygoo H, Ephritikhine G. (2009) Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol 183: 88–94 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. (2000) Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol 3: 224–228 [PubMed] [Google Scholar]
- O’Neal D, Joy KW. (1973) Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Arch Biochem Biophys 159: 113–122 [DOI] [PubMed] [Google Scholar]
- Orsel M, Boivin K, Roussel H, Thibault C, Krapp A, Daniel-Vedele F, Meyer C. (2005) Functional genomics of plant nitrogen metabolism. Leister D, , Plant Functional Genomics. Haworth Press, Binghamton, NY, pp 431–450 [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. (2010) Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ 33: 1486–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples MB, Freney JR, Mosier AR. (1995) Minimizing gaseous losses of nitrogen. Bacon PE, , Nitrogen Fertilizer in the Environment. Marcel Dekker, Inc, New York, pp 565–606 [Google Scholar]
- Pracharoenwattana I, Zhou W, Keech O, Francisco PB, Udomchalothorn T, Tschoep H, Stitt M, Gibon Y, Smith SM. (2010) Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J 62: 785–795 [DOI] [PubMed] [Google Scholar]
- Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C. (2010) The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo FM. (2004) Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: structure and regulation of their expression by physiological and stress conditions. Plant Sci 166: 971–982 [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH. (2007) ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS ONE 2: e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Sheen J. (2005) Sugar sensing and signalling networks in plants. Biochem Soc Trans 33: 269–271 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Baier M, Holman R, May AG, Bevan MW. (2006) Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J 46: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Rouached H, Secco D, Arpat B, Poirier Y. (2011) The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W, Lauerer M, Schulze E, Caboche M, Stitt M. (1997) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J 11: 671–691 [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP. (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA. (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18: 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Stewart GR. (1985) Nitrate assimilation and translocation by higher plants: comparative physiology and ecological consequences. Physiol Plant 64: 133–140 [Google Scholar]
- Smith FA, Raven JA. (1979) Intracellular pH and its regulation. Annu Rev Plant Physiol Plant Mol Biol 30: 289–311 [Google Scholar]
- Stitt M, Krapp A. (1999) The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ 22: 583–621 [Google Scholar]
- Strahm BD, Harrison RB. (2006) Nitrate sorption in a variable-charge forest soil of the Pacific Northwest. Soil Sci 171: 313–321 [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester-Bradley R, Kindred DR. (2009) Analysing nitrogen responses of cereals to prioritize routes to the improvement of nitrogen use efficiency. J Exp Bot 60: 1939–1951 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tercé-Laforgue T, Dubois F, Ferrario-Méry S, de Crecenzo MA, Sangwan R, Hirel B. (2004) Glutamate dehydrogenase of tobacco is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol 136: 4308–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Reich PB, Knops J, Wedin D, Mielke T, Lehman C. (2001) Diversity and productivity in a long-term grassland experiment. Science 294: 843–845 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Turano FJ. (1998) Characterization of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Physiol Plant 104: 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Lauter FR, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB. (2000) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J 21: 167–175 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM. (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Osakabe Y, Mizoi J, Nakashima K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J Biol Chem 285: 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33: W701– W704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, Zhang F, Li WX. (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Büchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG. (2004) The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett 576: 169–173 [DOI] [PubMed] [Google Scholar]