Abstract
Mitochondrial complex II (succinate dehydrogenase [SDH]) plays roles both in the tricarboxylic acid cycle and the respiratory electron transport chain. In Arabidopsis (Arabidopsis thaliana), its flavoprotein subunit is encoded by two nuclear genes, SDH1-1 and SDH1-2. Here, we characterize heterozygous SDH1-1/sdh1-1 mutant plants displaying a 30% reduction in SDH activity as well as partially silenced plants obtained by RNA interference. We found that these plants displayed significantly higher CO2 assimilation rates and enhanced growth than wild-type plants. There was a strong correlation between CO2 assimilation and stomatal conductance, and both mutant and silenced plants displayed increased stomatal aperture and density. By contrast, no significant differences were found for dark respiration, chloroplastic electron transport rate, CO2 uptake at saturating concentrations of CO2, or biochemical parameters such as the maximum rates of carboxylation by Rubisco and of photosynthetic electron transport. Thus, photosynthesis is enhanced in SDH-deficient plants by a mechanism involving a specific effect on stomatal function that results in improved CO2 uptake. Metabolic and transcript profiling revealed that mild deficiency in SDH results in limited effects on metabolism and gene expression, and data suggest that decreases observed in the levels of some amino acids were due to a higher flux to proteins and other nitrogen-containing compounds to support increased growth. Strikingly, SDH1-1/sdh1-1 seedlings grew considerably better in nitrogen-limiting conditions. Thus, a subtle metabolic alteration may lead to changes in important functions such as stomatal function and nitrogen assimilation.
Succinate:ubiquinone oxidoreductase (succinate dehydrogenase [SDH]; EC 1.3.5.1), commonly referred to as complex II, plays an essential role in mitochondrial metabolism both as a member of the electron transport chain and the tricarboxylic acid (TCA) cycle. This membrane-associated complex catalyzes the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol. SDH has been well characterized in bacteria and heterotrophic eukaryotes (Lemire and Oyedotun, 2002; Yankovskaya et al., 2003). In these organisms, complex II contains only four polypeptides: two peripheral membrane proteins, a flavoprotein (SDH1) that contains the succinate-binding site and an iron-sulfur protein (SDH2), and two small integral membrane proteins (SDH3 and SDH4) anchoring the SDH1-SDH2 subcomplex to the matrix side of the inner membrane (Yankovskaya et al., 2003). Interestingly, it has been shown that plant complex II may contain additional subunits of unknown function along with the four classical subunits (Eubel et al., 2003; Millar et al., 2004).
Few gene functional analyses have been employed to evaluate in plants the physiological role of complex II and their constituent subunits, which are all encoded in the nuclear genome in Arabidopsis (Arabidopsis thaliana; Figueroa et al., 2001, 2002; Millar et al., 2004). For instance, the absence of SDH2-3, which is one of the three genes encoding the iron-sulfur subunit and is specifically expressed in the embryo during seed development (Elorza et al., 2006), slows down seed germination, pointing to a role of a SDH2-3-containing complex II at an early step of germination (Roschzttardtz et al., 2009). The flavoprotein is encoded by two genes, designated SDH1-1 (At5g66760) and SDH1-2 (At2g18450). However, SDH1-2 is significantly expressed only in roots, albeit at a very low level (less than 10% of SDH1-1 in the same tissue), and its disruption has no effect on the growth and development of homozygous mutant plants, leaving SDH1-1 as the only gene that encodes for a functional flavoprotein (León et al., 2007). Molecular and genetic characterization of heterozygous SDH1-1/sdh1-1 mutant plants demonstrate that SDH1-1 is essential for pollen development and important for normal embryo sac development, explaining the reduced seed set of the heterozygous mutant plants and why no homozygous mutant plants could be obtained (León et al., 2007). Although these studies have clearly demonstrated the essential role of complex II in gametophyte development, its importance in other tissues such as photosynthetically active leaves remains unknown.
In recent years, evidence has been accumulating in support of a major role for mitochondria in photosynthetic metabolism (Raghavendra and Padmasree, 2003; Noguchi and Yoshida, 2008; Nunes-Nesi et al., 2008, 2011). A growing body of evidence suggests an important role for both the TCA cycle and the mitochondrial electron transport chain in the maintenance of optimal rates of photosynthesis. Indeed, photosynthetic performance can be modulated by modifications in these mitochondrial pathways. For instance, a wide range of mutant, antisense, or silenced plants with deficient expression of enzymes from the TCA cycle have been analyzed. Thus, tomato (Solanum lycopersicum) plants with reduced expression of aconitase or malate dehydrogenase showed an enhanced photosynthetic performance (Carrari et al., 2003; Nunes-Nesi et al., 2005). In contrast, tomato plants with less succinyl-CoA ligase or citrate synthase, or Arabidopsis plants with less mitochondrial isocitrate dehydrogenase, have no change in photosynthesis (Lemaitre et al., 2007; Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008), while tomato plants with less fumarase showed a decrease in photosynthesis (Nunes-Nesi et al., 2007). The reasons for these intriguing, quite diverse effects remain somewhat unclear at a mechanistic level. However, the reduced photosynthetic activity found in fumarase antisense plants was shown to be related to an impairment of stomatal function (Nunes-Nesi et al., 2007), and we have recently found that antisense repression of the iron-sulfur subunit of SDH in tomato results in a combined increase in stomatal conductance, photosynthetic rate, and growth (Araújo et al., 2011b). Few defects in the respiratory chain have been reported. The best-characterized respiratory chain mutant is a Nicotiana sylvestris mitochondrial mutant, CMSII, lacking a functional complex I (Gutierres et al., 1997). This leads to impaired photosynthesis and slower growth; however, plants attain biomass similar to that of wild-type plants and undergo reproductive development, although they are partially male sterile (Gutierres et al., 1997; Dutilleul et al., 2003a). Mutant plants were acclimated to the complex I defect, and this acclimation includes enhanced activity of nonphosphorylating NAD(P)H dehydrogenases, which bypass complex I (Sabar et al., 2000; Dutilleul et al., 2003a, 2003b). An Arabidopsis mutant lacking the 18-kD iron-sulfur subunit of complex I has also been described and shown to be affected in cold acclimation (Lee et al., 2002). Homozygous mutant plants grow more slowly than wild-type plants; nevertheless, these plants eventually reach heights similar to those of wild-type plants and are fully fertile. Altogether, these results indicate that a defect in complex I in plants is not lethal. In contrast, loss of complex II leads to a more severe phenotype, at least in gametophyte development (León et al., 2007), and this may be related to the dual role of SDH in both the TCA cycle and the respiratory chain.
Here, we analyze Arabidopsis SDH-deficient plants, compromised in the expression of the flavoprotein subunit of SDH, that were previously described by León et al. (2007). We demonstrate that a mild reduction in mitochondrial SDH activity has a positive impact on photosynthetic performance. Our results suggest that this effect is fairly specific, with detailed characterization revealing that the major effect is in stomatal function. Furthermore, mildly SDH-deficient plants grew better under nitrogen-limiting conditions, suggesting improved nitrogen assimilation.
RESULTS
Enhanced Growth of SDH-Deficient Plants
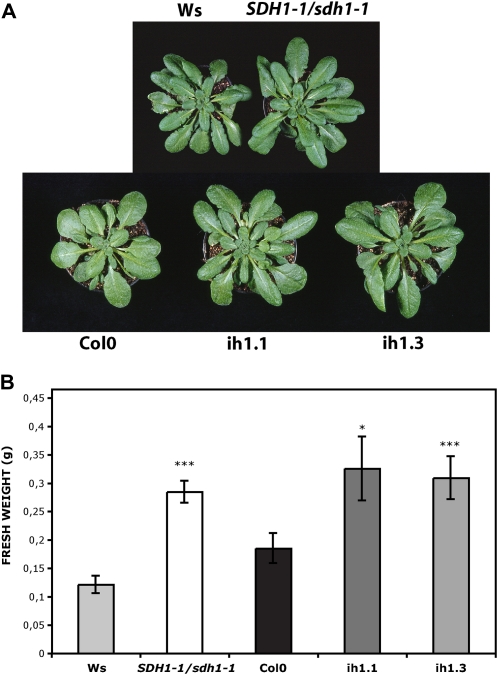
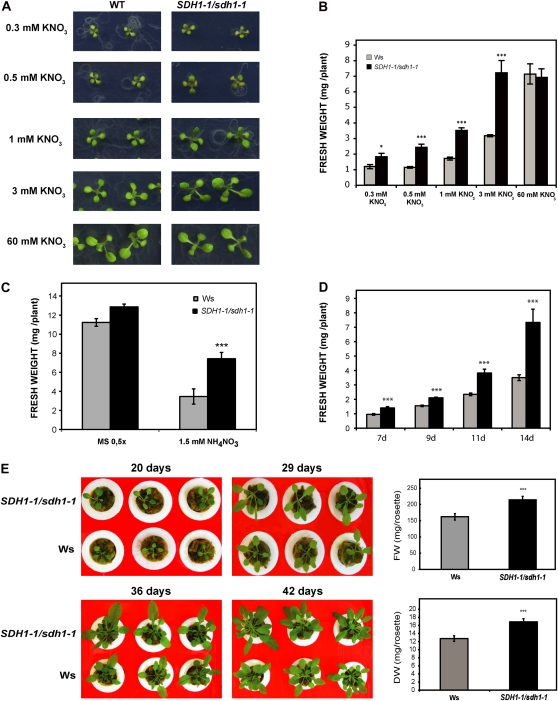
We have previously described heterozygous SDH1-1/sdh1-1 mutant plants and plants with down-regulation of SDH1-1 expression by RNA interference (RNAi; León et al., 2007). The heterozygous mutant plants showed consistent reductions in SDH1-1 mRNA (approximately 50% in flowers and approximately 25% in seedlings) and SDH activity (28%–34% in seedlings). SDH1-1 expression was also only partially silenced in the RNAi plants, suggesting that a drastic reduction may lead to unviable plants. To favor vegetative growth and obtain large leaves suitable for measuring photosynthetic parameters, we grew SDH1-1/sdh1-1 mutant plants and RNAi plants alongside their respective wild-type controls in short-day conditions (8 h of light/16 h of dark). A clear increase in the growth of the aerial part of mutant and RNAi plants was observed (Fig. 1A). Determination of shoot biomass confirmed that mutant and RNAi plants accumulate more shoot matter, 2.3- and 1.7-fold respectively, than their wild-type controls (Fig. 1B). Similar results were obtained in two different experiments with silenced plants and four different experiments with mutant plants, with increases in biomass ranging from 60% to 350%. Increases in fresh weight were paralleled by similar increases in dry weight (Supplemental Fig. S1). This enhanced growth phenotype was observed throughout growth (Supplemental Fig. S2). Furthermore, mutant plants attained higher biomass when sown directly in soil (Supplemental Fig. S3). However, it is worth mentioning that, in spite of this enhanced growth, seed yield was compromised, as described previously (León et al., 2007). Indeed, about 60% of the embryo sacs carrying the mutated sdh1-1 allele failed to complete their development, explaining why siliques contain fewer seeds (approximately 33% reduction) and are shorter than those of wild-type plants (León et al., 2007).
Figure 1.
SDH-deficient Arabidopsis plants showed an increase in size. Plants were grown for 2 weeks on plates and then 4 weeks in soil in short-day conditions. A, Photograph of representative 6-week-old plants. B, Biomass of 6-week-old shoots from wild-type (Ws, light gray bar; Col0, black bar), SDH1-1/sdh1-1 (white bar), RNAi ih1.1 (dark gray bar), and RNAi ih1.3 (gray bar) plants. Values are presented as means ± se of 11 individual plants per line. Asterisks indicate values that were determined by t test to be significantly different from the wild type (*** P < 0.01, * P < 0.05).
SDH Deficiency Results in Enhanced Photosynthesis That Correlates with Increases in Stomatal Conductance
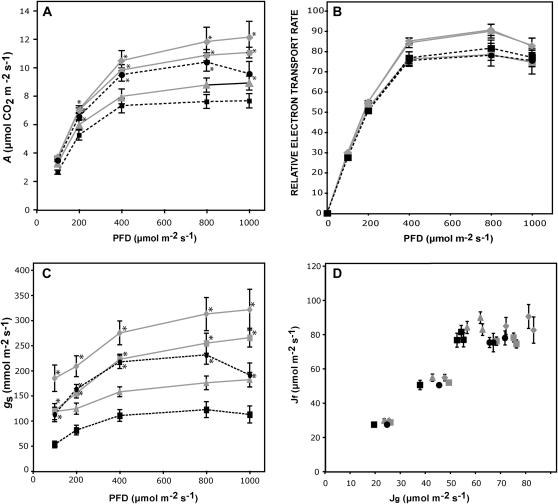
Given that mild SDH deficiency generated an increase in plant growth, we asked whether this phenomenon is associated with an increase in photosynthetic rates. First, gas exchange of fully expanded leaves was measured under photon flux densities (PFDs) that ranged from 100 to 1,000 μmol m−2 s−1. Compared with their respective wild-type controls, mutant and RNAi plants showed significantly higher CO2 assimilation rates (Fig. 2A). At saturating light levels, CO2 assimilation rates (A) were around 30% higher in the mutant and RNAi plants when compared with their respective controls. This was not due to an enhanced chloroplast electron transport, since relative electron transport rates did not change significantly between wild-type and mutant or RNAi plants (Fig. 2B). Moreover, nonphotochemical quenching indicated no differences between lines (data not shown). Interestingly, mutant and RNAi plants were characterized by a clear increase in stomatal conductance (gs; Fig. 2C; increases of around 80% for mutant plants and 45% and 75% for the two RNAi lines), and there was a strong positive correlation between A and gs (Pearson correlation of 0.84, P < 10−4, n = 25). No significant differences were found in dark respiration between mutant plants, RNAi plants, and their respective controls (data not shown). We also plotted electron transport rate determined from fluorescence against electron transport rate calculated from gas exchange, as described by von Caemmerer and Farquhar (1981; Fig. 2D). Altogether, these results suggest that photosynthesis is enhanced in SDH-deficient plants by a mechanism that improved CO2 uptake via the stomata.
Figure 2.
Effect of SDH deficiency on photosynthesis. A, CO2 assimilation rate at PFDs ranging from 100 to 1,000 μmol m−2 s−1. B, In vivo chlorophyll fluorescence was measured as an indicator of the electron transport rate by use of a LI-COR fluorometer at different light intensities. C, gs as a function of light intensity. D, Electron transport rates determined from fluorescence as in B (Jf) were plotted against electron transport rates calculated from gas exchange (Jg) according to von Caemmerer and Farquhar (1981). Values are presented as means ± se of determinations on six individual plants per genotype. All determinations were performed on fully expanded leaves from 6-week-old plants grown for 2 weeks on plates and 4 weeks in soil. Broken lines, heterozygous SDH1-1/sdh1-1 plants (black circles) are compared with their corresponding Ws wild-type plants (black squares); continuous lines, ih1.1 (gray squares) and ih1.3 (gray diamonds) partially silenced plants are compared with Col0 wild-type plants (gray triangles). In A to C, asterisks indicate values that were determined by t test to be significantly different (P < 0.05) from the wild type.
To further characterize the photosynthetic response in the SDH-deficient plants, we evaluated the response of A to the internal CO2 concentration (Ci) at 700 μmol photons m−2 s−1. These A/Ci curves indicate that SDH1-1/sdh1-1 mutant plants and RNAi plants resemble wild-type plants in their A responses (Supplemental Fig. S4). The maximum rate of carboxylation (Vcmax), rate of photosynthetic electron transport (Jmax), triose phosphate use, and Jmax/Vcmax were calculated for each genotype using the fitting model developed by Sharkey et al. (2007). No significant differences were found between SDH-deficient and wild-type plants (Table I). Gas-exchange parameters were also evaluated under natural growth conditions inside the greenhouse (Supplemental Table S1). SDH-deficient plants exhibited in these conditions A and gs that were significantly higher than those in the wild type, and importantly, these increases are associated with higher internal-to-ambient CO2 concentration (Ci/Ca) ratios. When taken together, our results are consistent with improved CO2 uptake in SDH-deficient plants independently of the mesophyll photosynthetic capacity to fix CO2 at a given Ci. Interestingly, we have recently observed similar effects by decreasing the iron-sulfur subunit of SDH in tomato (Araújo et al., 2011b). Further support for unaffected photosynthetic machinery was obtained by the evaluation of CO2 assimilation at saturating concentrations of 14CO2. Under these conditions, it is expected that CO2 uptake via the stomata would not be limiting; consistently, we found no significant differences either in total assimilation or in radiolabel incorporation into starch, sugars, amino acids, or organic acids (Supplemental Fig. S5).
Table I. Parameters derived from A/Ci curves.
Vcmax, Jmax, triose phosphate use (TPU), and Jmax/Vcmax ratio were computed using the fitting model developed by Sharkey et al. (2007). Values presented are means ± se of five individual plants per genotype, and no significant differences were found by t test between mutant SDH1-1/sdh1-1 and RNAi (ih1.1, ih1.3) plants and their respective wild-type controls (Ws and Col0).
| Parameter | Col0 | ih1.1 | ih1.3 | Ws | SDH1-1/sdh1-1 |
| Vcmax (μmol m−2 s−1) | 48.62 ± 7.50 | 50.64 ± 5.09 | 52.07 ± 3.65 | 47.75 ± 3.75 | 50.07 ± 4.42 |
| Jmax (μmol m−2 s−1) | 73.14 ± 9.24 | 70.02 ± 5.04 | 73.12 ± 3.93 | 70.43 ± 4.32 | 67.26 ± 3.56 |
| TPU (μmol m−2 s−1) | 5.02 ± 0.58 | 4.92 ± 0.35 | 5.05 ± 0.32 | 4.98 ± 0.31 | 4.87 ± 0.35 |
| Jmax/Vcmax | 1.54 ± 0.06 | 1.41 ± 0.09 | 1.42 ± 0.07 | 1.49 ± 0.07 | 1.37 ± 0.11 |
Enhancement of Stomatal Conductance Is Associated with Increases in Stomatal Aperture and Number
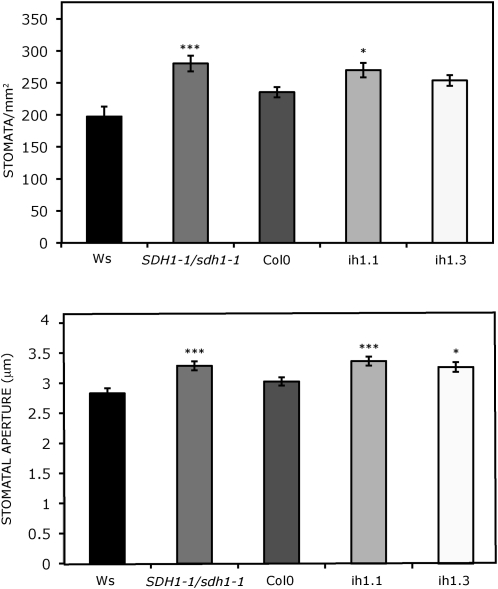
In order to analyze whether increases in stomatal conductance are related to changes in stomatal aperture and/or density, we evaluated these parameters in our mutant and RNAi SDH-deficient plants. Interestingly, we found an increase in stomata number that is statistically significant when comparing the mutant SDH1-1/sdh1-1 line and one of the RNAi lines (ih1.1) with their respective wild-type controls (Fig. 3). Furthermore, significant increases in stomatal aperture were observed in mutant and RNAi plants (Fig. 3), suggesting an important role for complex II in stomatal function. Increases of 35% to 40% and 15% to 18% were observed for stomata number and aperture, respectively, in different experiments with mutant and Wassilewskija (Ws) plants.
Figure 3.
Number and aperture of stomata in fully expanded leaves of mutant and silenced plants. Stomata frequency (stomata mm−2 leaf surface area) and aperture (μm) were determined. Values presented are means ± se of measurements made on 16 to 18 samples (stomata frequency) and on 78 to 80 stomata (aperture) from four individual plants per genotype. Plants were grown for 2 weeks under axenic conditions and then for 4 weeks in soil. Asterisks indicate values that were determined by t test to be significantly different from the wild type (* P < 0.05, *** P < 0.01).
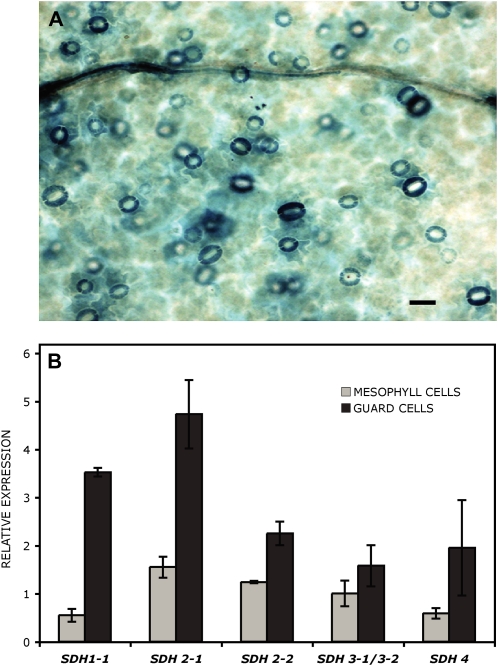
Consistent with an important role of complex II in guard cells, we observed a high SDH1-1 promoter activity in these cells. Indeed, strong GUS staining in guard cells was revealed when 0.8 kb of the SDH1-1 promoter was fused to the GUS reporter gene (Fig. 4A). Furthermore, data in the existing large expression databases confirmed that SDH1-1 expression is higher in guard cells than in mesophyll cells and that the same holds true for genes encoding other SDH subunits (Fig. 4B; data from Leonhardt et al. [2004] and Winter et al. [2007] at http://bar.utoronto.ca).
Figure 4.
SDH genes are highly expressed in guard cells. A, GUS expression in guard cells from Arabidopsis transgenic plants carrying a SDH1-1 promoter fusion. Histochemical localization of GUS activity is shown for leaves of 2-week-old seedlings grown under axenic conditions. The SDH1-1 promoter (788 bp) drives high expression in guard cells. Bar = 25 μm. B, Expression of complex II genes in mesophyll and guard cells. Data used to generate the digital northern blot were obtained using the Arabidopsis eFP browser at http://bar.utoronto.ca (Winter et al., 2007). For each gene, expression was normalized with respect to the median value across essentially all wild-type samples in the AtGenExpress tissue data set (Schmid et al., 2005).
Metabolic and Transcript Profiling of SDH-Deficient Plants
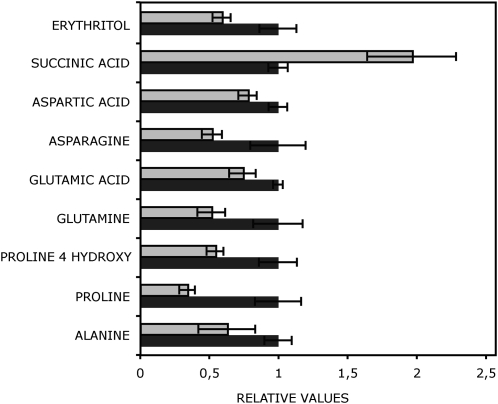
We compared the levels of metabolites in the mutant SDH1-1/sdh1-1 plants with those in wild-type plants grown in parallel using 6-week-old leaves and an established gas chromatography-mass spectroscopy protocol (Lisec et al., 2006). We were able to reliably identify 36 different metabolites (Supplemental Table S2). Those that are significantly different between mutant and wild-type plants are shown in Figure 5. As expected for SDH inhibition, we found increased levels of succinate. In contrast, no statistically significant changes were observable in the levels of other TCA cycle intermediates (malate, citrate), not even in fumarate, despite its being a reaction product of SDH (Supplemental Table S2). This may be related to the fact that mutant plants have only a mild reduction in SDH activity or the fact that fumarate may also be generated from malate during the operation of an anticlockwise phase of the TCA cycle (Sweetlove et al., 2010). Statistically significantly decreased levels, however, were found in the mutant plants for seven amino acids: Glu, Asp, Gln, Asn, Ala, Pro, and 4-Hyp (Fig. 5; Supplemental Table S2). These amino acids are key metabolites in nitrogen metabolism and are directly connected to TCA cycle intermediates or pyruvate (Glu, Asp, Ala) or derived from Glu (Gln, Pro) and Asp (Asn). Among the other identified metabolites, only erythritol level was found to be decreased significantly in mutant plants.
Figure 5.
Relative metabolite content of leaves from 6-week-old SDH1-1/sdh1-1 mutant plants. Data are normalized with respect to the mean response calculated for the wild type. The full data set from this metabolic profiling experiment is available as Supplemental Table S2. Values are presented as means ± se of determinations on six individual plants grown for 2 weeks on plates and then for 4 weeks in soil. The metabolites shown are those for which values in the mutant (gray bars) were determined by t test to be significantly different (P < 0.05) from the wild type (Ws; black bars).
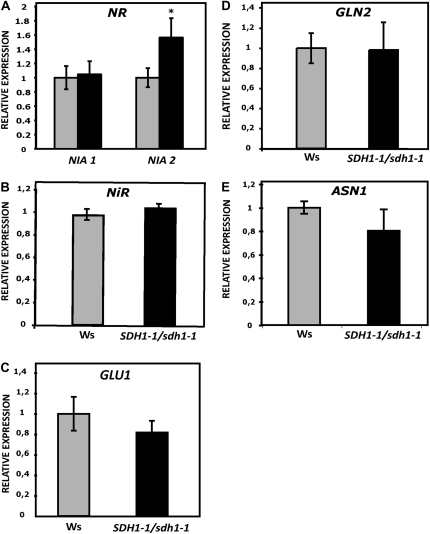
The observed decreases in amino acid levels could result from a slowing in their synthesis, which in turn may be the consequence of a slower functioning of the TCA cycle, source of the required carbon skeletons, and/or a deficiency in nitrogen assimilation. Our data argue against these possibilities, since mutant plants grew better (Fig. 1; Supplemental Figs. S1–S3) and no significant differences were found between wild-type and mutant plants for dark respiration, 14CO2 incorporation into starch, sugars, amino acids, and organic acids (at saturating [CO2]; Supplemental Fig. S5), and TCA cycle metabolite levels (malate, citrate, fumarate; Supplemental Table S2). Furthermore, neither the protein content nor the chlorophyll content was significantly altered in mutant plants (Supplemental Fig. S6), and analysis of leaf mRNA levels for genes in the nitrate assimilation pathway by real-time quantitative reverse transcription-PCR (qRT-PCR) showed that expression of these genes did not decrease in mutant plants (Fig. 6). While the levels of the transcripts for the plastid enzymes Gln synthetase (GLN2), ferredoxin-dependent Glu synthase (Fd-GOGAT; GLU1), and nitrite reductase (NiR) and for the cytosolic Asn synthetase (ASN1) did not differ significantly between wild-type and SDH1-1/sdh1-1 plants, transcript levels for one of the two NIA genes encoding nitrate reductase (NR) showed a modest increase in leaves from 6-week-old mutant plants when compared with wild-type plants (Fig. 6). However, this increase in NIA2 mRNA did not result in an increase in NR activity (Supplemental Fig. S7).
Figure 6.
Expression of genes in the nitrate assimilation pathway. Transcript levels in leaves of 6-week-old plants were determined by qRT-PCR for genes encoding NR (NIA1 and NIA2), NiR, GLN2, ASN1, and Fd-GOGAT1 (GLU1). Expression in mutant plants (black bars) is given relative to wild-type plants (set to 1; gray bars). Values are shown as means ± se from six biological replicates. The asterisk indicates a value that was determined by t test to be significantly different from the wild type (P < 0.05).
Altogether, these results strongly suggest that nitrogen metabolism was not impaired in mutant plants and that the decrease in amino acid levels may be due to a higher flux to proteins and other nitrogen-containing compounds (e.g. chlorophyll) to support increased growth.
Transcriptomes of wild-type and mutant SDH1-1/sdh1-1 leaves from 6-week-old plants were compared by DNA microarray analysis. Consistent with the hypothesis that these mildly SDH-deficient plants show only subtle modifications in their metabolism, as revealed by photosynthesis analysis (Fig. 2; Supplemental Figs. S4 and S5) and metabolite profiling (Supplemental Table S2), very limited effects on gene expression were found in the mutant plants (Table II). Only 16 genes were affected, five genes being up-regulated and 11 genes being down-regulated, and regulation of three genes (At2g15042, At2g18440, At4g15630) was confirmed by qRT-PCR (Supplemental Fig. S8).
Table II. Identification of Arabidopsis genes differentially expressed in the SDH1-1/sdh1-1 mutant.
DNA microarray analysis was performed with RNA obtained from leaves of 6-week-old Arabidopsis plants. Relative gene expression was calculated as the ratio of transcript levels in the SDH1-1/sdh1-1 mutant to those in the wild type.
| GeneChip IDa | Arabidopsis Genome Initiative No. | Relative Expression | Description | GO Biological Processb |
| 265893_at | At2g15042 | 3.92 | Protein binding | Unknown |
| 265330_at | At2g18440 | 2.44 | Gut15 (gene with unstable transcript 15) | Unknown |
| 257334_at | AtMg01370 | 2.35 | ORF111D, hypothetical protein | Unknown |
| 249726_at | At5g35480 | 2.15 | Unknown protein | Unknown |
| 263288_at | At2g36130 | 1.99 | Peptidyl-prolyl cis-trans-isomerase, putative/cyclophilin, putative/rotamase, putative | Protein folding |
| 245304_at | At4g15630 | 0.22 | Integral membrane family protein | Plasma membrane |
| 245003_at | AtCg00280 | 0.42 | PSBC, chloroplast gene encoding a CP43 subunit of the PSII reaction center. | Photosynthesis light reaction |
| 261518_at | At1g71695 | 0.47 | Peroxidase 12 (PER12, P12, PRXR6) | Response to oxidative stress |
| 245024_at | AtCg00120 | 0.48 | ATPA, encodes the ATPase α-subunit | ATP synthesis response to cold, defense response to bacterium |
| 265111_at | At1g62510 | 0.48 | Protease inhibitor/seed storage/lipid transfer protein family protein | Lipid transport |
| 249378_at | At5g40450 | 0.49 | Unknown protein | Unknown |
| 259640_at | At1g52400 | 0.54 | BGL1, BGLU18 (β-glucosidase 18) | Defense response to fungus |
| 252983_at | At4g37980 | 0.54 | ELI3-1 (elicitor-activated gene 3-1) | Plant-type hypersensitive response, response to bacterium |
| 244939_at | AtCg00065 | 0.54 | RPS12_RPS12A, chloroplast gene encoding ribosomal protein S12. | Translation |
| 259431_at | At1g01620 | 0.56 | PIP1;3,TMP-B, PIP1C (plasma membrane intrinsic protein 1C water channel) | Water transport, response to salt stress, response to water deprivation |
| 254609_at | At4g18970 | 0.66 | GDSL-motif lipase/hydrolase family protein | Lipid metabolic process |
Affymetrix probe set number.
Functional categories were identified using Gene Ontology (GO) from The Arabidopsis Information Resource (www.arabidopsis.org).
SDH Deficiency Results in Better Growth of Seedlings in Nitrogen-Limiting Conditions
We analyzed the growth of SDH1-1/sdh1-1 mutant and wild-type seedlings in nitrogen-limiting conditions. A modified Murashige and Skoog (MS) medium without nitrogen was supplemented with variable KNO3 concentrations ranging from 0.3 to 60 mm (Fig. 7, A and B), and growth was evaluated after 15 d. Notably, SDH1-1/sdh1-1 plants grew better than wild-type plants under a 0.3 to 3 mm nitrate regime: although the developmental stage was similar, as judged by the time of appearance and the number of true leaves, mutant plants clearly showed increased size (Fig. 7A) and significantly higher biomass (Fig. 7B). When mutant and wild-type seedlings were grown on 60 mm KNO3 (Fig. 7, A and B) or half-concentrated MS medium (Fig. 7C) containing 9.4 mm potassium nitrate and 10.3 mm ammonium nitrate, no significant differences were found between the genotypes. Similar results were obtained in several experiments when dry weight was determined (Supplemental Fig. S9A). Consistently, the biomass of mutant SDH1-1/sdh1-1 seedlings grown on 3 mm KNO3 was 1.7- to 2.7-fold higher than that of wild-type plants (11 experiments in which either fresh weight or dry weight was determined). Better growth of SDH1-1/sdh1-1 plants was also observed following the provision of 3 mm KNO3 in the absence of Suc (Supplemental Fig. S9B) and, additionally, when KNO3 was replaced by 1.5 mm NH4NO3 (Fig. 7C). Furthermore, enhanced growth was significant from 7 d onward (Fig. 7D), and differences in size were observed throughout growth, with plants grown hydroponically in the presence of 3 mm KNO3 (Fig. 7E). In some experiments, both shoot (rosette) and root weights of plants grown on 3 mm KNO3 were separately determined: similar increases in biomass were observed for roots and shoots of the mutant SDH1-1/sdh1-1 plants (Supplemental Fig. S10, A and B), while no significant differences in the rosette-root ratio were observed between genotypes. Increases in root biomass were mainly due to higher numbers of lateral roots (i.e. root branching appeared to be significantly increased in the mutant plants; Supplemental Fig. S10, C to E). Therefore, our results demonstrate that mild SDH-deficient plants have a better performance in nitrogen-limiting conditions, consistent with improved nitrogen assimilation and use.
Figure 7.
Effect of SDH deficiency on growth under low-nitrogen conditions. A and B, Photographs (A) and biomass (B) of 15-d-old wild-type (WT; light gray bars) and SDH1-1/sdh1-1 mutant (black bars) seedlings grown under axenic conditions on medium containing different concentrations of potassium nitrate. Eight independent shoot pools were weighed for each genotype and condition, and values (fresh weight per plant) are presented as means ± se. C, Biomass of 15-d-old shoots grown on medium containing NH4NO3. Seedlings were grown on solid medium containing either 1.5 mm NH4NO3 or 9.4 mm KNO3 + 10.3 mm NH4NO3 (0.5× MS). Fresh weights of eight biological replicates, each containing six to 25 shoots, were determined per genotype. D, Growth kinetics was analyzed by growing wild-type and SDH1-1/sdh1-1 mutant seedlings in solid medium containing 3 mm KNO3 and 1% Suc for the indicated times (in days). Fresh weights of eight biological replicates, each containing three to 20 shoots, were determined per genotype and time of growth. E, Improved growth of SDH-deficient plants grown hydroponically. Stratified seeds were sown on 0.5× MS (with 1% Suc) and, after 11 d in a 16-h/8-h day/night cycle, seedlings were transferred and grown hydroponically at 22°C under an 8-h-light/16-h-dark cycle. Twenty-day-old plants, 29-d-old plants, 36-d-old plants, and 42-d-old plants were photographed, and images of three representative plants per genotype are shown. Rosette fresh weight (FW) and dry weight (DW) of individual 42-d-old plants were determined: values presented are means ± se of 12 individual plants per line. Results show that the size increase in mutant plants is maintained throughout growth. Asterisks indicate values that were determined by t test to be significantly different from the wild type (* P < 0.05, *** P < 0.01).
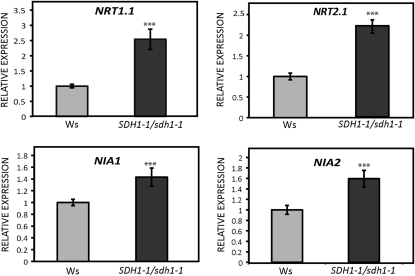
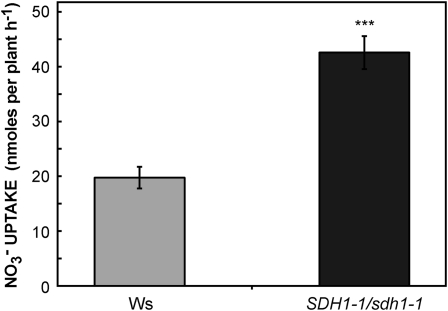
In order to understand the enhanced growth of mutant seedlings in nitrogen-limiting conditions, we analyzed the expression of key genes. We used qRT-PCR to measure the root transcript levels of NIA1 and NIA2, which encode the two isoforms of NR, and NRT1.1 and NRT2.1, encoding nitrate transporters involved in nitrate uptake by roots (Forde, 2000; Tsay et al., 2007). Interestingly, the transcript levels of NRT1.1 and NRT2.1 were 2.5- and 2-fold higher in the roots of 15-d-old seedlings grown on 3 mm KNO3, and NIA1 and NIA2 expression was also slightly increased (Fig. 8). Furthermore, nitrate uptake was evaluated using K15NO3 and 2-week-old seedlings grown on 3 mm KNO3. Nitrate uptake by SDH1-1/sdh1-1 mutant seedlings was higher than in the wild type (Fig. 9). Thus, the increase in biomass occurring in mutant seedlings grown in nitrogen-limiting conditions when compared with wild-type plants correlated with higher nitrogen uptake and assimilation.
Figure 8.
Expression of genes encoding nitrate transporters and nitrate reductase. Transcript levels for genes encoding nitrate transporters (NRT1.1 and NRT2.1) and nitrate reductase (NIA1 and NIA2) were determined by qRT-PCR in the roots of 15-d-old seedlings grown under axenic conditions on 3 mm KNO3. Expression in mutant SDH1-1/sdh1-1 seedlings (black bars) is given relative to wild-type seedlings (gray bars; set to 1). Values were normalized using clathrin as an internal control and are shown as means ± se from six biological replicates. Asterisks indicate values that were determined by t test to be significantly different from the wild type (*** P < 0.01).
Figure 9.
Higher nitrate uptake in SDH1-1/sdh1-1 mutant plants. Wild-type (gray bar) and SDH1-1/sdh1-1 mutant (black bar) seedlings were grown for 2 weeks on solid medium containing 3 mm KNO3 and 1% Suc. Nitrate uptake was then measured by incubating seedlings for 2 h in a solution with the same medium (3 mm KNO3) containing 20% K15NO3. Values are means ± se of six replicates, each containing 15 to 31 seedlings. Asterisks indicate a value that was determined by t test to be significantly different from the wild type (*** P < 0.01).
DISCUSSION
Mild SDH Deficiency and Stomatal Function
SDH1-1 is the gene encoding the functional flavoprotein of Arabidopsis mitochondrial complex II or SDH and is an interesting target for reverse genetics analysis to gain insight into the role of complex II in plants. We have previously established that sdh1-1 is a gametophytic mutation and that complex II is essential for gametophyte development (León et al., 2007). Therefore, only heterozygous SDH1-1/sdh1-1 and partially silenced SDH1 plants, with modest reductions in SDH1-1 expression, were viable. Here, we have characterized how this mild deficiency of mitochondrial complex II affects photosynthetic tissue, where the mitochondrial role is relatively unknown, and plant growth.
A clear increase in the growth of mutant and silenced plants was observed (Fig. 1; Supplemental Figs. S1–S3), and our results point to a specific effect on stomatal function. On the one hand, SDH-deficient plants showed higher CO2 assimilation rates, which correlated very well with higher stomatal conductance (Fig. 2). Furthermore, these changes in CO2 assimilation and stomatal conductance were caused by increases in both stomatal aperture and density (Fig. 3). On the other hand, very few additional significant alterations were detected in the mutant and silenced plants. The lack of differences in chloroplast electron transport rate (and nonphotochemical quenching), dark respiration, biochemical parameters such as the maximum rates of carboxylation by Rubisco and of photosynthetic electron transport (Table I; Supplemental Fig. S4), CO2 assimilation and distribution into starch, sugars, amino acids, and organic acids at saturating concentrations of CO2 (Supplemental Fig. S5), and TCA cycle metabolite levels (Supplemental Table S2) reveal that neither the photosynthetic machinery nor the TCA cycle is compromised. Moreover, few metabolites (mainly amino acids; Supplemental Table S2) and very few transcripts (Table II) were found to be different between mutant and wild-type plants, confirming that only subtle changes occur in the heterozygous mutant plants. Altogether, our results suggest that enhanced photosynthesis and growth in SDH-deficient plants is specifically mediated by a mechanism that improved CO2 uptake via the stomata. It is important to point out that our experiments were performed under controlled conditions that preclude water stress. Indeed, water loss from excised leaves from SDH1-1/sdh1-1 mutant and RNAi plants resulted in 27% to 28% fresh weight loss after 180 min, whereas in leaves from wild-type plants (Ws and Columbia [Col0]), fresh weight loss was only 22% to 23% (data not shown). Thus, SDH deficiency may have a negative impact on water loss by transpiration and water use efficiency; however, further work is required to analyze, for instance, the sensitivity of mutant stomata to low humidity and other environmental factors.
Quite diverse effects on photosynthetic performance have been observed on the down-regulation of various steps of the TCA cycle in tomato (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007; Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008; Araújo et al., 2011b). While the reasons for these differences are currently unclear in several cases, results obtained by down-regulation of fumarase and SDH in tomato (Nunes-Nesi et al., 2007; Araújo et al., 2011b) are especially relevant in the context of the data presented here. The fumarase-deficient tomato plants exhibited reduced biomass, CO2 assimilation, and stomatal conductance (Nunes-Nesi et al., 2007), which are diametrically opposite to what we found in tomato (Araújo et al., 2011b) and Arabidopsis (this work) SDH-deficient plants. It is highly interesting that similar results were obtained by inhibiting two different complex II subunits, the iron-sulfur subunit in tomato and the flavoprotein in Arabidopsis. This finding provides very strong support for a specific role of SDH in stomatal function and photosynthetic efficiency in plants. Further experiments in the antisense tomato plants suggest that these effects on stomatal aperture may be mediated by changes in the level of organic acids (e.g. fumarate, malate) in the apoplast (Araújo et al., 2011a, 2011b); thus, these data open the possibility that organic acids potentially synthesized in the mitochondrion may be involved in the regulation of stomatal aperture.
Nitrogen Assimilation in SDH-Deficient Plants
Our metabolic profiling analysis revealed that the levels of organic acids were unaffected in mutant SDH1-1/sdh1-1 whole leaves, with the exception of the SDH substrate succinate (Fig. 5; Supplemental Table S2). As outlined above, this may be due, at least in part, to the fact that mutant plants have only a mild reduction in SDH activity. Additionally, in the case of fumarate, the product of the SDH reaction, unaltered levels may not be unexpected, since this organic acid is present at a high concentration in Arabidopsis leaves (Chia et al., 2000), where it may represent a transient storage form of fixed carbon, and it has been reported recently that it is likely produced from malate by a cytosolic fumarase (Pracharoenwattana et al., 2010). In contrast, decreases in key amino acids of nitrogen metabolism were observed (Fig. 5). Although nitrogen deficiency typically results in decreased levels of Gln, which is the first amino acid formed during ammonium assimilation, and of many other amino acids, it also leads to decreased levels of protein and other nitrogen-containing structural components like chlorophyll, decreased nitrate reductase activity, inhibition of growth, and changes in plant architecture, including preferential root growth (Tschoep et al., 2009, and refs. therein). Therefore, the decreases in amino acids observed in our mutant plants do not appear to be the result of an impairment in nitrogen assimilation, since these plants grew better (Fig. 1; Supplemental Figs. S1–S3), and neither the protein content nor the chlorophyll content was altered (Supplemental Fig. S6). Moreover, leaf mRNA levels for genes in the nitrate assimilation pathway did not decrease in mutant plants (Fig. 6), and nitrate reductase activity did not differ between mutant and wild-type plants (Supplemental Fig. S7). It has to be pointed out that such an inverse relationship between the amino acid levels and growth or the nitrogen status is not without precedent, since Tschoep et al. (2009) found, using a soil-based growth system that allows a mild but sustained restriction of growth by nitrogen, that this condition did not alter protein or chlorophyll content and concomitantly led to decreases in growth and increases in amino acids. Thus, we interpreted the decrease in amino acid levels as the result of a higher synthesis of proteins and other nitrogen-containing compounds to support increased growth.
To test if nitrogen assimilation is altered in the mutant SDH1-1/sdh1-1, we analyzed the growth of wild-type and mutant seedlings in nitrogen-limiting conditions. Notably, mutant plants grew better, showing increased size and biomass (Fig. 7; Supplemental Figs. S9 and S10). Consistently, transcript levels of the nitrate transporters NRT1.1 and NRT2.1 were significantly elevated in roots of mutant seedlings grown under nitrogen-limiting conditions (Fig. 8), and nitrate uptake by the SDH1-1/sdh1-1 mutant was higher than in the wild type (Fig. 9). Altogether, our results indicate that a mild reduction in SDH led to enhanced nitrate uptake and assimilation and better growth under limiting nitrogen, an agronomically important trait. Although not directly comparable to our results, data obtained by growing plants at elevated CO2 concentrations may be relevant (Stitt and Krapp, 1999; Matt et al., 2001). Within this context, plants growing on nitrate under elevated CO2 led to an increase in nitrate uptake and assimilation and improved nitrogen use efficiency, which paralleled the increased rate of photosynthesis. Moreover, slight increases in NIA transcripts, minor effects on NR activity, and no effects on GLN transcripts or activity were found at elevated CO2, resembling our results on the SDH1-1/sdh1-1 plants (Fig. 6; Supplemental Fig. S7). However, these results contrast with those recently published by Bloom et al. (2010), who showed that enhanced CO2 inhibits nitrate assimilation in wheat (Triticum aestivum) and Arabidopsis; hence, this interaction remains far from fully understood. Further work is required to understand the relationship between the effects of SDH deficiency on stomatal function and nitrogen assimilation, a plausible hypothesis being that an increased supply of carbon has altered the utilization of nitrogen to maintain the carbon/nitrogen balance.
Since carbon and nitrogen assimilation has a large influence on plant growth and crop yields, attempts to increase the efficiency of these processes are manifold. Unfortunately, as yet, in only a few cases have significant improvements in carbon and nitrogen assimilation been achieved (Miyagawa et al., 2001; Sinclair et al., 2004; Yanagisawa et al., 2004; Nunes-Nesi et al., 2010). The results demonstrating that mild SDH-deficient plants grew better under nitrogen-limiting conditions are similar to those obtained in one successful example of metabolic engineering, in which transgenic Arabidopsis lines overexpressing the maize (Zea mays) Dof1 (for DNA-binding with one finger 1) transcription factor showed improved growth under nitrogen-limiting conditions (Yanagisawa et al., 2004). Thus, subtle modulation of metabolic pathways may be considered as a possible way to endow plants with desired characteristics.
CONCLUSION
We have characterized Arabidopsis plants with a mild deficiency in mitochondrial complex II (SDH) and found that these plants grew better than their wild-type counterparts. CO2 assimilation is improved through the enhancement of stomatal aperture and density, and this higher photosynthetic performance appears to lead to better nitrogen assimilation, maintaining the carbon/nitrogen balance to support the biosynthesis of macromolecules and growth. Although the detailed molecular mechanisms linking SDH deficiency with stomatal biogenesis and function and nitrogen metabolism are far from fully understood and deserve further work, it is striking that such a subtle metabolic alteration led to changes in very important functions associated with plant growth potential.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Heterozygous mutant SDH1-1/sdh1-1 plants and plants with partial down-regulation of SDH1-1 expression by RNAi (ih1.1 and ih1.3) have been described previously (León et al., 2007). In all experiments, these mutant and silenced plants were grown in parallel with their control wild-type Arabidopsis (Arabidopsis thaliana) ecotypes, Ws and Col0, respectively. Seeds were cold treated for 48 h at 4°C in darkness and then sterilized for 10 min with a solution of 10% (v/v) commercial bleach. After rinsing thoroughly with sterile distilled water, seeds were sown on one-half-concentrated MS medium, 1% (w/v) Suc, solidified with 0.8% (w/v) agar and supplemented or not with 50 mg L−1 kanamycin. For most experiments (Figs. 1–6; Tables I and II; Supplemental Tables S1 and S2; Supplemental Figs. S1, S2, and S4–S8), after 2 weeks in a 16-h/8-h day/night cycle, seedlings were transferred to soil and grown for an additional 4 weeks at 22°C under an 8-h-light/16-h-dark cycle (short-day conditions).
For growth under low-nitrogen conditions (Figs. 7–9; Supplemental Figs. S9 and S10), seeds were treated as described and sown on agar plates containing MS medium without nitrogen, 1% (w/v) Suc, and different concentrations of potassium nitrate. Growth was carried out at 22°C for 15 d under a 16-h-light/8-h-dark cycle. For NO3− uptake analyses, seedlings grown on 3 mm KNO3 in these conditions were subsequently incubated in a solution with the same medium containing 20% K15NO3. After 2 h, seedlings were washed, dried at 70°C for 48 h, weighed, and sent for isotopic analysis to the Cornell Isotope Laboratory. In the experiment of Figure 7E, seedlings were transferred and grown hydroponically at 22°C under an 8-h-light/16-h-dark cycle (Gibeaut et al., 1997). Growth medium was slightly modified to contain 3 mm KNO3 and 1.5 mm CaCl2 [replacing 1.5 mm Ca(NO3)2].
Homozygous transgenic plants carrying a SDH1-1 promoter fragment (788 bp) and the SDH1-1 5′ untranslated region (116 bp) fused to GUS were obtained using standard protocols. Briefly, the 5′ upstream sequence was obtained by PCR amplification of genomic DNA between a 5′ forward primer (5′-TAAGAATTCCACATTAATAAGCCAGGCC-3′) containing an EcoRI site and a 3′ reverse primer (5′-AATCCATGGCGTCTTGTTGTTGAT-3′) containing an NcoI site encompassing the SDH1-1 ATG initiation codon. This PCR product was cloned into pGEM-T plasmid (Promega), and the DNA fragment obtained by digestion with BamHI and NcoI was ligated into pCAMBIA 1381 (CAMBIA). Construct structure was verified by DNA sequencing and introduced into Agrobacterium tumefaciens GV3101 by electroporation. A. tumefaciens-mediated transformation of Arabidopsis plants was accomplished using the floral dip protocol (Clough and Bent, 1998).
Measurements of Photosynthetic Parameters
Fluorescence emission and gas-exchange measurements were made on intact, fully expanded leaves of 6-week-old plants grown as described (2 weeks under axenic conditions and then 4 weeks in soil) with an open-flow gas-exchange system (LI-COR; model LI-6400; http://www.licor.com/). Dark respiration was determined using the same gas-exchange system. Measurements were performed at a leaf chamber temperature of 22°C and a vapor pressure deficit of 1.55 ± 0.27 (sd) kPa. In the experiment of Figure 2, irradiances ranged from 100 to 1,000 μmol photons m−2 s−1, and the leaf chamber CO2 concentration was 400 μmol mol−1. In the experiment of Supplemental Figure S4, A/Ci curves were performed at a PFD of 700 μmol photons m−2 s−1. Measurements started at 350 μmol CO2 mol−1, and once the steady state was reached (within 3–5 min), CO2 concentration was gradually lowered to 50 μmol mol−1 and then increased stepwise up to 2,000 μmol mol−1, exactly as described by Long and Bernacchi (2003). Estimates of Vcmax, Jmax, and triose phosphate use were calculated for each A/Ci curve using the fitting model of Sharkey et al. (2007).
The 14C-labeling pattern of Suc, starch, amino acids, and organic acids was performed by illuminating leaf discs (10 mm diameter) from the same plants in an oxygen electrode chamber (Hansatech; www.hansatech-instruments.com) containing a saturated level of 14CO2 at a PFD of 700 μmol photons m−2 s−1 photosynthetically active radiation at 22°C for 30 min, and subsequent fractionation was performed exactly as described by Lytovchenko et al. (2002).
Analysis of Stomatal Numbers and Apertures
Stomatal numbers and apertures were determined from the same or similar leaves as used for gas-exchange analysis. Negative impressions were taken from the abaxial surface of 6-week-old leaves 2 h after the beginning of the light period with dental silicone. Positive images were then obtained using nail polish and viewed with a light microscope (Olympus IX81) equipped for differential interference contrast. To quantify stomata frequency, 16 to 18 samples were analyzed for each genotype (around 0.04 μm2 each sample). To evaluate stomatal aperture, 78 to 82 stomata were scored for each genotype.
Histochemical GUS staining was performed exactly as described (Elorza et al., 2004). Samples were visualized using a Nikon Eclipse 80i microscope equipped with differential interference contrast (Nomarski) optics.
Metabolic Profiling
Plants were grown for 2 weeks under axenic conditions and then for 4 weeks in soil, as described. Leaf samples from these 6-week-old plants were taken at the middle of the day, immediately frozen in liquid nitrogen, and stored at –80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of the extraction buffer, as described by Lisec et al. (2006). The levels of metabolites were quantified by gas chromatography-mass spectroscopy as described by Roessner et al. (2001). Data analysis was performed using the TagFinder program and included recent additions to the Max Planck Institute mass spectral libraries (Schauer et al., 2005).
Chlorophyll content was determined as described (Arnon, 1949) using pigment extracts from around 0.1 g of leaf tissue. Protein extracts were prepared as described (Roschzttardtz et al., 2009), and total protein content was assessed by the method of Bradford (1976).
Expression Analysis by Real-Time qRT-PCR
Plants were grown for 2 weeks under axenic conditions and then for 4 weeks in soil, as described. Total RNA was isolated from leaves of these 6-week-old plants harvested in liquid nitrogen using the TRIzol reagent according to the manufacturer’s protocol (Invitrogen) and treated with DNase I (Promega). cDNA synthesis was performed on 2 μg of RNA, using oligo(dT) as primer and the ImProm-II RT system (Promega). To evaluate gene expression, the following primer pairs, designed with the AmplifX software (http://ifrjr.nord.univ-mrs.fr/AmplifX-Home-page), were used: for NRT1.1, 5′-TGGTCGAGCTCAAACGTCTT-3′ (forward) and 5′-ATTAACGCTTCGCCGATACC-3′ (reverse); NRT2.1, 5′-CTTGAAGCTCCACACAGCA-3′ (forward) and 5′-ATCCACAACGTCCACAACCT-3′ (reverse); for NIA1, 5′-AAGGCAAAGGCAACTTCCTGGT-3′ (forward) and 5′-TCATCCTCGGTTCTGTTTGCGT-3′ (reverse); for NIA2, 5′-ACATGGCGGCCTCTGTAGAT-3′ (forward) and 5′-CCTGGAACGGGAGGTTTGTA-3′ (reverse); for NiR, 5′-CCTTCCTCCTCCACCAAACC-3′ (forward) and 5′-AACAGAGGCGGTGGATTCG-3′ (reverse); for GLU1, 5′-GCCTGAGGAAGCAACGATAG-3′ (forward) and 5′-CTCCTGTCATACCAGCAGCA-3′ (reverse); for GLN2, 5′-GCCTTTAGGTTGGCCTGTTG-3′ (forward) and 5′-GTCACGCCCCCAAATCTTG-3′ (reverse); for ASN1, 5′-GGGATTGATGCGATAGAGGGA-3′ (forward) and 5′-CGGAGAAACCATCTTGACC-3′ (reverse); and for clathrin, 5′-AATACGCGCTGAGTTCCCTT-3′ (forward) and 5′-AGCACCGGGTTCTAACTCAA-3′ (reverse). qRT-PCR assays were performed on one-tenth of the cDNA using the Mx3000P QPCR System (Stratagene; www.genomics.agilent.com) according to the manufacturer’s instructions and the SensiMix Plus SYBR kit provided by Quantace (www.bioline.com). Clathrin transcript was used as an internal control to normalize expression values, which were calculated taking into account the amplification efficiency of each primer pair.
Determination of NR Activity
The NR activity was measured following the protocol described by Scheible et al. (1997) in leaves from plants grown for 2 weeks on plates and 4 weeks in soil.
Microarray Hybridization and Data Analysis
Rosette leaves from 6-week-old (4 weeks in soil) Arabidopsis plants were collected in the middle of the light period and frozen in liquid nitrogen. Three independent biological replicates were performed for microarray experiments on the Arabidopsis ATH1 Affymetrix gene chip. Total RNA was extracted by the TRIzol method and treated with DNase I. The concentration and purity of RNA samples were determined with a NanoDrop spectrophotometer (NanoDrop Technologies). First- and second-strand cDNA syntheses (from 0.5 μg of RNA) and biotin labeling of antisense RNA were performed using the GeneChip 3′ IVT Express Kit, following the protocols of the manufacturer (Affymetrix). Amplified antisense RNA was quantified with the NanoDrop spectrophotometer, and 15 μg was fragmented and used to hybridize the ATH1 genome array for 16 h at 45°C. Hybridization, washing, and staining of the chips were carried out in the Affymetrix EukGE-WS2v5-450 Fluidics Station.
Arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G, and their quality was assessed using the Affymetrix quality controls. CEL files were imported into R software for statistical computing (http://www.r-project.org), and data quality was analyzed using the simpleaffy package from the Bioconductor Web site (http://www.bioconductor.org/). Cutoff values were determined according to Affymetrix. Arrays were normalized in R using Gene Chip Robust Multiarray Averaging from the Bioconductor affy package. Statistically significant differences (P < 0.05) between mutant and wild-type plants were determined using RankProduct from Bioconductor and 1,000 permutations.
Statistical Analysis
Student’s t tests were performed using the algorithm embedded into Microsoft Excel. The term “significant change” is employed in the text when it is supported by a value of P < 0.05.
The microarray data reported in this publication have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under experiment E-MEXP-2992.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fresh weight and dry weight of 6-week-old shoots.
Supplemental Figure S2. Improved growth of SDH-deficient plants.
Supplemental Figure S3. Biomass of rosette leaves from 6-week-old plants grown in soil.
Supplemental Figure S4. Rate of net CO2 assimilation as a function of internal CO2 concentration.
Supplemental Figure S5. Effect of SDH deficiency on photosynthetic carbon partitioning at the onset of illumination of leaves from 6-week-old plants.
Supplemental Figure S6. Chlorophyll and protein contents of leaves from mutant and wild-type plants are similar.
Supplemental Figure S7. NR activities of leaves from mutant and wild-type plants are similar.
Supplemental Figure S8. Validation of DNA microarray data for three selected genes.
Supplemental Figure S9. Growth of SDH1-1/sdh1-1 seedlings under low-nitrogen conditions in the presence or absence of Suc.
Supplemental Figure S10. Increased root growth of SDH-deficient plants grown under low-nitrogen conditions.
Supplemental Table S1. Gas-exchange parameters of wild-type and SDH-deficient plants.
Supplemental Table S2. Relative metabolite contents of wild-type and sdh1-1/SDH1-1 leaves.
Acknowledgments
We are very grateful to Luis León (Pontificia Universidad Católica de Chile) for helping us to analyze Affymetrix data and to Dr. Elena Vidal (Pontificia Universidad Católica de Chile) for her valuable help and suggestions in performing the nitrogen experiments.
References
- Araújo WL, Nunes-Nesi A, Fernie AR. (2011a) Control of stomatal aperture: a renaissance of the old guard. Plant Signal Behav 6: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011b) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplast: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Burger M, Rubio Asensio JS, Cousins AB. (2010) Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 328: 899–903 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR. (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DW, Yoder TJ, Reiter WD, Gibson SI. (2000) Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 211: 743–751 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G. (2003a) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, de Paepe R. (2003b) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, León G, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X. (2004) Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol 136: 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, Roschzttardtz H, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X. (2006) A nuclear gene for the iron-sulfur subunit of mitochondrial complex II is specifically expressed during Arabidopsis seed development and germination. Plant Cell Physiol 47: 14–21 [DOI] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP. (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Araya A, Jordana X. (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Jordana X. (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46: 241–250 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R. (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK. (2002) A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre T, Urbanczyk-Wochniak E, Flesch V, Bismuth E, Fernie AR, Hodges M. (2007) NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol 144: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire BD, Oyedotun KS. (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553: 102–116 [DOI] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X. (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Long SP, Bernacchi CJ. (2003) Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J Exp Bot 54: 2393–2401 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A, Sweetlove L, Pauly M, Fernie AR. (2002) The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M. (2001) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- Millar AH, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP. (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S. (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19: 965–969 [DOI] [PubMed] [Google Scholar]
- Noguchi K, Yoshida K. (2008) Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8: 87–99 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Araújo WL, Fernie AR. (2011) Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Sulpice R, Gibon Y, Fernie AR. (2008) The enigmatic contribution of mitochondrial function in photosynthesis. J Exp Bot 59: 1675–1684 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Zhou W, Keech O, Francisco PB, Udomchalothorn T, Tschoep H, Stitt M, Gibon Y, Smith SM. (2010) Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J 62: 785–795 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K. (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Fuentes I, Vásquez M, Corvalán C, León G, Gómez I, Araya A, Holuigue L, Vicente-Carbajosa J, Jordana X. (2009) A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol 150: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y. (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scheible W, Lauerer M, Schulze E, Caboche M, Stitt M. (1997) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J 11: 671–691 [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL. (2007) Fitting photosynthetic carbon dioxide response curves for C(3) leaves. Plant Cell Environ 30: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR. (2008) Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Purcell LC, Sneller CH. (2004) Crop transformation and the challenge to increase yield potential. Trends Plant Sci 9: 70–75 [DOI] [PubMed] [Google Scholar]
- Stitt M, Krapp A. (1999) The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ 22: 583–622 [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR. (2007) Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD. (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S. (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700–704 [DOI] [PubMed] [Google Scholar]