Abstract
A new regulatory pathway involved in plant response to oxidative stress was revealed using the iron-induced Arabidopsis ferritin AtFER1 as a model. Using pharmacological and genetic approaches, the DownSTream (DST) cis-acting element in the 3′-untranslated region of the AtFER1 mRNA was shown to be involved in the degradation of this transcript, and oxidative stress triggers this destabilization. In the two previously identified trans-acting mutants (dst1 and dst2), AtFER1 mRNA stability is indeed impaired. Other iron-regulated genes containing putative DST sequences also displayed altered expression. Further physiological characterization identified this oxidative stress-induced DST-dependent degradation pathway as an essential regulatory mechanism to modulate mRNA accumulation patterns. Alteration of this control dramatically impacts plant oxidative physiology and growth. In conclusion, the DST-dependent mRNA stability control appears to be an essential mechanism that allows plants to cope with adverse environmental conditions.
Keywords: ferritin, iron, mRNA stability, oxidative stress
Introduction
Aerobic organisms are constantly exposed to reactive oxygen species (ROS) that originate from both intracellular metabolism and exogenous sources. Although transient changes in the levels of ROS can have important signalling functions, elevated ROS levels can overcome the cellular anti-oxidant defenses and cause severe damage to nucleic acids, proteins and lipids (Rouault and Klausner, 1996; Touati, 2000; Theil, 2007; Toledano et al, 2007). Cellular redox fluctuations can trigger a wide spectrum of responses, mainly by implementing changes in gene expression patterns (Allen and Tresini, 2000). These changes are promoted primarily by transcriptional and post-transcriptional mechanisms. The transcriptional regulation of eukaryotic gene expression by ROS has been extensively studied and many signalling pathways have been identified (Allen and Tresini, 2000). There is increasing evidence that the expression of ROS-regulated genes is also modulated by post-transcriptional events in yeast and animal systems.
Apart from the regulatory mechanisms involving small RNAs, RNA-binding proteins (RBPs) are the main type of trans-acting factors, which recognize specific cis-elements in the target mRNA that are implicated in the regulation of mRNA stability (Valencia-Sanchez et al, 2006; Keene, 2007). Some RBPs associate with RNA sequences that are widely present in messenger RNAs, such as the 5′-cap structure or the 3′-poly(A) tail. Other RBPs interact with specific specialized mRNA sequences present in the 5′- and 3′-untranslated regions (UTRs) and affect changes in mRNA stability. Three mRNA decay regulatory pathways have been studied in detail, and the cis-elements were named AU-rich element (ARE; Shaw and Kamen, 1986; Chen and Shyu, 1995), iron responsive element (IRE; Casey et al, 1988; Rouault, 2006) and DownSTream (DST) element (McClure et al, 1989; Newman et al, 1993).
The DST cis-element was first identified in soybean plants in the 3′-UTR of the unstable SAUR (auxin-inducible small auxin-up) transcript (McClure et al, 1989). This downstream sequence, conserved among different plant species (Newman et al, 1993), is about 40 bp long and consists of two highly conserved sequences separated by a variable sequence (McClure et al, 1989). A repeat of this cis-element confers mRNA instability when inserted in the 3′-UTRs of different reporter genes in plants, but not in mammalian cells (Newman et al, 1993; Feldbrügge et al, 2002). A genome-wide study in Arabidopsis showed that the DST sequence was enriched in unstable transcripts (Narsai et al, 2007). Three trans-acting components of the DST pathway, named DST1–3, were isolated by a forward genetic approach (Johnson et al, 2000; Pérez-Amador et al, 2001; Lidder et al, 2004). Molecular and phenotypic analysis of the dst mutants showed deregulation of functional subsets of genes including some circadian clock-related genes (Pérez-Amador et al, 2001). However, to date, the only DST sequence shown to be functional is the one found in the SAUR gene (McClure et al, 1989). In addition, SAUR mRNA decay was not promoted by auxin, and phenotypes associated with auxin production, or perception, were not observed in the mutants altered in the DST pathway (Johnson et al, 2000), suggesting that DST-triggered mRNA degradation is not involved in auxin signalling.
Deciphering the signal that controls mRNA decay and degradation is of primary importance for our understanding of post-transcriptional regulation of biological processes. Using the Arabidopsis thaliana ferritin gene AtFER1 as a model, we present evidence that AtFER1 mRNA half-life is strongly decreased in response to pro-oxidant treatments such as excess iron (Fe) and hydrogen peroxide (H2O2). This oxidative stress-dependent degradation process involves a functional DST cis-element identified in the 3′-UTR region of the transcript. In the two mutants dst1 and dst2 that are defective in trans-acting factors, the AtFER1 mRNA decay was not promoted by Fe treatment, confirming that DST is involved in the Fe-mediated oxidative stress-induced AtFER1 mRNA degradation. By analysing transcriptome data, we identified additional genes that are upregulated in response to Fe excess, contain a DST sequence in their 3′-UTR and have their mRNA abundance altered in dst1 and dst2 mutants. By studying fitness of dst1 and dst2 mutants, we show that their growth is altered under high-Fe nutritional conditions, and that their response to oxidative stress is impaired. Together, our results reveal that the DST-dependent mRNA degradation mechanism is essential for the proper response of plants to Fe and to oxidative stress.
Results
AtFER1 mRNA stability is modulated by Fe treatment and ROS
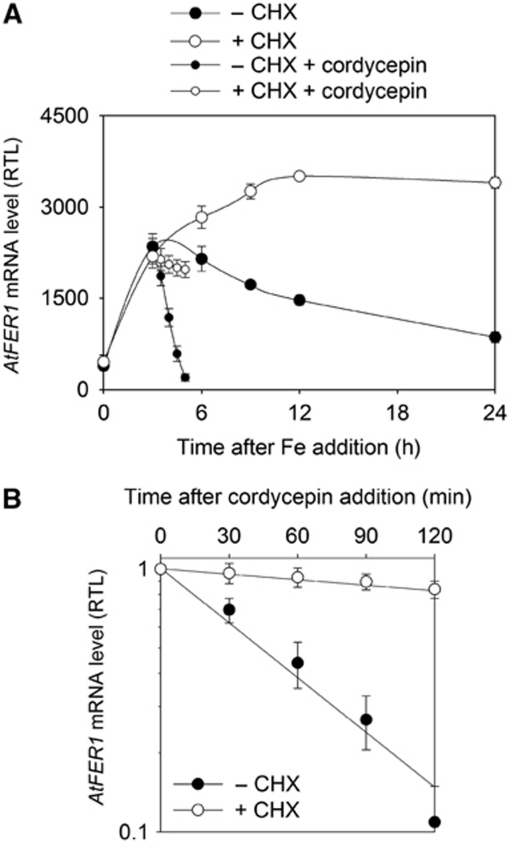
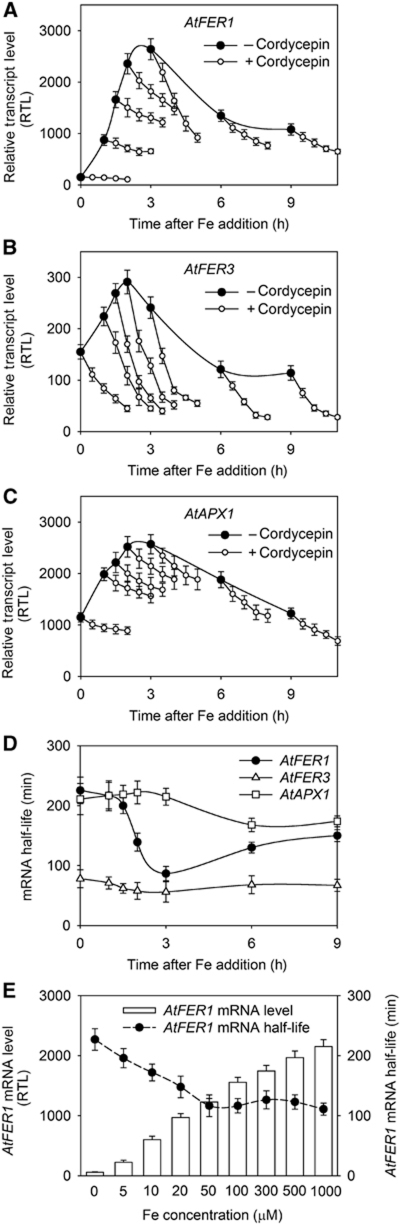
Fe was added to the culture medium of Arabidopsis cells cultivated in suspension, and AtFER1 mRNA accumulation was determined using quantitative qRT–PCR. Fe treatment led to a transient increase in AtFER1 mRNA abundance, reaching a maximum after 3–6 h, followed by a decrease in abundance at the later points of the time course (Figure 1A). In contrast, when cells were pre-treated for 1 h with the protein synthesis inhibitor cycloheximide (CHX), the increase in AtFER1 transcript abundance was not transient, but continued up to 15 h (Figure 1A). This result, in agreement with our previous observations obtained by northern blot (Arnaud et al, 2006), suggests that de novo protein synthesis is necessary either to decrease AtFER1 transcription rate or to increase AtFER1 mRNA turnover. In order to discriminate between these two possibilities, AtFER1 mRNA stability was determined using cordycepin, an inhibitor of transcription. Cells were treated or not with CHX for 1 h, and with Fe for 3 h prior to cordycepin addition, and AtFER1 mRNA abundance was monitored every 30 min for 2 h (Figure 1A). AtFER1 mRNA half-life was determined using a logarithmic regression curve according to Gallie et al (1991), in which the AtFER1 transcript amount was normalized relative to the abundance before the cordycepin treatment (Figure 1B). When cells were not pre-treated with CHX, AtFER1 mRNA abundance quickly decreased after inhibition of transcription, corresponding to an estimated half-life of 60 min. By contrast, when cells were pre-treated with CHX, the AtFER1 mRNA was much more stable, with an estimated AtFER1 mRNA half-life over 5 h. In order to determine whether Fe may affect AtFER1 mRNA stability, we next determined the transcript degradation rates as well as the half-lives at different time points after Fe addition. Fe treatment led to a transient increase of the AtFER1 degradation rate (Figure 2A). The highest degradation rate was observed 3 h after Fe addition, when the transcript abundance reached its maximum. AtFER1 mRNA half-life ranged from about 220 min in the absence of Fe treatment to about 90 min 3 h after Fe addition. By contrast, AtFER3 and AtAPX1, two other transcripts known to transiently accumulate in response to Fe (Petit et al, 2001; Fourcroy et al, 2004), did not exhibit such modulation of their degradation rate (Figure 2B and C). Based on the mRNA degradation patterns observed in Figure 2A–C, transcript half-lives were calculated at each time points of the kinetics (Figure 2D). AtFER1 mRNA half-life was quickly and strongly decreased between 2 and 6 h whereas AtFER3 and AtAPX1 decay was only slightly modulated. For all subsequent experiments, a 3-h iron treatment was selected for AtFER1 mRNA half-life determination, since it represents the most contrasted condition (when compared with the half-life obtained in control condition). These results indicate that AtFER1 mRNA stability is significantly and specifically modulated in response to Fe and that the process requires newly synthesized protein(s).
Figure 1.
Cycloheximide affects AtFER1 mRNA stability. (A) Effect of cycloheximide on the relative AtFER1 mRNA abundance after Fe treatment. Arabidopsis cells were grown in liquid medium for 7 days. Cells were treated (+CHX) or not (−CHX) with 100 μM cycloheximide for 1 h before Fe addition (Fe-citrate 300 μM). Cells were collected before, and 1, 3, 6, 9, 12 and 24 h after the Fe addition. Three hours after Fe addition, cordycepin was added, and cells were recovered every 30 min for 2 h. AtFER1 mRNA relative accumulation was determined by qRT–PCR using Sand transcript (At2g28390) as a reference. (B) Effect of cycloheximide on AtFER1 half-life. Relative AtFER1 mRNA abundance after cordycepin treatment as measured in (A) was plotted in a logarithmic scale. The relative half-life was calculated according to Gallie et al (1991). Values and standard errors were obtained from three independent experiments.
Figure 2.
AtFER1, but neither AtFER3 nor AtAPX1 mRNA stabilities are modulated by Fe. AtFER1 (A), AtFER3 (B) and AtAPX1 (C) mRNA degradation rate during the time course after Fe addition. Cells were collected before (0), and 1, 1.5, 2, 3, 6 and 9 h after treatment with 300 μM Fe-citrate. Total RNA was extracted and AtFER1, AtFER3 and AtAPX1 mRNA abundance was determined as described in Figure 1. Cordycepin was added at each time point mentioned above, and samples were recovered every 30 min for 2 h. (D) Half-life values were calculated from data presented in (A–C), according to Gallie et al (1991) and averages from three biological replicates ±s.d. are shown. (E) Effect of Fe concentration on AtFER1 mRNA accumulation and relative half-life. Cells were treated for 3 h with various concentrations of Fe, and cordycepin was added. AtFER1 mRNA accumulation was determined before cordycepin addition, and relative half-life was determined as described in the legend of Figure 1, and averages from three biological replicates ±s.d. are shown.
The effect of Fe concentration on AtFER1 mRNA accumulation and stability was then investigated in a range from 10 μM to 1 mM. For all Fe concentrations tested, the maximum of AtFER1 mRNA accumulation and its highest degradation rate were obtained 3 h after treatment (data not shown). Therefore, AtFER1 mRNA half-life was determined at this time point. AtFER1 mRNA stability continuously decreased in response to increasing the Fe concentration of the culture medium until reaching a plateau at 50 μM; higher Fe concentrations did not affect the stability of this transcript more dramatically (Figure 2E) while AtFER1 mRNA accumulation continued to increase in response to increasing Fe concentrations up to 1 mM. Therefore, these results indicated that the modulation of the AtFER1 mRNA decay is a response independent of the initial amount of mRNA.
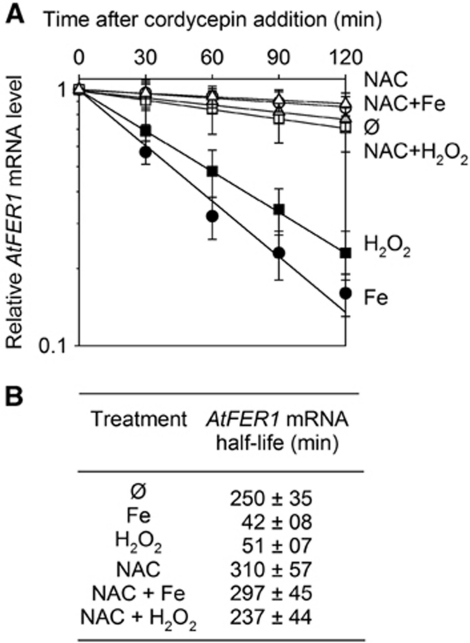
Fe can cause the accumulation of ROS. Therefore, we aimed to determine whether AtFER1 mRNA degradation was due to Fe itself or to its mediation of oxidative stress. When cells were treated with H2O2, AtFER1 mRNA stability was affected to the same extent as after Fe treatment (Figure 3A and B), suggesting that the observed effect of Fe on AtFER1 stability could be mediated by ROS. In order to test this hypothesis, cells treated with Fe or H2O2 were pre-treated with N-acetylcysteine (NAC), an anti-oxidant compound (Lobréaux et al, 1995). NAC pre-treatment completely prevented the AtFER1 mRNA destabilization in response to Fe and H2O2 (Figure 3A and B). Together, these data indicate that an Fe-mediated oxidative stress leads to a specific AtFER1 mRNA destabilization. This post-transcriptional control of AtFER1 expression likely enables the transcript to quickly return to its basal level after being transiently overaccumulated in response to Fe treatment.
Figure 3.
AtFER1 mRNA degradation requires an oxidative step. Cells were treated (NAC) or not (∅) for 1 h with 1 mM N-acetylcysteine before adding 300 μM Fe-citrate (Fe) or 5 mM hydrogen peroxide (H2O2) or water as control (∅). Three hours later, transcription was stopped by cordycepin addition. (A) AtFER1 mRNA relative abundance was measured as described in the legend of Figure 1. (B) AtFER1 relative half-life was calculated from the data obtained in (A). Values and standard errors were obtained from three independent experiments.
A DST-dependent destabilization pathway is involved in AtFER1 degradation
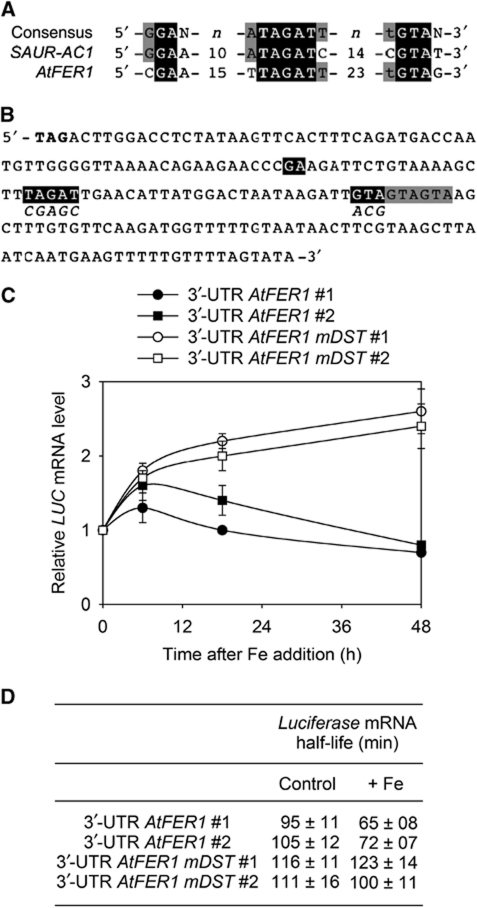
In order to search for putative post-transcriptional regulatory cis-elements, a sequence analysis of the AtFER1 mRNA was performed. We found regions in the 3′-UTR of the transcript that exhibit similarity with the DST sequence previously identified in SAUR transcripts (McClure et al, 1989; Sullivan and Green, 1996). In SAUR transcripts, an element called DST (for DownSTream) has been characterized (McClure et al, 1989). The element consists of a central ATAGAT motif flanked by a 5′-GGA motif and a 3′-GTA motif (Figure 4A). We identified in the 3′-UTR of AtFER1 the presence of a TAGAT motif surrounded by several GAA and GTA motifs (Figure 4A and B). Interestingly, such sequence similarity was not detected in the 3′-UTR of AtFER3 and AtAPX1 mRNAs, which are not destabilized by Fe.
Figure 4.
AtFER1 transcript degradation in response to Fe requires the DST cis-acting element present in its 3′-UTR. (A) Sequence alignment of DST sequence of AtSAUR-AC1 (At4g38850) with the DST found in the 3′-UTR of AtFER1. Numbers refers to the numbers of nucleotides between the indicated sequences. Nucleotides in black correspond to those found in the consensus of DST sequences (Sullivan and Green, 1996). (B) Sequence of the 3′-UTR of AtFER1 mRNA. Nucleotides in bold correspond to the stop codon. Nucleotides potentially belonging to a DST element are shown in black. Nucleotides in italic correspond to the mutations introduced in the 3′-UTR AtFER1 mDST constructs. (C) LUC mRNA abundance after Fe addition to transgenic plants. LUC is under the control of the strong 35S-CaMV promoter. The 3′-UTR of AtFER1 containing (3′-UTR AtFER1mDST) or not (3′-UTR AtFER1) mutations in the putative DST motif was introduced downstream of LUC. Two representatives from five independent transgenic lines are presented. For comparing transgenic lines, LUC mRNA abundance was fixed to one before Fe addition. Values and standard errors were obtained from three independent experiments. (D) LUC mRNA half-life in the transgenic lines analysed. Before Fe addition (control), or 3 h after 300 μM Fe-citrate addition (+Fe), cordycepin was added. LUC mRNA half-life was determined as described in the legend of Figure 3. Values and standard errors were obtained from three independent experiments.
To investigate the involvement of this DST-like sequence in AtFER1 destabilization, transgenic plants expressing the reporter gene Luciferase (LUC) under the control of the strong and constitutive 35S-CaMV promoter were used. In control plants (3′-UTR AtFER1), the 3′-UTR of AtFER1 mRNA sequence was introduced downstream of the LUC coding sequence. To test the functionality of the AtFER1 DST-like sequence, a PCR-mutagenized version of the 3′-UTR of AtFER1 mRNA was obtained by mutation of the TAGAT motif and of the closest GTA motif (3′-UTR AtFER1 mDST). Plants were grown in liquid medium and LUC transcript accumulation was measured by qRT–PCR over a period of 48 h after Fe addition. In order to compare the response of the different transgenic lines, the relative amount of LUC mRNA detected before Fe treatment was set to one for all lines. Fe addition led to an increase of the LUC transcript accumulation during the first 6 h in all lines. However, while this increase was transient in control plants, the reporter mRNA continued to overaccumulate in 3′-UTR AtFER1 mDST lines (Figure 4C). Thus, mutation in the DST cis-element of AtFER1 led to LUC mRNA overaccumulation after Fe addition. To strengthen the relationship between the DST sequence found in the 3′-UTR of AtFER1 and the modulation of AtFER1 mRNA decay, we measured the half-life of the LUC transcripts, both in control condition and 3 h after Fe addition. The LUC mRNA half-life of the 3′-UTR AtFER1 lines was significantly reduced after 3 h of Fe treatment. By contrast, the decay of LUC remained unchanged in the lines containing the mutated DST (Figure 4D). Taken together, our data suggest that the DST-like sequence identified in the AtFER1 3′-UTR is likely functional and involved in the control of AtFER1 mRNA stability in response to Fe.
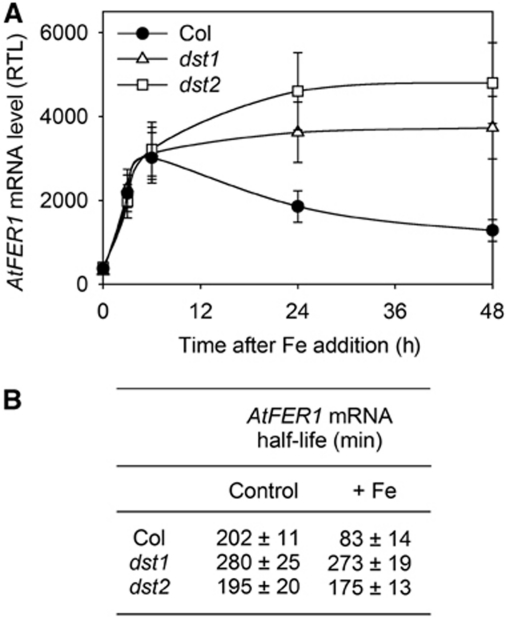
AtFER1 mRNA stability was also studied in two trans-acting mutants, named dst1 and dst2, previously identified in a genetic screen aimed at isolating factors involved in the DST-dependent mRNA degradation pathway (Johnson et al, 2000; Pérez-Amador et al, 2001; Lidder et al, 2004). AtFER1 mRNA accumulation in response to Fe was measured in Col, dst1 and dst2 grown in liquid medium (Figure 5A). As previously shown in Arabidopsis cells (Figure 1A), Fe led to a transient AtFER1 mRNA accumulation in Col seedlings, with a maximum between 3 and 6 h. In dst1 and dst2 mutants, AtFER1 transcript overaccumulated in the latter times after Fe addition (Figure 5A). In both dst1 and dst2, AtFER1 mRNA half-life was not affected by Fe (Figure 5B). This result demonstrates that the overaccumulation of AtFER1 transcript in dst mutants resulted from an impairment of its destabilization. In the same samples, AtFER3 and AtAPX1 half-life was not affected by the dst mutations. As expected (Johnson et al, 2000), SAUR-AC1 stability was affected in dst1 and dst2 (not shown). However, SAUR-AC1 accumulation was not modulated in response to Fe (Supplementary Figure S1), suggesting that oxidative stress activates only some of the potential targets of the DST pathway. Half-life values obtained for LUC-3′-UTR AtFER1 (Figure 4D) are different from those determined for the endogenous AtFER1 transcript. This difference suggests that additional elements present in AtFER1 mRNA sequence may have also a function in the DST-dependent degradation process. Such an hypothesis has also been raised for SAUR transcript (Newman et al, 1993; Gil and Green, 1996; Feldbrügge et al, 2002).
Figure 5.
DST1 and DST2 are trans-acting factors involved in AtFER1 mRNA degradation in response to Fe. (A) AtFER1 mRNA accumulation after Fe addition in Col, dst1 and dst2 seedlings. AtFER1 relative accumulation was determined as described in the legend of Figure 1. Mean values±s.d. are shown (n=3). (B) AtFER1 mRNA half-life in Col, dst1 and dst2 seedlings. Before Fe addition (control), or 3 h after 300 μM Fe-citrate addition (+Fe), cordycepin was added. AtFER1 mRNA half-life was determined as described in the legend of Figure 3.
Thus, AtFER1 undergoes a new post-transcriptional level of regulation of its expression. The DST-like sequence identified in the AtFER1 3′-UTR is necessary and sufficient to control the transient accumulation of the transcript in response to Fe. Furthermore, the two trans-acting factors DST1 and DST2 are involved in this process. Importantly, these results identify oxidative stress as an inducer of the DST-dependent AtFER1 mRNA destabilization. Accordingly, we refer to the oxidative stress-responsive pathway as the ‘oxDST mRNA degradation pathway’ hereafter.
Identification of targets regulated by the oxDST mRNA degradation pathway
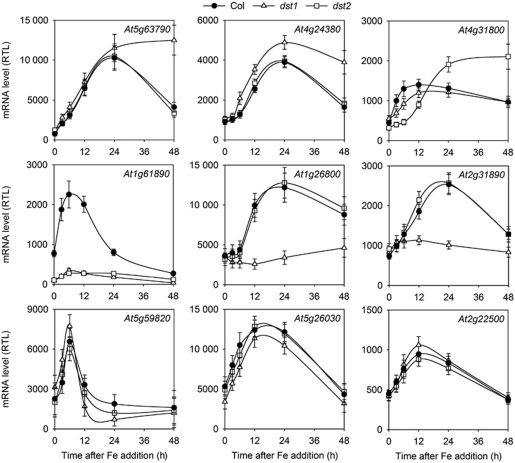
To get further insight into the molecular importance of the oxDST pathway, we searched for additional genes regulated by this mechanism. We screened among transcripts containing potential DST-like sequences in their 3′-UTR for those regulated in response to Fe. First, Arabidopsis transcripts containing putative DST sequences were searched using the Patmatch software (TAIR). We used the only two transcripts shown to contain functional DST sequences, AtSAUR-AC1 and AtFER1, to define an Arabidopsis DST consensus (Figure 4A). This bioinformatic analysis of the Arabidopsis genome revealed 1719 genes containing potential DST-like sequences (Supplementary Table 1). Then, to select for the Fe-regulated genes, we used publicly available transcriptome data obtained on Arabidopsis cell cultures by our group (http://www.catma.org/). Among the 1719 genes containing potential DST-like sequences, 13 genes were strongly regulated by Fe in Arabidopsis cells (Supplementary Table 1). Among these 13 genes (Table I), 9 of them are expressed in Arabidopsis seedlings and were further selected for mRNA abundance analysis in Col, dst1 and dst2 plants in response to Fe excess treatment (Figure 6). All selected transcripts accumulated in response to Fe in control plants. Three genes (At5g59820, At5g26030 and At2g22500) exhibited a similar profile of mRNA accumulation in the three genotypes. However, three other genes, At5g63790, At4g24380 and At4g31800 overaccumulated in dst1 or dst2 in the later time points of the kinetic analysis, similar to AtFER1. Additionally, the accumulation of three transcripts in response to Fe was totally abolished in dst1 (At1g26800) or in dst2 (At2g31890), or in both mutants (At1g61890). From these experiments, additional genes potentially regulated by the oxDST mRNA degradation pathway were identified. Furthermore, our results suggest that DST1 and DST2 can act collectively as shown in the case of AtFER1, but each of them can also individually affect specific gene subsets.
Table 1. Genes found to be regulated by iron in transcriptome data and to contain a putative DST sequence.
| Locus | Name | Location | Activity | Biological function |
|---|---|---|---|---|
| At1g26800 | Zn Finger | Nucleus (?) | Transcription factor activity (?) | Unknown |
| At1g61890 | MATE | Mitochondria | Transport | Drug transmembrane transporter |
| At2g22500 | UCP5 | Mitochondria | Transport | Dicarboxylic acid transporter |
| At2g31890 | RAP | Nucleus | Transcription factor activity | Iron homeostasis (?) |
| At3g09350 | FES1A | Cytoplasm | HSP70 binding protein | Response to stress |
| At3g28210 | SAP12/PMZ | Unknown | Transcription factor activity (?) | Response to stress |
| At3g54810 | BME3 | Nucleus | Transcription factor activity | Circadian clock |
| At5g26030 | FC1 | Chloroplast | Ferrochelatase | Haeme biosynthesis |
| At4g24380 | Hydrolase | Unknown | Unknown | Folate biosynthesis |
| At4g31800 | WRKY18 | Nucleus | Transcription factor activity | Response to stress |
| At5g59820 | ZAT12 | Nucleus | Transcription factor activity | Response to stress |
| At5g63790 | ANAC102 | Nucleus | Transcription factor activity | Response to stress |
| At4g16370 | OPT3 | Membrane | Oligopeptide/metal transport | Iron homeostasis |
| AGI, name, location, activity and biological function of the corresponding proteins are indicated as provided on TAIR site (http://www.arabidopsis.org). | ||||
Figure 6.
Expression of nine candidate genes in Col, dst1 and dst2. Genes regulated by Fe were selected from public transcriptome databases (http://www.catma.org/). Among these genes, those exhibiting a putative DST in their 3′-UTR were searched using the Patmatch program (TAIR). Transcript accumulation was determined as described in the legend of Figure 3. Mean values±s.d. are shown (n=3).
The oxDST mRNA decay control is crucial for plant fitness
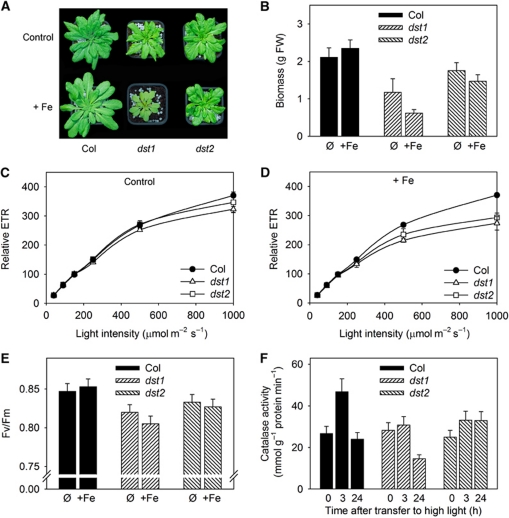
The oxDST mRNA degradation pathway is controlled by Fe and ROS (Figures 3, 4 and 5). Ferritin was shown to be involved in the coordination of Fe metabolism and the response to oxidative stress (Ravet et al, 2009). Interestingly, some of the genes affected by alteration in the oxDST pathway are transcription factors related to oxidative stress responses. ANAC 102 (At5g63790) belongs to the NAC transcription factors and is involved in low oxygen responses in Arabidopsis (Christianson et al, 2009). In addition to its involvement in pathogen responses, WRKY 18 (At4g31800) has recently been associated with abiotic stress responses (Chen et al, 2010). The RAP gene (At2g31890) encodes a MYC transcription repressor that may be related to Fe metabolism (Loulergue et al, 1998). Therefore, the oxDST pathway may participate in the establishment of plant responses to Fe overload and oxidative stress. To test this hypothesis, the fitness of dst1 and dst2 plants was evaluated on regular soil or on soil enriched with 2 mM Fe-EDDHA. On control soil, only dst1 growth was decreased, suggesting that a mutation in that gene affects plant fitness even in absence of harsh treatment (Figure 7A). However, increasing Fe availability in the soil dramatically affected growth of both mutants. Measurement of shoot biomass revealed that Fe addition to the soil is significantly deleterious when the oxDST pathway is altered (Figure 7B). In contrast, this treatment is beneficial for improving wild-type plant growth, in agreement with previous reports (Ravet et al, 2009). In order to try to understand how the oxDST pathway affects Arabidopsis biomass, the photosynthetic efficiency was evaluated using chlorophyll fluorescence measurements. The electron transport rate (ETR) was measured under different light intensities and Fe nutrition conditions (Figure 7C and D). In control conditions, no statistically significant difference was measured in the dst mutants compared with wild type, although ETR values were slightly decreased in dst1 (Figure 7C). By contrast, in Fe-enriched soil, ETR was significantly lower in both mutants when compared with Col, especially under high light conditions known to induce oxidative damage in the chloroplast (Figure 7D). In addition, maximum quantum yield of the PSII (Fv/Fm parameter) was lower in dst1 and, to a lower extent, in dst2, especially when plants were grown on Fe-enriched soil (Figure 7E). These observations reinforced the idea that plants altered in the oxDST pathway are more sensitive to oxidative stress.
Figure 7.
Physiological function of Fe-induced DST-dependent mRNA degradation. (A) Plants were grown on soil and irrigated with water (control: ∅) or a 2-mM Fe-EDDHA solution (+Fe). Plants are 35 days old and are representative of each genotype and condition. (B) Shoot biomass (FW) of plants shown in (A). Values presented are means and standard errors from 10 plants. (C, D). Relative electron transport rate (ETR) was determined on 28-day-old plants at different light intensities using a fluorometer camera device. Plants were grown on soil and irrigated with water (C) or a 2-mM Fe-EDDHA solution (D). Means and standard deviation from 6 to 8 biological replicates are shown. (E) Maximum quantum yield of PSII (Fv/Fm) parameter was determined on the same plants used for the ETR presented in (C) and (D). Mean values and standard deviation from 6 to 8 biological replicates are shown. (F) Evolution of catalase activity after exposure to high light (800 μmol photons m−2 s−1). Plants were grown for 21 days on soil and irrigated with water. They were transferred during various time under high light conditions. Total protein extract from leaves were prepared and used for catalase activity determination. Values and standard errors were obtained from 6 to 8 biological replicates.
The ability of the dst mutants to properly respond to oxidative stress was further studied by exposing plants to high light conditions (Figure 7F). The responses of plants to high light treatment are well documented. One of the major effects of such a treatment is to promote a strong oxidative stress. Consequently, plants activate various enzymatic and non-enzymatic anti-oxidant responses. Among them, catalase activity is known to be strongly induced, and is therefore a good marker of the plant responses to high light exposure (Fernández and Strand, 2008; Li et al, 2009). As expected, high light treatment led to a transient increase of catalase activity in control plants (Figure 7F). Interestingly, the catalase activity was not increased in the two dst mutants under high light and the catalase activity appeared even lower in dst1 when compared with Col and dst2 after 24 h of exposure. Together, these data demonstrate that the oxDST mRNA degradation pathway is crucial for establishing appropriate responses to oxidative stress, and that alteration of the pathway reduced photosynthesis, growth and ultimately plant fitness.
Discussion
The results from this study identify the oxDST pathway as an essential regulatory mechanism to modulate mRNA accumulation patterns in response to oxidative stress, and reveal that alteration of this control dramatically impacts plant physiology. Therefore, the oxDST-dependent mRNA stability control appears to be an important mechanism that allows plants to cope with adverse environmental conditions. These results are of significance because they describe a trigger of molecular events leading to the degradation of specific mRNAs in response to oxidative stress, and demonstrate the physiological importance of sequence-specific mRNA decay mechanisms in plants.
In animals, the sequence-specific mRNA decay systems, IRE and ARE, play a central role in Fe homeostasis and oxidative stress responses. RBPs interact with specific sequences present in the UTRs of the target transcripts, thus regulating mRNA stability and translation (Klausner et al, 1993; Harrison and Arosio, 1996; Hentze et al, 2004). The post-transcriptional control of Fe homeostasis in animals in response to Fe availability occurring through the IRE system enables (i) to regulate the stability of the transferrin receptor mRNA through IRE sequences found in the 3′-UTR of the transcript and (ii) to link this regulation to the control of the ferritin mRNA translation mediated by an IRE sequence located in the 5′-UTR of the transcript. In plants, we previously showed that this IRE system does not regulate ferritin expression despite the conservation of the RBP (Arnaud et al, 2007). ARE-like elements have been observed in several plant transcripts, suggesting that the mechanism could be functional in plants (Chen and Stern, 1991; Ohme-Takagi et al, 1993; Chen et al, 1995). However, the only evidence of a functional ARE in plants concerns the plastid-encoded PetD mRNA (Chen et al, 1995), which is a prokaryotic-type transcript. In contrast, ARE-mediated regulation of nuclear-encoded plant genes has never been reported so far. The destabilizing DST element, originally identified in the 3′-UTR of the soybean SAUR-AC1 mRNA, has been shown to be conserved among different plant species (Newman et al, 1993), but the cis-element failed to promote mRNA decay in mammalian cells (Feldbrügge et al, 2002). Recently, a large-scale analysis of elements overrepresented in the 3′-UTR of unstable Arabidopsis transcripts confirmed that the DST motifs fall into that category (Narsai et al, 2007).
In this report, we demonstrate that the mutations in the trans-acting factors DST1 and DST2 genes impair plant response to oxidative stress mediated by high light and Fe excess treatment. At the molecular level, Fe and H2O2 promote the mRNA decay of AtFER1, the most abundant ferritin transcript in Arabidopsis, and potentially other ROS-related transcripts. Therefore, the DST pathway probably evolved specifically in plants, where it may fulfill a similar function to other systems (ARE for instance) described in other eukaryotic lineages.
The plant-specific DST pathway(s)
The DST destabilizing element was identified in the 3′-UTR of the auxin-related unstable SAUR-AC1 mRNA and made it possible to identify trans-acting factors involved in the corresponding mRNA degradation process. This led to the molecular and phenotypic characterization of the corresponding dst1 and dst2 mutants (Johnson et al, 2000). However, SAUR-AC1 stability was not modulated by auxin treatment (Gil and Green, 1996) and the dst1 and dst2 mutants did not reveal any developmental or auxin-related phenotype (Pérez-Amador et al, 2001; Gutierrez et al, 2002). Only a discrete molecular phenotype related to circadian clock-regulated genes was observed (Lidder et al, 2005). The biological function of the DST pathway, therefore, remained to be elucidated. Here, we report convergent data for the activation by ROS of an mRNA degradation pathway that depends on the DST cis/trans-acting elements identified so far. Also, phenotypic analyses of the trans-acting mutants dst1 and dst2 revealed for the first time the major importance of the DST-dependent mRNA degradation pathway for plant fitness.
Previous transcriptome analysis of the dst1 mutant had already identified specific mRNAs that are overaccumulated when compared with wild-type plants (Pérez-Amador et al, 2001; Lidder et al, 2005). Interestingly, AtFER1 was not detected as a potential target of dst1 in this screen. Our experiments showed that in the absence of oxidative treatments, AtFER1 mRNA does not fall into the category of the unstable transcripts with a half-life over 3–4 h, both in Arabidopsis cells and in seedlings (Figure 1). In the same condition (i.e., in the absence of oxidative stress), eight of the nine other DST-containing mRNAs tested in our search for oxDST pathway candidate genes were not affected by the dst mutations (Figure 6). Additionally, SAUR-AC1 mRNA stability was not affected by oxidative stress. Taken together, it appears that the DST pathway can differentially affect mRNA stability depending on the conditions experienced by the plants, some DST-containing mRNAs being constitutively processed (e.g., SAUR-AC1), others being conditionally destabilized (e.g., AtFER1). These results indicate that the DST-dependent mRNA degradation is involved in different signalling pathways. DST1 and DST2 do not appear to be involved in a specific pathway since they act both in the presence and in the absence of stimuli. Therefore, they may represent the core machinery of this mRNA degradation pathway. In addition, DST1 and DST2 are not strictly acting collectively because some of the mRNAs we tested are affected by only one of the mutations in the oxDST pathway (Figure 6). The accumulation of circadian-regulated genes was also differentially affected by the dst1 mutation (Lidder et al, 2005). Thus, DST1 and DST2 could be part of a selective machinery, mobilized in different ways depending on effectors applied, proteins recruited and transcripts targeted. These new insights into the DST regulatory mechanisms prompted us to propose an integrative model (Figure 8) describing our present view of the overall process.
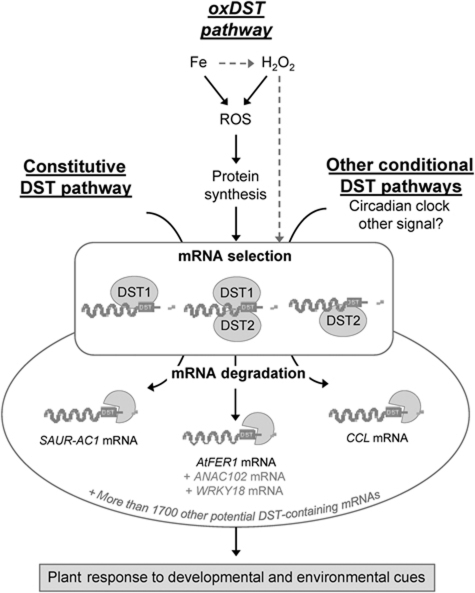
Figure 8.
A model for DST-mediated mRNA degradation pathways in plant. In the current knowledge, DST-dependent mRNA degradation relies on the presence of a conserved DST cis-element present in the 3′-UTR of target transcripts, and on two trans-acting factors DST1 and DST2. So far, three independent pathways are known to control the entry of DST-containing mRNAs for degradation. The first pathway (named constitutive DST pathway) involving the DST mRNA decay mechanism has been shown to constitutively degrade the AtSAUR-AC1 transcript, an unstable mRNA. Further genetic insights identified a second DST pathway controlling the proper oscillation of the accumulation of the circadian-regulated transcript CCL. Therefore, this pathway can be considered as a conditional DST pathway since clock is likely a signal controlling CCL half-life modulation. Finally, the present report demonstrates that the major ferritin transcript AtFER1 is processed only under oxidative stress conditions. In this third pathway (named oxDST pathway), Fe activates the entry of AtFER1 mRNA in the degradation pathway through ROS and the process involves an unstable protein. H2O2 also modulates AtFER1 decay, suggesting that not only Fe can promote this ROS-mediated AtFER1 decay. However, H2O2 can be produced in response to Fe through the Fenton reaction, or may trigger AtFER1 destabilization by an alternative pathway. Thus, the H2O2-mediated degradation of AtFER1 through the oxDST pathway is more hypothetical at that time (grey arrows). In addition to AtFER1, this study also revealed ANAC102 and WRKY18 as two other potential direct targets of the oxDST pathway. However, the direct link between their decay modulation and the DST factors remains to be investigated (grey labelling). In the present model, the pathways are distinct beside a common dependence regarding DST1 and DST2 factors. In one hand, AtSAUR-AC1 is constantly destabilized and its decay is not affected by iron. On the other hand, AtFER1 is clock regulated but independently to the DST factors. Thus, several DST pathways can be distinguished. In regard to the differential involvement of DST1 and DST2 depending on the transcript and the pathway considered, it appears that DST factors likely act as components of a selective machinery for mRNA degradation, in which they would be involved separately or collectively.
The importance of the oxDST mRNA decay in response to oxidative stress
Our results provide a line of evidence that AtFER1 mRNA is conditionally destabilized through the DST pathway, making it the second validated DST cis-element in plants after the one identified in SAUR-AC1. Interestingly, the process is activated by Fe and H2O2. In vivo, ROS production is catalysed by cellular Fe through the Fenton reaction, and thus Fe homeostasis is tightly coordinated with the redox status of the cell. Plants are particularly prone to redox imbalance due to the photosynthetic activity in the chloroplast, and ferritins are Fe-storage proteins found in plastids where they exert an essential control on ROS accumulation (Ravet et al, 2009).
However, the oxDST mediated mRNA decay is not a general response observed in genes regulated by Fe or ROS. For instance, another ferritin mRNA, AtFER3, does not contain any DST and its decay is not promoted by Fe or ROS. Interestingly, we identified Zat12 as a DST-like containing gene regulated by Fe treatment. The transcription factor ZAT12 plays major functions in oxidative stress response, and in particular it controls AtAPX1 transcription (Rizhsky et al, 2004). However, based on APX1 and ZAT12 expression data, we consider that ZAT12 mRNA is unlikely to be a functional target of the oxDST pathway.
Nevertheless, our search for candidate genes of the oxDST pathway has revealed other transcripts, which overaccumulate in response to Fe in the dst mutants, similar to AtFER1. These mRNAs could represent additional direct targets of the oxDST pathway. Among them are two transcription factors involved in abiotic and biotic stress responses, namely ANAC102 and WRKY18 (Christianson et al, 2009; Chen et al, 2010). Other transcripts did not accumulate in response to Fe in the dst mutants. Such alteration could illustrate general downstream effects due to an impairment of the transcriptional activation of the responses. As an ultimate consequence, plants deficient in the DST machinery are sensitive to Fe nutrition, exhibit altered responses to high light treatment, and photosynthesis and growth are reduced. The molecular mechanisms underlying these altered responses remain to be determined. The Arabidopsis catalase genes do not contain putative DST (Supplementary Table 1), and the observed reduced photosynthetic efficiency is an integrative parameter that is probably altered due to the direct and indirect deregulation of several genes.
Conclusion and future challenges
By nature, aerobic organisms have evolved complex mechanisms to cope with oxygen. Adjustments in gene expression profiles, collectively at the transcriptional and post-transcriptional levels, decisively influence the overall response of the organism. Plants are photosynthetic and sessile organisms. In that context, mRNA decay control is of particular interest because it provides a quick means to adjust RNA levels and to target specific subsets of transcripts. The exponentially growing number of reports describing small RNA-mediated post-transcriptional mechanisms reveals the significance of that additional level of control of gene expression, especially in response to environmental cues. Here, we demonstrate that the sequence-specific DST mRNA degradation is also crucial in plants, where it is required to cope with oxidative stress.
Examples of transcript stability modulation in response to ROS are well documented in yeast and animal systems. Mechanisms such as those involving IRE or ARE have been investigated in detail at the molecular level, likely because they are of major importance to understand the implication of ROS in pathological processes such as neurodegenerative disorders (Finkel and Holbrook, 2000; Bokov et al, 2004). In plants, the eukaryotic mRNA degradation machinery is well documented (Belostotsky and Sieburth, 2009). However, the process by which a signal will coordinate the conditional degradation of specific mRNA subsets is poorly understood. In this report, we show that (i) Fe and ROS activate AtFER1 mRNA degradation, and (ii) this process occurs in a DST-dependent manner. Furthermore, mutants in the DST pathway: (iii) exhibit altered accumulation of ROS-related mRNAs and (iv) are sensitive to Fe nutrition and have altered responses to high light. Thus, our study considerably reassesses the significance of such post-transcriptional regulatory mechanisms. In addition, ROS emerged as an activator of these processes, revealing the complexity of the DST-mediated pathway(s) (Figure 8).
The next important challenge will be to identify the DST trans-acting factors and other RBPs that may be recruited for mRNA selection processing. Interestingly, a recent report on the exoribonuclease XRN4 indicates that AtFER1 could be overaccumulated in the corresponding mutant (Rymarquis et al, 2011). It is likely that the ultimate steps of DST mRNA degradation involve such nucleolytic processes. More important is the nature of the DST factors. The genes corresponding to the dst1 and dst2 EMS mutants have not yet been identified, although the genetic mapping of the dst1 mutation has been initiated (Johnson et al, 2000). Identification of these factors will permit us to gain a better understanding how the pathway is activated and how the selectivity of targeted mRNA is achieved among the potentially broad range of DST-containing mRNAs.
Materials and methods
Cell culture
Arabidopsis thaliana L. (Columbia ecotype) suspension cells were grown at 24°C under continuous light (100 μmol photons m−2 s−1) on a rotating table (60 rev per minute) in a medium containing 20 mM KNO3, 1.2 mM CaCl2, 450 μM MgSO4, 375 μM KH2PO4, 60 μM Na2HPO4, 40 μM NaH2PO4, 40 μM MnSO4, 30 μM H3BO3, 25 μM glycine, 10 μM ZnSO4, 5 μM Fe(III)-EDTA, 4 μM nicotinic acid, 2.5 μM pyridoxine-HCl, 1.5 μM KI, 1.2 μM thiamine-HCl, 300 nM Na2MoO4, 100 nM ANA, 30 nM CoCl2, 30 nM CuSO4, 0.1 g l−1 casein hydrolysate, 0.1 g l−1 myo-inositol, 15 g l−1 sucrose, pH 5.7 (Arnaud et al, 2006). Cells were subcultured with a 1/10-dilution factor once a week. Experiments were carried out 1 week after subculture.
Plant material and growth conditions
For molecular analyses, seeds from Arabidopsis thaliana L. (Columbia ecotype), dst1 and dst2 mutants (Johnson et al, 2000) and transgenic lines were surface sterilized and grown in 10 ml half-strength Murashige and Skoog liquid medium (Sigma), pH 5.7, supplemented with 1% sucrose, 0.5 g l−1 MES and 50 μM Fe(III)-EDTA (Arnaud et al, 2007). Seedlings were grown under continuous light (100 μmol photons m−2 s−1) on a rotating table (60 rev per minute). The medium was renewed after 1 week of culture and plants were grown for three additional days before treatments.
For physiological analyses, plants were sown on soil (Neuhaus Humin Substrat N2, Klasman-Deilmann GmbH) in pots as single plant. Plants were grown in controlled growth chambers at 20°C with a relative humidity of 70%, under short-day conditions (8 h light/16 h dark photoperiod) at two light intensities, 250 and 800 μmol photons m−2 s−1 corresponding to control and high light conditions, respectively.
Chemical treatments
Plants grown on Fe-enriched soil were irrigated with 2 mM Fe-EDDHA (Sequestrene, Fertiligene). All other chemicals were purchased from Sigma-Aldrich. Cells and seedlings grown in liquid medium were treated with 300 μM Fe-citrate (Lobréaux et al, 1995). H2O2, CHX, cordycepin, NAC were used at 5 mM, 100 μM, 400 μM and 1 mM final concentrations, respectively.
Plasmid constructs and plant transformation
All constructs were made using standard recombinant DNA techniques (Sambrook and Russell, 2000) and sequenced prior to their use. An NheI/XbaI fragment containing the LUC+ coding sequence from pSP-LUC+ (Promega) was subcloned in SpeI/XbaI-digested pBluescript (Stratagene) to obtain pBS-LUC+. The 35S promoter sequence from pCAMBIA 1301 (Cambia) was subcloned in SalI/NcoI-digested pBS-LUC+ generating pBS-35SLUC+. The AtFER1 3′-UTR was amplified with thermostable Pfu DNA polymerase (Promega) on Arabidopsis thaliana L. (Columbia ecotype) genomic DNA using 3′-AtFer1F and 3′-AtFer1R primers (Supplementary Table 2). Mutagenesis of the DST sequence of AtFER1 3′-UTR was made using the two-step PCR method as described previously (van Wuytswinkel and Briat, 1995) using primers 3′-AtFER1FmutA, 3′-AtFER1FmutF, 3′-AtFER1RmutA and 3′-AtFER1RmutB primers (Supplementary Table 2). In both constructs, external primers introduced XbaI and SacI sites at the 5′ and 3′ ends of the amplified fragment, respectively. DNA fragments were cloned in XbaI/SacI-digested pBS-35SLUC+ to obtain pBS-35S∷LUC-3′UTR AtFER1 and pBS-35S∷LUC-3′UTR AtFER1mDST. The constructs were excised by KpnI/SacI digestion and subcloned into pBIBHygro binary vector. The resulting plasmids were introduced into Agrobacterium tumefaciens MP90 strain (Hofgen and Willmitzer, 1988). These bacteria have been used to transform plants using floral dip method (Clough and Bent, 1998).
RNA isolation and qRT–PCR analysis
Total RNA from cells and seedlings was extracted using TRI reagent (Euromedex). Total RNA was treated with DNAse (Promega) prior to reverse transcription using M-MLV Reverse Transcriptase (Promega) and oligo dT(23) (Promega). Quantitative qRT–PCR was performed in a Light-Cycler® (Roche Diagnostics) using the LightCycler FastStart DNA MasterPLUS SYBR GREEN I (Roche). PCR amplification of the various genes analysed in this study was performed using 3′-UTR gene-specific primers listed in Supplementary Table 2.
Data analysis was performed using Light-Cycler 3 data-analysis software (Roche). Relative transcript levels (RTL=1000 × 2−ΔCt) were evaluated by calculation of the difference between the crossing time of the target gene and the threshold crossing time of the control gene (ΔCt) for the respective templates (Arrivault et al, 2006). Gene expression was monitored in technical duplicate and biological triplicate, and results presented were standardized using the Sand gene (Czechowski et al, 2005; Remans et al, 2008).
mRNA half-life determination
Transcript half-life was determined after treatment by the transcription inhibitor cordycepin. Samples were collected every 30 min for 2 h after transcription inhibition. Total RNA was extracted and mRNA relative abundance was determined by qRT–PCR. The values were normalized relative to the value prior cordycepin treatment, plotted as a function of time and subjected to a regression analysis. The regression slope (k) was used to calculate half-life according to the equation t1/2=ln2/k (Gallie et al, 1991).
Photosynthesis measurements
Chlorophyll fluorescence measurements were performed on 28-day old plants using a fluorometer camera device (Photon System Instruments, Brno, Czech Republic). Relative ETR was used to evaluate the photosynthetic efficiency (ETR=light intensity × Photosystem II efficiency). Photosystem II efficiency (Psi PSII) was determined according to Genty et al (1989) using the following equation: Psi PSII=(Fm−Ft)/Fm where Ft is the fluorescence determined at different light intensities ranging from 12 to 1000 μmol photons m−2 s−1 and Fm is the maximal dark-adapted fluorescence measured with a 0.8-ms pulse of 6000 μmol photons m−2 s−1 of actinic light. Photochemical efficiency of PSII, expressed as the maximum quantum yield of PSII (Fv/Fm), was determined using the following equation: Fv/Fm=(Fm−F0)/Fm where F0 is the initial minimal fluorescence on dark-adapted leaves.
Catalase activity
Catalase activity was determined in crude leaf extract at 20°C as previously described (Cakmak and Marschner, 1992). Plants were grown for 21 days under moderate light intensity (250 μmol photons m−2 s−1), then transferred under high light (800 μmol photons m−2 s−1) for 24 h. Protein concentration was determined according to Schaffner and Weissmann (1973) using BSA as standard.
CATMA arrays
High-throughput mRNA analyses have been performed on Arabidopsis cell suspension treated or not with Fe, using the Complete Arabidopsis Transcriptome MicroArray (CATMA) technology and data are publicly available at http://www.catma.org/. Two individual experiments have been performed. Experimental and data analysis procedures are described at ftp://urgv.evry.inra.fr/CATdb/GEO/. The files GEO-Soft-no27-iron-signaling-exp24.txt.gz and GEO-Soft-rs05-15-iron-signaling-exp105.txt.gz can be downloaded for further experimental details.
Bioinformatic analyses
DST sequences were searched in 3′-UTR of Arabidopsis genes using the TAIR9 3′-UTR data set (TAIR, http://www.arabidopsis.org). Bioinformatic search for the three conserved motifs was performed by using the Patmatch software (TAIR) and GAA-n-TAGAT-n-GTA as an algorithm based on the Arabidopsis DST sequence consensus that we established in this report (see Results). The stringency of the screen was optimized by fixing to n=50 nucleotides the maximum spacing between the different motifs.
Supplementary Material
Acknowledgments
This work was funded by Institut National de la Recherche Agronomique and Centre National de la Recherche Scientifique, by the Action Concertée Incitative ‘Biologie Cellulaire Moléculaire et Structurale’ number BCMS166 from the Ministère de l’Education Nationale de l’Enseignement Supérieur et de la Recherche and by the ANR-Blanche DISTRIMET No. 25383 from the Agence Nationale de la Recherche. The work of KR and NA was supported by a thesis fellowship from the Ministère de l’Education Nationale de l’Enseignement Supérieur et de la Recherche. GK research is supported by an European-FP7-International Outgoing Fellowships (Marie Curie) (AtSYSTM-BIOL; PIOF-GA-2008- 220157). We thank Pr Pamela Green for the gift of dst1 and dst2 mutants, Pr Elizabeth Pilon-Smits and Pr Marinus Pilon for discussions and critical reading of the manuscript.
Author contributions: KR, GR, NA, E-BD JB and FG performed experiments; KR, NA, GK J-FB and FG designed research; KR, GK and FG analysed the data; KR, J-FB and FG wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen RG, Tresini M (2000) Oxidative stress and gene regulation. Free Radic Biol Med 28: 463–499 [DOI] [PubMed] [Google Scholar]
- Arnaud N, Murgia I, Boucherez J, Briat JF, Cellier F, Gaymard F (2006) An iron-induced nitric oxide burst precedes ubiquitin-dependent protein degradation for Arabidopsis AtFer1 ferritin gene expression. J Biol Chem 281: 23579–23588 [DOI] [PubMed] [Google Scholar]
- Arnaud N, Ravet K, Borlotti A, Touraine B, Boucherez J, Fizames C, Briat JF, Cellier F, Gaymard F (2007) The iron-responsive element (IRE)/iron-regulatory protein 1 (IRP1)-cytosolic aconitase iron-regulatory switch does not operate in plants. Biochem J 405: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Belostotsky D, Sieburth L (2009) Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 12: 96–102 [DOI] [PubMed] [Google Scholar]
- Bokov A, Chaudhuri A, Richardson A (2004) The role of oxidative damage and stress in aging. Mech Ageing Dev 125: 811–826 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Marschner H (1992) Magnesium Deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol 98: 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J, Hentze M, Koeller D, Caughman S, Rouault T, Klausner R, Harford J (1988) Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science 240: 924–928 [DOI] [PubMed] [Google Scholar]
- Chen C-YA, Shyu A-B (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Stern DB (1991) Specific ribonuclease activities in spinach chloroplasts promote mRNA maturation and degradation. J Biol Chem 266: 24205–24211 [PubMed] [Google Scholar]
- Chen Q, Adams C, Usack L, Yang J, Monde R, Stern D (1995) An AU-rich element in the 3′ untranslated region of the spinach chloroplast petD gene participates in sequence-specific RNA-protein complex formation. Mol Cell Biol 15: 2010–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M, Arizti P, Sullivan ML, Zamore PD, Belasco JG, Green PJ (2002) Comparative analysis of the plant mRNA-destabilizing element, DST, in mammalian and tobacco cells. Plant Mol Biol 49: 215–223 [DOI] [PubMed] [Google Scholar]
- Fernández A, Strand Å (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Vansuyt G, Kushnir S, Inze D, Briat JF (2004) Iron-regulated expression of a cytosolic ascorbate peroxidase encoded by the APX1 gene in Arabidopsis seedlings. Plant Physiol 134: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Feder JN, Schimke RT, Walbot V (1991) Post-transcriptional regulation in higher eukaryotes: the role of the reporter gene in controlling expression. Mol Gen Genet 228: 258–264 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais J, Baker N (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochem Biophys Acta 990: 87–92 [Google Scholar]
- Gil P, Green PJ (1996) Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J 15: 1678–1686 [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275: 161–203 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC (2004) Balancing acts: molecular control of mammalian iron metabolism. Cell 117: 285–297 [DOI] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Perez-Amador MA, Lidder P, Green PJ (2000) Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc Natl Acad Sci USA 97: 13991–13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD (2007) Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol 72: 157–165 [DOI] [PubMed] [Google Scholar]
- Klausner RD, Rouault TA, Harford JB (1993) Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72: 19–28 [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer B, Niyogi K (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138: 2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidder P, Johnson MA, Sullivan ML, Thompson DM, Pérez-Amador MA, Howard CJ, Green PJ (2004) Genetics of the DST-mediated mRNA decay pathway using a transgene-based selection. Biochem Soc Trans 32: 575–577 [DOI] [PubMed] [Google Scholar]
- Lobréaux S, Thoiron S, Briat JF (1995) Induction of ferritin synthesis in maize leaves by an iron-mediated oxidative stress. Plant J 8: 443–449 [Google Scholar]
- Loulergue C, Lebrun M, Briat JF (1998) Expression cloning in Fe2+ transport defective yeast of a novel maize MYC transcription factor. Gene 225: 47–57 [DOI] [PubMed] [Google Scholar]
- McClure B, Hagen G, Brown C, Gee M, Guilfoyle T (1989) Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Taylor CB, Newman TC, Green PJ (1993) The effect of sequences with high AU content on mRNA stability in tobacco. Proc Natl Acad Sci USA 90: 11811–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Amador M, Lidder P, Johnson M, Landgraf J, Wisman E, Green P (2001) New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13: 2703–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Briat JF, Lobréaux S (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT–PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227: 1343–1349 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Rouault TA (2006) The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2: 406–414 [DOI] [PubMed] [Google Scholar]
- Rouault TA, Klausner R (1996) Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem Sci 21: 174–177 [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ (2011) Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2000) Molecular Cloning, 3-Volume Set: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schaffner W, Weissmann C (1973) A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56: 502–514 [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R (1986) A conserved AU sequence from the 32 untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667 [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ (1996) Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA 2: 308–315 [PMC free article] [PubMed] [Google Scholar]
- Theil EC (2007) Coordinating responses to iron and oxygen stress with DNA and mRNA promoters: the ferritin story. Biometals 20: 513–521 [DOI] [PubMed] [Google Scholar]
- Toledano MB, Kumar C, Le Moan N, Spector D, Tacnet F (2007) The system biology of thiol redox system in Escherichia coli and yeast: differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett 581: 3598–3607 [DOI] [PubMed] [Google Scholar]
- Touati D (2000) Iron and oxidative stress in bacteria. Arch Biochem Biophys 373: 1–6 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- van Wuytswinkel O, Briat JF (1995) Conformational changes and in vitro core-formation modifications induced by site-directed mutagenesis of the specific N-terminus of pea seed ferritin. Biochem J 305: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.