Abstract
Marfan syndrome (MFS) is a heritable connective tissue disorder caused by mutations in the gene coding for FIBRILLIN-1 (FBN1), an extracellular matrix protein. MFS is inherited as an autosomal dominant trait and displays major manifestations in the ocular, skeletal, and cardiovascular systems. Here we report molecular and phenotypic profiles of skeletogenesis in tissues differentiated from human embryonic stem cells and induced pluripotent stem cells that carry a heritable mutation in FBN1. We demonstrate that, as a biological consequence of the activation of TGF-β signaling, osteogenic differentiation of embryonic stem cells with a FBN1 mutation is inhibited; osteogenesis is rescued by inhibition of TGF-β signaling. In contrast, chondrogenesis is not perturbated and occurs in a TGF-β cell-autonomous fashion. Importantly, skeletal phenotypes observed in human embryonic stem cells carrying the monogenic FBN1 mutation (MFS cells) are faithfully phenocopied by cells differentiated from induced pluripotent-stem cells derived independently from MFS patient fibroblasts. Results indicate a unique phenotype uncovered by examination of mutant pluripotent stem cells and further demonstrate the faithful alignment of phenotypes in differentiated cells obtained from both human embryonic stem cells and induced pluripotent-stem cells, providing complementary and powerful tools to gain further insights into human molecular pathogenesis, especially of MFS.
Marfan syndrome (MFS) is a heritable dominant disorder of fibrous connective tissue, caused by mutations in the gene encoding fibrillin-1 on chromosome 15 (1, 2). MFS shows striking pleiotropism and clinical variability (3, 4). Cardinal pathological features occur in three systems—skeletal, ocular, and cardiovascular (4–8)—and share overlapping features with congenital contractural arachnodactyly, which is caused by a mutation in the FIBRILLIN-2 (FBN2) gene (9). FBN1 mutations are detected in the majority of the patients fulfilling the clinical criteria, but also in incomplete phenotypes, referred to as type 1 fibrillinopathies (10). FBN1 is an extracellular matrix glycoprotein containing 43 calcium-binding EGF-like domains and 78 cysteine-containing TB motifs (11, 12). Mutations in FBN1 are the etiology of many phenotypes observed in MFS. The most common mutations found in FBN1 in MFS are missense mutations (56%), mainly substituting or creating a cysteine in a calcium-binding EGF-like domain. Other mutations are frame-shift, splice, and nonsense mutations (13). There are only a few reports of patients with marfainoid features and a molecularly proven complete deletion of a FNB1 allele (14–16). Most of FBN1 deletions are associated with a severe or classical Marfan phenotype (17–20). Although the molecular pathogenesis of MFS was initially attributed to a structural weakness of the fibrillin-rich microfibrils within the ECM, more recent results have documented that many of the pathogenic abnormalities in MFS are the result of alterations in TGF-β signaling (18, 19). Mutations in other genes have been reported to cause MFS-related disorders, such as TGF-β receptor-I and -II in MFS type 2 and Loeys-Dietz syndrome, and myosin heavy chain (MYH)11 and actin/alpha2 smooth muscle/aorta (ACTA2) in familial thoracic aortianeurysms and dissections (21, 22).
To date, by necessity most knowledge of MFS has been obtained by extrapolation of studies in the mouse Fbn1 null/transgenic models (2, 23–27). However, with the derivation of human embryonic stem cells carrying a common FBN1 mutation, as well as human induced pluripotent-stem (iPS) cells from MFS patients, we now have a unique opportunity to examine key features of this syndrome on a human genome background. Moreover, we can address whether phenotypes observed following reprogramming of somatic cells to pluripotency are legitimately reflected in pluripotent stem cells directly obtained from human MFS embryos. Below, we describe our studies that used human MFS embryonic stem cells and iPS cells to unveil a unique skeletogenic phenotype featuring impaired osteogenic differentiation and the ability to undergo chondrogenesis in the absence of exogenous TGF-β. Importantly, our study demonstrates that phenotypes observed in MFS embryonic stem cells are phenocopied reliably in MFS reprogrammed iPS cells.
Results
Derivation of Human Marfan Embryonic Stem Cells and iPS Cells from an MFS-Specific Patient.
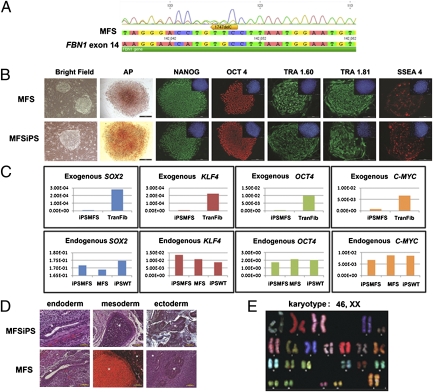
In the routine clinical practice of in vitro fertilization, embryos are sometimes tested via preimplantation genetic diagnosis for common disorders; genetic testing occurs at the eight-cell stage before blastocyst formation. We obtained a human blastocyst carrying a FBN1 mutation, following preimplantation genetic diagnosis, and derived a human embryonic stem cell line (referred to as MFS cells) via standard derivation conditions on mouse embryonic fibroblast feeder cells. The embryos and the MFS cells were both shown to carry a frame-shift mutation (c.1747delC) in the 5′ region (exon 14) of the FBN1 gene that results in a stop codon (in exon 15) at the amino acid position 624 (Fig. 1A).
Fig. 1.
Characterization of MFS and MFSiPS cells. (A) DNA sequencing analysis of MFS cells showing a mutation in FBN1 exon 14. (B) Cell morphology of representative MFS and iPS clones by phase contrast (bright field), alkaline phosphatase (AP) staining, and immunofluorescence staining for pluripotent markers: NANOG, OCT-4, TRA-1–60, TRA-1–81, and SSEA-4. (Insets) Nuclear counterstaining performed with DAPI. (Scale bars, 100 μm.) (C) qPCR for the expression of exogenous and endogenous SOX2, KLF4, OCT4, and C-MYC genes. (D) Differentiation of MFS and MFSiPS cells, teratomas containing cells from three germ layers (endoderm, mesoderm, and ectoderm) developed from MFSiPS and MFS cells injected into the dorsal flank of nude mice. Endoderm (gut epithelium), mesoderm, (cartilage), and ectoderm (neuroectoderm) are indicated by white asterisks. (Scale bars, 100–250 μm.) (E) Spectral karyotyping analysis of MFSiPS cells. TransFib, parental transduced fibroblasts.
Control WT human embryonic stem cells (referred to as WT cells) were derived from a blastocyst that does not carry FBN1 mutation donated for research.
Human iPS cells (MFSiPS cells) were generated from fibroblasts obtained from a patient with MFS that harbored a FBN1 splice-site mutation (c.3839–1 g > t) that causes skipping of exon 31, (proband FB1121), leading to a severe neonatal clinical phenotype (28). A second MFSiPS cell line was generated from fibroblasts obtained from a different MFS patient (proband FB1592) harboring a FBN1 frame-shift mutation (c.1642del3ins20bp) previously characterized (29) (Fig. S4). The iPS cell derivation was as previously described (30).
MFS fibroblasts and control WT fibroblasts from healthy male (WTiPS) were transduced with the retroviral vectors harboring the human reprogramming genes SOX2, OCT4, KLF4, and c-MYC. Two clones, collectively referred to as MFSiPS cells, were then selected for detailed analysis, with both giving similar results. MFSiPS cells exhibited morphology similar to MFS embryonic stem cells, were alkaline phosphatase-positive (ALPL), and were immunoreactive for NANOG, OCT4, TRA1.60, TRA1.81, and SSEA4 (Fig. 1B). Expression levels of endogenous and exogenous human pluripotency-associated genes SOX2, KLF4, OCT4, and C-MYC were also analyzed by quantitative real-time PCR (qPCR) (Fig. 1C) MFSiPS cells were successfully cultured in an undifferentiated state for more than 25 passages. Pluripotency of MFSiPS and WTiPS cells was assessed by differentiation into cell types of all three embryonic germ layers, and by teratoma formation in vivo (Fig. 1D). Moreover, spectral karyotype analysis showed normal karyotype of MFSiPS cells (Fig. 1E).
Impaired Osteogenic Differentiation in MFS Embryonic Stem Cells.
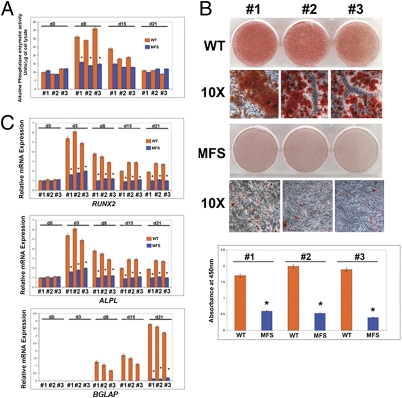
To identify skeletogenic phenotypes of embryonic MFS stem cells (MFS cells), we evaluated their osteogenic differentiation. Cells positive for CD73, one of the mesenchymal cellular markers, representative of cells differentiating toward both osteogenic and chondrogenic fates (31–34), were isolated and analyzed. Alkaline phosphatase enzymatic activity and Alizarin red staining revealed a striking impairment in osteogenic differentiation of MFS cells compared with WT human embryonic stem cells (Fig. 2 A and B). This impairment was further supported by qPCR analysis of osteogenic markers RUNX2, ALPL, and osteocalcin (BGLAP) (Fig. 2C).
Fig. 2.
Impairment of osteogenic differentiation in MFS cells. (A) Alkaline phosphatase enzymatic activity is significantly reduced in MFS cells. (B) Alizarin red staining reveals lack of mineralization in MFS cells. Histogram below represents quantification of staining. #1, #2, and #3 represent three independent isolations of CD73+ cells. (C) qPCR showing down-regulation of specific osteogenic markers in MFS cells. The relative mRNA level in each sample was normalized to its GAPDH content. Values are given as relative to GAPDH expression.
Enhanced Activation of TGF-β Signaling in MFS Cells Inhibits Osteogenic Differentiation.
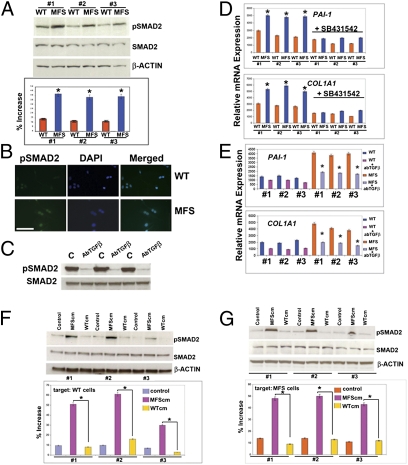
A potential explanation for the impairment of osteogenesis might be enhanced activation of TGF-β and downstream signaling. Therefore, we investigated the extent of SMAD2 phosphorylation in MFS cells and corresponding WT controls. Immunoblotting and immunofluorescence analyses revealed a greater endogenous phosphorylated SMAD2 in MFS cells (Fig. 3 A and B), which could be blocked by treatment with pan–TGF-β–neutralizing antibody (Fig. 3C). Moreover, a higher than normal activation of TGF-β signaling in the MFS cells was further indicated by the up-regulation of TGF-β1–induced ECM markers PAI-1 and collagen (COL1A1) (35–38) in MFS cells compared with WT controls (Fig. 3D). These differences were abrogated by treatment with either SB431542, a selective inhibitor of endogenous TGF-β signaling with no effect on bone morphogenetic protein (BMP) signaling (39), or a pan–TGF-β–neutralizing antibody (Fig. 3 D and E). Moreover, ELISA and immunoblotting analyses detected higher levels of active TGF-β1 in the MFS conditioned-medium compared with WT conditioned-medium (Fig. S2 A–C), but qPCR analyses showed normal steady-state levels of TGF-β transcripts (Fig. S2D). The presence of abundant levels of active TGF-β in the conditioned-medium of MFS cells was also confirmed by its ability to promote a strong phosphorylation of SMAD2, either when applied to WT or MFS cells (Fig. 3 F and G). Notably, conditioned-medium collected from WT control cells failed to activate SMAD2 phosphorylation (Fig. 3 F and G). These findings suggested that MFS cells are engaged in active autocrine TGF-β signaling.
Fig. 3.
Enhanced activation of TGF-β1 signaling in MFS cells. (A) immunoblotting analysis showing a more intense phosphorylation of SMAD2 in MFS cells. To assess for the total amount of endogenous SMAD2 and to control for equal loading and transfer of the samples, the membrane was reprobed with anti-SMAD2 and anti–β-actin antibody. Histogram below represents quantification of phosphorylated SMAD2 protein obtained by the ImageJ program. The relative intensity of each band was normalized to its respective β-actin loading controls. (B) Immunofluorescence using anti-pSMAD antibody detects strong staining in MFS cells compared with WT. DAPI nuclear counterstaining. (Scale bar, 50 μm.) (C) Treatment with pan–TGF-β–neutralizing antibody (1.2 μg/mL) dramatically decreases the endogenous phosphorylated SMAD2 in MFS cells. (D) qPCR analysis reveals up-regulation of PAI-1 and COL1A1 expression in MFS cells; treatment with SB431542 (10 μM) inhibits the up-regulation. (E) Up-regulation of PAI-1 and COL1A1 expression is abrogated by pan–TGF-β–neutralizing antibody. (F) Treatment of WT cells with serum-free conditioned-media collected from MFS cells (MFScm) induces heavy phosphorylation of SMAD2 protein, whereas conditioned-media collected from WT cells (WTcm) does not. Control represents untreated cells. Control for loading and transfer of the samples and densitometry analysis of pSMAD2 bands were performed as above. (G) Similar results are observed on treated MFS cells. Asterisks indicate statistically significant differences: *P < 0.05.
Inhibition of TGF-β Signaling Rescues the Osteogenic Differentiation in MFS Cells.
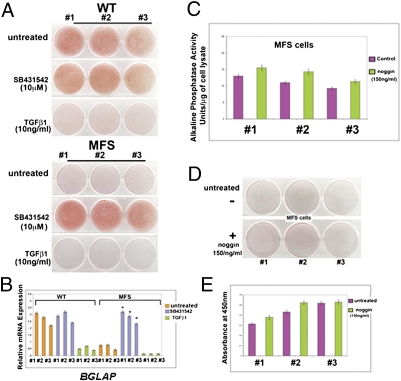
Having established significant differences in TGF-β signaling activation between MFS and WT cells, we sought to verify whether inhibition of this signaling could effectively rescue the osteogenic differentiation in MFS cells. For this purpose, MFS cells and their corresponding WT controls were treated during the osteogenic assay with 10 μM SB-431542. Treatment with SB-431542 rescued the phenotype, leading to robust osteogenic differentiation of MFS cells to a similar degree as observed in untreated controls, as indicated by Alizarin red staining of mineralized ECM and qPCR analysis of BGLAP (Fig. 4 A and B). In contrast, TGF-β1 treatment inhibited osteogenesis in both MFS and WT cells (Fig. 4 A and B). Of note, treatment with noggin, an inhibitor of BMP signaling, did not restore the osteogenic differentiation in MFS cells (Fig. 4 C–E). Collectively, the results above demonstrated that MFS cells are unable to differentiate along the osteogenic lineage because of an enhanced autocrine TGF-β signaling.
Fig. 4.
Inhibition of TGF-β signaling rescues the osteogenic differentiation in MFS cells. (A) Treatment with SB431542 rescues the osteogenic differentiation of MFS cells. In contrast, TGF-β1 inhibits dramatically osteogenic differentiation. (B) qPCR of BGLAP, a late osteogenic marker. Values are presented as above. *P < 0.05. (C) Alkaline phosphatase enzymatic activity at day 10. (D) Alizarin red staining at day 21 showing that noggin treatment does not rescue the osteogenic differentiation in MFS cells. (E) Alizarin red staining quantification.
MFS Cells Undergo Chondrogenic Differentiation Without Requirement for Exogenous TGF-β.
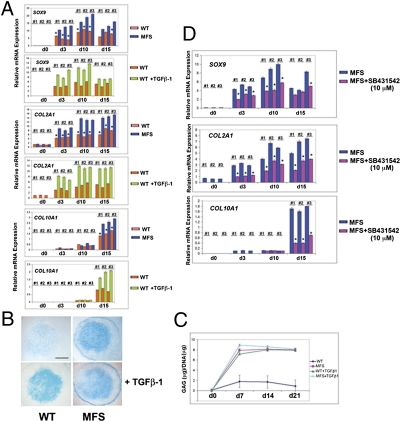
TGF-β signaling has previously been shown to promote mesenchymal cell differentiation toward chondrocytes (40–42). Therefore, we compared chondrogenic differentiation of MFS and WT cells. We implemented a high-density micromass culture system of MFS and WT cells in the presence or absence of exogenous TGF-β1. Expression analysis of signature genes of chondrogenesis indicated that MFS micromasses could differentiate efficiently toward the chondrogenic lineage even in absence of exogenous TGF-β1, whereas chondrogenesis was greatly impaired in WT micromasses in the absence of exogenous TGF-β1 (Fig. 5A). This striking difference in chondrogenesis was further confirmed by Alcian blue staining and proteoglycans synthesis (Fig. 5 B and C). Importantly, treatment with SB-431542 impeded the chondrogenic ability of MFS cells in absence of exogenous TGF-β1 (Fig. 5D), thus strengthening the role of active TGF-β signaling in determining phenotypic differences between MFS and WT cells.
Fig. 5.
MFS cells undergo chondrogenesis without requirement of exogenous TGF-β1 protein. (A) qPCR analysis of chondrogenic markers indicates that MFS cells undergo chondrogenesis in absence of exogenous TGF-β1, whereas WT cells adopt chondrogenic fate only in presence of exogenous TGF-β1 (2 ng/mL). *P < 0.05. (B) Alcian blue staining reveals strong staining for proteoglycans in MFS micromasses cultured in absence of TGF-β1. (Scale bar, 250 μm.) (C) Quantification of GAGs confirm the chondrogenic differentiation of MFS micromasses in absence of TGF-β1. (D) qPCR showing that SB431542 treatment prevents MFS cells to undergo chondrogenesis in absence of exogenous TGF-β1. *P < 0.05.
MFSiPS Cells Recapitulate Osteogenic Phenotype of MFS Cells.
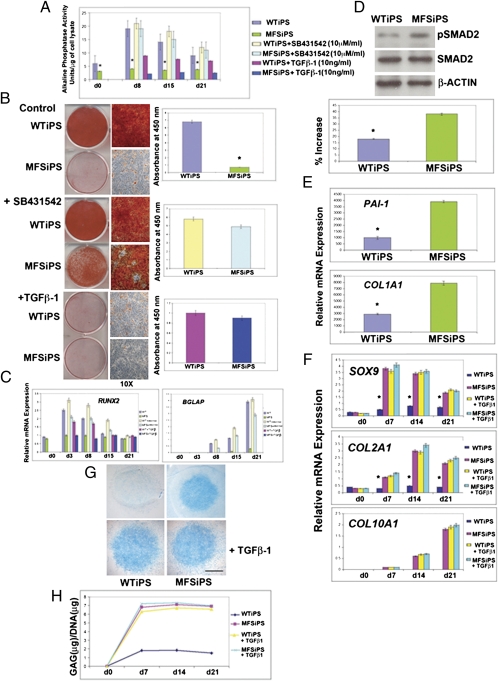
Much of controversy surrounds the potential use of iPS cells as a model for human disease given potential differences of iPS and embryonic stem cells (43, 44). Here we examined whether MFSiPS cells phenocopy observations described above for MFS embryonic stem cells. For this purpose, we differentiated and isolated cells positive for CD73 and examined osteogenesis in detail. We observed distinct inhibition of osteogenic differentiation in MFSiPS cells of the same magnitude as we observed with MFS embryonic stem cells, as indicated either by ALPL activity, Alizarin red staining, or quantitative gene expression analysis (Fig. 6 A–C and Fig. S4 E and F). Similarly, SB-431542 treatment promoted osteogenic differentiation, but TGF-β1 was inhibitory (Fig. 6 A–C and Fig. S4 E and F), thus indicating enhanced activation of TGF-β signaling also in MFSiPS-derived cells.
Fig. 6.
MFSiPS cells phenocopy MFS embryonic stem cells. (A) Alkaline phosphatase enzymatic assay detects significantly lower levels of activity MFSiPS. (B) Alizarin red staining showing poor mineralization of extracellular matrix in MFSiPS cells. SB431542 treatment rescues osteogenic differentiation of MFSiPS, whereas addition of TGF-β1 (2 ng/mL) inhibits osteogenesis. (C) qPCR analysis of osteogenic markers RUNX2 and BGLAP. (D) Higher levels of endogenous phosphorylated SMAD2 detected by immunoblotting analysis in MFSiPS cells. Histogram below showing quantification of phosphorylated SMAD2. Control for loading and transfer of the samples were determined as described in Fig. 3. (E) qPCR analysis indicates up-regulation of PAI-1 and COL1A1 expression in MFSiPS cells. (F) qPCR of chondrogenic markers in MFSiPS and WTiPS micromass cultures with or without exogenous TGF-β1. (G) Alcian blue staining indicates that MFSiPS are able to differentiate without TGF-β1 supplement. (Scale bar, 250 μm.) (H) Quantification of GAGs confirming the chondrogenic differentiation of MFSiPS micromass cultured without exogenous TGF-β1. Asterisks indicate statistically significant differences: *P < 0.05.
Enhanced Activation of TGF-β Signaling in MFSiPS Cells.
We next examined activation of TGF-β signaling in MFSiPS cells. Immunoblotting and immunofluorescence analyses of phosphorylated SMAD2 (Fig. 6D, and Figs. S3A and S4G), as well as expression of PAI-1 and COL1A1 genes (Fig. 6E), indicated enhanced activation of TGF-β signaling also in MFSiPS cells, similarly to MFS. Pan–TGF-β–neutralizing antibody blocked both the endogenous phosphorylated SMAD2 and up-regulation of PAI-1 and COL1A1 genes (Fig. S3 B and C). ELISA detected higher levels of active TGF-β1 in the medium of MFSiPS than WTiPS cells (Figs. S1, S3D, and S4H).
MFSiPS Cells Undergo Chondrogenic Differentiation Without Requirement for Exogenous TGF-β.
Given the striking similarity of the MFSiPS phenotype to that of MFS cells, we hypothesized that these two pluripotent stem cells sources should exhibit the same chondrogenic phenotypes. Examination of micromass cultures of MFSiPS with and without exogenous TGF-β1 indicated that MFSiPs cells shared the ability to differentiate without TGF-β1 supplementation, an ability confirmed by Alcian blue staining and synthesis of glycosaminoglycans (GAGs) (Fig. 6 F–H and Fig. S4 I–M). Congruent with observations in MFS cells, we also observed that treatment of MFSiPS cells with SB-431542 inhibited chondrogenesis in the absence of exogenous TGF-β1 (Fig. S3 E and F).
Discussion
The present study highlights the ability of MFS patient-induced iPS cells to faithfully phenocopy the skeletogenic phenotype identified in human MFS embryonic stem cells. To our knowledge, this report of human embryonic stem cells with a monogenic disease and its phenotype recapitulation using patient-specific fibroblast-derived iPS cells is unique.
To date, over 600 mutations have been published in the Universal Marfan databases, but only a minority are recurring mutations (45). Understanding the mechanisms by which genetic variations contribute to syndromes, like MFS, is a central goal of human genetics and will facilitate the development of preventive strategies and treatments. Indeed, iPS cells offer promise for defining the functional effects of multiple genetic variations observed in MFS probands.
Several recent reports have uncovered intricate genomic differences (e.g., genetic and epigenetic alterations) between iPS cells and their ESC counterparts, therefore fading the glitter of iPS cells and stimulating controversy about their future (43, 46–50). In our study we demonstrate a tight correlation between embryonic stem cells and iPS cell MFS phenotypes.
Mutations in FBN1 are associated with increased activity and bioavailability of TGF-β1 (27, 51), which is suspected to be the basis for phenotypical similarities of FBN1 mutations in MFS and mutations in the receptors for TGF-β in MFS-related diseases. Observations of increased activity and bioavailability of TGF-β1 have been obtained from studies carried out exclusively on MFS mouse models (27, 51). The present article reveals increased activation of TGF-β and enhancement of its mediated signaling in human MFS cells. Our results demonstrate higher levels of active TGF-β and enhanced phosphorylation of endogenous SMAD2 in human embryonic MFS stem cells and MFSiPS cells compared with WT control cells. Remarkably, high levels of active TGF-β and enhanced phosphorylation of endogenous SMAD2 were sustained throughout the osteogenic differentiation (Fig. S4). Moreover, we show the biological consequences of activation of TGF-β signaling in the skeletogenic context of MFS. Our data point toward a molecular and unique mechanism underlying MFS whereby enhanced TGF-β signaling inhibits osteogenic differentiation while promoting chondrogenesis in a TGF-β1 cell-autonomous fashion. These findings are strongly supported by the fact that either treatment with SB 431542, an effective inhibitor of ALK4, -5, and -7, but not of ALK1, -2, -3, or -6 (39) or pan–TGF-β–neutralizing antibody abrogate the observed skeletogenic phenotypes upon decreased activation of phosphorylated SMAD2. In contrast, noggin, an inhibitor of BMP-mediated signaling, does not rescue the osteogenic differentiation. The impairment of osteogenic differentiation observed in MFS and MFSiPS cells is supported by clinical studies showing osteopenia and bone fracture susceptibility in MFS patients (52–54).
The in vitro chondrogenic differentiation of mesenchymal cells requires the complex involvement of growth factors and cell–cell and cell–matrix interactions, similar to developmental chondrogenesis in vivo (55). The chondroinductive effect of TGF-β1 has been well established in embryonic and adult mesenchymal cells (40–42). Several reports have demonstrated the critical roles of intracellular signaling cascades activated by TGF-β family members in promoting cartilage-specific gene expression (56). By using micromass cell cultures we demonstrate that MFS cells are able to differentiate along the chondrogenic lineage without requirement of any exogenous TGF-β1, whereas control WT cells do not. These results reflect the enhanced activation of TGF-β in MFS cells. It is tempting to speculate that abundant level of active TGF-βs in MFS patients might trigger high proliferation of chondrocytes in the growth plate or enrichment of chondroprogenitor cells.
Of note, disproportionate overgrowth of the long endochondral bones (dolichostenomelia) is often the most striking and immediately evident manifestation of MFS skeletal defects (6, 7, 57). Therefore, in the light of our data one could hypothesize that the enhanced activation of TGF-βs may favor the longitudinal overgrowth of long-bones of MFS patients. Our hypothesis is strongly supported by the fact that dolichostenomelia was one of the dominant clinical manifestations reported in the MFS patient from which we have derived iPS cells (28).
Using iPS technology, we have created a human MFS “model” phenocopying human embryonic MFS stem cells, strongly supporting the hypothesis that these are bona fide disease phenotypes resulting from the FBN1 mutations and not simply sporadic cell-culture artifacts. Indeed, MFSiPS cells provide a unique and useful tool to further dissect molecular aspects of MFS, as well as characterize novel targets that may yield exciting opportunities for screening MFS novel treatments.
From a translational medicine perspective, the use of an iPS strategy will allow reprogramming of MFS adult fibroblasts harboring different mutations in FBN1 gene, enabling comprehensive molecular investigations aimed at elucidating the mechanisms underlying the clinically observed pathological variability and helping to pave the way to personalized therapeutic interventions.
Methods
Cell Lines Derivation, Characterization, and Culture Conditions.
Derivation of Marfan human embryonic stem cells, MFSiPS cells, teratoma formation and karyotype are described in detail in SI Methods.
Osteogenic Differentiation, Reverse Transcription (RT), and Quantitative Real-Time PCR (qPCR)
Osteogenic differentiation assays, RT, and qPCR were previously described (58, 59). Additional details are available in SI Methods.
Further methods are described in SI Methods.
Supplementary Material
Acknowledgments
The authors thank Ha Nam Nguyen for the generation of retrovirus. This work was supported by the National Institutes of Health Grants, NIH-U01 HL100397, NIH-U01 HL099776, RC1 HL100490, and RC2 DE020771 (to M.T.L.); and California Institute Regenerative Medicine Grant RL1-00662-1 (to M.T.L. and E.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113442109/-/DCSupplemental.
References
- 1.Dietz HC, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 2.Pereira L, et al. Targetting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet. 1997;17:218–222. doi: 10.1038/ng1097-218. [DOI] [PubMed] [Google Scholar]
- 3.Dietz HC, Loeys B, Carta L, Ramirez F. Recent progress towards a molecular understanding of Marfan syndrome. Am J Med Genet C Semin Med Genet. 2005;139C:4–9. doi: 10.1002/ajmg.c.30068. [DOI] [PubMed] [Google Scholar]
- 4.Robinson PN, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–787. doi: 10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKusick VA. The defect in Marfan syndrome. Nature. 1991;352:279–281. doi: 10.1038/352279a0. [DOI] [PubMed] [Google Scholar]
- 6.Pyeritz RE, McKusick VA. The Marfan syndrome: Diagnosis and management. N Engl J Med. 1979;300:772–777. doi: 10.1056/NEJM197904053001406. [DOI] [PubMed] [Google Scholar]
- 7.Pyeritz RE. The Marfan syndrome. Annu Rev Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- 8.McKUSICK VA. The cardiovascular aspects of Marfan's syndrome: A heritable disorder of connective tissue. Circulation. 1955;11:321–342. doi: 10.1161/01.cir.11.3.321. [DOI] [PubMed] [Google Scholar]
- 9.Gelb BD. Marfan's syndrome and related disorders—More tightly connected than we thought. N Engl J Med. 2006;355:841–844. doi: 10.1056/NEJMe068122. [DOI] [PubMed] [Google Scholar]
- 10.Hayward C, Brock DJ. Fibrillin-1 mutations in Marfan syndrome and other type-1 fibrillinopathies. Hum Mutat. 1997;10:415–423. doi: 10.1002/(SICI)1098-1004(1997)10:6<415::AID-HUMU1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 11.Pereira L, et al. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum Mol Genet. 1993;2:1762. doi: 10.1093/hmg/2.10.1762. [DOI] [PubMed] [Google Scholar]
- 12.Ramirez F, Pereira L. The fibrillins. Int J Biochem Cell Biol. 1999;31:255–259. doi: 10.1016/s1357-2725(98)00109-5. [DOI] [PubMed] [Google Scholar]
- 13.Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum Mol Genet. 1995;4(Spec No):1799–1809. doi: 10.1093/hmg/4.suppl_1.1799. [DOI] [PubMed] [Google Scholar]
- 14.Adès LC, et al. FBN1, TGFBR1, and the Marfan-craniosynostosis/mental retardation disorders revisited. Am J Med Genet A. 2006;140:1047–1058. doi: 10.1002/ajmg.a.31202. [DOI] [PubMed] [Google Scholar]
- 15.Faivre L, et al. De novo 15q21.1q21.2 deletion identified through FBN1 MLPA and refined by 244K array-CGH in a female teenager with incomplete Marfan syndrome. Eur J Med Genet. 2010;53:208–212. doi: 10.1016/j.ejmg.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson S, et al. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: A potential modifier of phenotype? Hum Mol Genet. 2003;12:2269–2276. doi: 10.1093/hmg/ddg241. [DOI] [PubMed] [Google Scholar]
- 17.Blyth M, Foulds N, Turner C, Bunyan D. Severe Marfan syndrome due to FBN1 exon deletions. Am J Med Genet A. 2008;146A:1320–1324. doi: 10.1002/ajmg.a.32229. [DOI] [PubMed] [Google Scholar]
- 18.Dietz HC, et al. Four novel FBN1 mutations: Significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics. 1993;17:468–475. doi: 10.1006/geno.1993.1349. [DOI] [PubMed] [Google Scholar]
- 19.Liu W, Schrijver I, Brenn T, Furthmayr H, Francke U. Multi-exon deletions of the FBN1 gene in Marfan syndrome. BMC Med Genet. 2001;2:11. doi: 10.1186/1471-2350-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mátyás G, et al. Large genomic fibrillin-1 (FBN1) gene deletions provide evidence for true haploinsufficiency in Marfan syndrome. Hum Genet. 2007;122:23–32. doi: 10.1007/s00439-007-0371-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhu L, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 22.Guo DC, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 23.Carta L, et al. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem. 2006;281:8016–8023. doi: 10.1074/jbc.M511599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Judge DP, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest. 2004;114:172–181. doi: 10.1172/JCI20641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldadah ZA, Brenn T, Furthmayr H, Dietz HC. Expression of a mutant human fibrillin allele upon a normal human or murine genetic background recapitulates a Marfan cellular phenotype. J Clin Invest. 1995;95:874–880. doi: 10.1172/JCI117737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charbonneau NL, et al. In vivo studies of mutant fibrillin-1 microfibrils. J Biol Chem. 2010;285:24943–24955. doi: 10.1074/jbc.M110.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neptune ER, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, et al. Mutant fibrillin-1 monomers lacking EGF-like domains disrupt microfibril assembly and cause severe marfan syndrome. Hum Mol Genet. 1996;5:1581–1587. doi: 10.1093/hmg/5.10.1581. [DOI] [PubMed] [Google Scholar]
- 29.Schrijver I, et al. Premature termination mutations in FBN1: Distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet. 2002;71:223–237. doi: 10.1086/341581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrne JA, Nguyen HN, Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS ONE. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuli R, et al. Characterization of multipotential mesenchymal progenitor cells derived from human trabecular bone. Stem Cells. 2003;21:681–693. doi: 10.1634/stemcells.21-6-681. [DOI] [PubMed] [Google Scholar]
- 32.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, et al. A subpopulation of mesenchymal stromal cells with high osteogenic potential. J Cell Mol Med. 2009;13(8B):2436–2447. doi: 10.1111/j.1582-4934.2009.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bühring HJ, et al. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–271. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- 35.Keeton MR, Curriden SA, van Zonneveld AJ, Loskutoff DJ. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 36.Laping NJ, et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 37.Ignotz RA, Massagué J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem. 1986;261:4337–4345. [PubMed] [Google Scholar]
- 38.Penttinen RP, Kobayashi S, Bornstein P. Transforming growth factor beta increases mRNA for matrix proteins both in the presence and in the absence of changes in mRNA stability. Proc Natl Acad Sci USA. 1988;85:1105–1108. doi: 10.1073/pnas.85.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 40.Mello MA, Tuan RS. Effects of TGF-beta1 and triiodothyronine on cartilage maturation: in vitro analysis using long-term high-density micromass cultures of chick embryonic limb mesenchymal cells. J Orthop Res. 2006;24:2095–2105. doi: 10.1002/jor.20233. [DOI] [PubMed] [Google Scholar]
- 41.Kawaguchi J, Mee PJ, Smith AG. Osteogenic and chondrogenic differentiation of embryonic stem cells in response to specific growth factors. Bone. 2005;36:758–769. doi: 10.1016/j.bone.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 42.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 43.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: Facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panopoulos AD, Ruiz S, Izpisua Belmonte JC. iPSCs: Induced back to controversy. Cell Stem Cell. 2011;8:347–348. doi: 10.1016/j.stem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Faivre L, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: An international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 51.Habashi JP, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohlmeier L, Gasner C, Marcus R. Bone mineral status of women with Marfan syndrome. Am J Med. 1993;95:568–572. doi: 10.1016/0002-9343(93)90351-o. [DOI] [PubMed] [Google Scholar]
- 53.Kohlmeier L, Gasner C, Bachrach LK, Marcus R. The bone mineral status of patients with Marfan syndrome. J Bone Miner Res. 1995;10:1550–1555. doi: 10.1002/jbmr.5650101017. [DOI] [PubMed] [Google Scholar]
- 54.Moura B, et al. Multidisciplinary Marfan Syndrome Clinic Group Bone mineral density in Marfan syndrome. A large case-control study. Joint Bone Spine. 2006;73:733–735. doi: 10.1016/j.jbspin.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 55.DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 2000;8:309–334. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 56.Tuli R, et al. Transforming growth factor-beta-mediated chondrogenesis of human mesenchymal progenitor cells involves N-cadherin and mitogen-activated protein kinase and Wnt signaling cross-talk. J Biol Chem. 2003;278:41227–41236. doi: 10.1074/jbc.M305312200. [DOI] [PubMed] [Google Scholar]
- 57.Giampietro PF, Raggio C, Davis JG. Marfan syndrome: Orthopedic and genetic review. Curr Opin Pediatr. 2002;14:35–41. doi: 10.1097/00008480-200202000-00006. [DOI] [PubMed] [Google Scholar]
- 58.Quarto N, et al. Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res. 2010;25:1680–1694. doi: 10.1359/jbmr.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quarto NWD, Wan DC, Longaker MT. Molecular mechanisms of FGF-2 inhibitory activity in the osteogenic context of mouse adipose-derived stem cells (mASCs) Bone. 2008;42:1040–1052. doi: 10.1016/j.bone.2008.01.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.