Abstract
Proteasomes execute the degradation of most cellular proteins. Although the 20S core particle (CP) has been studied in great detail, the structure of the 19S regulatory particle (RP), which prepares ubiquitylated substrates for degradation, has remained elusive. Here, we report the crystal structure of one of the RP subunits, Rpn6, and we describe its integration into the cryo-EM density map of the 26S holocomplex at 9.1 Å resolution. Rpn6 consists of an α-solenoid-like fold and a proteasome COP9/signalosome eIF3 (PCI) module in a right-handed suprahelical configuration. Highly conserved surface areas of Rpn6 interact with the conserved surfaces of the Pre8 (alpha2) and Rpt6 subunits from the alpha and ATPase rings, respectively. The structure suggests that Rpn6 has a pivotal role in stabilizing the otherwise weak interaction between the CP and the RP.
Keywords: 26S proteasome, cryoelectron microscopy, PSMD11, S9, PCI domain
Protein degradation is of vital importance for the maintenance of protein homeostasis, for the removal of misfolded proteins, and for the control of numerous regulatory processes (1, 2). In eukaryotic cells, the main pathway for protein degradation is the ubiquitin-proteasome system (3). It has the capability of degrading almost any protein, and yet it acts with exquisite specificity. The ubiquitin system selects proteins and marks them for destruction, whereas the 26S proteasome is the executioner of proteolysis. Malfunctions of the system have been implicated in a variety of diseases (2).
The 26S proteasome is a molecular machine of approximately 2.5 MDa built from two copies each of 34 canonical subunits and several proteasome interacting proteins, which are present in substochiometric amounts (4–6). The 26S holocomplex comprises two subcomplexes: The barrel-shaped core particle (CP) that harbors the proteolytically active sites and sequesters them from the cellular environment, and the regulatory particles (RPs) that bind to one or both ends of the CP. Their role is to prepare substrates for degradation; this preparation includes the recognition of polyubiquitylated proteins, their deubiquitylation and unfolding and, eventually, assistance in their translocation into the CP through the gate in the α-ring of the CP.
The CP, which is a stack of four seven-membered rings (α1–7; β1–7; β1–7; α1–7), is structurally well characterized; it is highly conserved from archaea to mammals, and crystal structures are available for CPs from several species (7–9). In contrast to the CP, the structure of the RP is only dimly understood. So far, all attempts to crystallize the RP alone or in association with the CP have been unsuccessful. Recently, EM single particle analysis has provided a map of the 26S holocomplex at medium resolution (9.1 Å), which provides a platform for the integration of high-resolution structures of the constituent subunits (10).
The RP is composed of a core of 19 different subunits, which can dissociate into a “base” and a “lid” subcomplex (11). The base is thought to form the proximal part of the RP, which associates with the α-rings of the CP, whereas the lid forms the distal end. The base comprises a heterohexameric AAA-ATPase module (Rpt1–Rpt6) and the non-ATPase subunits Rpn1 and Rpn2 (11). The often substoichiometric subunits Rpn10 and Rpn13 are also commonly assigned to the base subcomplex (5). The lid part of the RP is composed of the Rpn3, Rpn5–Rpn9, and Rpn11–Rpn12 subunits (11). The lid subunits can be classified into two groups according to their predicted domain structure: Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, and Rpn12 are predicted to share a C-terminal module present in proteasome, COP9/signalosome, and eIF3 subunits (PCI module), whereas Rpn8 and Rpn11 subunits have an MPN (Mpr1, Pad1 N-terminal) domain in common (12). Functionally, Rpn10 and Rpn13 serve as polyubiquitin receptors, whereas Rpn11 has deubiquitylation activity (4, 5). The PCI module was proposed to have a structural role and is composed of an N-terminal helix bundle and a winged-helix subdomain (13–15).
The PCI subunit Rpn6 was found to be an essential component of the 26S proteasome in Saccharomyces cerevisiae (16), Trypanosoma brucei (17), Plasmodium falciparium (18), and Drosophila melanogaster (19). Upon conditional knock-out in S. cerevisiae, only partially assembled complexes lacking all the lid subunits were found, and the cells were arrested in G2/M phase (20). Similarly, a temperature-sensitive Rpn6 mutant strain of S. cerevisiae yielded only partially assembled complexes at the restrictive temperature, suggesting a critical role of Rpn6 for assembly (21).
Here, we present the crystal structure of Rpn6 from D. melanogaster. The distinctive shape of this subunit and the prevalence of α-helices allowed us to fit the structure into the 9.1 Å cryo-EM map of the 26S proteasome of Schizosaccharomyces pombe with high confidence. The hybrid structure reveals highly conserved contact interfaces between Rpn6 and subunits of both the CP and RP.
Results and Discussion
Crystallization and Structure Solution.
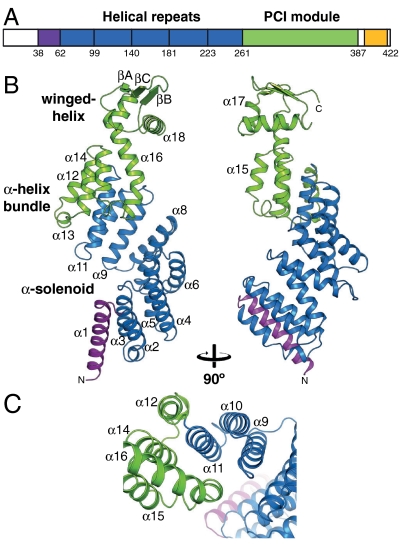
Rpn6 of D. melanogaster was expressed as a soluble 6xHis-tag fusion protein in Rhodococcus erythropolis (22). The 6xHis-tag was cleaved for biochemical analysis. Size-exclusion chromatography suggested that Rpn6 is monomeric in physiological buffer. Crystals from the 422-residue full-length protein showed only weak diffraction to approximately 9 Å resolution. To find a better construct for crystallization, we performed a limited proteolysis experiment using Proteinase-K. Mass spectrometry analysis of the most prominent SDS-PAGE fragment bands showed that the N-terminal region up to residue 29 is most sensitive to protease cleavage, indicating that it is flexibly linked. At higher protease concentrations, the protein is furthermore nicked at position 337, which maps to the PCI module (Fig. 1A). The Rpn6 construct comprising residues 30–422 yielded hexagonal crystals diffracting to 2.5 Å resolution. The crystal structure was solved by Gd-MAD at 3.0 Å resolution (Table S1). The model comprises residues 38–390 (Fig. 1B); the remaining residues were not resolved in the electron density and are presumably disordered.
Fig. 1.
Crystal structure of Rpn6. (A) Domain structure of Rpn6. The purple region denotes a capping helix; the yellow region is predicted to be α-helical. (B) Ribbon representation of Rpn6, colored by domain structure. Two views related by 90° rotation are shown. N and C termini and selected secondary structure elements are indicated. (C) Detailed view of the interface between the solenoid fold and the PCI module.
Structure Overview.
The crystal structure of Rpn6 consists of an α-helical solenoid followed by the PCI module (Fig. 1B). The overall shape is that of a right-handed suprahelical turn with approximate dimensions of 100 Å × 45 Å (height × width). The solenoid contains a slightly elongated N-terminal capping helix and five double-helix repeats with structural similarity to tetratricopeptide repeats (TPR). However, the helices are approximately one turn longer than in canonical TPR units; i.e., each repeat contains approximately 40 residues compared to 34 for TPRs. A conserved sequence signature for the Rpn6 repeats could not be detected. The hydrophobic final helix of the solenoid, α11, forms the central hub of a helix bundle, contacting helices α12, α14, and α16 of the PCI module (Fig. 1C). This interaction enables additional contacts between α9 and α16 that reinforce the solenoid-PCI module interface, strongly suggesting that the orientation of the two domains is rigidly fixed to form the right-handed suprahelical configuration. Thus, there is no discrete N-terminal boundary of the PCI module, which supports the conclusion of prior bioinformatic analyses of PCI-protein sequences (14). The winged-helix subdomain of Rpn6 has an elongated first helix, α16, which is markedly kinked in the center. Its N-terminal segment forms part of the helix bundle (Fig. 1C). The three-stranded antiparallel β-sheet of the PCI module is located at the tip of the suprahelical structure. α18, the so-called recognition helix in canonical winged-helix transcription factors, is arranged perpendicular to the long axis of the protein. In DNA complex structures, this helix is placed into the major groove of DNA (23). Whether the corresponding structure in Rpn6 serves such a function is unknown, but might be worth further investigation. The 26S proteasome has been implicated to play a role in transcription (24) and DNA double-strand repair (25), which could require physical association of the 26S proteasome to nucleic acid. Comparison with the other known structures of PCI module proteins, Csn7 and eIF3κ (13, 15), indicated that the winged-helix subdomains are less divergent than the N-terminal helix bundles (Fig. S1). The elongated helices in the Rpn6 helix bundle (helices α12, α14, α16) appear to ensure a rigid connection to the α-solenoid; these elongations are absent in Csn7 and eIF3κ. In addition, the proximal part of the helical bundle subdomain in all three structures appears to function as a buttress for the winged-helix domain.
Rpn6 Surface Conservation.
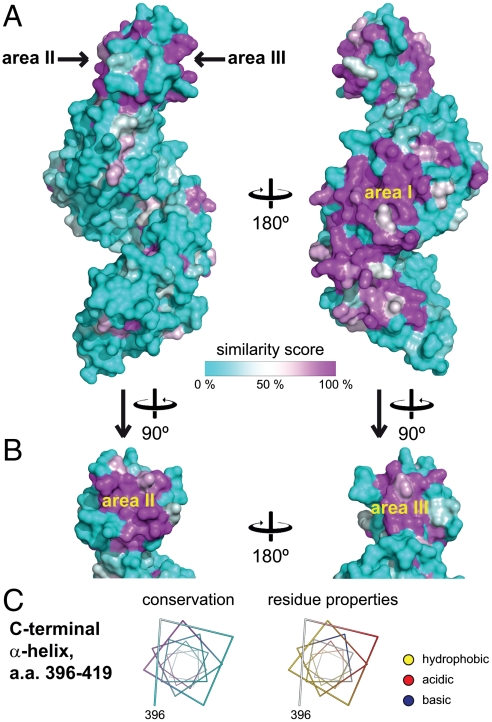
To identify functionally important regions, we performed an extensive sequence alignment of 21 putative Rpn6 sequences (Fig. S2) and mapped the similarity score onto the surface of the crystal structure (Fig. 2A). In the solenoid segment, a large continuous area of increased surface conservation was found on the convex outer face between helix α8 and α10 (region I, Fig. 2A). This area has few surface charges (Fig. S3B). The adjacent loop connection between helices α6 and α7 (residues 158–162) is also highly conserved. On the concave face, the adjacent residues Lys82, Lys84, Lys87, Arg90, and Phe124 are almost invariant. All these areas face approximately in the same direction, while there is essentially no surface conservation on the opposite (convex) side (Fig. 2A), strongly suggesting that the former is involved in contacts with other subunits of the 26S proteasome complex, while the latter is exposed to solvent.
Fig. 2.
Surface analysis of Rpn6. (A) Surface conservation mapped onto the surface of Rpn6. On the left, the same orientation as in Fig. 1A is shown. The similarity score from a multiple alignment of 21 related sequences (Fig. S2) was mapped onto the molecular surface of Rpn6. A cyan-white-magenta color gradient indicates increasing surface conservation. Regions I, II, and III are indicated. (B) Side views on the winged-helix subdomain. (C) The predicted C-terminal helix. The helix is represented as a helical wheel, and residue properties are indicated. (Left) Conservation is represented using the same color scheme as in panel A. (Right) Hydrophobic side chains are indicated in yellow. Positively and negatively charged functional groups are colored blue and red, respectively. The rest of the surface is shown in white.
Surface conservation in the PCI module of Rpn6 is limited to two smaller areas located at the flanks on the β-sheet (regions II and III, Fig. 2B). Region II includes the end of helix α16, strand βA and the connecting linker. Region III is composed of helix α18 and strand βB. Both are predominantly hydrophobic, implying that they might serve as protein–protein interfaces (Fig. S3). Interestingly, region II and region III of adjacent Rpn6 chains contact each other in the crystal lattice. The alignment of the β-sheets creates a continuous β-ribbon that traverses the crystals along the 6-fold screw axis (Fig. S4A), suggesting that the six PCI subunits in the lid might be arranged similarly within the complex. The buried surface area of approximately 460 Å2 on each partner is probably too small for a stable interaction consistent with our finding that Rpn6 is monomeric in solution. This observation suggests that other interactions must contribute to complex formation. A likely candidate for this additional interface is a conserved region at the C-terminus (residues 396–419) that was disordered in the crystal structure. In agreement, the sequence alignment suggests that this segment is flexibly attached to the PCI module via a poorly conserved linker (Fig. S2). Secondary structure prediction strongly suggests that the respective region forms an amphipathic α-helix (4). Mapping conservation and surface properties on this predicted helix shows that conservation is limited to the hydrophobic face (Fig. 2C), suggesting that it is involved in interactions with other subunits, probably in a coiled-coil conformation. Intriguingly, all proteasomal PCI subunits were predicted to comprise such a helical segment C-terminal to the PCI module (4).
Interactions of Rpn6 Within the Lid.
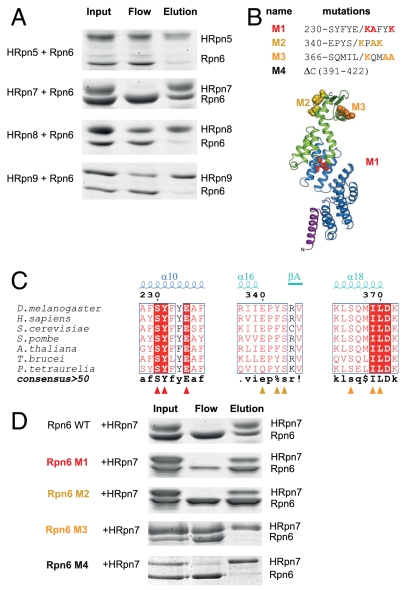
To test whether or not the PCI subunits interact with each other in the lid subcomplex, we incubated Rpn6 separately with 6xHis-tagged Rpn5, Rpn7, or Rpn9 of D. melanogaster, followed by Ni-affinity precipitation (Fig. 3A). Under the conditions tested, only Rpn6 and Rpn7 formed a stable binary complex. To analyze this interaction in more detail, mutations were introduced into Rpn6 (Fig. 3B). At the center of region I, we replaced the highly conserved peptide sequence 230-SYFYE-234 with KAFYK, yielding mutant M1 (Fig. 3C). Similarly, Rpn6 mutants M2 and M3 were generated by analogous substitutions of conserved peptide motifs in regions II and III, respectively. Finally, we removed the putative C-terminal α-helix by truncation at position 391 (M4). Interaction analysis of these mutant Rpn6 forms with 6xHis-tagged Rpn7 clearly showed that both an intact region III and the putative C-terminal helix are required for the interaction (Fig. 3D). This finding strongly suggests that the observed interaction is specific. Rpn6 and Rpn7 are thus likely to be in direct contact with each other in the lid, probably employing a bipartite interface.
Fig. 3.
Binary interaction of Rpn6 and Rpn7. (A) Probing for direct interactions of Rpn6 with lid particle subunits. Purified Rpn6 was incubated individually with His-tagged Rpn5, Rpn7, Rpn8, or Rpn9 from D. melanogaster. The Coomassie-stained SDS-PAGE gels show the initial mixtures, unbound proteins, and proteins precipitated with Ni-affinity resin. (B) Location of Rpn6 mutations in the structure. The respective amino acid residue substitutions are indicated. Mutated residues are shown in space-filling mode. (C) Excerpts from Rpn6 sequence alignment showing the mutated regions. (D) Both the PCI module interface region III and the C-terminal helix of Rpn6 are required for the interaction with Rpn7. His-tagged Rpn7 was incubated with either wild-type Rpn6(30–422) or Rpn6(30–422) mutants M1, M2, M3, or M4 and analyzed as described for panel A.
Location of Rpn6 in the 26S Proteasome.
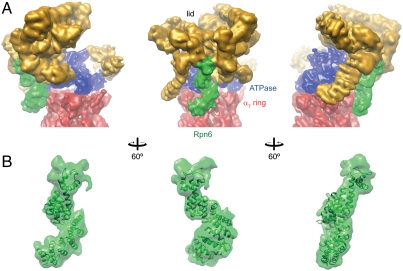
Finally, we fitted the crystal structure of Rpn6 into the 9.1 Å cryo-EM density of the 26S proteasome from S. pombe, which is the highest-resolution map available so far (10) (Fig. 4A). An exhaustive six-dimensional real-space search yielded a single solution, with high confidence (Fig. S5). The size of Rpn6 (49 kDa), its distinctive shape, and the prevalence of α-helices enabled the high-precision fit into the map (Fig. S6). We estimate that the accuracy of the fit considerably exceeds the resolution of the map (9 Å), probably by an order of magnitude. In the resulting model, Rpn6 forms a protrusion that is located at the outer rim of the lid particle, reaching down to the ATPase and alpha rings with its α-solenoid segment (Fig. 4A). This interface appears to be the most extensive direct contact between the lid and core particles (a second contact formed by a protrusion to the left of Rpn6 appears weaker). There is additional density at the N-terminus of the Rpn6 model that might correspond to residues 1–37, most of which were not included in the crystallization construct. For Rpn6 of S. pombe, an additional pair of helices was predicted for this segment and included in our homology model (Fig. 4B). Regions on Rpn6 with high surface conservation match almost perfectly with the areas buried in the complex, while the rather poorly conserved face projects toward the solvent (compare Figs. 2A and 4A).
Fig. 4.
Location of Rpn6 in the 26S proteasome. (A) Rpn6 density within the 9 Å cryo-EM density of the 26S proteasome from S. pombe. Three views are shown. The lid, base, and core subcomplex densities are indicated in gold, blue, and red, respectively. Density ascribed to Rpn6 is colored green. The core particle is clipped off at the β7 ring. (B) Detailed view of the Rpn6 fitted into the EM envelope. Density assigned for Rpn6 was segmented from the map. The homology model of Rpn6 from S. pombe including the predicted N-terminal helices α(-1) and α0 are included. Similar orientations as in panel A are shown.
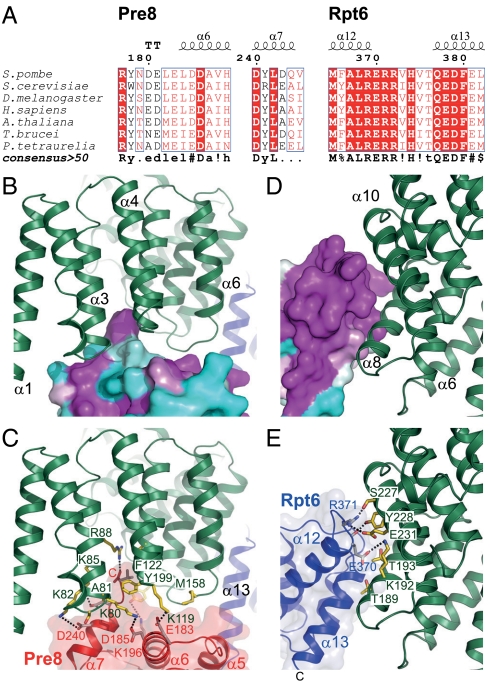
The subunits contacted by the solenoid domain of Rpn6 were previously assigned as Rpt6 and Pre8 (alpha2) using the 9.1 Å cryo-EM density of the 26S proteasome from S. pombe and cross-linking data (10). For both, high-confidence molecular models are available (7, 26–28). Both subunits share a conspicuous surface conservation in the Rpn6 contact areas, which is indicative of coevolution of the interface residues (Fig. 5 A–E). Interestingly, conditional mutation of Rpt6 in S. cerevisiae resulted in the same G2/M phase transition arrest as the Rpn6 deletion (20, 29). Closer inspection reveals that the CP subunit Pre8 is in proximity to the N-terminus of α3 (residues 79–85), the loop between α4 and α5 (118–122), and the loop between α6 and α7 (157–160) (Fig. 5 B and C). Together, these elements form an extensive, conserved interface. Under the reasonable assumption that the peptide backbones in the complex are similar to those in the individual crystal structures, a tentative assignment of molecular interactions is possible: The small side chains of Ser79Rpn6 and Ala81Rpn6 (S. pombe numbering, add 2 for D. melanogaster) enable tight contacts to α7 of Pre8. The adjacent residues Lys80Rpn6, Lys82Rpn6, and Lys85Rpn6 are in hydrogen-bonding distance to Glu183Pre8/Asp185Pre8, Asp240Pre8, and Asp243Pre8, respectively. Because of the proximity to Asp159Rpn6 and Asp160Rpn6, the C-terminal Val245Pre8 might rearrange to form a salt bridge with its carboxylate group to Arg88Rpn6. The Val245Pre8 side chain would then point into a hydrophobic pocket formed by Phe122Rpn6, Ala126Rpn6, and Arg88Rpn6. In an alternative scenario, the side chains of Phe122Rpn6 and Met158Rpn6 might rearrange locally and engage in contacts with two hydrophobic pockets located between helices α7 and α6, and at the loop connection between α5 and α6 of Pre8, respectively. Moreover, Asp159Rpn6 is in hydrogen-bonding distance to Arg177Pre8 and His189Pre8; Lys119Rpn6 may form a salt bridge with Glu183Pre8. The exposed side chain of residue Tyr199Rpn6, which is located in the strongly conserved loop between helices α8 and α9 (residues 186–202), could reach toward the highly conserved Lys196Pre8.
Fig. 5.
Putative interactions of Rpn6 with Pre8 and Rpt6. (A) Excerpts from Pre8 and Rpt6 sequence alignments for the Rpn6 contact regions. (B and C) Detailed view of the interactions with Pre8. (Upper) The similarity score of an extensive alignment of Pre8 sequences mapped onto the homology model surface. Rpn6 is shown as a green ribbon. (Lower) Putative key interactions at the interface. Both proteins are shown in ribbon representation. Selected side chains are shown as sticks. Putative hydrogen bonds are indicated by dashed lines. (D and E) Detailed view of the interactions with Rpt6.
The Rpt6–Rpn6 interface involves Rpn6 helices α8 and α10 (Fig. 2, area I), which are located opposite to Rpt6 helices α12 and α13 (i.e. helices 3 and 4 in its four-helical bundle subdomain) (Fig. 5 D and E). Specifically, the conserved helix α8 of Rpn6 aligns with helix α12 in Rpt6. Residues Thr234Rpn6, Ser227Rpn6, Tyr228Rpn6, and Glu231Rpn6 (the latter three mutated in Rpn6-M1, Fig. 3) at the groove between helices α8 and α10 of Rpn6 cradle the highly conserved C terminus of Rpt6 helix α12, extending the contact area. While Tyr228Rpn6 is placed for contacting the backbone at Rpt6 residue 370, the Arg371Rpt6 side chain is in hydrogen-bonding distance to both Ser227Rpn6 and Glu231Rpn6. Glu370Rpt6 could form hydrogen bonds with Lys192Rpn6 and Asn196Rpn6.
Interestingly, surface conservation on Rpn6 extends toward the lid beyond the observed contact area—for example, Ala186Rpn6 is extending the conserved edge of helix α8 continuing from Thr189Rpn6 and Thr193Rpn6. This conservation suggests that Rpn6 could accommodate different conformational states of the ATPase ring. “Wobbling” or “wagging” motions of the active ATPase ring relative to the CP have been proposed (30, 31), and ATP-dependent structural changes involve binding of ubiquitin conjugates (32). In addition, there is structural evidence for a wagging motion of the whole RP (30).
The tip of the PCI module of Rpn6 is part of a horseshoe structure with six radially projecting protrusions that are included in the lid density (Fig. 4A). The contact points within the horseshoe coincide with the conserved regions II and III at the flanks of the β-sheet in Rpn6 (Fig. 4B). Region III, which was implicated in direct interactions of Rpn6 with Rpn7 (Fig. 3D), is situated to the right (Fig. 4B), suggesting that this density corresponds to Rpn7. Furthermore, yeast-two hybrid assays indicate a physical interaction of Rpn6 and Rpn5 via their PCI modules (21), suggesting that the density to the left of Rpn6 represents Rpn5. It is thus tempting to speculate that the horseshoe represents the density for the six proteasome PCI subunits, arranged like the Rpn6 chains in the crystal (Fig. S4A). According to this hypothesis, the interacting winged-helix subdomains form the inner rim, and the N-terminally adjacent α-helical bundles and solenoids form the protrusions. At the current resolution, the density for the C-terminal helix of Rpn6 cannot be assigned with confidence.
Arrangement of the PCI Subunits in the RP.
Our mutational analysis showed that the winged-helix subdomain in the PCI module of Rpn6 is important for interactions with the PCI subunit Rpn7, consistent with the proposed function as a PCI:PCI interaction module (14). In addition to an intact winged-helix motif, the conserved C-terminal helix of Rpn6 is required for the interaction with Rpn7. This requirement for an additional contact might explain why we were not able to identify a second PCI binding partner of Rpn6. Rpn5, Rpn6, and Rpn9 form a subcomplex together with Rpn8 and Rpn11 (33), suggesting that one of the latter non-PCI subunits is required for the attachment of Rpn6 to Rpn5 in addition to the subunit II–III interface. The assembly pathway of the lid suggests that the PCI subunits Rpn3 and Rpn7 form a dimer, and PCI subunit Rpn12 attaches to the Rpn3/Rpn7 dimer after its binding to the Rpn6/Rpn5/Rpn8/Rpn9/Rpn11 pentamer (33). Thus, Rpn6 and Rpn7 followed by Rpn3 and Rpn12 could form the right end of the horseshoe, perhaps stabilized by coiled-coil interactions of their C-terminal helices; the interaction of Rpn6 with Rpn5 would require Rpn8 and Rpn11, resulting in the second subcomplex. The sequence of PCI subunits in the horseshoe structure would thus be (from the left) Rpn9-Rpn5-Rpn6-Rpn7-Rpn3-Rpn12 (Fig. S4B). Such a model is consistent with native MS analysis of the COP9/signalosome, in which each subunit of the lid subcomplex has a homolog (34). Thus, the lid and COP9/signalosome architectures might be evolutionarily conserved.
Conclusions
In our hybrid structure, Rpn6 contacts at least four subunits from three functional units of the proteasome, the lid, the ATPase, and the proteolytic core particle. Interactions with Rpn6 thus appear to reinforce the contacts between the lid and base and also between the regulatory and core particles. This Rpn6 role is consistent with increased occurrence of partially assembled proteasome particles in the temperature-sensitive rpn6-2 mutant of budding yeast (21). Interestingly, this mutant harbors mutations both at the interface to the alpha ring, F132L (residue F122 in S. pombe) and the lid subunits Rpn5 and Rpn7, L377P (residue L365 in D. melanogaster). The latter mutation would presumably interrupt helix α18, compromising the structural integrity of the winged-helix subdomain.
Because of the symmetry mismatch between the heptameric alpha ring and the hexameric AAA-ATPase, their contacts appear rather sparse and weak, thus enabling relative motions. Indeed, symmetry mismatches have often evolved to allow for motions of macromolecules during their functional cycle (35). Thus, Rpn6 appears to have a pivotal role in holding the complex together by acting as an additional clamp between RP and CP. Monomeric Rpn6 might also be functionally important. The reported interactions of Rpn6 with the ubiquitin ligase regulatory complex COP9/signalosome probably control its own degradation (19, 36), which might in turn regulate the assembly and activation of 26S proteasomes through the availability of monomeric Rpn6. Such a regulation is consistent with the critical role of Rpn6 for the integrity of the 26S proteasomes complex.
Materials and Methods
Detailed experimental procedures are given in SI Materials and Methods. Briefly, Rpn6 from D. melanogaster was expressed as a His6-tag fusion protein including a tobacco etch virus (TEV) protease site in R. erythropolis (L-88) cells and purified by Ni2+-immobilized metal affinity chromatography, TEV cleavage, Mono-Q anion exchange chromatography, and Superose-12 size-exclusion chromatography. Crystals were grown using 100 mM Tris-HCl pH 7.5 200 mM Li2SO4 and 12% PEG-3350 as a precipitant. The Rpn6 crystal structure was solved by multiwavelength anomalous dispersion using gadolinium(3+), using diffraction data acquired at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. The exact position and orientation of Rpn6 in the 9.1 Å electron density map of the 26S proteasome was determined by an exhaustive six-dimensional search procedure.
Supplementary Material
Acknowledgments.
We thank the staff of the Joint Structural Biology Group at the European Synchrotron Radiation Facility, Grenoble, France, of the Max-Planck-Institute of Biochemistry Crystallization Facility and the Max-Planck-Institute of Biochemistry Core Facility for their excellent support; and Johannes Söding for valuable discussions. Materials for the R. erythropolis expression system were kindly provided by Khalid Ibrahim Sallam and Noriko Tamura, Sapporo, Japan. This work was supported in part by funding from the European Union 7th Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648). F.F. is grateful to a Career Development Award from the Human Frontier Science Project. K.L. was supported by continuous mentorship from Prof. Haim J. Wolfson as well as a fellowship from the Clore Foundation Ph.D. Scholars program.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3TXM and 3TXN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117648108/-/DCSupplemental.
References
- 1.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 4.Förster F, Lasker K, Nickell S, Sali A, Baumeister W. Toward an integrated structural model of the 26S proteasome. Mol Cell Proteomics. 2010;9:1666–1677. doi: 10.1074/mcp.R000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K. The proteasome: Overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 7.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 8.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 9.Unno M, et al. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 10.Bohn S, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci USA. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 12.Kim T, Hofmann K, von Arnim AG, Chamovitz DA. PCI complexes: Pretty complex interactions in diverse signaling pathways. Trends Plant Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- 13.Dessau M, et al. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell. 2008;20:2815–2834. doi: 10.1105/tpc.107.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Z, et al. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 16.Saito A, et al. cDNA cloning and functional analysis of p44.5 and p55, two regulatory subunits of the 26S proteasome. Gene. 1997;203:241–250. doi: 10.1016/s0378-1119(97)00524-6. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Wang CC. Functional characterization of the 11 non-ATPase subunit proteins in the trypanosome 19 S proteasomal regulatory complex. J Biol Chem. 2002;277:42686–42693. doi: 10.1074/jbc.M207183200. [DOI] [PubMed] [Google Scholar]
- 18.Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci USA. 2011;108:4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lier S, Paululat A. The proteasome regulatory particle subunit Rpn6 is required for Drosophila development and interacts physically with signalosome subunit Alien/CSN2. Gene. 2002;298:109–119. doi: 10.1016/s0378-1119(02)00930-7. [DOI] [PubMed] [Google Scholar]
- 20.Santamaria PG, Finley D, Ballesta JP, Remacha M. Rpn6p, a proteasome subunit from Saccharomyces cerevisiae, is essential for the assembly and activity of the 26 S proteasome. J Biol Chem. 2003;278:6687–6695. doi: 10.1074/jbc.M209420200. [DOI] [PubMed] [Google Scholar]
- 21.Isono E, Saito N, Kamata N, Saeki Y, Toh EA. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima N, Tamura T. A novel system for expressing recombinant proteins over a wide temperature range from 4 to 35 degrees C. Biotechnol Bioeng. 2004;86:136–148. doi: 10.1002/bit.20024. [DOI] [PubMed] [Google Scholar]
- 23.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 24.Collins GA, Tansey WP. The proteasome: A utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Krogan NJ, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 26.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Förster F, et al. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 30.Walz J, et al. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 31.Saeki Y, Tanaka K. Unlocking the proteasome door. Mol Cell. 2007;27:865–867. doi: 10.1016/j.molcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 34.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Zwickl P, Baumeister W, Steven A. Dis-assembly lines: The proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]
- 36.Kwok SF, Staub JM, Deng XW. Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol. 1999;285:85–95. doi: 10.1006/jmbi.1998.2315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.