Abstract
Plant metabolic engineering is commonly used in the production of functional foods and quality trait improvement. However, to date, computational model-based approaches have only been scarcely used in this important endeavor, in marked contrast to their prominent success in microbial metabolic engineering. In this study we present a computational pipeline for the reconstruction of fully compartmentalized tissue-specific models of Arabidopsis thaliana on a genome scale. This reconstruction involves automatic extraction of known biochemical reactions in Arabidopsis for both primary and secondary metabolism, automatic gap-filling, and the implementation of methods for determining subcellular localization and tissue assignment of enzymes. The reconstructed tissue models are amenable for constraint-based modeling analysis, and significantly extend upon previous model reconstructions. A set of computational validations (i.e., cross-validation tests, simulations of known metabolic functionalities) and experimental validations (comparison with experimental metabolomics datasets under various compartments and tissues) strongly testify to the predictive ability of the models. The utility of the derived models was demonstrated in the prediction of measured fluxes in metabolically engineered seed strains and the design of genetic manipulations that are expected to increase vitamin E content, a significant nutrient for human health. Overall, the reconstructed tissue models are expected to lay down the foundations for computational-based rational design of plant metabolic engineering. The reconstructed compartmentalized Arabidopsis tissue models are MIRIAM-compliant and are available upon request.
Current challenges in using plants as factories for bio-energy and nutraceuticals require predesigned and efficient strategies for metabolic engineering (1). Currently, plant metabolic engineering mostly involves trial-and-error approaches, without the utilization of computational modeling procedures to rationally design genetic modifications. The marginal contribution played by metabolic modeling in plants until now stands in marked contrast to its prominent success in microbial metabolic engineering (2, 3). Metabolic network reconstructions were manually reconstructed for dozens of bacterial species (3), and automated approaches recently generated draft models for a total of 130 bacteria (4). A modeling approach, called constraint-based modeling (CBM), serves to analyze the function of such large-scale metabolic networks by solely relying on simple physical-chemical constraints, overcoming the problem of missing enzyme kinetic data (3). Applications of CBM for large-scale microbial networks has proven to be highly successful in predicting metabolic phenotypes in metabolic engineering and many other applications (3).
The reconstruction of metabolic network models for multicellular eukaryotes is significantly more challenging than that for bacteria, because of the larger size of the networks, the subcellular compartmentalization of metabolic processes, and the considerable variation in tissue-specific metabolic activity. The reconstruction of plant metabolic networks is further complicated by the metabolome size and high-level complexity of metabolism because of extensive secondary metabolism. A significant step forward in plant metabolic modeling has been made with the publication of large-scale Arabidopsis network models (5, 6), which relied on genomic and biochemical data extracted from the literature. Although both models were shown to carefully reproduce several experimental metabolic phenotypes, their scope is limited: (i) Both networks do not account for tissue-specific metabolism, but rather piece together the entire set of metabolic reactions that can take place within different tissues. (ii) Both models focus only on primary metabolism and, hence, cannot be used for metabolic engineering of many target metabolites of interest that are mostly present in secondary metabolism. (iii) Both models include very partial experimental localization data that is limited only for central metabolism.
In this work we present a computational pipeline for the reconstruction of genome-scale, subcellular compartmentalized metabolic network models for multiple Arabidopsis tissues and cell cultures. The validity of the model reconstruction steps is demonstrated via computational tests (cross-validation tests and simulations of known metabolic functionalities), and via comparison with experimental datasets (i.e., metabolomics and flux measurements under various compartments and tissues). The utility of the derived models was demonstrated in the prediction of measured fluxes in metabolically engineered seed strains. It is further demonstrated how the model can be used to computationally design metabolic engineering strategies for vitamin E overproduction, a significant nutrient for human health. The reconstructed Arabidopsis models are MIRIAM-compliant (7) (Dataset S1), are available in Datasets S2, S3, and S4, and upon request in a standard System Biology Markup Language format.
Results and Discussion
Reconstruction of Genome-Scale, Subcellular Compartmentalized Metabolic Network Models for Arabidopsis Tissues.
The reconstruction of the tissue-specific Arabidopsis metabolic models involves a computational pipeline consisting of three major steps (Fig. 1): (i) Global model reconstruction: involving the assembly of known Arabidopsis reactions from various databases and automatic gap-filling. (ii) Compartmentalized model reconstruction: involving the integration of the global model derived in the previous step with various experimental data sources for enzyme subcellular localization. (iii) Tissue-specific model reconstruction: involving the integration of the compartmentalized model derived in the previous step with tissue-specific protein-expression data. Below, each reconstruction step is further elaborated; the full details are provided in SI Text.
Fig. 1.
Arabidopsis model reconstruction computational pipeline. Stage 1: Global model reconstruction. Assembly of known Arabidopsis reactions followed by automatic gap-filling. Stage 2: Compartmentalized model reconstruction. Integration of the global model with various experimental localization data sources and utilization of network-based localization prediction method. Stage 3: Tissue-specific models reconstruction. Integration of the compartmentalized model with tissue-specific protein-expression data to create a separate model for 10 different tissues.
Step 1: Global model reconstruction.
Data on traditional metabolic pathways, including the participating metabolites and reactions, stoichiometric coefficients, and gene-to-reaction mapping was extracted from AraCyc (8) and KEGG-Arabidopsis (Kyoto Encyclopedia of Genes and Genomes) databases (9). All reactions were checked for correct atom balancing (using the method described in ref. 10) for proton and oxygen balancing. An extensive literature search was performed to identify 71 metabolites that are transported across the extracellular membrane to define the model boundaries (Table S1). Additional data on key cellular biomass constituents was extracted from the literature (Table S1). Data on reaction directionality was extracted from KEGG, complemented by a set of heuristic biochemical rules (11). The above datasets were used to compile a draft network model consisting of 1,363 unique reactions and 1,078 metabolites.
To address the problem of dead-end reactions, we implemented a gap-filling algorithm that, in accordance with previous computational approaches (12), aims to bridge network gaps by searching for a minimal set of reaction directionality relaxations and reaction additions (preferably from other plants), such that the core set of reactions in the draft model could be activated (i.e., carry nonzero metabolic flux under a stoichiometric steady-state assumption) (SI Text) (13). The application of the gap-filling algorithm resulted in 191 reaction directionality relaxations and the addition of 31 known plant reactions, as well as 223 reactions not known previously to occur in plants (Datasets S2 and S4). To assess the performance of the gap-filling approach, we applied a standard cross-validation test, repeating the gap-filling procedure given four-fifths of the known Arabidopsis reactions, aiming to predict the missing one-fifth. The resulting enrichment of the predicted missing reactions with known Arabidopsis reactions is highly significant (hyper-geometric P value < 10−14), with a precision of 0.61 and recall of 0.42 (Table S2).
To further validate the actual existence in Arabidopsis of the reactions identified in the gap-filling procedure, we performed a BLAST search of the known enzyme sequences catalyzing these reactions in other organisms against the Arabidopsis genome (Dataset S2). Reassuringly, we found that the resulting BLAST E-scores show significantly higher sequence similarity for the set of identified gap-filling reactions in comparison with a random set of reactions from the same source (Wilcoxon P value < 0.05), testifying for the ability of the gap-filling approach to correctly identify missing reactions in Arabidopsis. As an example, the prediction of the gap-filling method enabled the activation of the cyanide metabolism, which is known to function in Arabidopsis (14). Specifically, the mandelonitrile lyase reaction (transforming 4-hydroxymandelonitrile to hydrogen cyanide) and its downstream reactions are blocked in the draft network, as a pathway for synthesizing 4-hydroxymandelonitrile is currently unknown in Arabidopsis. Our gap-filling analysis suggested it may be synthesized from the aldoxime product (15), p-hydroxyphenylacetaldoxime, via the newly added reaction catalyzed by 4-hydroxyphenylacetaldehyde oxime monooxygenase (a cytochrome P450-type enzyme), which is known to participate in dhurrin biosynthesis in the Sorghum bicolor plant. A Blast search of the known enzyme-coding gene from Sorghum against the Arabidopsis genome identified several Arabidopsis genes with high E-score similarity, all belonging to the cytochrome P450 family.
Following previous model reconstructions (13, 16), we further validated the global model reconstruction using a set of 176 simulations of known metabolic functions, such as amino acids, as well as secondary metabolites biosynthesis and degradation (SI Text and Table S3). Although 98% of the simulations were successful, demonstrating that this model is indeed highly functional, the ones that failed were used to manually identify and add several missing reactions.
Step 2: Compartmentalized model reconstruction.
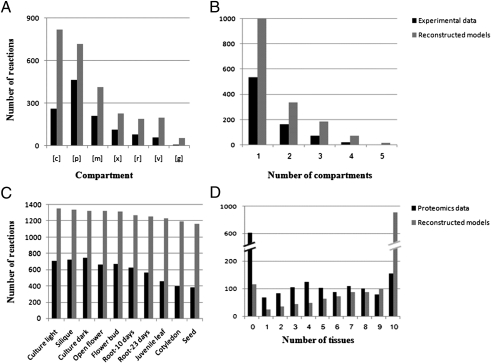
To extend the genome-scale Arabidopsis model reconstructed in the above step to account for subcellular compartmentalization of metabolic processes, we used a variant of our previously developed pertaining method (17). Given the known localization of a subset of the enzymes in the network, the method predicts the most likely localization of the remaining enzymes based on a parsimony principle of minimizing the number of metabolite transmembrane transport required to activate the enzymes with a known localization in the corresponding compartments. As input, we considered the known localization of 49% of the reactions taken from the SUBA database (the Arabidopsis Subcellular Database) (18) (Dataset S4), aiming to predict the localization of the remaining reactions (SI Text). The analysis resulted in a compartmentalized model (Table 1), with each enzyme localized to the cytosol, plastid, mitochondrion, endoplasmic reticulum (ER), peroxisome, vacuole, and golgi-apparatus (Fig. 2A). A detailed list of the predicted intracellular transporters is provided in Table S4. A high percentage of the reactions are predicted to be localized in multiple compartments, in accordance with the experimental data used for the model reconstruction, with 21%, 11%, and 4% of the reactions localized to two, three, or a higher number of compartments, respectively (Fig. 2B). To assess the performance of the compartmentalized model reconstruction step, we used a cross-validation test (SI Text). The resulting prediction accuracy was significantly high (hyper-geometric P value < 10−13), with a precision of 0.47 and recall of 0.50 (Table S2). To further validate predictions of enzyme subcellular localizations, we used recently published metabolomic data by Krueger et al. (19), which describes subcellular distribution of dozen of compounds from leaves of Arabidopsis. Comparing the measured metabolites in leaves to those computationally predicted to belong to the leaves model showed a significantly high prediction accuracy (hyper-geometric P value = 0.03), with a precision of 0.62 and recall of 0.72.
Table 1.
Statistics on the reconstructed metabolic network models
| Model | Arabidopsis reactions | Gene-reaction associations | Arabidopsis metabolites | Compartmentalized reactions | Intracellular transporters |
| Compartmentalized model | 1,363 | 1,065 | 1,078 | 1,194 | 772 |
| Seed model | 989 | 798 | 872 | 948 | 595 |
| Silique model | 1,143 | 934 | 1,037 | 1,057 | 655 |
Here we focus on the smallest (seed) and largest (silique) reconstructed tissue models.
Fig. 2.
Compartmentalized (A and B) and tissue-specific (C and D) model properties. (A) Reactions’ distribution in the compartments, based on experimental localization data (black) and on the reconstructed compartmentalized model (gray). c, cytoplasm; p, plastid; m, mitochondrion; x, peroxisome; r, ER; v, vacuole; g, Golgi-apparatus. (B) Multiple localizations’ distribution, based on experimental localization data (black) and on the reconstructed compartmentalized model (gray). (C) Reactions’ distribution in the tissues, based on tissue-specific proteomics data (black) and on the reconstructed tissue models (gray). (D) Multiple tissue assignment distribution, based on tissue-specific proteomics data (black) and on the reconstructed tissue models (gray).
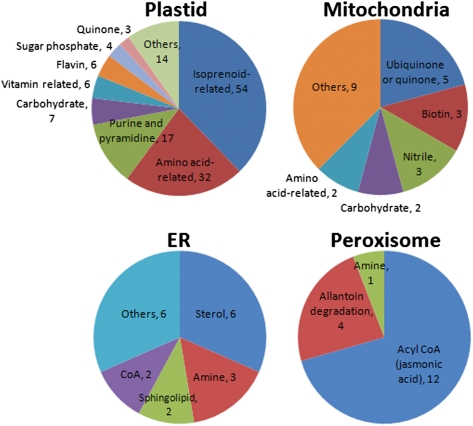
We subsequently used the metabolic network model to examine classes of metabolites that are uniquely assigned to a particular compartment (Fig. 3). The relatively large size of the plant metabolome and extensive transport of metabolites between compartments makes such analysis at the network-scale of particular interest. A large portion of the unique metabolites detected in the plastids belong to the isoprenoids pathway including chlorophylls, carotenoids, diterpene, geranylgeranyl diphosphate, vitamin E, and cytokinines. Other known plastidic metabolites, uniquely localized in our model, include amino acid-related compounds, and thiamine (vitamin B1) (20). Myo-inositol and nonstandard amino acids, such as homoserine and glutamylcysteine, were predicted to be also uniquely present in the plastids. In the mitochondria, quinone and ubiquinones (21), biotin-related compounds (22), and nitriles were the most dominant, as expected, and in the ER most metabolites were fatty acid-related, including sterols, sphingolipids, and CoAs (23). In the peroxisome, CoA metabolites were prominent, particularly those associated with jasmonic acid biosynthesis (24). Another interesting localization is that of allantoin catabolism intermediates, allantoate, ureidoglycine, and ureidoglycolate, in peroxisome, in accordance with previous findings in other plant species (25).
Fig. 3.
Subcellular localization of classes of metabolites (and their size represented by the proceeding numbers) predicted to be localized in a single compartment.
Step 3: Tissue-specific model reconstruction.
To reconstruct metabolic network models for specific Arabidopsis tissues, the compartmentalized model was integrated with tissue-specific protein expression data (26) from juvenile leaves, open flowers, flower buds, 10-d roots, 23-d roots, siliques, seeds, and cotyledons, as well as cell cultures grown in light and in dark, using our previously developed method (13) (SI Text). Briefly, for each tissue, a core set of reactions was determined using the tissue-specific proteomics data (Dataset S4). Then, a search for a minimal set of reactions from the compartmentalized model was performed, such that the core set of tissue-specific reactions could be activated. The proteomic data provided tissue-localization data for 63% of the reactions in the model. Using this data as input for the tissue-model reconstructions enabled the prediction of tissue assignment of 92% of the reactions in the resulting tissue models (Fig. 2D) (i.e., obtaining a reassuringly high reaction coverage). As much as 942 reactions are predicted to participate in all reconstructed tissue models, including reactions in primary metabolic pathways, such as glycolysis, pentose-phosphate pathway, fatty acid metabolism, nucleotide metabolism, and amino acid metabolism. The reconstructed metabolic network model for a cell culture grown in light consists of the highest number of reactions, followed by the network models of silique, cell culture grown in dark conditions, and flower. The reconstructed network model of the seed is the smallest, reflecting the more specialized metabolic nature of this tissue (Fig. 2C and Table 1).
To assess the performance of this reconstruction step, we first applied a cross-validation test for all tissues (SI Text), obtaining significantly high accuracy results, with high precision/recall values (Table S2). As an additional validation to tissue models, we used gene-expression data measured in the same Arabidopsis tissues (for which the tissue models were reconstructed) (27). Focusing on proteins that were not detected via proteomics in various tissues, we found that the expression level of their coding genes is significantly higher, specifically in tissues in which they are predicted to function (based on the reconstructed tissue models), compared with their expression level in other tissues (Wilcoxon signed rank test P value < 10−4). This finding directly testifies for the predictive performance of the tissue-model reconstruction approach, and hence for the plausibility of the reconstructed tissue models.
To further validate the prediction of metabolite distribution in the different tissues, we performed a targeted metabolomics analysis for three isoprenoid compounds, chlorophyll b, lutein, and violaxanthin, across seven tissues (leaf, cotyledon, open flower, flower bud, 10-d root, seed, and silique) (Fig. 4). We found that all three metabolites are detectable in all measured tissues (other than root) in accordance with the reconstructed models for these tissues, which include these metabolites in all tissue models other than the one for root (with the exception of lutein in silique and violaxanthin in seed). In marked contrast, using strictly the proteomic data (used as input to reconstruct the tissue-models) to infer metabolite presence in different tissues, resulted in a significantly lower match to the metabolomics data, wrongly predicting that chlorophyll b and lutein would not be present in many other tissues. These results highlight the added value of our tissue models, demonstrating how the integration of proteomic data from various tissues with a metabolic network models enable to lead to improved prediction accuracy of metabolic behavior.
Fig. 4.
Experimental support for predicted tissue specificity of three isoprenoids metabolites. The graphs shows the results of HPLC-targeted metabolomics across various Arabidopsis tissues, and the tables denote the presence of the metabolites in the different tissues, as inferred strictly from proteomic data (i.e., based on the presence of substrate using enzymes), and based on the presence of these metabolites in the reconstructed tissue models.
Global View of the Models Generated for Different Arabidopsis Tissues.
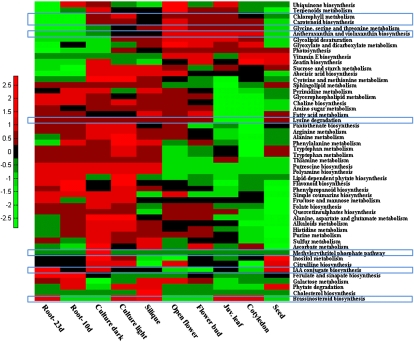
Examination of pathway activity profiles in different tissues demonstrated known tissue-specific metabolic profiles, as well as highlighting new, yet unknown profiles (Fig. 5). For example, the biosynthesis of the carotenoids antheraxanthin and violaxanthin through the xanthophyll cycle is known to be activated under excessive light conditions, as part of a photo-protection mechanism (28), and accordingly showed higher representation in the “light” tissues and cell cultures. Our models suggests that the methylerythritol 4-phosphate (MEP) pathway, which generates the precursors for multiple isoprenoids including the carotenoids (29), is controlled differently than the downstream carotenoid biosynthesis because it is showing constant representation in all tissues and a higher representation in the dark culture model. This finding is in accordance with the recently reported activation of the general MEP pathway in “dark”-grown Arabidopsis seedlings (30). Carotenoid biosynthesis, on the other hand, is highly represented in the light tissues as well as in the dark-culture model (31). In a different example, chlorophyll metabolism is mostly underrepresented in roots, a tissue that typically lacks chlorophyll. The lysine degradation pathway showed an interesting metabolic profile, because it is more represented in mature vs. younger tissues, in accordance with the recently reported role of lysine in senescence metabolism (32). Indole-3-acetic acid (IAA) biosynthesis is enriched in the root tissue model, in accordance with the observation that aerial tissues are not the only source of the IAA, with primary root tip showing high IAA synthesis capacity (33). Brassinosteroids were proposed to be synthesized most actively in young developing organs, especially in silique and apical shoots (34). In accordance with these findings, and although indicated to exist only in siliques based on the proteomics data, highest brassinosteroid biosynthesis representation in the models is seen for the silique model, followed by the young tissues of cotyledon and juvenile leaf models, as well as the root-tissue model. Finally, by clustering the different tissues according to pathway activity profiles, a separation is observed between the dark tissues and cultures from the light tissues and culture, a separation that is not evident in the proteomics data-based clustering (SI Text and Fig. S1).
Fig. 5.
The pathway-content of the reconstructed metabolic network models for the various tissues and cell-cultures. Red (green) entries represent pathways that consist of a high (low) number of reactions in a corresponding tissue; that is, above (below) the mean number of pathway reactions across all tissues. Pathways discussed in the text are highlighted via blue squares.
Validating the Tissue Model's Ability to Correctly Predict Flux Measurements.
To validate our model's ability to correctly predict metabolic flux reroutes following genetic modifications, which is essential for metabolic engineering, we used recently measured flux data (through 13C labeling) in culture-developing embryos following the knockdown of the plastidic pyruvate kinase (PK) (35). The knockdown of PK (converting phosphoenolpyruvate into pyruvate), was shown to markedly effect the formation of fatty-acids while increasing starch accumulation. To predict metabolic changes following the PK knockdown, we used the reconstructed seed model, as previous studies have shown similar metabolic behavior between seeds and culture developing embryos (35). First, the WT flux distribution was predicted by integrating the measured fluxes with the seed model through a standard optimization approach (Materials and Methods). Then, the effect of the PK knockdown was predicted via two computational approaches: minimization of metabolic adjustment (MOMA) (36) and regulatory on/off minimization (ROOM) (37), the potential use of which in metabolic engineering in plants was recently highlighted (38). With both methods, the predicted fluxes following the knockdown were found to be significantly correlated with the measured flux following the knockdown, with a Spearman correlation of 0.57 (P value = 0.006) for MOMA and 0.52 (P value = 0.005) for ROOM (Table S5). Specifically, a decrease in flux was predicted for the initial plastidic and cytosolic reaction of fatty acid biosynthesis, in accordance with experimental measurements.
Discussion
In this work we present the reconstruction of fully compartmentalized tissue-specific models of Arabidopsis thaliana, a model plant belonging to the Brassicaceae family. This family comprises many plants important to industry and nutrition. Despite monocot-dicot evolutionary divergence, the genetic make-up of the Arabidopsis model plant contains similarities to the genetic make-up of cereals (39), making its study highly useful to closely related plants, as well as to cereal researchers, where metabolic engineering is commonly used for quality-trait improvement. To demonstrate the potential utility of the resulting models in the prediction of metabolic engineering strategies, we use it to computationally predict genetic manipulations that are expected to increase vitamin E content, a significant nutrient for human health. Specifically, we applied computational methods for predicting 71 target enzymes, the knockout of which should increase vitamin E accumulation, some of which were already found to be connected to vitamin E biosynthesis in an intricate manner (SI Text and Table S6). It should be noted, however, that further experimental validation should be performed to reaffirm these predictions.
The described model reconstruction pipeline allows an efficient, semiautomatic model construction via integration of different “omics” data, and is generic and fast, and capable of processing a large variety of data sources. However, several limitations should be noted: (i) As the approach hinges upon different molecular data, its accuracy depends on the quality of the latter. (ii) Several assumptions are used in the reconstruction process, such as the assumption of minimal addition of reactions and directionality relaxation to resolve network gaps, and the assumption of minimal number of metabolite transmembrane transport used in the localization assignment. These assumptions, although commonly used for similar purposes (12, 17), may in some cases be oversimplified and lead to false identification of gap-filling solutions. (iii) The resulting models do not explicitly account for transcriptional and metabolic regulation, data that is still unavailable for most Arabidopsis metabolic functions, and thus cannot accurately predict some of the organism's functions.
A number of future refinements and extensions to the computational pipeline presented here may improve the quality of the resulting network models. First, the assignment of reactions’ directionality can be improved via complete thermodynamic analysis (40), which is significantly more complicated in Eukaryote than in bacteria because of the presence of subcellular compartments with varying pH levels, ionic strength, and membrane potentials, the exact quantification of which is difficult. Performing a more limited analysis, testing directionality feasibility of the model's reactions (following ref. 41), has found that the activation of 93% of the reactions is indeed feasible, even after elimination of futile cycles (the rest were annotated in Dataset S2, accordingly). Second, newly developed metabolomic analysis platforms are now beginning to allow steady-state and flux profiling of previously nondetected compounds in different compartments and tissues (42) that will improve consistency and accuracy of the reconstructed models. Third, we demonstrated flux predictions in WT vs. mutant strains using the minimization of metabolic adjustment objective function. Further work is also required to improve the prediction of fluxes through the use of alternative objective functions that may be better suited for specific plant tissues (43).
The computational pipeline described herein can be straightforwardly used to reconstruct updated versions of the Arabidopsis models when new data becomes available, as well as tissue models for a variety of other plants that would significantly expand our understanding of plant metabolism in a tissue-specific manner. Such reconstructed tissue models, in addition to those presented here for Arabidopsis, open up future opportunities for metabolic engineering of increased production rates of various nutraceuticals important for human health.
Materials and Methods
Predicting Flux Measurements.
The reconstructed seed model was applied to predict changes in metabolic flux rates between WT and a PK knockdown mutant strain (pkpβ1pkpa). The predictions were compared with a set of flux measurements, including four exchange fluxes (uptake/secretion of metabolites) and 49 inner fluxes (35). Quadratic programming was applied to first find a feasible flux distribution, satisfying stoichiometric mass-balance and reaction directionality constraints, that is as close as possible to the measured WT fluxes. Two computational approaches were used: the MOMA method (36), which minimizes the Euclidean norm of the flux differences, and the ROOM (37) method, which minimizes the total number of significant flux changes. Then, effect of the knockdown was predicted by constraining the flux through PK reaction, and using each minimization method again to search for a feasible flux distribution that undergoes a minimal redistribution with respect to the flux configuration of the WT. Spearman correlation was computed between the log fluxes predicted by the seed model following the knockdown, with the measured changes in flux.
Experimental Validation of Predicted Isoprenid Tissue Distribution.
A. thaliana plants (ecotype Col-0) were grown on soil in (22 °C; 70% humidity) with 18/6 h of light/dark cycles in growth rooms. Leaves, cotyledons, flower buds, open flowers, seeds, siliques, and roots tissues were analyzed. Apart from dry seeds (50 mg), 100 mg of each tissue were collected in three biological replicates, according to Baerenfaller et al. (26). Isoprenoid extraction and analysis was performed using HPLC coupled to photodiode array, detector, as previously described (44).
Supplementary Material
Acknowledgments
We thank Nathan Lewis, Gad Galili, Arren Bar-Even, Keren Yizhak, and Raphy Zarecki for their help and fruitful comments. Work in the A.A. laboratory was supported by the Minerva Foundation and the European Research Council project Systems Analysis of Plant Metabolism through the Integration of Heterogeneous Data from Genetics, Informatics and Metabolomics (FP7 program). T.S. is supported by a grant from the Israel Science Foundation; A.A, E.R, and T.S are supported by a joint grant from the Ministry of Science and Technologies for an integrated study of plant metabolism; and S.M.-O. is supported by the Ministry of Science and Technology. A.A. is the incumbent of the Adolpho and Evelyn Blum Career Development Chair of Cancer Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100358109/-/DCSupplemental.
References
- 1.Aharoni A, Jongsma MA, Bouwmeester HJ. Volatile science? Metabolic engineering of terpenoids in plants. Trends Plant Sci. 2005;10:594–602. doi: 10.1016/j.tplants.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Burgard AP, Pharkya P, Maranas CD. Optknock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 3.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 5.Poolman MG, Miguet L, Sweetlove LJ, Fell DA. A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 2009;151:1570–1581. doi: 10.1104/pp.109.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira Dal'Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010;152:579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Novère N, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 8.Mueller LA, Zhang P, Rhee SY. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma H, Zeng AP. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- 12.Orth JD, Palsson BO. Systematizing the generation of missing metabolic knowledge. Biotechnol Bioeng. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: Application to human liver metabolism. Mol Syst Biol. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzfeld Y, et al. beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kliebenstein DJ, Rowe HC, Denby KJ. Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 2005;44:25–36. doi: 10.1111/j.1365-313X.2005.02508.x. [DOI] [PubMed] [Google Scholar]
- 16.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mintz-Oron S, Aharoni A, Ruppin E, Shlomi T. Network-based prediction of metabolic enzymes’ subcellular localization. Bioinformatics. 2009;25:i247–i252. doi: 10.1093/bioinformatics/btp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35(Database issue):D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger S, et al. A topological map of the compartmentalized Arabidopsis thaliana leaf metabolome. PLoS ONE. 2011;6:e17806. doi: 10.1371/journal.pone.0017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lunn JE. Compartmentation in plant metabolism. J Exp Bot. 2007;58:35–47. doi: 10.1093/jxb/erl134. [DOI] [PubMed] [Google Scholar]
- 21.Crane FL. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion. 2007;7(Suppl):S2–S7. doi: 10.1016/j.mito.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Picciocchi A, Douce R, Alban C. The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory protein components. J Biol Chem. 2003;278:24966–24975. doi: 10.1074/jbc.M302154200. [DOI] [PubMed] [Google Scholar]
- 23.Moreau P, et al. The plant ER-Golgi interface: A highly structured and dynamic membrane complex. J Exp Bot. 2007;58:49–64. doi: 10.1093/jxb/erl135. [DOI] [PubMed] [Google Scholar]
- 24.Reumann S, Ma C, Lemke S, Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells XE, Lees EM. Ureidoglycolate amidohydrolase from developing French bean fruits (Phaseolus vulgaris [L.].) Arch Biochem Biophys. 1991;287:151–159. doi: 10.1016/0003-9861(91)90400-d. [DOI] [PubMed] [Google Scholar]
- 26.Baerenfaller K, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 27.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 28.Saga G, et al. Mutation analysis of violaxanthin de-epoxidase identifies substrate-binding sites and residues involved in catalysis. J Biol Chem. 2010;285:23763–23770. doi: 10.1074/jbc.M110.115097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips MA, León P, Boronat A, Rodríguez-Concepción M. The plastidial MEP pathway: Unified nomenclature and resources. Trends Plant Sci. 2008;13:619–623. doi: 10.1016/j.tplants.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009;60:424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Colors in the dark: A model for the regulation of carotenoid biosynthesis in etioplasts. Plant Signal Behav. 2009;4:965–967. doi: 10.4161/psb.4.10.9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepansky A, et al. Lysine catabolism, an effective versatile regulator of lysine level in plants. Amino Acids. 2006;30:121–125. doi: 10.1007/s00726-005-0246-1. [DOI] [PubMed] [Google Scholar]
- 33.Ljung K, et al. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada Y, et al. Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 2003;131:287–297. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonien J, Schwender J. Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 2009;151:1617–1634. doi: 10.1104/pp.109.144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci USA. 2005;102:7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libourel IG, Shachar-Hill Y. Metabolic flux analysis in plants: From intelligent design to rational engineering. Annu Rev Plant Biol. 2008;59:625–650. doi: 10.1146/annurev.arplant.58.032806.103822. [DOI] [PubMed] [Google Scholar]
- 39.Ewing R, Poirot O, Claverie JM. Comparative analysis of the Arabidopsis and rice expressed sequence tag (EST) sets. In Silico Biol. 1999-2000;1:197–213. [PubMed] [Google Scholar]
- 40.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys J. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schellenberger J, Lewis NE, Palsson BO. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys J. 2011;100:544–553. doi: 10.1016/j.bpj.2010.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldberg L, Venger I, Malitsky S, Rogachev I, Aharoni A. Dual labeling of metabolites for metabolome analysis (DLEMMA): A new approach for the identification and relative quantification of metabolites by means of dual isotope labeling and liquid chromatography-mass spectrometry. Anal Chem. 2009;81:9257–9266. doi: 10.1021/ac901495a. [DOI] [PubMed] [Google Scholar]
- 43.Feist AM, Palsson BO. The biomass objective function. Curr Opin Microbiol. 2010;13:344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adato A, et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 2009;5:e1000777. doi: 10.1371/journal.pgen.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.