Abstract
For much of the time since their discovery, the sirtuin family of deacetylase enzymes has been associated with extensions in lifespan. This longevity promoting capacity in numerous model systems has enabled the sirtuins to gain `celebrity status' in the field of aging research. However, the mechanisms underpinning these changes remain incompletely defined. A general phenotype long associated with aging is the dysregulation of biological systems, which partly occurs via the accumulation of damage over time. One of the major sources of this damage is oxidative stress, which can harm both biological structures, and the mechanisms with which they are repaired. It is now becoming clear that the beneficial lifespan effects of sirtuins, along with many of their other functions, are closely linked to their ability to regulate systems that control the redox environment. Here we investigate the links between sirtuins and their oxidative/redox environment, and review the control mechanisms which are regulated by the activity of sirtuin deacetylase proteins.
Keywords: SIRT1, SIRT3, sirtuins, mitochondria, redox stress, resveratrol, metabolism
Introduction to sirtuin biology
The free radical hypothesis governing the pathophysiology of disease advocates that excess oxidative stress, and/or the diminished capacity to control reactive species, plays a role in disease initiation and progression [1–4]. In parallel, members of the NAD-dependent sirtuin deacetylase family are increasingly recognized as pivotal mediators in the regulation of nuclear, cytosolic and mitochondrial quality control programs – mechanisms which enhance cellular homeostasis and diminish degenerative disease susceptibility. Taken together, these concepts suggest that the control of redox stress, and the biological effects of sirtuin-dependent protein deacetylation, may be intertwined. To date, the majority of studies have suggested caloric-dependent factors in the control of sirtuin biology (recently reviewed [5, 6]). However, evidence is emerging that several sirtuin isoforms are activated during redox stress and may modulate crucial responses, including the adaptation to hypoxia and ameliorating reactive oxygen species-induced pathologies.
As background, the Sir2 (silence information regulator 2) or sirtuin family of class III deacetylases differ from class I and II histone deacetylases (HDACs) by their protein sequences and in that they are NAD+- rather than Zn-dependent enzymes [7]. Seven mammalian homologues of Sir2 have been identified, and are designated as SIRT1 through SIRT7 [8]. SIRT1, SIRT2 and SIRT3 are classified as subclass I enzymes, which show closest homology to yeast Sir2, and exhibit the most robust deacetylase activity. SIRT4 and SIRT5 are assigned to subclasses II and III, respectively; while SIRT6 and SIRT7 are classified as subclass IV enzymes [8]. Sirtuins appear to also catalyze ADP-ribosyl transferase activity, although this biology has been less well studied [9]. There is also recent work to suggest that SIRT5 functions as a `desuccinylase' and/or a `demalonylase' rather than a deacetylase, which may explain a lack of functional targets for SIRT5 compared to SIRT3 [10]. The mammalian sirtuins are found in distinct subcellular compartments, and emerging evidence shows that they regulate specific biological functions, via the deacetylation of target proteins, within these restricted subcellular locations. The biological effects of sirtuin-mediated lysine residue deacetylation include the direct modulation of transcription, and substrate-specific diverse effects on cell growth, aging, stress-tolerance and metabolism [11, 12]. This review will focus on how redox signaling controls sirtuin abundance and activity and, conversely, how sirtuins regulate the response to changes in redox potential and oxygen concentration through substrate-specific deacetylation events.
Nuclear and Cytosolic Sirtuins
SIRT1 is the most widely characterized protein in the sirtuin family, and as such a great deal of information has been amassed regarding its regulation over the last decade. The potential role of SIRT1 in a number of vital physiological pathways has made it a prime target for pharmacological intervention (see [13, 14]). One major area of SIRT1 research has been its role in caloric restriction (CR), which has long been known to increase lifespan in a variety of organisms. CR was first shown to have an effect on sirtuin activity in yeast and worms, where loss or gain of the SIRT1 homologue led to a reduction or increase, respectively, in lifespan [15, 16]. These effects were mediated in a large part by the availability of the sirtuin activator, NAD+, which was increased during CR and is required for the function of the sirtuin protein [17]. Later work demonstrated that CR may affect sirtuin function through several NAD+- linked redox pathways in yeast: CR may increase nicotinamide levels, a specific inhibitor of SIRTs [18]; or reduce the levels of cellular NADH, which acts as a competitive inhibitor of sirtuin activity [19]. The extent to which NAD+ is involved in the overall control of SIRT function in mammals is, however, less well established. Finally, CR has been shown to increase the protein expression of a number of SIRTs, including SIRT1, SIRT3 and SIRT5 [20–22], which may further enhance their overall deacetylase activity.
The ability of SIRT1 homologues to increase lifespan in various organisms led to the search for novel pharmacological activators of sirtuins. One of the best characterized is resveratrol, a weak polyphenolic antioxidant found in red wine. Resveratrol improves the enzyme kinetics of SIRT1 by increasing its ability to bind and deacetylate substrates, such as p53, in an NAD+ [13]. Addition of resveratrol to yeast activated the SIRT1 homologue, Sir2, leading to increased DNA stability and lengthening lifespan by 70% [13]. Further work by the same group extended these findings to metazoans, showing that resveratrol increased lifespan in fruit flies and worms by 20–30% in a sirtuin manner [23]. More recent studies have identified a group of small-molecule activators of SIRT1, which have a 1000-fold increase in potency compared to resveratrol. Treatment of mice and rat diabetes-model animals with these compounds, which include SRT1720, SRT1460 and SRT2183, led to an improvement in overall metabolic function and a reduction in diabetic progression [14]. While the authors related these results to an upregulation of mitochondrial function, based on the ability of SIRT1 to deacetylate and activate PGC-1α (see below; [14]), there has been some controversy regarding whether these compounds are in fact direct activators of SIRT1, and therefore what mechanism is behind this improvement [24]. While the role of SIRT1 in increasing lifespan in higher animals remains an open question [25], it would appear from knockout and over-expression studies (see below) that SIRT1 may be involved in ameliorating some of the phenotypes linked to aging diseases, such as diabetes, obesity and cancer (reviewed in detail here:[26]).
SIRT2, SIRT6 and SIRT7 are the other sirtuins that reside in the nucleus and cytosol [11]. SIRT2 is the only isoform predominantly localized in cytoplasm, although it does transiently reside in the nucleus during phases of mitosis [27]. Multiple target proteins that are deacetylated by SIRT2 have been characterized [12]. SIRT6 appears to possess nuclear ADP-ribosyltransferase activity, and it functions as a classical histone deacetylase [28, 29]. SIRT7 associates with the nucleolus, and only a limited number of substrates for this deacetylase have been identified [11]. The biology relevant to redox and hypoxic stress associated with these sirtuins will be discussed later in this review.
The role of redox signaling and oxygen in the regulation of SIRT1
In contrast to the emerging characterization of substrates responsive to SIRT1 deacetylation, our knowledge pertaining to the molecular control of SIRT1 itself, and especially in response to redox stress and differing oxygen levels, is less well established. The gene transcript encoding SIRT1 is induced in response to mild oxidative stress [30], although the transcriptional regulatory program controlling this regulation has not been delineated. Furthermore, in response to hypoxia the SIRT1 promoter is directly trans-activated by HIF transcription factors [31]. Emerging data also shows that the SIRT1 protein is modulated by numerous regulatory events in response to altered redox states. At pathological levels, H2O2 and redox stress associated with environmental toxins diminish SIRT1 protein levels, via increased proteosomal degradation [32]. The activity of SIRT1 is additionally modulated by post-translational modifications that alter deacetylase activity. For example, oxidative stress has been shown to promote the desumoylation and inactivation of SIRT1, resulting in increased susceptibility to apoptosis [33].
Redox Targets of SIRT1
A large number of sirtuin deacetylation substrates have now been identified in the human proteome, and several of these are related to maintaining redox homeostasis within the cell. One of the first major targets of SIRT1 to be identified was the tumor suppressor p53. As a transcription factor, p53 is involved in activating a suite of pro- and anti-oxidant genes, such as sestrins, mitochondrial manganese superoxide dismutase and glutathione peroxidase 1 (reviewed in [34] ). SIRT1 binds and deacetylates p53 at Lys382, which attenuates its transcriptional activity [35, 36]. The cellular localization of p53 in response to reactive oxygen species (ROS) has been shown to be dependent on SIRT1 deacetylation, and may help to regulate the switch from antioxidant protection to apoptosis. Over-expression of SIRT1 attenuated the transcriptional activity of p53 in response to exogenous hydrogen peroxide treatment, which prevented the induction of a p53-mediated apoptosis program [37]. At lower ROS levels, the SIRT1-p53 interaction appears to be responsible for regulating the induction of an antioxidant program. In wild-type mouse embryonic stem cells, the removal of culture medium antioxidants led to the translocation of p53 to mitochondria and subsequent apoptosis. However in SIRT1−/− cells, this endogenous increase in ROS led to the nuclear translocation of p53, which the authors speculated would result in the induction of an antioxidant response [38].
A second pathway that relies on sirtuins to activate an antioxidant response involves the forkhead box O (FOXO) transcription factor family. Both SIRT1 and SIRT2 have been shown to deacetylate and activate FOXO3a in response to oxidative stress [39, 40]. This is significant, as FOXO3a is a transcriptional activator of the SOD2 gene which encodes the mitochondrial-localized MnSOD antioxidant protein [41]. A second target of SIRT1-FOXO3a is catalase, an important enzyme that protects against damage caused by excess hydrogen peroxide. Again, the action of SIRT1 appears to be bi-directional, with low levels of hydrogen peroxide leading to the upregulation of FOXO3a-induced catalase, and higher levels leading to the switch to FOXO3a-mediated apoptosis [42]. In the heart, pressure overload and related oxidative stress leads to an upregulation in SIRT1, and mimicking this increase using transgenic over-expression of SIRT1 in mice leads to the induction of protective mechanisms, such as increased catalase expression [43]. However, high levels of SIRT1 led to increased oxidative stress, apoptosis and cardiac hypertrophy. As such, these sirtuins appear to act as ROS sensors, with the ability to induce protective mechanisms in response to low-level stresses, and signaling for apoptosis when the stress becomes too great.
Given the requirement for NAD+ in sirtuin function, mechanisms which regulate the intracellular NAD+:NADH ratio have a powerful role in SIRT1 regulation. One of the most closely studied is the interaction between cellular NAD+ levels (particularly the NAD+ salvage pathway) and AMP-activated kinase (AMPK), a key energy homeostasis enzyme. One of the first studies to link AMPK function and SIRT1 activity looked at glucose metabolism during muscle development, and found that reduced levels of glucose led to the activation of SIRT1[44]. Reducing the glucose available to myoblasts led to the activation of AMPK and, consequently, the induction of NAD+ salvage pathway enzyme Nampt, which increased intracellular NAD+ levels and activated SIRT1 [44]. These results were further extended by studies demonstrating that AMPK, acting as a metabolic fuel sensor, could stimulate transcriptional activity downstream of SIRT1 [45]. A reduction in cellular energy stores leads to the phosphorylation and activation of AMPK, which leads to an increase in available NAD+ within the cell. This stimulates SIRT1 to activate several transcriptional activators, such as FOXO proteins and the peroxisome proliferator-activator receptor-γ coactivator 1α (PGC-1α), leading to an upregulation of genes involved in catabolism and mitochondrial biogenesis [46]. The activation of AMPK in this manner can also occur through SIRT1, using positive feedback mechanisms. LBK1, an upstream kinase that phosporylates and activates AMPK under nutrient stress conditions, has been shown to be acetylated on multiple lysine residues. Stimulation or over-expression of SIRT1 leads (either directly or indirectly) to the deacetylation of LBK1, which promotes its translocation from the nucleus to the cytoplasm, allowing it to phosphorylate AMPK [47], [48]. As such, there appears to multiple levels of metabolic regulation occurring through the AMPK-SIRT1 axis, and many of these steps require further elucidation.
As noted above, SIRT1 has the ability to interact with and deacetylate PGC-1α, a major transcriptional coactivator involved in cellular metabolism and mitochondrial biogenesis. The functional role of this deacetylation, however, appears to depend greatly upon the tissue type and metabolic conditions in which it takes place. It has been demonstrated that in the liver, SIRT1 is activated in response to fasting, which leads to the deacetylation of PGC-1α [49]. Acting through this pathway, PGC-1α deacetylation can both inhibit glycolytic genes and induce the expression of those involved with gluconeogenesis [49]. A contemporary study showed that in the adrenal PC12 cell line, SIRT1 could directly interact with and deacetylate PGC-1α, and that this led to a decrease in both PGC-1α transcriptional activity and related mitochondrial oxidative metabolism [50]. Conversely, induction of SIRT1 activity using resveratrol, and subsequent PGC-1α deacetylation, led to an increase in mitochondrial function in numerous oxidative tissue types (including muscle and brown adipose tissue), indicating that deacetylation was responsible for an increase in PGC-1α transcriptional activity [51]. In skeletal muscle, fasting induced SIRT1 activity and PGC-1α deacetylation, which led to an increase in mitochondrial fatty-acid oxidation [52]. More recent work has indicated that SIRT1-dependent deacetylation of PGC-1α is required for the induction of mitochondrial biogenesis and activity in response to adiponectin in muscles [53] and Fibroblast Growth Factor 21 in adipocytes [54], and therefore is crucial to the function of these anti-diabetic metabolic regulators. Loss of PGC-1α activity has been linked to a down-regulation of mitochondrial antioxidant proteins, such as MnSOD, indicating that SIRT1 may have a direct effect on cellular redox states through this pathway [55].
Following the discovery of resveratrol, and its effect on health and lifespan, there were several avenues of research that opened up to elucidate the mechanisms involved. One pathway that was of interest to the cardiovascular field was the effect of resveratrol on the endothelial nitric oxide synthase gene, eNOS, which when uncoupled leads to ROS production and quenching of nitric oxide (NO). Previous work had shown that resveratrol was an agonist of the estrogen receptor [56], and that estrogens led to the upregulation of the eNOS gene involved in the synthesis of cardioprotective NO (reviewed in [57]. Addition of resveratrol to endothelial HUVEC cells led to a time- and concentration-dependent increase in eNOS expression and cellular NO levels [57], indicating a direct effect on the NO pathway. Similar research showed that calorie restriction, another known SIRT1 activator, was capable of enhancing eNOS expression and increasing mitochondrial biogenesis through transcription factors such as PGC-1α [58]. To square the circle, further research demonstrated that SIRT1 interacted with and deacetylated eNOS in vivo, leading to an upregulation of eNOS activity and intracellular NO levels [59]. As such, it would appear that SIRT1 has an important cardioprotective effect on vascular tone in an eNOS-dependent manner.
SIRT1 in hypoxia
As with other aspects of redox stress, non-mitochondrial sirtuins play a prominent role in the response to hypoxia. The transcriptional response to hypoxia is regulated, in a large part, by the Hypoxia Inducible Factor (HIF) family of proteins, the best characterized of which are HIF1α and HIF2α (reviewed in [60]). HIF proteins are constitutively expressed, however under normoxic conditions they are kept at low levels by continual hydroxylation and proteasomal degradation. A change in the cellular environment from normoxia to hypoxia attenuates this proteolysis, stabilizing the HIF subunits and allowing them to transcriptionally upregulate key hypoxia-response genes. Both HIF1α and HIF2α have been shown to be targets of SIRT1 deacetylation, however this interaction leads to very different outcomes for these two subunits. HIF2α forms a complex with SIRT1 under hypoxic conditions, and is deacetylated at three lysine residues (K385, K685, and K741) in the carboxy terminus [61]. This deacetylation upregulates HIF2α transcriptional activity, and leads to an increase in the abundance of HIF2α-dependent hypoxia-related proteins such as erythropoietin [61]. Recent work by the same group has shown that SIRT1 protein levels are increased at the initiation of hypoxia, and that this induction is regulated by HIF2α transcriptional activity [31].
In contrast to HIF2α, SIRT1 has been shown to act as a repressor of HIF1α activity. Under normal conditions SIRT1 can bind to, and deacetylate, HIF1α. This deacetylation prevents HIF1α from interacting with the transcriptional coactivator p300, thereby blocking its transcriptional activity [62]. However, under hypoxic conditions, the repression of HIF1α by SIRT1 is lifted indirectly through changes in the metabolic environment. The reduction in oxygen levels during hypoxia leads to a shift in the NAD+:NADH ratio, and results in a decrease in the NAD+ available to stimulate SIRT1. This NAD+-dependent loss of SIRT1 activity allows HIF1α to remain acetylated, thereby facilitating its hypoxic transcriptional activity [62]. Interestingly, HIF1α activity has also been shown to be regulated by SIRT6. HIF1α transcriptionally regulates a number of genes involved in the shift to glycolysis under low oxygen conditions. These genes, along with HIF1α itself, are upregulated in SIRT6-deficient cells, leading to an increase in glucose uptake and glycolysis [63]. In wild-type cells, SIRT6 acts as a H3K9 histone deacetylase, blocking the HIF1α-dependent transcription of multiple glycolytic genes, and co-repressing HIF1α in a deacetylase-dependent manner [63]. As such, it would appear that while SIRT1 can regulate the activity of HIF subunits in general, SIRT6 may act as a glucose metabolism-specific regulator of hypoxia-related genes.
The redox phenotype in SIRT1 knockout and transgenic mice
The composite of data discussed would suggest that SIRT1 should play an adaptive role in protection against redox stressors. A major limit to testing this hypothesis has been the production of SIRT1−/− mice, as this deletion leads to high levels (~75%) of embryonic lethality. Nevertheless, in vivo studies have also been performed with either heterozygous SIRT1+/− mice, tissue-specific knockout mice, or following over-expression of SIRT1 using transgenic technology. Here mice have been studied in the context of degenerative and redox stress diseases. Heterozygous SIRT1 knockout mice show increased susceptibility to oxidative stress in the kidney [64] and the combined SIRT1+/− p53+/− mice show greater tumor susceptibility compared to p53 haploinsufficiency alone [65]. Tissue-specific deletion of SIRT1, including in the liver and brain, has been shown to negatively alter fat metabolism and increase susceptibility to diet-induced obesity [66, 67]. Limited copy number over-expression of SIRT1 has been shown to protect against redox stress in the brain, heart and kidneys, to ameliorate hepatic hepatosteatosis that results from diet induced obesity, and to ameliorate colon cancer [43, 68–70]. SIRT1 is also implicated as being necessary to enable the innate adaptive reprogramming to resist ischemia-reperfusion injury, as a component of the ischemic preconditioning program [71].
Additional nuclear and cytosolic sirtuins and redox biology
Although the role of SIRT2 has not been as well characterized as SIRT1 or SIRT3, it does play a regulatory role in modulating redox stress tolerance. SIRT2 is mainly localized in the cytosol, however it can undergo translocation to the nucleus during certain events, such as mitosis [72]. Indeed, other sirtuins such as SIRT3 have been shown to translocate under different cellular conditions [73], although this may be the result of using an over-expression vector with an incomplete murine SIRT3 [74]. Redox stress has been shown to result in the upregulation of both SIRT2 transcript and protein levels [40, 75]. Functional characterization of this process suggests that redox-stress upregulation of SIRT2 is associated with increased cell death, in parallel with the induction of the pro-apoptotic protein Bim [40], and that overexpression of SIRT2 similarly promotes neurodegeneration [76]. In contrast, basal levels of SIRT2 under low-stress conditions upregulate mitochondrial MnSOD via FOXO3a deacetylation, with the subsequent attenuation of reactive oxygen species levels [40]. Genetic knockdown of SIRT2 is associated with upregulation of the cytosolic chaperone 14-3-3ζ, which in turn sequesters the pro-apoptotic mitochondrial protein BAD in the cytosol and augments tolerance against anoxia-reoxygenation induced cell death [75]. A `dose-dependent' function of SIRT2 under redox stress conditions is further supported by data showing that a SIRT2 inhibitor rescues α-synuclein-mediated toxicity in a cellular model of Parkinson's disease [77], and that SIRT2 knockdown is protective in neuronal and cardiac cells [75, 77].
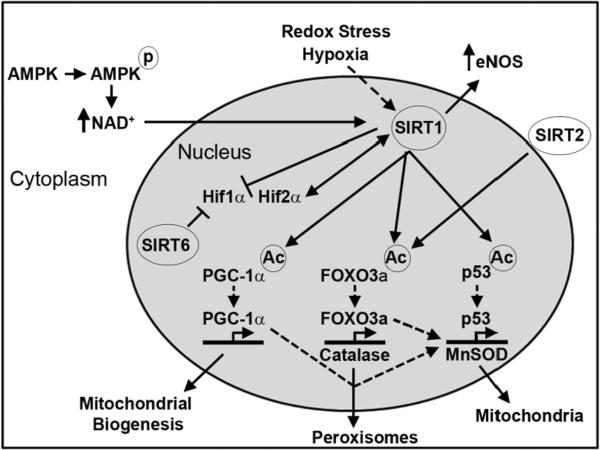
The role of SIRT6 in biological control under redox and hypoxic stress has not been well characterized, although mice lacking SIRT6 are predisposed to accelerated senescence. This includes the development of degenerative features, telomerase shortening and a lifespan limited to approximately 4 weeks [78]. As noted above, SIRT6 additionally modulates hypoxic signaling through corepression of HIF1α during the control of glucose homeostasis [63]. SIRT7 is the least-well studied sirtuin, although the genetic knockout of this gene does have a robust phenotype, with a 50% reduction in lifespan and the development of degenerative features and an inflammatory cardiomyopathy [79]. In the absence of SIRT7, p53 exhibits robust acetylation, which is postulated to diminish resistance to genotoxic and oxidative stress [79]. The role of SIRT7 in ameliorating oxidative stress is also evident in that it blunts cell proliferation under oxidative stress conditions [80]. A summary of the role played by non-mitochondrial sirtuins in redox biology can be found in Figure 1.
Figure 1.
Of the non-mitochondrial sirtuins, SIRT1 is the most widely characterized deacetylase. In common with the other sirtuins, SIRT1 is activated by NAD+; levels of which can be increased following AMPK activation. Redox stress and hypoxia may, directly or indirectly, increase SIRT1 levels or activate its deacetylase activity (broken arrow). In terms of oxidative stress, one of the main roles of SIRT1 is to deacetylate and activate transcription factors involved in upregulating antioxidant proteins, such as MnSOD and catalase. In addition, SIRT2 has also been shown to activate the transcription factor FOXO3a through deacetylation. The activity of SIRT1, along with SIRT6, can inhibit HIF1α activity; however, SIRT1 has the opposite effect on HIF2α. Finally, SIRT1 deacetylates and activates the eNOS, leading to an increase in intracellular cardioprotective NO.
Mitochondrial sirtuins
The role of redox signaling and oxygen in the regulation of SIRT3
Of the three family members found in the mitochondria, SIRT3 functions as the major sirtuin deacetylase [81]. An initial controversy regarding the localization of the murine SIRT3 [82] has now been resolved, with the identification and functional characterization of its mitochondrial localization sequence [74, 83, 84]. The number of SIRT3 substrates identified is increasing at a prodigious pace, and their role in numerous biological programs is still being explored. Here we focus on those substrates that modulate redox-stress directly, and discuss their role in redox-stress associated pathology. SIRT4 and SIRT5 are also enriched in the mitochondria, although fewer substrates of these two sirtuins have been described. As mentioned previously, SIRT5 may function to remove succinyl and/or malonyl, rather than acetyl, groups from mitochondrial targets [10]. However, where they are relevant to the role of sirtuin biology in redox stress, these data have been included. As numerous SIRT3 mitochondrial targets play a role in the control of ROS generation and amelioration (see below), studies have begun to explore whether ROS levels themselves regulate SIRT3. Both SIRT3 transcript and protein levels are induced by the generation of mitochondrial ROS [85]. Whether SIRT3 is modulated at the post-translational level, in response to redox signaling, has not been explored. However, prolonged high fat feeding results in evidence of hepatic redox stress, and an associated reduction in SIRT3 protein levels [86]. SIRT3 levels are also induced in parallel with mitochondrial biogenesis, where the transcriptional co-activator PGC-1α and transcription factor ERRα, respectively, transactivate the SIRT3 gene promoter [87].
Redox Targets of SIRT3
Mitochondria are the major source of cellular oxidative stress, with ROS formation occurring during normal function of the electron transport chain (ETC). Any reduction in mitochondrial function, through processes such as metabolic disease progression or aging, can augment oxidative stress [88]. Mitochondria have therefore developed numerous biological programs to combat this innate oxidative stress and maintain functional homeostasis. One such program utilizes reduced glutathione, via the generation of NADPH by isocitrate dehydrogenase 2 (IDH2). IDH2 uses NADP+ as a substrate in the TCA cycle, during the decarboxylation of isocitrate to α-ketoglutarate, which yields NADPH. This, in turn, is used for the regeneration of reduced glutathione to confer antioxidant properties [89]. IDH2 was initially shown to be differentially acetylated in a nutrient dependent manner in a proteomics screen by Kim et al. [90]. A role for SIRT3 in this process was identified by Schlicker et al., who showed that in the presence of NAD+ purified SIRT3, but not SIRT5, deacetylated IDH2 and increased its activity [91]. The sites of SIRT3 deacetylation (K211 and K212) found by Schlicker et al. were different to those identified in the initial proteomics screen (K75 and K241), and it is unclear how the latter two residues are regulated [90, 91]. Recently, this work has been linked to age-related hearing loss (AHL). It was previously shown that CR attenuates AHL by slowing neuronal loss in the cochlea, which correlated with increased SIRT3 expression levels [89]. Further work demonstrated that CR prevented AHL in 12 month old mice and attenuated spiral ganglion neuronal loss. However, these benefits were completely abolished in SIRT3 knockout animals, suggesting that the process was SIRT3-dependent [89]. CR also ameliorated oxidative damage to DNA in various organs, including the brain and liver, and promoted hair cell survival in the cochlea of aged mice. To provide a mechanism for these SIRT3-dependent benefits, Someya et al. showed that GSH/GSSG ratios were increased, and that NADPH levels were elevated in liver, brain and the inner ear following CR. CR also decreased the acetylation levels of IDH2, increasing its activity, in a SIRT3-dependent manner [89]. Confirming previous results, they further demonstrated that SIRT3 directly deacetylates and augments the activity of IDH2, and that overexpression of SIRT3 or IDH2 was sufficient to increase NADPH levels and attenuate oxidative stress-mediated cell death [89]. Linking CR, SIRT3 and IDH2 supports the concept that oxidative stress is a major component of aging, and that nutrient status can regulate the cellular response to degenerative pathologies.
Another major mitochondrial defense against oxidative stress is superoxide dismutase (SOD). Humans express three forms of SOD, which converts superoxide anion (O2−) to hydrogen peroxide (H2O2), limiting ROS damage to nucleic acids, proteins and lipids. SOD1 and SOD3 are localized in the cytosol and extracellular space, using copper and zinc as prosthetic groups, respectively. SOD2 contains manganese (MnSOD) and localizes to the mitochondria [92]. MnSOD, like IDH2, has been linked to age-related disorders such as cancer and cardiovascular disease [93–95]. SIRT3 was first shown to regulate SOD2 by modulating FOXO3a activity in the mitochondria, which led to subsequent FOXO3a-mediated SOD2 expression [95, 96]. It is not clear if these results were due to an over-expression artifact leading to the nuclear localization of SIRT3 [74]. Nevertheless, more recent evidence has directly tied SIRT3 to MnSOD function. Qiu et al. showed that caloric restriction lowers oxidative stress in a SIRT3 dependent manner [97]. SIRT3 overexpression in mouse embryonic fibroblasts (MEFs) lowered ROS levels in an SOD2-dependent manner, suggesting that acetylation status regulates SOD2 activity. They further showed that SIRT3 interacts with SOD2, and that SIRT3-mediated deacetylation of SOD2 increased its activity, which provided protection against oxidative stress [97]. SOD2 was previously identified as an acetylated protein at K68 and K130 [90, 98]. However, there is some uncertainty regarding which acetylated lysine residue regulates SOD2 activity. Qui et al. showed that mutating K68 and K130 did not alter total acetylation levels of SOD2. However acetylation of K53 and K89, two lysine residues near the active site of SOD2, regulated its activity [97]. While a second study by Tao et al. confirmed that SIRT3 regulates SOD2 activity, and subsequent protection against ionizing radiation (see below), this group identified K122 as the crucial lysine [99]. Fasting wild-type mice for 36 hours promoted deactylation of SOD2, which was blocked in SIRT3 knockout animals. Hyperacetylation of SOD2 in SIRT3-deficient mice led to lower enzymatic activity and elevated ROS levels [99]. Further complicating the post-translational regulation of SOD2, Chen et al. recently showed that acetylation of K68 does indeed regulate SOD2 activity, and that SIRT3 deacetylates SOD2 at this site [85]. The differences observed in the site-specific regulation of SOD2 by SIRT3 may reflect cell type, species or stress conditions, or may reflect a higher-order regulation that requires changes in several lysine residues. As Chen et al. state, it is interesting to note that the lysine to arginine mutation (which mimics deacetylation) increased SOD2 activity for K53R, K89R and K122R mutants, while in the K68R mutant it does not [85]. Although the precise site(s) of regulation remains unclear, it is clear ROS levels are tightly controlled by SIRT3 by increasing the activity of two major ROS regulating enzymes, IDH2 and SOD2.
Indirect SIRT3 and SIRT5 targets for regulation of mitochondrial oxidative stress
MnSOD and IDH2 represent SIRT3 targets that directly regulate mitochondrial ROS. However, other targets of SIRT3 and SIRT5 have indirect effects on oxidative stress. As mentioned previously, the electron transport chain generates ROS, and SIRT3 has been shown to deacetylate complexes I, II, V and most recently III [86, 100, 101]. SIRT5 is also implicated in the deacetylation of cytochrome c, although this has not been functionally characterized [91]. Secondly glutamate dehydrogenase (GDH), which can regulate glutamate oxidative stress and generate NAPDH, is deacetylated by SIRT3 [81] (and inhibited by SIRT4 mediated ADP-ribosylation) [102]. A final example of indirect regulation of ROS levels by mitochondrial sirtuins occurs during fasting, when protein catabolism is augmented to provide TCA cycle intermediates. Protein catabolism leads to the generation of ammonium, which can result in oxidative stress [103]. To prevent damage, ammonium is converted to urea for subsequent elimination in the urea cycle. SIRT3 and SIRT5 regulate the mitochondrial localized reactions of the urea cycle, with SIRT5 acting on carbamoyl-phosphate synthase 1 (CPS-1), and SIRT3 deacetylating ornithine transcarbamoylase (OTC) [20, 104]. Both deacetylation reactions promote urea cycle function, leading to the elimination of the oxidative stress-promoting ammonium.
Biological roles of SIRT3 in redox-associated pathology
Oncogenesis
Numerous studies have highlighted a pivotal role for SIRT3 in cancer, even going as far as referring to it as a tumor suppressor. However, the ability of SIRT3 to act as a tumor suppressor is directly related to its role in regulating ROS levels. SIRT3 knockout cells display a propensity towards carcinogenesis, with increased ROS, nuclear and mitochondrial genomic instability and diminished contact inhibition [93]. Combining the expression of oncogenes with deletion of SIRT3 further promotes a cancer phenotype, which can be rescued by overexpression of SOD2. SIRT3 knockout mice also have a higher incidence of breast cancer, while human breast cancer samples exhibit lower levels of SIRT3 [93].
Cancer cells display a characteristic metabolic profile, and SIRT3 appears to play a role in preventing the switch to this phenotype. In cancer cells, hexokinase II activity is greatly upregulated, which promotes increased glycolysis and attenuates oxidative phosphorylation – classic indicators of the Warburg effect (reviewed in [105]). Hexokinase II binds to VDAC in the outer mitochondrial membrane, and utilizes ATP to generate glucose-6-phosphate (G6P). G6P is shuttled through glycolysis, generating pyruvate and lactic acid; or through the pentose phosphate pathway to generate NADPH, which is used to drive anabolic reactions. Some of the G6P is also used to generate citrate in the mitochondria, which enters the cytosol to allow phospholipid and cholesterol synthesis. SIRT3 knockout MEFs show elevated levels of glycolytic and pentose phosphate pathway metabolites, increased glucose uptake and lactate production, and lower levels of TCA cycle intermediates – all suggesting a metabolic profile similar to cancer cells [106]. SIRT3 has been shown to deacetylate cyclophilin D (CypD), which limits its interaction with the mitochondrial permeability transition pore (mPTP). Importantly, this deacetylation also prevents hexokinase II from binding to VDAC and the mPTP, limiting its ability to use ATP and augment glycolysis [107]. Shulga et al. speculate that this would limit the transition to a Warburg phenotype, thereby attenuating tumor growth [107]. Other studies have also found that SIRT3 drives oxidative phosphorylation and augments ATP generation [74, 100].
Metabolic reprogramming of cancer cells relies on HIF-1α, and the loss of SIRT3 leads both to its stabilization, and increased levels of HIF-1α gene targets, including hexokinase II [106]. As noted above, SIRT1 can deacetylate HIF-1α. Although SIRT3 does not directly interact with HIF-1α, HIF-1α is stabilized in SIRT3 knockouts via an indirect mechanism. The increased ROS levels found in SIRT3 knockouts increase HIF-1α stabilization promoting cancer cell metabolism [106]. Bell et al. also showed the effect of SIRT3 on HIF-1α stability, along with the ability of SIRT3 to attenuate tumor growth using xenograft models [108]. SIRT3 also appears to regulate p53 activity, by altering growth arrest and apoptosis in a complex manner involving a similar mechanism to HIF-1α, namely elevated ROS levels affecting p53 activity [109–111]. Although the majority of data shows that SIRT3 can act as a tumor suppressor, SIRT3 may promote tumorigenesis in some forms of cancer. SIRT3 expression levels were elevated in oral squamous cell carcinoma (OSCC); while inhibition and down regulation of SIRT3 slowed growth, promoted apoptosis and increased susceptibility to radiation and chemotherapy [112]. These conflicting roles of SIRT3 as tumor suppressor or promoter may be due to its role in regulating p53, with SIRT3 inhibiting the ability of p53 to arrest growth [110]. Overall it appears that SIRT3 can regulate cellular energetics, and possibly substrate selection (e.g. lipids; see below), by increasing mitochondrial oxidative phosphorylation through deacetylation of various electron transport chain targets, which results in attenuated ROS levels and glycolytic metabolism.
Lipotoxicity
Obesity and over-nutrition can lead to diabetes, the metabolic syndrome and non-alcoholic fatty liver disease (NAFLD), along with an increase in circulating free fatty acids, i.e. lipotoxicity. Lipotoxicity generates a pro-inflammatory state with increased oxidative stress, promoting insulin resistance systemically and diminishing insulin secretion by the pancreas (reviewed in [113]). Concurrent with this pro-inflammatory state, mitochondrial function and biogenesis, through defects in PGC-1α, also diminish with obesity and diabetes – further promoting ROS generation [3]. Interestingly, expression of SIRT3 leads to an upregulation of PGC-1α, and consequently increases the transcription of its biogenesis gene targets [87, 114, 115].
Nutritional status greatly alters mitochondrial protein acetylation levels, with fasting and high fat diets leading to the hypo- and hyper-acetylation of mitochondrial targets, respectively [90, 116]. Hirschey et al. showed that fasting increases SIRT3 expression and lowers acetylation levels in mouse livers. Although phenotypically normal, SIRT3 knockout animals had higher levels of triglycerides and β-oxidation intermediates, suggesting a defect in lipid processing. They showed that long chain acyl-CoA dehydrogenase (LCAD) is hyperacetylated at K42 in SIRT3 knockout mice, leading to lower activity, and that SIRT3 deacetylates LCAD directly [117]. Hallows et al. also found LCAD to be a SIRT3 target [20]. It has also been shown that SIRT3 prevents lipid accumulation in HepG2 cells by acting alongside AMPK, which phosphorylates acetyl-CoA carboxylase (ACC) and inhibits lipid synthesis. SIRT3 appears to regulate AMPK activity, however the mechanism of the cross-talk between these two nutrient sensing proteins is currently unknown [118]. On the opposite end of the nutritional spectrum, six weeks of high fat diet (HFD) led to hyperacetylation of numerous protein targets in mouse livers involved in metabolic pathways [116]. Under these conditions, SIRT3 activity was diminished, while oxidative stress was elevated, further supporting a role of SIRT3 in regulating oxidative stress [116]. SIRT3 knockout animals exposed to HFD showed even higher levels of acetylation and diminished mitochondrial respiration [116]. The absence of SIRT3 similarly increased susceptibility to lipotoxicity in primary hepatocytes, and this phenotype was ameliorated by either the reconstitution of SIRT3, or via the administration of the antioxidant N-acetylcysteine [86]. It appears that SIRT3 may alleviate the pro-inflammatory state induced by lipotoxicity through a variety of mechanisms, including altering mitochondrial biogenesis, aiding oxidative stress systems and regulating lipid accumulation. SIRT3 may therefore represent a potential new target to combat lipotoxicity and its concurrent disease states.
Cardiac hypertrophy
Sirtuins and protein deacetylation have also been linked to cardiovascular health [119]. The renin-angiotensin system (RAS) and Angiotensin II (Ang II) are common targets in the treatment of cardiovascular disease, including hypertension, renal disease, hypertrophy and Marfan syndrome. Ang II is pro-inflammatory and increases ROS generation [120]. Similar to CR, knockout of Ang II Receptor (AT1R) in mice increased lifespan by 26%, and AT1R knockouts have lower levels of cardiac fibrosis, hypertrophy and attenuated aortic damage [121]. AT1R deficient mice also showed lower levels of oxidative stress, and increased mitochondrial density when compared to controls. As a possible mechanism explaining this anti-aging phenotype, it was shown that AT1R knockouts had higher levels of both SIRT3 and the NAD+ generating enzyme nicotinamide phosphoribosyltransferase, Nampt. Conversely, AngII treatment lowered the expression levels of Nampt and SIRT3, with these effects being blocked by candesartan, an AT1R blocker [121]. Further linking the sirtuins to promoting cardiovascular health, Miyazaki et al. showed that Sirt1 attenuates AT1R expression levels [122].
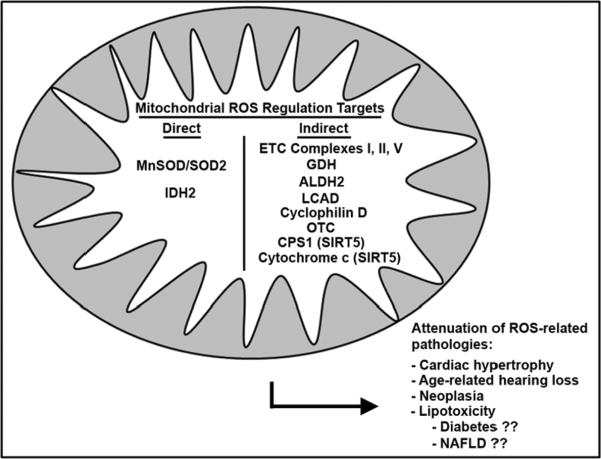
To directly investigate the role of SIRT3 in cardiac hypertrophy, Sundaresan et al. induced cardiac hypertrophy chemically and physically; both of which enhanced SIRT3 expression [94, 95]. SIRT3 knockout animals subjected to the same stimuli displayed an exacerbated hypertrophic response, suggesting that the augmented SIRT3 level seen in wild-type animals is a protective response used to combat the hypertrophic program. Indeed, overexpression of SIRT3 attenuated the hypertrophic response both in vivo and in vitro, preserving cardiac function [95]. Increasing exogenous levels of the SIRT3 substrate, NAD+, attenuated the hypertrophic response through SIRT3 deacetylation and activation LKB1 and AMPK. Hsu et al. also showed that elevated NAD+, as mediated by Nampt, induces a cardio-protective phenotype [123]. As mentioned previously, it is not clear if the SIRT3-mediated effect on nuclear and cytosolic targets, i.e. FOXO3a and LKB1, is a by-product of over-expression of the short form of SIRT3 that fails to localize to the mitochondria, or if SIRT3 acts on targets outside the mitochondria through unspecified mechanisms. An overview of the role of the mitochondrial sirtuins on redox stress biology is schematized in Figure 2.
Figure 2.
Mitochondrial localized SIRT3 and SIRT5 regulate oxidative stress utilizing both direct and indirect mechanisms. Deacetylating manganese superoxide dismutase (MnSOD/SOD2) and isocitrate dehydrogenase 2 (IDH2) augments their activity, providing a direct mechanism to regulate ROS. The deacetylation of various other mitochondrial enzymes indirectly attenuates oxidative stress, which includes regulating mitochondrial function, namely through deacetylation of cytochrome c and complexes of the electron transport chain (ETC I, II and V). Another indirect mechanism is through controlling ROS generating metabolic intermediate production by deacetylating aldehyde dehydrogenase (ALDH2), glutamate dehydrogenase (GDH), long chain acyl-dehydrogenase (LCAD), carbamoyl phosphate synthetase I (CPS-1) and ornithine transcarbamylase (OTC). Finally SIRT3 can regulate apoptosis and glucose metabolism through its actions on cyclophilin D. The cumulative effect of this ROS regulation is to attenuate various age-related pathologies including oncogenesis, cardiac hypertrophy, lipotoxicity and age-related hearing loss.
ALDH2 and acetaminophen - an exception to the redox-stress ameliorative role of SIRT3
Unlike the protective role of SIRT3 in modulating redox-stress described above [86, 89, 99], it has recently been found that SIRT3-mediated mitochondrial protein deacetylation exacerbated oxidative stress associated with acetaminophen-induced liver injury (AILI) [124]. The level of the lipid peroxidation adduct, trans-4-hydroxy-2-nonenal (4-HNE) was lower in SIRT3−/− mice, compared to SIRT+/+, in response to a toxic dose of acetaminophen, in parallel with less AILI in the SIRT3−/− mice. The resistance of SIRT3−/− mice to AILI was partly due to the acetylation of mitochondrial aldehyde dehydrogenase 2 (ALDH2), a dehydrogenase which functions to oxidize and detoxify aldehydes, including the lipid peroxidation product 4-HNE [125]. In contrast to the known anti-oxidant effect of SIRT3, via the deacetylation and activation of redox scavenger substrates, SIRT3-mediated deacetylation of ALDH2 does not change its enzyme activity directly. Rather, the deacetylation of ALDH2 facilitates the binding of the acetaminophen toxic metabolite, N-acetyl-p-benzoquinoneimine (NAPQI), directly to ALDH2, with a concomitant blunting of dehydrogenase activity [124]. Taken together, this finding suggests that the acetylation of mitochondrial proteins can also function as an allosteric inhibitor to the binding of reactive metabolites. This mechanism is proposed to function in the enhanced susceptibility to acetaminophen-induced injury under fasting and caloric restricted conditions [126, 127], nutrient states known to promote the activation of sirtuin enzymes.
Conclusions and future directions
The majority of the work carried out thus far on mammalian sirtuin proteins has focused on SIRT1 and SIRT3. SIRT1, as the major nuclear deacetylase, plays a pivotal role in the acetylation-based regulation of numerous proteins, including many that form the transcriptional response to changes in redox conditions. SIRT3, as the major mitochondrial deacetylase, acts as the in situ regulator of proteins which ameliorate damage in one of the major ROS-producing organelles in the cell. Our knowledge of the functional activity of these proteins has grown exponentially over the last few years, and will likely continue to uncover new pathways regulating the cellular response to oxidative stress. Table 1 summarizes all the ROS modulatory proteins that are substrates of the sirtuins, and describes the functional consequences of sirtuin activation. One of the major fundamental areas to be tackled in this field is how intracellular redox conditions affect the transcriptional control of SIRT1 and SIRT3 protein levels. Work in this area will help to determine the level of feedback control inherent in this system, and will establish how finely balanced these mechanisms are. Additionally, both in the redox field in particular, and the sirtuin field in general, future research effort will likely turn to the other sirtuins. While SIRTs 2,4,5,6 and 7 may have a more limited functional scope than SIRT1 and SIRT3, it is likely that their substrate-specific activity will be crucial in the regulation of many important pathways. As such, while they appear to be the forgotten members currently, the future may yield much more insight into their function within the cell.
Table 1.
| Redox-related Gene | Sirtuin | Localization | Result of Sirtuin Action | Reference(s) |
|---|---|---|---|---|
| p53 | SIRT1, SIRT7 | Nucleus | Mediates transcriptional activity depending on sirtuin expression level | 33–38,77 |
| FOXO3a | SIRT1, SIRT2 | Nucleus, cytoplasm | Activates transcriptional activity | 39,40 |
| LBK1 | SIRT1 | Nucleus | Activates LBK1, leading to AMPK phosphorylation | 47,48 |
| PGC-1α | SIRT1 | Nucleus | Activates transcriptional co-activation | 49 – 55 |
| eNOS | SIRT1 | Nucleus | Activates eNOS activity | 59 |
| HIF1α | SIRT1, SIRT6 | Nucleus | Repression of transcriptional activity | 62,63 |
| HIF2α | SIRT1 | Nucleus | Activates transcriptional activity | 31,61 |
| 14-3-3ζ | SIRT2 | Cytoplasm | Upregulates 14-3-3ζ, prevents anoxia-reoxygenation mediated apoptosis | 75 |
| IDH2 | SIRT3 | Mitochondria | Activates enzyme, reduces ROS | 89 – 91 |
| SOD2 | SIRT3 | Mitochondria | Activates enzyme, reduces ROS | 85, 90, 97–99 |
| NDUFA9 | SIRT3 | Mitochondria | Activates enzyme, upregulates ETC activity | 100 |
| SDHB | SIRT3 | Mitochondria | Activates enzyme, upregulates ETC activity | 101 |
| ATP5A | SIRT3 | Mitochondria | Activates enzyme, upregulates ETC activity | 86 |
| GDH | SIRT3, SIRT4 | Mitochondria | Activates enzyme (SIRT3), represses enzyme by ADP-ribosylation (SIRT4) | 81, 102 |
| OTC | SIRT3 | Mitochondria | Activates enzyme, reduces ammonia toxicity and ROS | 20 |
| CPS1 | SIRT5 | Mitochondria | Activates enzyme, reduces ammonia toxicity and ROS | 104 |
| CypD (PPIF) | SIRT3 | Mitochondria | Deactivates enzyme, prevents interaction with mPTP | 107 |
| LCAD | SIRT3 | Mitochondria | Activates enzyme, promotes lipid processing | 20,117 |
| ALDH2 | SIRT3 | Mitochondria | Deacetylation by SIRT3 allows NAPQI binding to ALDH2, reducing its activity | 124 |
Highlights
-
>
Sirtuin deacetylase enzymes are closely associated with extensions in lifespan.
-
>
A common phenotype in aging is the dysregulation of biological systems over time.
-
>
This can be caused by damage accumulation, e.g. from reactive oxygen species (ROS).
-
>
Recent research has linked the beneficial effects of sirtuins to the control of ROS.
-
>
We review these links, and examine the mechanisms which regulate sirtuin activity.
Acknowledgements
All the authors are funded by the Division of Intramural Research of the NHLBI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts: None
Reference List
- [1].Sugamura K, Keaney JF., Jr. Reactive oxygen species in cardiovascular disease. Free Radic Biol Med. 2011 doi: 10.1016/j.freeradbiomed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Whaley-Connell A, McCullough PA, Sowers JR. The role of oxidative stress in the metabolic syndrome. Rev Cardiovasc Med. 2011;12:21–29. doi: 10.3909/ricm0555. [DOI] [PubMed] [Google Scholar]
- [3].Pagel-Langenickel I, Bao J, Pang L, Sack MN. The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr Rev. 2010;31:25–51. doi: 10.1210/er.2009-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Naik E, Dixit VM. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J Exp Med. 2011;208:417–420. doi: 10.1084/jem.20110367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sack MN. Caloric excess or restriction mediated modulation of metabolic enzyme acetylation - proposed effects on cardiac growth and function. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- [8].Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem.Biophys.Res.Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- [9].Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- [10].Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- [12].Bao J, Sack MN. Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol Life Sci. 2010;67:3073–3087. doi: 10.1007/s00018-010-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- [14].Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- [16].Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- [17].Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- [18].Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de CR, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- [22].Geng YQ, Li TT, Liu XY, Li ZH, Fu YC. SIRT1 and SIRT5 activity expression and behavioral responses to calorie restriction. J Cell Biochem. 2011 doi: 10.1002/jcb.23315. [DOI] [PubMed] [Google Scholar]
- [23].Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- [24].Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J.Biol.Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J.Biol.Chem. 2007;282:19546–19555. doi: 10.1074/jbc.M702990200. [DOI] [PubMed] [Google Scholar]
- [28].Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J.Biol.Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- [29].Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, Zipp F, Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat.Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- [31].Chen R, Dioum EM, Hogg RT, Gerard RD, Garcia JA. Hypoxia increases sirtuin 1 expression in a hypoxia-inducible factor-dependent manner. J Biol Chem. 2011;286:13869–13878. doi: 10.1074/jbc.M110.175414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat.Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vaziri H, Dessain SK, Ng EE, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- [36].Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- [37].Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D. Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic Biol Med. 2006;40:2175–2182. doi: 10.1016/j.freeradbiomed.2006.02.014. [DOI] [PubMed] [Google Scholar]
- [38].Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- [40].Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- [41].Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- [42].Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Washida N, Tokuyama H, Hayashi K, Itoh H. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem Biophys Res Commun. 2008;372:51–56. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- [43].Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ.Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- [44].Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr.Opin.Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J.Biol.Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, Zang M. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J.Biol.Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- [50].Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J.Biol.Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- [51].Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [52].Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, Ogata H, Kubota N, Takamoto I, Hayashi YK, Yamauchi N, Waki H, Fukayama M, Nishino I, Tokuyama K, Ueki K, Oike Y, Ishii S, Hirose K, Shimizu T, Touhara K, Kadowaki T. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- [54].Chau MD, Gao J, Yang Q, Wu Z, Gromada J. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1alpha pathway. Proc Natl Acad Sci U S A. 2010;107:12553–12558. doi: 10.1073/pnas.1006962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B, Chen Y. PGC-1 alpha regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxid Redox Signal. 2010;13:1011–1022. doi: 10.1089/ars.2009.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gehm BD, McAndrews JM, Chien PY, Jameson JL. Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc Natl Acad Sci U S A. 1997;94:14138–14143. doi: 10.1073/pnas.94.25.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wallerath T, Deckert G, Ternes T, Anderson H, Li H, Witte K, Forstermann U. Resveratrol, a polyphenolic phytoalexin present in red wine, enhances expression and activity of endothelial nitric oxide synthase. Circulation. 2002;106:1652–1658. doi: 10.1161/01.cir.0000029925.18593.5c. [DOI] [PubMed] [Google Scholar]
- [58].Clementi E, Nisoli E. Nitric oxide and mitochondrial biogenesis: a key to long-term regulation of cellular metabolism. Comp Biochem.Physiol A Mol.Integr.Physiol. 2005;142:102–110. doi: 10.1016/j.cbpb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- [59].Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc.Natl.Acad.Sci.U.S.A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- [62].Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- [63].Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].He W, Wang Y, Zhang MZ, You L, Davis LS, Fan H, Yang HC, Fogo AB, Zent R, Harris RC, Breyer MD, Hao CM. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, Gardner K, Man YG, Hung MC, Finkel T, Deng CX. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol.Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, Souza DO, Gao Q, Diano S, Gao XB, Horvath TL. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010;30:11815–11825. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc.Natl.Acad.Sci.U.S.A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Sueyasu K, Washida N, Tokuyama H, Tzukerman M, Skorecki K, Hayashi K, Itoh H. Kidney-specific overexpression of Sirt1 protects against acute kidney injury by retaining peroxisome function. J Biol Chem. 2010;285:13045–13056. doi: 10.1074/jbc.M109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Nadtochiy SM, Redman E, Rahman I, Brookes PS. Lysine deacetylation in ischaemic preconditioning: the role of SIRT1. Cardiovasc Res. 2011;89:643–649. doi: 10.1093/cvr/cvq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem.Biophys.Res.Commun. 2008;366:174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- [74].Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr., Leclerc J, Nguyen P, Gius D, Sack MN. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J.Cell Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lynn EG, McLeod CJ, Gordon JP, Bao J, Sack MN. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett. 2008;582:2857–2862. doi: 10.1016/j.febslet.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Suzuki K, Koike T. Mammalian Sir2-related protein (SIRT) 2-mediated modulation of resistance to axonal degeneration in slow Wallerian degeneration mice: a crucial role of tubulin deacetylation. Neuroscience. 2007;147:599–612. doi: 10.1016/j.neuroscience.2007.04.059. [DOI] [PubMed] [Google Scholar]
- [77].Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- [78].Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- [79].Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ.Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- [80].Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J.Physiol Pharmacol. 2008;59(Suppl 9):201–212. [PubMed] [Google Scholar]
- [81].Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol.Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?--functional localization of an NAD+-dependent protein deacetylase. Biochem.J. 2008;411:e11–e13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–525. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cooper HM, Huang JY, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS.ONE. 2009;4:e4986. doi: 10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011 doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Giralt A, Hondares E, Villena JA, Ribas F, Diaz-Delfin J, Giralt M, Iglesias R, Villarroya F. Peroxisome Proliferator-activated Receptor-{gamma} Coactivator-1{alpha} Controls Transcription of the Sirt3 Gene, an Essential Component of the Thermogenic Brown Adipocyte Phenotype. J Biol Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- [89].Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol.Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- [91].Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J.Mol.Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- [92].Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, Troncoso J, Johnson B, Saffary R, Goh EL, Song H, Park BJ, Kim MJ, Kim S, Dawson VL, Dawson TM. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol.Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J.Clin.Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Jacobs KM, Pennington JD, Bisht KS, ykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int.J.Biol.Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- [98].Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- [99].Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc.Natl.Acad.Sci.U.S.A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- [103].Braissant O. Ammonia toxicity to the brain: effects on creatine metabolism and transport and protective roles of creatine. Mol Genet Metab. 2010;100(Suppl 1):S53–58. doi: 10.1016/j.ymgme.2010.02.011. [DOI] [PubMed] [Google Scholar]
- [104].Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [108].Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011 doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kawamura Y, Uchijima Y, Horike N, Tonami K, Nishiyama K, Amano T, Asano T, Kurihara Y, Kurihara H. Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J Clin Invest. 2010;120:2817–2828. doi: 10.1172/JCI42020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- [112].Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Andrikopoulos S. Obesity and type 2 diabetes: slow down!--Can metabolic deceleration protect the islet beta cell from excess nutrient-induced damage? Mol Cell Endocrinol. 2010;316:140–146. doi: 10.1016/j.mce.2009.09.031. [DOI] [PubMed] [Google Scholar]
- [114].Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J.Biol.Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- [115].Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11:55–62. doi: 10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- [119].Lu Z, Scott I, Webster BR, Sack MN. The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. Circ.Res. 2009;105:830–841. doi: 10.1161/CIRCRESAHA.109.204974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].de Cavanagh EM, Inserra F, Ferder L. Angiotensin II blockade: a strategy to slow ageing by protecting mitochondria? Cardiovasc Res. 2011;89:31–40. doi: 10.1093/cvr/cvq285. [DOI] [PubMed] [Google Scholar]
- [121].Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G. Disruption of the Ang II type 1 receptor promotes longevity in mice. J.Clin.Invest. 2009;119:524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Miyazaki R, Ichiki T, Hashimoto T, Inanaga K, Imayama I, Sadoshima J, Sunagawa K. SIRT1, a longevity gene, downregulates angiotensin II type 1 receptor expression in vascular smooth muscle cells. Arterioscler.Thromb.Vasc.Biol. 2008;28:1263–1269. doi: 10.1161/ATVBAHA.108.166991. [DOI] [PubMed] [Google Scholar]
- [123].Hsu CP, Hariharan N, Alcendor RR, Oka S, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through autophagy in cardiomyocytes. Autophagy. 2009;5:1229–1231. doi: 10.4161/auto.5.8.10275. [DOI] [PubMed] [Google Scholar]
- [124].Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011;12:840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Doorn JA, Hurley TD, Petersen DR. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem.Res.Toxicol. 2006;19:102–110. doi: 10.1021/tx0501839. [DOI] [PubMed] [Google Scholar]
- [126].Whitcomb DC, Block GD. Association of acetaminophen hepatotoxicity with fasting and ethanol use. JAMA. 1994;272:1845–1850. doi: 10.1001/jama.1994.03520230055038. [DOI] [PubMed] [Google Scholar]
- [127].Fernando WK, Ariyananda PL. Paracetamol poisoning below toxic level causing liver damage in a fasting adult. Ceylon Med J. 2009;54:16–17. doi: 10.4038/cmj.v54i1.467. [DOI] [PubMed] [Google Scholar]