Abstract
Animal mRNAs are regulated by hundreds of RNA binding proteins (RBPs). The identification of RBP targets is crucial for understanding their function. A recent method, PAR-CLIP, uses photoreactive nucleosides to crosslink RBPs to target RNAs in cells prior to immunoprecipitation. Here, we establish iPAR-CLIP (in vivo PAR-CLIP) to determine, at nucleotide resolution, transcriptome-wide binding sites of GLD-1, a conserved, germline-specific translational repressor in C. elegans. We identified 439 reproducible target mRNAs and demonstrate an excellent dynamic range of target detection by iPAR-CLIP. Upon GLD-1 knockdown, protein but not mRNA expression of the 439 targets was specifically upregulated, demonstrating functionality. Finally, we discovered strongly conserved GLD-1 binding sites nearby the start codon of target genes. These sites are functional in vitro and likely confer strong repression in vivo. We propose that GLD-1 interacts with the translation machinery nearby the start codon, a so far unknown mode of gene regulation in eukaryotes.
Introduction
Gene expression in eukaryotes is extensively regulated at the post-transcriptional level. Animal genomes encode hundreds of RNA binding proteins (RBPs) that modulate maturation, stability, transport, editing and translation of target RNAs (Martin and Ephrussi, 2009; Moore and Proudfoot, 2009; Sonenberg and Hinnebusch, 2009). Numerous RBPs are associated with specific human diseases (Lukong et al., 2008), but the biological function of most RBPs is not known. Individual RBPs can bind and regulate hundreds to thousands of target RNAs (Lebedeva et al., 2011; Mukherjee et al., 2011) and consequently regulate multiple cellular processes at the same time. To understand the function of RBPs it is thus necessary to identify their targets and binding sites at a transcriptome-wide level.
Recent methods to biochemically identify targets of RBPs include RNA immunoprecipitation (RIP) or UV crosslinking and immunoprecipitation (CLIP) approaches, followed by microarray profiling or sequencing of bound RNAs (Konig et al., 2010; Tenenbaum et al., 2000; Ule et al., 2003). One of the more recent methods, PAR-CLIP, uses photoreactive nucleosides to crosslink RBPs to target RNAs in living cells prior to immunoprecipitation and next-generation sequencing of bound RNAs (Hafner et al., 2010). PAR-CLIP has three main attractive features. First, labeling of nascent RNAs with photoreactive nucleosides increases UV crosslinking efficiency and thus enables comprehensive target identification. Second, crosslinked target sequences are enriched for specific nucleotide changes within or next to RBP binding sites. For example, when labeling with 4-thiouridine, specific thymidine to cytidine changes (termed T to C conversions in what follows) are observed at nucleotides which are in physical contact to parts of the RBP (Hafner et al., 2010). Therefore, PAR-CLIP makes it possible to identify RBP binding sites at nucleotide resolution. Third, and perhaps most importantly, the conversions serve as internal controls reflecting how well target RNAs were crosslinked to RBPs and allow to directly separate crosslinked target sequences from the background of non-target RNAs.
The vast majority of RIP- or CLIP-based studies were carried out in cell lines, which allow mostly mechanistic but limited functional insights (e.g.(Lebedeva et al., 2011; Mukherjee et al., 2011)) because the function of RBPs is often context dependent. For example, it is well known that very early animal development is driven by complex regulatory interactions between numerous maternally supplied RBPs and their targets. These regulatory networks must be studied in vivo. Another example is the large class of microRNAs (miRNAs), which guide an RNA binding complex to specific binding sites on target mRNAs to regulate mRNA turnover and translation. miRNAs are known to regulate the majority of animal genes but are thought to frequently function in a context dependent manner (e.g. (Didiano and Hobert, 2006)). A famous model organism for studying developmental biology, miRNAs, or celltype-specific regulators in general is the nematode C. elegans. Its transparency makes it also a candidate for UV crosslinking based approaches, and indeed it has been shown that it is possible to use CLIP to extract regulatory interactions between the miRNA effector complex and miRNA targets in C. elegans (Zisoulis et al., 2010). However, this approach does not permit to obtain binding sites at nucleotide resolution and it is unclear if it is efficient enough to detect regulatory interactions in specific tissues of the worm because the miRNA effector complex is believed to be active in almost all tissues. We therefore reasoned that PAR-CLIP might allow us to overcome these difficulties, opening the exciting possibility to obtain a snapshot of most or all in vivo RBP:mRNA interactions for a RBP of choice, even if only expressed in a single tissue, and to resolve these regulatory interactions at nucleotide resolution.
To design a proof of principle experiment for in vivo PAR-CLIP (termed “iPAR-CLIP” in what follows), we selected the KH-domain containing RBP GLD-1 (Germline development Defective) as a suitable and interesting candidate. GLD-1 is a relatively well studied, conserved RBP that has multiple crucial functions during C. elegans germ cell development (Lee and Schedl, 2010). The most prominent phenotype upon loss of GLD-1 is a defect in maintenance of meiotic prophase during oogenesis, resulting in the formation of a tumor in the proximal part of the hermaphrodite germline (Lee and Schedl, 2010). GLD-1 is believed to be exclusively expressed in the adult germline (Lee and Schedl, 2010) and is known to recognize a relatively well defined primary sequence motif (Galarneau and Richard, 2009; Ryder et al., 2004; Wright et al., 2011). This allows to computationally check if biochemically identified binding sites on mRNAs are indeed likely to be directly bound by GLD-1. Importantly, in addition to a RIP-CHIP experiment, which identified hundreds of likely GLD-1 targets (Wright et al., 2011), there are ~20 functionally validated targets which can serve as positive controls. All of these targets are translationally repressed by GLD-1 (Lee and Schedl, 2010; Wright et al., 2011). Moreover, the availability of a GLD-1 flag tagged rescue strain allowed us to explore iPAR-CLIP based on well established flag IPs and independent of GLD-1 antibodies (Schumacher et al., 2005). Finally, GLD-1 belongs to the GSG/STAR protein family that includes human/mouse QUAKING, SAM68 and Drosophila HOW. STAR proteins share a conserved region containing a maxi-KH RNA binding domain suggesting similar RNA binding specificities (Lee and Schedl, 2010).
As it is the case for most RBPs, endogenous GLD-1 binding sites are known to reside in the 3' untranslated regions (3' UTRs) of target mRNAs. For GLD-1, the only known exception is the GLD-1 target gna-2 which has a binding site in its 5' UTR (Lee and Schedl, 2004). This binding site has been proposed to protect gna-2 from nonsense-mediated mRNA decay (Lee and Schedl, 2004). Altogether, specific regulation of target mRNA translation by RBPs has been much more extensively studied in 3' UTRs than in 5' UTRs. Very recently, one eukaryotic RBP that binds to 5' UTRs of target mRNAs (e.g. (Gebauer et al., 1999; Gebauer et al., 2003)) has been shown to regulate translation initiation at open upstream reading frames (Medenbach et al., 2011). Indeed, it seems likely that mapping the position of a RBP binding site within the context and organization of target mRNAs will not only be important for understanding the mechanisms of gene regulation mediated by specific binding sites but also to understand their function. With the ability of iPAR-CLIP to pinpoint binding sites at nucleotide resolution, we were able to start to systematically explore the binding repertoire of GLD-1 and its functional consequences.
To establish iPAR-CLIP, we first needed to explore how well mRNAs expressed in different tissues can be labeled with photoreactive nucleosides. We then performed multiple iPAR-CLIP experiments to carefully establish technical and biological reproducibility. Our computational methods, motif analyses, and known GLD-1 targets allowed us to assess the quality of the 439 reproducible target genes harboring thousands of binding sites identified by iPAR-CLIP. To test if iPAR-CLIP targets are indeed expressed in the germline, we sequenced mRNAs in worms with and without the germline (“RNA-seq”). These data allowed us also to quantify the dynamic range of target detection, i.e. if iPAR-CLIP is biased to detect highly expressed targets. To test whether regulation by GLD-1 is direct and depends on the presence of GLD-1 binding sites identified by iPAR-CLIP, we performed gelshift assays and created transgenic reporters expressing mRNAs with wildtype or mutated GLD-1 binding sites. However, in practice these methods do not allow to test the entire set of 439 target genes in vivo. We therefore knocked down GLD-1 and used a high-throughput quantitative shotgun proteomics approach to quantify changes in protein expression for thousands of genes. If the 439 targets indeed represent functional GLD-1 targets then we would expect that they are, on average, up-regulated in protein but not in mRNA expression.
Results
Labeled transcripts are efficiently UV crosslinked to interacting proteins in C. elegans
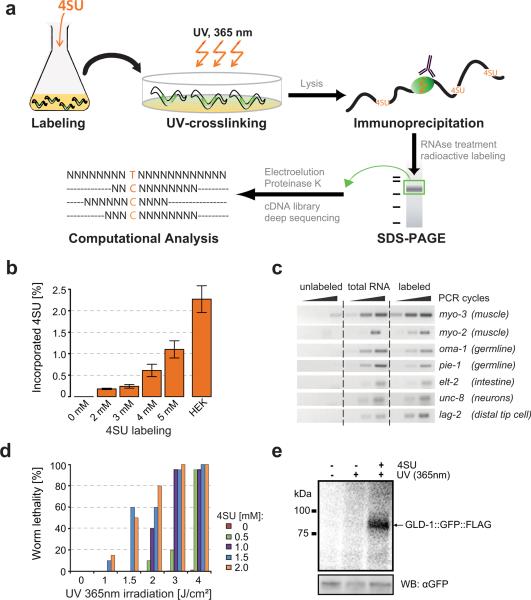
iPAR-CLIP relies on the labeling of newly transcribed C. elegans RNA with photoreactive nucleosides such as 4-thiouridine (4SU) or 6-thioguanosine (6SG). Labeled transcripts are subsequently UV crosslinked to proteins in living worms followed by immunoprecipitation and next-generation sequencing of bound RNAs (Figure 1a). 4SU or 6SG containing RNA can be thiol-specifically biotinylated and thereafter visualized on a membrane using streptavidin-antibody (described in (Dolken et al., 2008)). Using this assay we examined the incorporation of photoreactive nucleosides in C. elegans (Figure S1). The duration of the labeling, the concentration of photoreactive nucleosides and the amount of food in the medium influenced 4SU/6SG incorporation (Figure S1). Nevertheless, we were able to select parameters that allowed relatively efficient labeling while minimizing costs. Alternatively, C. elegans can be labeled on plates by adding photoreactive nucleosides to the food source (Figure S1e). For exact determination of 4SU incorporation rates into C. elegans total RNA we established a LC-MS/MS based method which allowed to quantitatively measure uridine and 4SU. Depending on the concentration of 4SU used, 0.2 – 1.1% of uridines were replaced by 4SU. Thus, worms can be labeled with an incorporation rate that is comparable to the incorporation rate used for PAR-CLIP in cell lines (Figure 1b). To test whether transcripts in different tissues are labeled, we separated labeled from non-labeled C. elegans RNA using streptavidin beads. Labeled RNA was eluted from the beads, reverse transcribed and the presence of tissue-specific transcripts was assayed by PCR. All tested transcripts (e.g. muscle-, germline-, intestine- and neuron-specific mRNAs) were detected in the labeled fraction. Compared to total RNA their relative quantities remained unchanged, indicating uniform labeling across tissues (Figure 1c).
Figure 1. in vivo PAR-CLIP.
(a) iPAR-CLIP methodology. Photoreactive nucleosides (4-thiouridine) were added to C. elegans L1 larvae in liquid culture. Alternatively, 6-thioguanosine (6SG) can be used. Adult worms expressing a GLD-1::GFP::FLAG fusion protein in the germline (marked in green) were irradiated with UV light (365nm). After immunoprecipitation of GLD-1 crosslinked RNA was partially digested and radiolabeled. Protein-RNA-complexes were size-separated on a denaturing gel. After gel elution, proteins were digested by Proteinase K and RNA was converted into a cDNA library for next generation sequencing. Binding sites of RNA binding proteins can be identified by a high incidence of conversions (T to C when using 4SU) in the sequence reads. (b) 4SU incorporation rates after labeling of worms with different concentrations of 4SU were measured by LC-MS/MS. For comparison, incorporation of 4SU in HEK293 cells labeled as described in (Hafner et al., 2010) is shown. Error bars due to technical variability range from the lowest to the highest incorporation rates measured in one sample. (c) PCRs showing labeling of transcripts specifically expressed in muscle (myo-2, myo-3), germline (oma-1, pie-1), intestine (elt-2), neurons (unc-8) or in a few cells including the distal tip cell (lag-2). Labeled: Total RNA of 4SU-labeled worms was biotinylated and isolated using streptavidin-beads. Unlabeled: same procedure using non 4SU labeled worms. (d) Adult worms labeled with different concentrations of 4SU were UV-irradiated with different doses of UV 365nm light. Dead and surviving worms were counted. (e) Phosphorimage of SDS-gel resolving 5′-32P-labeled RNA crosslinked to GLD-1::GFP::FLAG immunoprecipitates. IPs were prepared from lysates of worms grown in the presence or absence of 4SU and crosslinked with UV 365 nm light. For comparison, samples prepared from non-irradiated worms were included. Lower panel: immunoblot with anti-GFP antibody (loading control).
When 4SU-labeled and unlabeled worms were UV irradiated, 4SU-labeled worms died depending on the amount of 4SU used for labeling and the UV irradiation time (Figure 1d). Since unlabeled worms survived, we assumed that labeled worms die due to crosslinking of 4SU-containing transcripts to interacting proteins. Next, we 4SU-labeled and UV crosslinked adult worms that express a rescuing GLD-1∷GFP∷FLAG fusion protein (BS1080, provided by Tim Schedl). GLD-1 was immunoprecipitated using anti-FLAG antibody. Covalently bound RNA was partially digested, radioactively labeled and RNA-protein complexes were size-separated on a denaturing gel (Figure 1e). In 4SU labeled worms, RNA was efficiently UV crosslinked to GLD-1. The crosslinked RNA was recovered and converted into a cDNA library, which was then deep sequenced (Illumina).
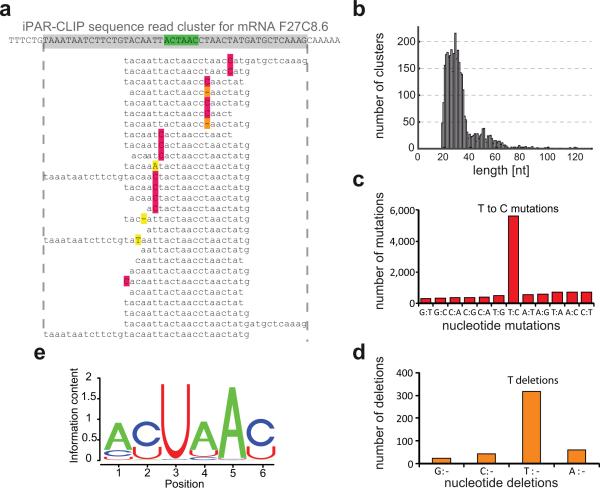
Sequence reads were mapped to the transcriptome (ModENCODE gene models (Gerstein et al., 2010)) and overlapping or adjacent sequence reads mapping with edit distance 0 or 1 were organized into sequence read clusters (Figure 2a). Identical reads were treated as a single read as they might be PCR amplification contaminations. Most clusters are between 18 and 70 nucleotides in length (Figure 2b). In the clustered sequence reads, T to C conversions are around 10 fold enriched over all other mutations, reflecting efficient 4SU-protein crosslinking (Figure 2c). At the same time, T deletions were up to 10 fold enriched over all other deletions (Figure 2d), consistent with previous observations (Kishore et al., 2011; Zhang and Darnell, 2011). We therefore considered T to C mutations as well as T deletions as T conversions which flag nucleotides that are directly crosslinked to the RBP and ranked the sequence read clusters by the total number of T conversions within each cluster.
Figure 2. Identification of GLD-1 target sites by iPAR-CLIP.
(a) Sequence reads obtained by iPAR-CLIP were mapped to C. elegans mRNAs and, after removing redundant reads, organized into sequence read clusters (GLD-1 binding motif highlighted in green, T to C conversions in red, T deletions in orange). (b) Length distribution of the identified sequence read clusters. In the sequence read clusters, T to C mutations (c) and T deletions (d) were ~10 fold enriched over other types of mutations or deletions. (e) GLD-1 binding motif (p value < 10−250) identified in the top 100 clusters using MEME.
iPAR-CLIP recovers all known GLD-1 targets and the known binding motif
Previously identified GLD-1 targets served as positive controls to test, whether iPAR-CLIP enables comprehensive identification of RNA-protein interactions. We compiled 18 targets from the literature that have been studied in detail and for all of these GLD-1 was shown to act as a translational repressor (Table S1). Using iPAR-CLIP, we were able to identify all of them. Moreover, we identify 8 mRNAs that were suggested as GLD-1 targets but not further validated (Table S2). Remarkably, we find 9 previously identified GLD-1 targets in our top 30 targets ranked by the number of T conversions (Table S3).
A motif search (MEME (Bailey and Elkan, 1994)) on the top 100 identified clusters resulted in a highly significant GLD-1 binding motif (p-value < 10−250) that is not identical but in agreement with previously identified binding sites (Figure 2e) (Ryder et al., 2004; Wright et al., 2011). However, our identified binding motif is almost identical to the recently published binding motif of QUAKING, the human ortholog of GLD-1 (Hafner et al., 2010). This result is explained by the observation that the RNA-binding domains of GLD-1 and QUAKING are highly conserved (Lee and Schedl, 2010).
Identified targets and binding sites are highly reproducible
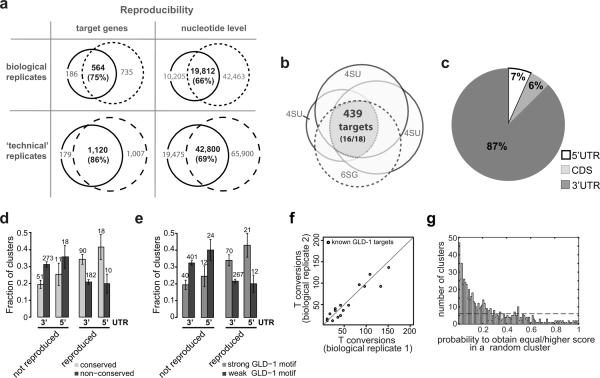
We considered it critical to assay biological and technical reproducibility of iPAR-CLIP. We performed three 4SU iPAR-CLIPs, including two biological (1 and 2) and two technical replicates (2 and 3). In two biological replicates we identify 750 (1) and 1,299 (2) putative GLD-1 targets when considering all clusters with at least two T conversions. In what we call `technical replicates', worms were combined for labeling, but UV crosslinked separately. In (3) the UV irradiation time was prolonged compared to (2). After UV crosslinking samples were again processed in parallel. In (3) we detect 2,127 putative targets, suggesting that the number of identified interactions increases with UV irradiation time.
564 of 750 targets identified in (1) were also found in (2), indicating a biological reproducibility of 75% for the smaller set of targets (Figure 3a). Biological reproducibility increases to 82% when only clusters with at least 3 T conversions are included. 1,120 of 1,299 targets identified in (2) were also identified in (3), suggesting a technical reproducibility of at least 86% (Figure 3a). Remarkably, reproducibility on nucleotide level in the same `technical replicates' is 69% (72%) for all clusters with at least two (three) conversions, demonstrating that not only targets but also binding sites can be reproduced well. Moreover, the number of T conversions in known GLD-1 targets strongly correlates in biological replicates (Figure 3f).
Figure 3. iPAR-CLIP is highly reproducible.
(a) 75% of the target genes and 66% of the nucleotides identified in iPAR-CLIP sequence read clusters could be reproduced in a biological replicate. Technical reproducibility on gene and nucleotide level is at least 86% and 69%, respectively. (b) Three 4SU iPAR-CLIPs and one 6SG iPAR-CLIP overlap in 439 targets, including 16 of 18 known GLD-1 targets. (c) For the 439 targets, 87% of the identified clusters were mapped to 3′UTRs, 7% to 5′UTRs. (d) Reproduced target sites, that were identified in all four iPAR-CLIP experiments, but not non-reproduced sites (identified in one iPAR-CLIP) are highly likely to be conserved (see Supplemental Methods). Error bars represent the standard deviation computed from binomial statistics and reflect the uncertainty due to the limited number of data points. (e) Reproduced, but not non-reproduced target sites are highly likely to contain strong GLD-1 binding motifs (see Supplemental Methods). (f) Number of T conversions in known GLD-1 targets in two biological replicates, normalized by the number of sequence reads. (g) Each cluster was tested for the presence of a GLD-1 binding motif and assigned a score. Plotted is the probability to obtain an equally high or higher score in a random cluster of same length and similar dinucleotide composition for the 630 identified clusters. 39% of the 630 identified clusters can be explained singly by the presence of GLD-1 motifs over background (dashed line).
439 genes are stable, reproducible GLD-1 targets
In addition to 4SU iPAR-CLIP experiments, we performed iPAR-CLIP using the photoreactive nucleoside 6-thioguanosine (6SG). We thereby wanted to account for any bias in the composition of crosslinked target sequences due to crosslinking of uridines, or due to the use of RNAse T1 (Kishore et al., 2011; Lebedeva et al., 2011). In PAR-CLIP experiments RNAse T1 is used to partially digest crosslinked target RNAs after immunoprecipitation (see Methods). Since RNAse T1 cuts after guanosines it might deplete G-rich target sequences. By using 6SG, G-containing target sequences will be crosslinked, stabilized and protected against digestion. Alternatively, a different RNAse without a nucleotide bias can be used (Kishore et al., 2011). Three 4SU iPAR-CLIPs and one 6SG iPAR-CLIP overlapped in 439 identified targets (Figure 3b and Table S4). The set of 439 targets contains 16 of 18 validated functional GLD-1 targets and is thus highly enriched for biologically relevant GLD-1 targets. Compared to all germline-expressed genes, GO-Term-Analysis for the 439 candidates results in the top-scoring categories `cell division' (37 genes; Bonferroni-corrected p-value < 1.9 · 10−7), `cytokinesis' (28; 4.5 × 10−6), `cell fate commitment' (15; 7.4 × 10−5), `embryonic development ending in birth or egg hatching' (165; 1.4 × 10−4) (Table S5). All of these terms are in agreement with the functional roles of GLD-1 in the C. elegans germline.
87% of the 630 sequence read clusters in 439 genes mapped to 3'UTRs and 7% mapped to 5'UTRs (Figure 3c). We observed that 5'UTR and 3'UTR binding sites that were manifestly conserved during nematode evolution (see Supplemental Methods) were more likely to be identified in multiple iPAR-CLIP experiments than non-conserved sites (Figure 3d). Thus, binding sites confirmed in separate experiments are more likely to be functional compared to sites that were only present in one experiment. Reproduced target sites in 5'UTRs and 3'UTRs are further more likely to contain strong GLD-1 binding motifs (see Supplemental Methods) than non-reproduced sites (Figure 3e).
Identified targets are highly enriched for the GLD-1 binding motif
Each of 630 clusters in 439 genes was tested for the presence of a GLD-1 binding motif and assigned a score. We calculated the probability to obtain an equally high or higher score in a random cluster of similar dinucleotide composition and show that GLD-1 sequence read clusters are highly enriched for the GLD-1 binding motif (Figure 3g) over background. In this calculation, 39% of the 630 identified clusters can be explained by the presence of GLD-1 motifs over background. However, the background model used in this calculation is exceedingly conservative. The reason is that clusters are depleted in G's because the GLD-1 binding motif does not contain G's. When using the much more balanced nucleotide composition of C. elegans 3' UTRs as background, ~60% of the 630 clusters can be explained by GLD-1 binding.
Identified targets are expressed in the germline and distributed over a wide range of mRNA expression levels
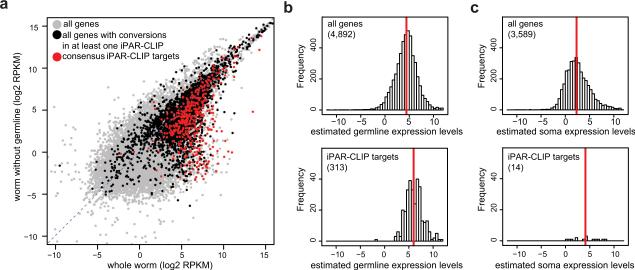
It is important to test whether iPAR-CLIP allows to specifically identify GLD-1 targets in the germline and to discriminate them from RNAs that are expressed in other celltypes but might encounter GLD-1 after lysis. To obtain a comprehensive list of transcripts expressed in the germline or soma of adult worms, we performed mRNA sequencing of the GLD-1∷GFP∷FLAG rescue strain used in our experiments and a glp-4 mutant strain that develops all somatic tissues but does not develop gonads at restrictive temperatures (Beanan and Strome, 1992). This strategy was developed previously (Reinke et al., 2000) but was carried out using microarrays and gene models that since then have been heavily modified. To estimate the expression of transcripts present in the C. elegans germline, expression levels of transcripts (computed as RPKMs, Supplemental Methods) in the worm without germline were weighted with the relative volume of germline/(germline + soma) and subtracted from those in the rescue strain, resulting in positive RPKM values only for germline-expressed genes (see Supplemental Methods). Using these datasets, we find that the vast majority of identified GLD-1 targets are expressed in the germline (Figure 4a). The identified targets are distributed over a wide range of expression levels. Compared to all mRNAs present in the germline, they show only a slight shift towards higher mRNA expression levels, indicating that iPAR-CLIP captures both highly and lowly expressed transcripts and thus has a good dynamic range of target detection (Figure 4b,c).
Figure 4. GLD-1 targets are expressed in the germline and distributed over a wide range of expression levels.
(a) mRNAs from wildtype worms and mutant worms that lack a germline were sequenced (RNA-seq). RPKM values reflect mRNA abundances (see Methods). The vast majority of the 439 reproducible GLD-1 targets (red) are expressed in the germline. GLD-1 target genes which are supported by at least one iPAR-CLIP replicate but not by all four (black) have less distinct germline expression. (b) RPKMs from mutant worms (glp-4) were weighted to account for the different proportions of somatic cells in the different samples and subtracted from wildtype RPKMs (Supplemental Methods). The resulting RPKMs quantify, if positive and significant (p-value < 0.01), expression levels in the germline (Supplemental Methods). Upper panel: histogram of germline expression levels (log units) for all genes with confident fold changes. Lower panel: For the 313 iPAR-CLIP targets for which we can confidently quantify germline expression, the detection range spans four orders of magnitude in expression levels and is only slightly shifted towards higher RPKM values compared to background expression. Red line: median (c) Soma expression levels were computed analogously (right panels). Only 14 out of 439 iPAR-CLIP targets were confidently quantified as exclusively expressed in somatic tissues and not expressed in the germline at detectable levels.
iPAR-CLIP identified functional GLD-1 targets
To test whether targets identified by iPAR-CLIP are functional, we knocked down GLD-1 by RNAi and measured changes in protein abundance compared to mock-treated worms. To accurately quantify protein expression in C. elegans we applied an in vivo stable isotope labeling method, the “SILAC worm”, that we and colleagues have recently established (Grün, Kirchner, Thierfelder et al, in preparation). The method allows for accurate and precise measurements of changes in protein abundances between any two C. elegans samples. In two measurements, we obtained SILAC ratios between GLD-1 RNAi and control sample for proteins corresponding to 3,874 unique gene loci, among them 2,795 germline expressed proteins and 217 of the 439 identified GLD-1 targets. Protein quantification by SILAC and mRNA quantification by RT-qPCR indicate that GLD-1 is around 2-fold downregulated after RNAi.
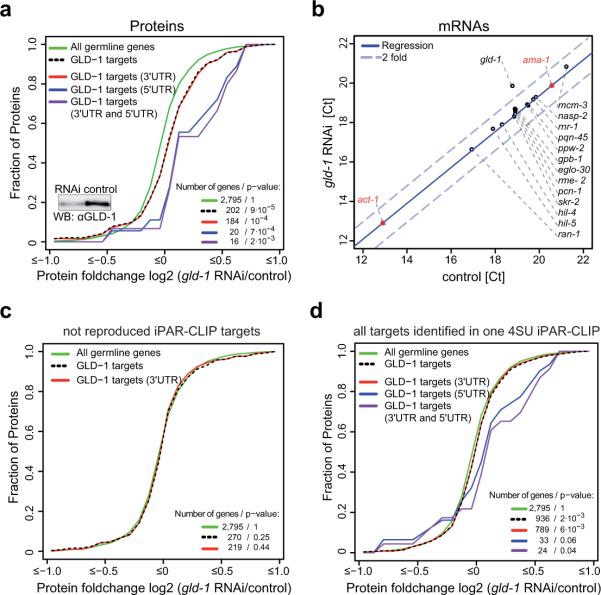
Upon knockdown of GLD-1, GLD-1 targets show a highly significant shift towards higher protein expression levels (p-value < 9 · 10−5) (Figure 5a). To test whether the change in protein expression is transcriptionally or post-transcriptionally driven, we performed RT-qPCRs for 14 randomly selected targets that are de-repressed upon GLD-1 knockdown, for gld-1 and two negative controls. With the exception of gld-1 mRNA, mRNA levels of the tested genes remain constant upon GLD-1 knockdown, indicating that GLD-1 regulates identified targets at the post-transcriptional level (Figure 5b).
Figure 5. Identified iPAR-CLIP targets are functional in vivo.
(a, c, and d) show cumulative fractions of fold-changes in protein expression after GLD-1 knockdown. Protein fold changes for altogether 3,874 genes were measured by using SILAC in C. elegans. (a) Out of the 439, protein fold changes for 202 germline-expressed, reproducible iPAR-CLIP targets were obtained. Upon GLD-1 knockdown, reproducible iPAR-CLIP targets show a highly significant shift (p value < 9 · 10−5) towards higher protein expression levels compared to all genes expressed in the germline. Changes in protein abundance averaged over targets that contain 3′UTR sites, targets that contain 5′UTR sites and targets that contain both 3′UTR and 5′UTR sites are shown. (b) RT-qPCRs for 14 targets that change on protein level upon GLD-1 knockdown (same samples as in (a)). RT-qPCRs for gld-1 itself and 2 negative controls (ama-1, act-1) were included. (c) In contrast to the reproducible set of 439 iPAR-CLIP targets, targets identified in only one of four iPAR-CLIP experiments are not de-repressed upon GLD-1 knockdown. (d) Changes in protein abundance upon GLD-1 knockdown averaged over all 2,127 putative targets identified in one iPAR-CLIP replicate.
In contrast, putative targets that could not be reproduced in any of the iPAR-CLIP replicates do not show any significant change in protein expression upon GLD-1 knockdown (Figure 5c). Similarly, we did not see a significant change in protein expression, when we as an extreme case averaged over all 2,127 transcripts detected in one iPAR-CLIP experiment (Figure 5d). These control experiments demonstrate that (a) the group of 439 genes is specifically up-regulated upon GLD-1 knockdown, and thus very likely directly regulated by GLD-1 (b) it is necessary to do iPAR-CLIP in replicates.
Surprisingly, we observed that targets containing 5'UTR sites in addition to 3'UTR sites show a more pronounced shift towards higher expression levels than all 3'UTR targets (Figure 5a). We measured protein fold changes for 20 targets that contain 5'UTR sites. Compared to all targets with binding sites in 3' UTRs, they show a significantly higher de-repression upon GLD-1 knockdown (p-value < 0.03, Figure S2a).
In vitro and in vivo validation of selected GLD-1 binding sites
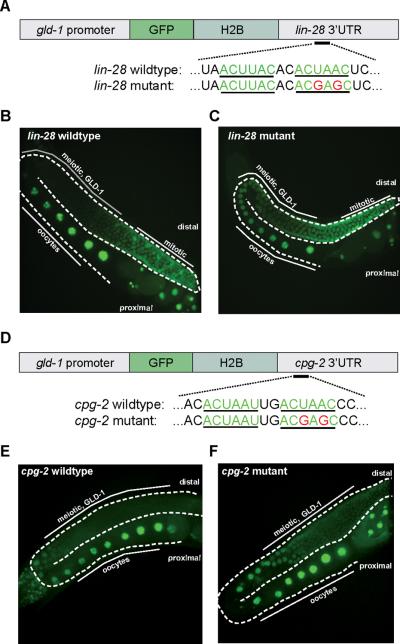
We used quantitative gelshift assays and transgenic reporters to test whether regulation of targets by GLD-1 is direct. In our iPAR-CLIP experiments, we identified lin-28 as a mid-range scoring target of GLD-1. lin-28 has not been described to be expressed in the germline and is not known to be a GLD-1 target. We first validated germline expression of the lin-28 mRNA (by RNA-seq and PCR on dissected gonads, data not shown). Next, we confirmed binding of GLD-1 to the identified target site in its 3'UTR in vitro (Figure 7b, Figure S3 and S4). For in vivo validation, we cloned the 3'UTR of lin-28 downstream of green fluorescent protein (GFP) fused to Histone H2B and used the gld-1 promoter to drive expression of the fusion protein throughout the germline (Figure 6a). We used the Mos1-mediated single copy insertion technique (mosSCI (Frokjaer-Jensen et al., 2008)) to integrate the constructs into the C. elegans genome. The reporter was expressed in the distal, mitotic region of the germline, in developing oocytes and embryos. GLD-1 protein is absent in these parts of the germline (Lee and Schedl, 2010). In the pachytene region, where GLD-1 is present, we did not observe GFP expression (Figure 6b). However, when we mutated one of two GLD-1 binding motifs in the lin-28 3'UTR, the reporter showed continuous expression throughout the germline (Figure 6c). The results show that GLD-1 can repress lin-28 in vivo and that the interaction is direct and depends on the presence of the GLD-1 binding motif. In addition to that, we designed a pgld-1∷GFP-H2B∷cpg-2 reporter construct. Since cpg-2 immunoprecipitates with GLD-1 (Lee and Schedl, 2001), it was suggested as a GLD-1 target but not further validated. In our iPAR-CLIP experiments, cpg-2 is one of the highest ranking targets (Table S3) and we show that mutation of one GLD-1 binding motif in the cpg-2 3'UTR causes de-repression of the reporter in vivo (Figure 6d–f).
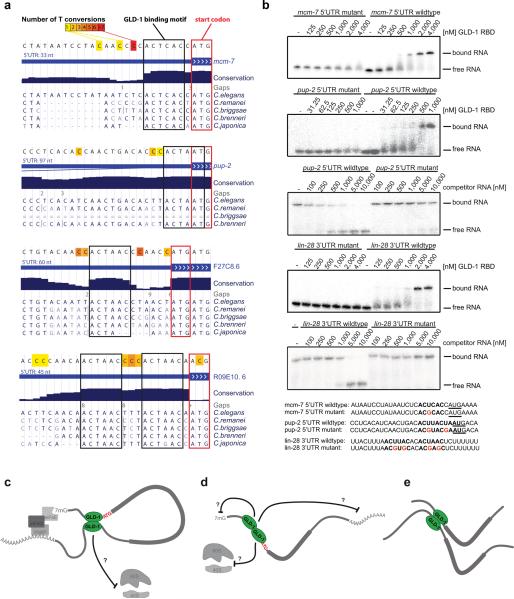
Figure 7. GLD-1 binds close to the start codon.
(a) Examples of 5′UTR binding sites with highly conserved GLD-1 binding motifs directly upstream of start codons. The number of T conversions is color-coded. Nucleotides surrounding binding sites are not conserved. (b) Gelshift assays demonstrating binding of the GLD-1 RNA binding domain (RBD) to 5′UTR target sites in vitro, depending on the GLD-1 binding motif. For direct titration experiments, radiolabeled RNA was incubated with increasing concentrations of GLD-1. For competition binding assays, unlabeled competitor RNA was titrated into labeled GLD-1 RNA complexes (Supplemental Methods). Mutant competitor RNAs did not outcompete wildtype RNAs in the tested range of concentrations, indicating that their affinity is at least one order of magnitude lower compared to wildtype sequences. (c–e) Models for GLD-1 dimer dependent translational regulation via the 5′UTR (see discussion).
Figure 6. Identified iPAR-CLIP binding sites are functional in vivo.
(a,d) Reporter constructs containing a gld-1 promoter fused to GFP::Histone 2B and lin-28 or cpg-2 3′UTRs with unaltered or mutated versions of the GLD-1 binding motif were introduced into C. elegans. (b,c,e,f) Mutation of the GLD-1 binding motif leads to de-repression of reporter constructs in the meiotic region of the germline where GLD-1 is expressed.
GLD-1 regulates targets by binding to highly conserved 5'UTR sites near start codons
Finally, we discovered that GLD-1 frequently binds to 5'UTR sites nearby (or overlapping with) the start codon of target genes. These sites are often highly and specifically conserved within nematode species, indicating that they are functionally important (Figure 7a and Figure S5). We performed gelshift assays using immunoprecipitated, full-length GLD-1 (Figure S4) or the GLD-1 RNA binding domain (Figure 7b) to confirm that GLD-1 binds to identified 5'UTR target sites near start codons in vitro. The affinity of GLD-1 for the tested 5'UTR sites and the in vivo validated lin-28 3'UTR target site are in the same order of magnitude (Figure S3). In contrast, the affinity of mutated GLD-1 target sites is reduced by at least one order of magnitude compared to the respective wildtype (Figure 7b). Moreover, T conversions in the iPAR-CLIP sequence reads appear directly next to and within start codons. In a systematic analysis using three 4SU iPAR-CLIP datasets, we computed the relative distances of T conversions to start codons, demonstrating that crosslinked nucleotides reside frequently proximal to start codons (Figure S6). Restricting the computation to 5' UTRs of at least 100 nt in length did not change the overall shape of the histogram (data not shown), suggesting that these sites cannot be explained by short 5' UTRs.
Taken together, our data suggest that GLD-1 binds to highly conserved target sites near start codons.
Discussion
iPAR-CLIP identifies binding sites of RBPs in C. elegans
We performed a proof of principle experiment and identified hundreds of target sites for the RBP GLD-1 at nucleotide resolution in the model organism C. elegans. We determined parameters at which C. elegans can be labeled by adding photoreactive nucleosides to worm cultures or on plates at a moderate cost per iPAR-CLIP. We did not observe changes in developmental speed or morphology of labeled worms for 4SU/6SG concentrations used in our experiments. Thus, we believe that iPAR-CLIP allows at least in principle to identify targets of RBPs in all tissues. However, our proof of principle experiment was carried out for an RBP expressed in the germline, which comprises about a third of adult cells. It is not clear how well iPAR-CLIP would work for RBPs that are expressed in only a few cells. Naively, it should be possible and sufficient to simply scale up the input material by increasing the number of worms or the concentration of photoreactive nucleosides used for labeling. We note that only at higher labeling concentrations (> 5mM) we observed a slightly retarded development but no other obvious phenotypes. In any case, we have shown that iPAR-CLIP has a good dynamic range of target detection covering expression levels over four orders of magnitude. Thus, at least relatively highly expressed targets in small tissues should be detectable.
iPAR-CLIP identified a large number of functional and direct GLD-1 targets
We identified 439 GLD-1 targets that were reproducible in four experiments with different photoreactive nucleosides and UV irradiation times. This target list, together with a large amount of additional detailed information, is provided as Supplemental Material or can be browsed in the context of the UCSC genome database using DORINA (http://dorina.mdc-berlin.de) (Anders et al., NAR 2011, in press).
We carried out several follow up experiments and analyses that showed that (a) the vast majority of the 439 targets is, as expected, expressed in the germline and target expression covers four orders of magnitude (Figure 4a–c), suggesting a good dynamic range of target detection (b) the GLD-1 binding motif is extremely significantly enriched in the target sites (p value ~ 0) and 40–60% of the target sites can be explained by the GLD-1 binding motif, showing that a large fraction of the targets are directly bound by GLD-1. These results can be further strengthened by considering a GLD-1 “affinity score” that was recently published based on classical RIP-CHIP experiments (Wright et al., 2011). 64% of the 439 genes contain binding sites with at least one of the top 30 high affinity nucleotide 7mer motifs present (data not shown). Further, reproducible target sites, but not non-reproducible sites, are highly likely to contain strong GLD-1 motifs (affinity score ≥ 1, see Supplemental Methods and Figure 3e). (c) quantitative gel shift assays validated binding sites (d) using transgenic reporters, we show for cpg-2, a high-ranking GLD-1 target, that mutation of the identified GLD-1 binding site causes de-repression in vivo. We could further validate lin-28, a mid-range scoring target of the 439, as a direct GLD-1 target by transgenic reporters (e) when knocking down GLD-1, a translational repressor, protein expression of the 439 targets was highly significantly up-regulated. However, mRNA expression of all tested targets remained constant, which makes indirect transcriptional effects extremely unlikely. (f) reproducible target sites, but not non-reproducible sites, are highly likely to be conserved (see below).
Taken together, these results show that the 439 targets are not only highly enriched in direct but also functional targets. We note that our GLD-1 knockdown was suboptimal in strength as we observed that GLD-1 in numerous worms was not efficiently knocked down, explaining the relatively small protein fold changes that we measure via SILAC. We further note that ~70% of our 439 targets can also be validated by RIP-CHIP experiments (Wright et al., 2011) (Figure S7). Interestingly, the difference between these two data sets comes mostly from genes which are expressed at low or mid-range levels (Figure S7). In any case, the power of iPAR-CLIP is perhaps most convincingly demonstrated by our ability to map binding sites at nucleotide resolution in vivo which allowed us for example to identify a set of targets with GLD-1 binding sites nearby the start codon (see below).
Reproducibility correlates with functionality
Our replicate experiments showed that the size of the set of target genes can significantly vary between iPAR-CLIP experiments. For example, when we prolonged the UV irradiation time, more RNA-protein interactions were captured. Nevertheless, genes from a smaller target set are typically also present in the larger set (75–86%, Figure 3a). An explanation for this scenario is that iPAR-CLIP is a highly sensitive method that captures transient interactions in addition to stable, biologically relevant targets. Several lines of evidence support this explanation and demonstrate that reproducible targets are more likely to be functional than others: (a) almost all previously known functional targets are contained in the set of 439 reproducible iPAR-CLIP targets (Figure 3b) (b) protein levels of the 439 targets were significantly up-regulated when knocking down GLD-1 (Figure 5a) but not so protein expression of the set of targets identified exclusively in a single iPAR-CLIP experiment (Figure 5c,d) (c) conserved target sites are more likely to be reproduced across different experiments than non-conserved sites (Figure 3d). It therefore appears to be important to carry out iPAR-CLIP in several replicate experiments.
GLD-1 binds to highly conserved 5'UTR sites near start codons
With the exception of a single case, all known GLD-1 binding sites have been identified in 3'UTRs. Surprisingly, we discovered that 7% of target sites for the 439 reproducible iPAR-CLIP target genes reside in 5'UTRs. We believe that these sites are functional due to the following reasons. (a) Targets that contain 5'UTR sites in addition to 3'UTR sites show on average a much stronger de-repression upon GLD-1 knockdown (Figure 5a). This difference in response to the knockdown between 3'UTR targets and targets with 5' UTR sites is statistically significant (p-value < 0.03) and cannot be explained with a higher number of binding sites for targets with 5' UTR sites (Figure S2). In fact, on average, targets with 5' UTR sites have fewer sites in 3' UTRs compared to targets with 3' UTR sites only (Figure S2c). Furthermore, while we could not confirm functionality when we averaged protein fold changes over all 2,127 transcripts detected in one iPAR-CLIP experiment, we see a significant shift (Figure 5d, p value < 0.04, 0.06) in protein expression upon GLD-1 knockdown if we restrict to targets with 5'UTR sites. These results also indicate that 5' and 3' UTR sites function via different mechanisms. (b) gel-shifts confirmed direct binding of GLD-1 to the 5' UTRs of mcm-7 and pup-2 (Figure 7b) (c) We found a strong correlation between detecting cross linking in GLD-1 binding sites in 5' UTRs and conservation of the GLD-1 binding motif within the binding site (Figure 3d) (d) As exemplary shown in Figure 7a, we often observed that nucleotides just outside of the conserved GLD-1 binding motif are mutated between different nematode species, indicating that 5' UTR GLD-1 binding sites are specifically deeply conserved (e) Intriguingly, we observed that binding sites in 5' UTRs often reside nearby the start codon (~20 nucleotides, Figure S6) and that the number of observed biochemical RNA-protein crosslink interactions, quantified by T conversions, peaks nearby the start codon. This effect cannot be explained by small 5' UTR sizes because it still holds true when restricting the analysis to 5' UTRs which are at least 100 nt long (data not shown).
In animals, little is known about RBPs that specifically bind sites in 5' UTRs to regulate translation. In Drosophila, Sex lethal (SXL) has recently been shown to bind downstream of upstream open reading frames (uORFs) to regulate initiation at the start codon of the uORF and thus to indirectly modulate translation of the target gene (Medenbach et al., 2011). For GLD-1, binding to the 5'UTR of a single target gene, gna-2 (also identified in our experiments), has been described (Lee and Schedl, 2004). gna-2 harbors two upstream open reading frames (uORFs). Binding by GLD-1 has been proposed to repress translation and at the same time protect gna-2 from non-sense mediated decay, likely by inhibiting translation of uORFs (Lee and Schedl, 2004). By that time, the exact GLD-1 binding site in the gna-2 5'UTR could not be identified. Using iPAR-CLIP we detected crosslinked nucleotides and a strong GLD-1 binding motif precisely overlapping the stop codon of the uORF nearby the start codon of the CDS. GLD-1 binding could thus directly influence both translation of the uORF and the main ORF. We carefully checked all 5' UTRs for which we discovered GLD-1 binding sites for the presence of uORFs. For five targets, including gna-2, we observe highly conserved GLD-1 binding motifs overlapping with stop codons of uORFs (Y97E10AL.2, T04C12.5, C05C10.5, T23G11.2, R09E10.6). Cooperation of GLD-1 with uORFs is thus likely one mechanism of target regulation. However, whereas nearly all identified 5'UTR targets show binding sites close to the start codon, only a small number of these contain uORFs: 26 of 30 genes with reproduced 5'UTR clusters show GLD-1 binding near the start codon and 20 of the 26 do not contain uORFs. The 5' UTR for eight of the 26 is longer than 100 nt. We thus speculate that GLD-1 can also regulate targets by interacting with the translation machinery nearby the start codon, a so far unknown mode of gene regulation in eukaryotes.
What could be mechanistic and functional consequences of GLD-1 binding nearby start codons? GLD-1 is thought to bind as a dimer (Lee and Schedl, 2010). Under this assumption, several distinct architectural consequences of GLD-1 binding to 5' UTRs arise (Figure 7c–e). Binding of GLD-1 as a dimer or oligomer to both 5'UTR and 3'UTR sites may stabilize or promote circularization of mRNAs while blocking translation initiation at the start codon (Figure 7c). Release of GLD-1 would then allow rapid initiation of translation. Conversely, accessing a binding site nearby the start codon of an actively translated and thus circularized target may be promoted by a site in the 3' UTR. A testable prediction of this model would be that disrupting binding to the 3' UTR site is expected to have an impact on binding the 5' UTR site (and vice versa). This scenario stands in contrast to GLD-1 binding as a dimer to 5' UTR sites only (Figure 7d) which may also be an efficient way to repress translation of GLD-1 targets. However, this mode of GLD-1 binding might be more appropriate for cases where a less rapid regulatory response upon changes in GLD-1 levels is wanted. We note that iPAR-CLIP identified cases of two nearby binding sites within the same 5' UTR as well as cases of orphan 5' UTR sites. Thus, at this point there is support for both models. As a third possibility GLD-1 could also help to bring together different target molecules (Figure 7e), a possible mechanism for regulating localization of RNA molecules.
Clearly, further experiments are needed to distinguish between these models and to also understand the cases where GLD-1 binds stop codons of uORFs. However, our results illustrate that a starting point for a detailed understanding of the function of GLD-1 interactions with RNA is the knowledge of the precise position of binding sites within the target mRNA and relative to binding sites of other interacting RBPs. We believe that we have shown that iPAR-CLIP makes it possible to explore this “post-transcriptional regulatory code” in vivo.
Methods
I. Experimental Procedures
Labeling of C. elegans with photoreactive nucleosides
Arrested L1 worms were typically grown in liquid culture supplemented with 2mM 4-thiouridine (4SU) or 6-thioguanosine (6SG) and harvested at the adult stage (Supplemental Methods).
Isolation of labeled RNA and dot-blot assays
Thiol-specific biotinylation, dot-blot assays and pull-down of labeled RNA using streptavidin-beads were carried out as described previously (Dolken et al., 2008) (Supplemental Methods).
In vivo PAR-CLIP
Synchronized L1 worms were grown in liquid culture supplemented with 2mM 4SU or 6SG. Living adult worms were transferred to NGM plates and crosslinked on ice using a Stratalinker (Stratagene) with customized 365nm UV-lamps (energy setting: 2J/cm2). Worms were lysed on ice and cleared lysates were treated with RNase T1 (1U/μl) for 15 min at 22°C. GLD-1∷GFP∷FLAG fusion proteins were immunoprecipitated for 1h at 4°C using anti-FLAG antibody (Supplemental Methods). Immunoprecipitates were treated with RNase T1 (100U/μl) for exactly 12 min at 22 °C. Subsequently, PAR-CLIP was carried out as described previously (Hafner et al., 2010). cDNA libraries were sequenced on a Genome Analyzer II (Illumina).
in vivo SILAC
In vivo SILAC was performed as described in (Grün, Kirchner, Thierfelder et al., in preparation). Briefly, worms were metabolically labeled with 15N 132C6-Lysine (Cambridge Isotope Laboratories, USA, hereafter referred to as “heavy” Lysine) by feeding them with the metabolically labeled lysine auxotroph Escherichia coli strain AT713. Worms were cultivated for one generation on peptone-free NGM plates supplemented with antibiotic-antimycotic (Invitrogen, USA), seeded with an excess of labeled bacteria. An unsynchronously growing population of L4 to adult stage worms was used as “heavy” reference for LC-MS/MS (Supplemental Methods).
II. Computational Analysis
Mapping and clustering of iPAR-CLIP sequence reads
After adapter removal and pre-processing, Illumina small RNA reads were mapped against all mRNA isoforms annotated by the ModEncode consortium (Gerstein et al., 2010) using the read alignment software BWA version 0.5.8a (Li and Durbin, 2009). Overlapping or immediately adjacent reads were grouped into iPAR-CLIP sequence read clusters (see Supplemental Methods).
Motif analyses
To define the GLD-1 binding site motif, MEME (Bailey and Elkan, 1994) version 4.4.0 was run on the top 100 clusters from one 4SU iPAR-CLIP library (Supplemental Methods).
Quantification of transcript expression
Reads of all samples were mapped to the transcriptome using TOPHAT (Trapnell et al., 2009). Expression of a gene locus was quantified with CUFFLINKS (Trapnell et al., 2010) given all mRNA isoforms annotated by the ModEncode consortium (Gerstein et al., 2010) as reference annotation by reads per kilobase of gene locus sequence per million mapped reads (RPKM) (Pepke et al., 2009) (Supplemental Methods).
Quantification of protein expression
Protein expression was quantified by SILAC ratios computed by MaxQuant (Cox and Mann, 2008) against “heavy” labeled wild-type L4 to adult stage worms as reference (Supplemental Methods). Protein fold changes between GLD-1 knockdown and control samples were computed by dividing the respective SILAC ratios.
Accession numbers
The sequencing data have been deposited in the GEO database under the accession number GSE33543.
Supplementary Material
Highlights
iPAR-CLIP identifies binding sites of RNA-binding proteins (RBPs) in C. elegans
iPAR-CLIP identified hundreds of direct targets of the germline-specific RBP GLD-1
Effects of GLD-1 knockdown on protein expression of targets confirm functionality
Conserved 5'UTR sites near start codons are bound by GLD-1 and functional in vitro
Acknowledgements
We are indebted to Tim Schedl for providing the BS1080 rescue strain and for helpful discussions. All other strains used in this project were provided by the Caenorhabditis Genetic Center, which is funded by the National Center for Research Resources. We are thankful to James Williamson for providing purified MBP∷GLD-1 STAR fusion protein. ACJ thanks Fabio Piano for support. We thank all members of the Rajewsky lab for discussions and support, in particular Toshiaki Kogame for injections, Andranik Ivanov for MEME runs, and Salah Ayoub and Lena von Oertzen for technical assistance. We acknowledge Claudia Langnick and Mirjam Feldkamp from the Wei Chen lab (MDC) for the sequencing runs. ACJ thanks Boehringer-Ingelheim Fonds for a PhD fellowship. NR thanks the NYU department of Biology for funding stays at NYU where he carried out part of the work. DG received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement N°HEALTH-F4-2010-241504 (EURATRANS). DM was supported by grant R01 HD046236. We thank Eric Miska for lin-28 discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
Author Contributions ACJ designed and performed the iPAR-CLIP experiments. NR designed and performed the computational experiments. DM and ACJ performed the transgenic experiments. MS conducted initial labeling experiments and contributed the GLD-1 knockdown and SILAC worms. GM provided SILAC runs and 4SU incorporation measurements, supervised by SK. DG provided RPKM/SILAC fold change and sequence conservation computations. NR conceived and supervised the project. ACJ and NR analyzed and interpreted the data and wrote the paper.
References
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings / … International Conference on Intelligent Systems for Molecular Biology; ISMB; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development (Cambridge, England) 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nature structural & molecular biology. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA (New York, N.Y. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature genetics. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC molecular biology. 2009;10:47. doi: 10.1186/1471-2199-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Corona D, Preiss T, Becker P, Hentze MW. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5' and 3'UTRs of msl-2 mRNA is independent of the poly(A) tail. The EMBO journal. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Grskovic M, Hentze MW. Drosophila sex-lethal inhibits the stable association of the 4SU ribosomal subunit with msl-2 mRNA. Mol Cell. 2003;11:1397–1404. doi: 10.1016/s1097-2765(03)00176-x. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science (New York, N.Y. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nature methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes & development. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes & development. 2004;18:1047–1059. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. C. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development. Advances in experimental medicine and biology. 2010;693:106–122. doi: 10.1007/978-1-4419-7005-3_8. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW. Translational Control via Protein-Regulated Upstream Open Reading Frames. Cell. 2011;145:902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr., Tuschl T, Ohler U, Keene JD. Integrative Regulatory Mapping Indicates that the RNA-Binding Protein HuR Couples Pre-mRNA Processing and mRNA Stability. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nature methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nature structural & molecular biology. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science (New York, N.Y. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. The EMBO journal. 2011;30:533–545. doi: 10.1038/emboj.2010.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature biotechnology. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nature structural & molecular biology. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.