Abstract
Psychiatric genetics research, as exemplified by the DISC1 gene, aspires to inform on mental health etiology and to suggest improved strategies for intervention. DISC1 was discovered in 2000 through the molecular cloning of a chromosomal translocation that segregated with a spectrum of major mental illnesses in a single large Scottish family. Through in vitro experiments and mouse models, DISC1 has been firmly established as a genetic risk factor for a spectrum of psychiatric illness. As a consequence of its protein scaffold function, the DISC1 protein impacts on many aspects of brain function, impacting both neurosignalling and neurodevelopment. DISC1 is a pathfinder for understanding psychopathology, brain development, signaling and circuitry. Though much remains to be learnt and understood, potential targets for drug development are starting to emerge, and in this review, we will discuss the 10 years of research that has helped us understand key roles of DISC1 in psychiatric disease.
DISC1: on the path to pathfinder
In the original Scottish family, a remarkable 70% of those with the DISC1 gene disruption [1] had a psychiatric diagnosis of schizophrenia (SZ), bipolar disorder (BP) or major depressive disorder (MDD) (Fig. 1), and all tested had deficits in the amplitude of the event-related potential P300 that characterizes SZ and BP [2]. This broad spectrum of disorder is consistent with recent epidemiological evidence, which points to a substantial sharing of genetic risk between SZ and BP and thus, a blurring of diagnostic and genetic boundaries for these disorders [3, 4]. Subsequent DISC1 genetic studies have provided evidence for both regulatory and coding mutations in DISC1 that are associated with SZ, BP, MDD, schizoaffective disorder and autism, and with quantitative variation in behavior and brain function within the normal range (reviewed in [5–8]). Multiple, ultra-rare mutations, apparently unique to patients, have also been identified [9, 10]. Although these follow on studies have been crucial in establishing a general role for DISC1 in risk of psychiatric illness, its status as a translational pathfinder rests less on the genetic evidence and more on our growing understanding of DISC1 biology. In this review, we will summarize what the past 10 years have taught us about the biology and function of DISC1.
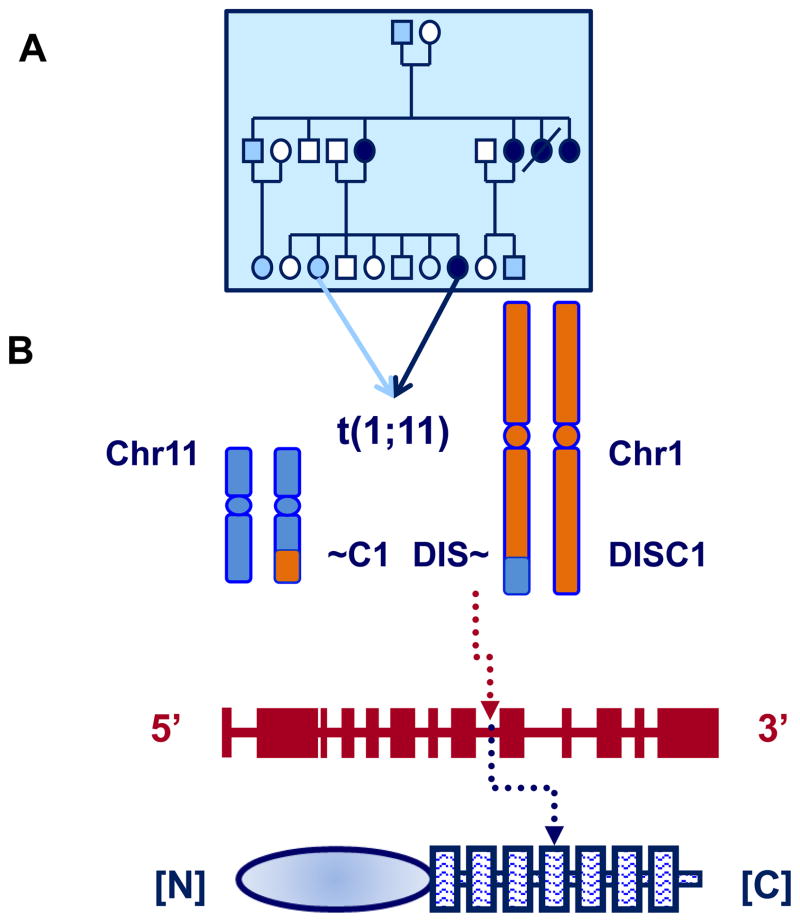
Figure 1. The Scottish family, DISC1 gene disruption and native protein.
Panel A shows an illustrative sub-portion of the Scottish family tree whose members carry the t(1;11) translocation. Those individuals represented with white shapes have neither the t(1;11) translocation nor a major psychiatric diagnosis. Those individuals represented by light blue shapes carry the t(1;11), but at the time of clinical assessment no major psychiatric diagnosis. Those with a dark blue fill also carry the t(1;11) and have a psychiatric diagnosis of schizophrenia, bipolar disorder or recurrent major depression.
Panel B is a visualisation of the balanced translocation between chromosomes 1 (blue) and 11 (orange). This translocation results in a disruption of DISC1 between exons 9 and 10 (c).
(c) Is a schematic of the DISC1 genome structure (exons shown as vertical lines) (top) and the native protein structure (below), with the disordered N-terminal [N] head domain (left) and the C-terminal [C] coiled coil domain (right). The translocation site is indicated by a broken arrow.
DISC1: where, when and how?
In 2005, critical advances in understanding the role of DISC1 came from two independent, but complementary, directions. Millar et al [11] described a different chromosomal rearrangement, this time disrupting the phosphodiesterase 4B (PDE4B) gene in two cousins, one with SZ and the other psychosis. Moreover, they showed that PDE4B interacted dynamically with DISC1, likely modulating cAMP levels in a protein kinase A phosphorylation-dependent fashion [11]. PDE4 activity is linked to memory formation in the fly (cognate dunce mutation), to mood disorder in the mouse (via knock out studies) and is a direct target for rolipram, a non-selective PDE4 inhibitor with antipsychotic and antidepressant activity in pre-clinical models [12]. Kamiya et al [13] reported that in utero application of short hairpin loop RNA oligonucleotides (shRNA) targeting mouse Disc1 resulted in reduced migration of neurons out of the sub-ventricular zone to the cortical plate, accompanied by altered cell polarity and reduced dendritic arborisation. These studies demonstrated for the first time a direct role for DISC1 both in neurosignalling and in early brain development, in harmony with current concepts of schizophrenia [14].
DISC1 is widely expressed not only in the brain, most highly during fetal neurogenesis and in the adult hippocampus, but also in other tissues. DISC1 shows little sequence similarity to other known proteins and the three-dimensional structure is unsolved, but bioinformatic analysis predicts multiple C-terminal coiled coil domains and a disordered N-terminal domain [5, 8]. Moreover, DISC1 forms dimers, octamers and higher order states, a process that is influenced by the common Ser704Cys amino acid sequence variant. This variant impacts on protein binding, at least for the interacting partner NUDEL1, and may have pathological implications, as insoluble aggregates of DISC1 are associated with chronic psychiatric disease [15, 16]. Critically, biochemical and expression studies establish DISC1 as a multifunctional, neurodevelopmentally regulated scaffold, or ‘hub’, protein [5, 7, 8, 17, 18]. It is these features of DISC1 which explain the connection between psychiatric genetics and neuroscience, setting out a pathway for translation of therapies from the bench to the clinic. Interested readers are directed to Bradshaw and Porteous [7] and Soares et al. [8] for recent detailed reviews of the genetic, biochemical and biophysical evidence supporting these connections.
Genetics and biology of the DISC1 pathway
The scaffold function of DISC1 helps resolve the genetics paradox. The set of DISC1 interacting partners are enriched for proteins known to have a role in neurodevelopment, neurosignalling, cytoskeletal, centrosomal and synaptic function [17, 8]. Through an interaction with the amyloid precursor protein APP, DISC1 is potentially involved in neurodegeneration [19], and high molecular pathway connectivity links DISC1 to the causal Huntington’s Disease protein, HTT [18]. Thus, DISC1 has the potential to simultaneously affect a wide range of plausible risk processes in disease susceptibility [5, 8, 20, 21]. Genetic association studies point to the effect of common, non-coding variants in DISC1 and genes encoding several DISC1 partners, including PDE4B, PDE4D, NDE1, NDEL1, LIS1, FEZ1, PCM1 and TNIK (reviewed in [7]). The interplay between single nucleotide polymorphism (SNP) variants within DISC1 affects the risk of SZ and BP [22], and several papers have demonstrated a genetic interplay between DISC1 and NDE1 [23]. An epistatic interaction between the DISC1 Ser704Cys variant and NDEL1 affects the risk for SZ and manifests at the cellular level as altered neurite extension [24]. Finally, epistasis occurs between DISC1 and CIT as well as NDEL1 and CIT, as validated in part by functional brain imaging measures of hippocampal engagement and working memory in healthy controls [25]. Moreover, common cis-variants of DISC1 alter the expression levels of the DISC1 gene by up to 20% of normal [26]. These modest reductions of DISC1 gene expression levels may exert subtle, but pervasive effects on neurodevelopment, neurophysiology and neural circuitry through the scaffold function of DISC1. Common, non-coding variants in DISC1, PDE4B, PDE4D and NDE1 are transcriptional modulators of cAMP signaling, cytoskeletal, synaptogenic, neurodevelopmental and sensory perception proteins [26]. This set of regulated proteins is significantly enriched for current targets of psychiatric drug development [26]. Thus, there is a combined impact of rare, highly penetrant variants and of common, low penetrant gene variants within the DISC1 pathway affecting the manifestation of psychiatric illness, cognition and brain function. Of further consideration is the recent evidence for as many as 50 different DISC1 isoforms, with dramatic differences in their pre- and post-natal brain expression profiles, and for the impact of common DISC1 genetic variants on relative isoform abundance [27]. Follow on studies from the same group have confirmed that when expressed in vitro the predicted truncated version of DISC1 binds to native DISC1 [28]. As predicted [8], these putative truncated forms of DISC1 do not bind NDEL1 under the conditions used, nor did they bind PDE4B, but binding to FEZ1 and GSK3β was retained [28]. These results lend weight to earlier evidence from the same group for reduced expression of NUDEL, FEZ1 and LIS1 in schizophrenia, which was potentiated by the common DISC1 Ser704Cys polymorphism [29]. Replication of the experimental findings with respect to short isoforms of DISC1 protein in post-mortem materials and in vivo models will now be important as they may point to important mechanisms for DISC1-mediated developmental regulation. It is premature to judge the net impact of genetic variation within the DISC1 pathway on variation within the normal range of human behaviour, or on the elevated risk and spectrum of mental illness, but given the number of conditions that can arise as a consequence of DISC1 mutations the concept of DISCopathies may have utility and validity. Moreover, biological studies lend significant weight to the proposition that DISC1 participates in key neuronal processes (Fig. 2).
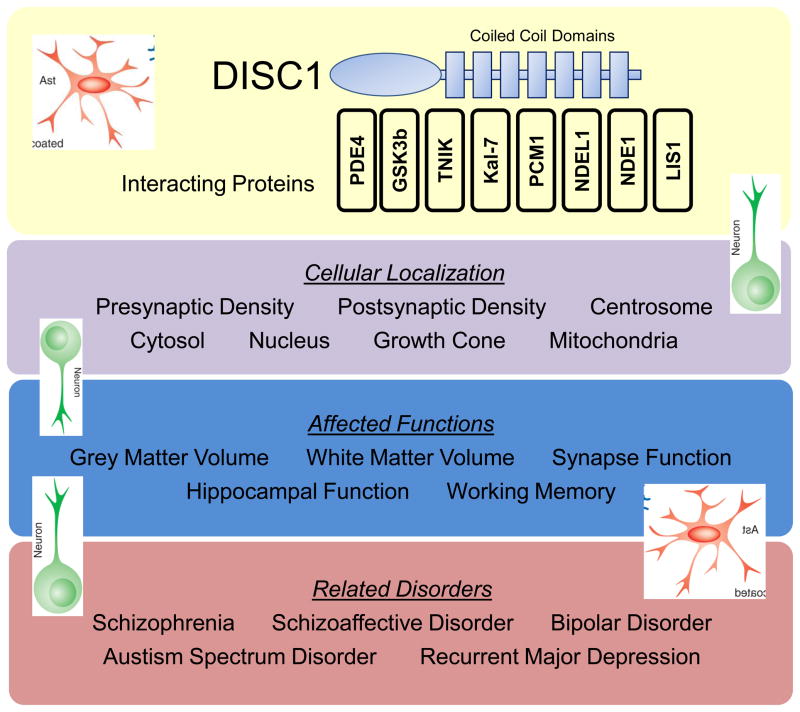
Figure 2. DISC1: from psychopathology to structure and function.
The schema is a conceptual hierarchy linking DISC1 and its binding partners (yellow box) to their cellular localization (purple box), the functions affected by their loss or mutation (blue box), and the psychiatric disorders shown to be linked to DISC1 function (red box).
Modeling DISC1-mediated brain circuitry and human psychopathology in the laboratory mouse
With the notable exception referred to above [27–30], access to well-characterized sample sets of fetal and adult human brain tissue is a major limitation in biological psychiatry. Fortunately, many different laboratory mouse models of Disc1 are now available that can at least partially compensate for the lack of human samples and help model the role of DISC1 [6, 31, 32]. Interested readers are referred to the recent article by Kelly and Brandon [32] who provide a comprehensive review of mouse models for the study of DISC1 and DISC1 interactors. Each model provides insights into cellular and molecular mechanisms, but a key feature worth emphasizing is how the observed behavioral phenotypes can be related back to the spectrum of psychotic and mood disorder observed in the original Scottish pedigree [2] (Fig. 1). Thus, deficits in both schizophrenia-associated phenotypes (e.g. prepulse inhibition) and those related to mood disturbance (e.g. forced swim test) were observed in a model overexpressing a truncated form of DISC1 [33]. Similarly, depression-related phenotypes were prominent in an ENU-induced amino acid substitution variant Gln31Leu, whereas a Leu100Pro substitution mutant showed more schizophrenia-associated abnormalities [34]. The Leu100Pro mutation has also been recently shown to modulate the developmental pattern of NRXN1 and NRXN3 gene expression [35], two genes linked primarily to autism, but also to schizophrenia.
Understanding how DISC1 influences neuronal circuitry depends upon being able to modulate DISC1 expression in both a temporally and a spatially specific manner. Conditional knockout mice are not yet available, but modulation of DISC1 in a temporally and spatially specific manner has been achieved by generation of transgenic mice over expressing a truncated form of DISC1 under the control of an inducible CaMKII promoter [36–38]. The caveat for this mouse model being that the impact is limited to pyramidal neurons in the cortex and both pyramidal and granule neurons in the hippocampus. Transient knock down of Disc1, limited by direct injection of sHRNA constructs to the ventricular zone at the mid-embryonic stage, was shown to affect the prenatal and perinatal pyramidal neuron lineage in the prefrontal cortex [39]. Histological abnormalities were observed perinatally, but neither robust neurochemical nor behavioral phenotypes. Schizophrenia is typically diagnosed in late adolescence or early adulthood, so it was intriguing to note that in the Disc1 knockdown mouse model phenotypes relevant to schizophrenia, including disturbed dopaminergic neurotransmission and interneuron deficits in the cortex, plus several behavioral changes, such as deficits in prepulse inhibition and working memory, became apparent in young adulthood [39].
DISC1 biology and neurodevelopment
Having demonstrated, at least in part, the validity of these experimental constructs in the mouse as models for human DISC1 variations, it is important to tease apart the mechanisms of DISC1 action (Fig. 2). Biochemical studies have confirmed DISC1 interaction with LIS1, NDEL1, NDE1 and PCM1, all components of the microtubule-associated motor complex [5, 7, 31, 32]. These proteins are also involved in neurodevelopment processes, consistent with the early demonstration in mice that DISC1 plays a fundamental role in corticogenesis, especially radial neuronal migration and dendritic arborization [13]. In support of this, the DISC1 interacting proteins PCM1, BBS, CAMDI, Dixdc1 and APP are all involved in radial neuronal migration in the cortex [19, 40–42]. Moreover, DISC1 was elegantly shown to determine the proliferation and fate of neural progenitors through interaction with GSK3β and regulation of β-catenin activity [43]. More recently, Ishizuka et al. [44] reported that in mice, phosphorylation of DISC1 at Ser710 acts as a switch between progenitor proliferation and post-mitotic neuronal migration: unphosphorylated DISC1 regulates canonical Wnt signaling via an interaction with GSK3β and Ser710 phosphorylation enhances DISC1-BBS1 binding, possibly sequestering DISC1 from modulating Wnt / β-catenin activity. The emerging picture is of multiple DISC1 protein interactions regulating and coordinating neurodevelopmental processes in the cerebral cortex. DISC1 regulates neuronal migration and dendritic arborization in the CA1 and the dentate gyrus of the hippocampus [21, 45–49]. Whereas knockdown of Disc1 generally leads to delayed migration and less arborization, the opposite is true of newborn neurons in the adult dentate gyrus, suggesting that the key action of DISC1 is to regulate the tempo of migration [45]. That said, these demonstrations of cellular function have yet to conclusively identify the underlying molecular mechanisms.
DISC1 biology and neurosignalling
Cyclic AMP signalling regulates multiple brain processes, and by hydrolysing cAMP, the phosphodiesterases (PDEs) play a critical role. The Gln31Leu and Leu100Pro amino acid substitution mutations in the mouse [34] are components of PDE4 binding sites [50], reduce the strength of the DISC1/PDE4B interaction, and for the Gln31Leu mice only, reduce PDE4B cAMP hydrolysis activity in the brain, consistent with altered DISC1/PDE4 function relating directly to their reported behavioural abnormalities [34, 50]. A modulatory role of another key DISC1 interactor, GSK3β, has also been implicated in cAMP-dependent activation of PDE4 [51]. Indeed, in mice carrying the Gln31Leu and Leu100Pro mutations, PDE4 and GSK3 activities synergise to modulate behavior [52, 53]. Moreover, both DISC1 mutations reduce the strength of the interaction with GSK3, as with PDE4 [52, 53]. Thus the phenotypes of mice carrying the Gln31Leu and Leu100Pro mutations are likely due, in part, to dysregulated pathways resulting from altered DISC1/PDE4 and DISC1/GSK3 interaction and function. It is critical for understanding the functions of DISC1 in psychiatric illnesses that these pathways are identified. Altered β-catenin and Wnt signaling has already been implicated [43]. Another clue is provided by the identified interaction between DISC1/PDE4 and the LIS1/NDE1/NDEL1 complex [54]. Within this complex, DISC1/PDE4 modulates phosphorylation of NDE1 at position Thr131 by cAMP-dependent protein kinase A (PKA) [55]. Thr131 is located in the binding interface for LIS1 and PDE4 [56]. Phosphorylation of this site reduces LIS1 binding, but promotes NDEL1 binding, and by implication, regulates the function of the complex as a whole [55].
DISC1 is expressed in dendritic spines [54] where it associates with the post-synaptic density [34] and the pre-synapse [57] (Fig. 2). DISC1 determines the tempo of spine and synapse development, the rate of acquisition of intrinsic excitability, and the development of functional synapses in the adult mouse brain [45]. In cultured mouse neurons, DISC1 modulates spine density and the number of miniature excitatory postsynaptic currents, with simultaneous effects upon cell surface expression of the GluR1 AMPA receptor subunit [58]. Moreover, mice carrying the Gln31Leu and Leu100Pro mutations display significantly reduced spine density [59] suggesting that dysregulated synapse development may contribute to their behavioural phenotypes.
The underlying mechanisms likely involve several of the DISC1 binding partners also identified at the post synaptic density (PSD), including PDE4 [34] and the LIS1/NDE1/NDEL1 complex [54, 56], but we currently have the most information about the interaction with Kal-7 [58] and TNIK [57] (Fig. 2).
Kal-7 is a GDP/GTP exchange factor for Rac1 that modulates dendritic spines in response to NMDA receptor activity. DISC1 complexes simultaneously with Kal-7 and the PSD scaffold protein PSD-95 to modulate Rac1 activity, and there is evidence that these associations are regulated by NMDA receptor signalling [58]. Thus, it has been proposed that DISC1 modulates NMDA receptor-dependent Rac1 activity via Kal-7 to regulate spine function, and that this may be a pathological mechanism in schizophrenia [58].
TNIK modulates Wnt signaling [60] and binds Rap2 to control neuronal properties including spine density [61]. DISC1 binds and inhibits TNIK. In neurons, this decreases the levels of the PSD proteins GluR1, stargazin and PSD-95 [57]. Moreover, TNIK knockdown decreases expression of these proteins and additionally reduces expression of GluR2, GluR3 and the NMDA receptor subunit NR2B, in parallel decreasing the amplitude of AMPA receptor miniature excitatory postsynaptic currents [57]. By contrast, DISC1 knockdown increases expression of GluR1, GluR2, GluR3 and TNIK, but reduces PSD-95 and stargazin levels. DISC1 and TNIK are thus essential for dynamic regulation of crucial post-synaptic proteins, including glutamate receptors. Interestingly, in a cell culture model measuring outputs via micro electrode arrays, TNIK, but not DISC1, knockdown profoundly dysregulated neural network development [62].
NMDA receptor hypofunction is widely held to be a significant contributory pathological factor in schizophrenia [63]. The behaviour and pharmacological responses of mice engineered to have reduced expression of the NMDA receptor subunit NR1 make them an interesting model for schizophrenia [64]. There is emerging evidence that NMDA receptor function is directly linked to DISC1 as these mice display reduced dendritic spine density and decreased synaptic DISC1 expression [65]. Moreover, NMDA receptor antagonists decrease DISC1 expression, inducing overextended migration of newborn neurons, an abnormality that is rescued by DISC1 overexpression [66]. Given that we now have a raft of evidence supporting the relevance of the DISC1 pathway to the etiology of major mental illness, including an emerging evidence base for an influence on the NMDA receptor, does this mean that DISC1 is a good target for drug development?
Drugging DISC1: the challenges and opportunities
Those who suffer from major mental illness are poorly served by current drug treatments. In the last 50 years there have been few, if any, fundamental innovations in antipsychotics or antidepressants; many patients respond poorly, if at all, and many of these medications have serious side effects [67]. To date, translational research in schizophrenia has relied heavily on pharmacological models of dysregulated dopaminergic or glutamatergic signalling [68], but genetic models are coming to the fore, including a mouse model that overexpresses the striatal D2-receptor [69] and a model with a partial NR1 knockout that results in reduced NMDA expression [64] and has now been shown to have reduced synaptic DISC1 [65]. That said, the construct validity of these models is modest. The neurodevelopmental, circuitry and behavioral abnormalities reported in a range of Disc1 mouse models suggests they could have great potential for translational studies of major mental illness. Disc1 mutant mice display a range of behavioural phenotypes that model relevant aspects of schizophrenia and mood disorder, which can be rescued by treatment with classical anti-psychotics and anti-depressants [34]. Critically, early work demonstrates that relevant phenotypes can be modulated by PDE4 or GSK3 inhibition, which we know are mechanistically linked to the DISC1 pathway and thus offer novel therapeutic mechanisms and targets [34, 53]. These latter insights are essential as we contemplate screening for new clinically useful compounds across a wide spectrum of unmet mental health needs, and critically, show the absolute need to understand the biology of the DISC1 pathway in great detail (see below). Furthermore, the developmental nature of schizophrenia and related conditions raises the question of when to treat and how to prevent their appearance [14]. At least partial answers may come soon from refined conditional ‘knock out’ and ‘knock in’ studies in Disc1 mutant mouse models. But, more fundamentally, which, or indeed how many, of the functional consequences of DISC1 offer the best and most tractable targets for drug development or intervention?
DISC1 is not ‘druggable’ in the conventional sense: it does not fall into any of the ‘preferred’ classes of targets, overwhelmingly ion channels, G-protein coupled receptors, metabolic enzymes and kinases. But the phosphodiesterases continue to be a major target for drug development in relation to CNS, cardiovascular and inflammatory disorders [12]. The trick here will be to find a way in which the DISC1-PDE4 interaction can be selectively modulated both temporally and spatially, to circumvent the emetic and other limiting downsides of direct inhibition of PDE4 [70]. The identification of GSK3β as a DISC1 interactor [43] highlights again the Wnt signaling pathway as a target for drug development, as does the link from DISC1 through Girdin to the AKT1/mTor pathway [44,46]. This link between DISC1 and the AKT1/mTOR pathway brings into focus the role of lithium (or lithium-like molecules) in the treatment of bipolar disorder. Thus, for PDE4, GSK3β and AKT1/mTOR there is the attractive possibility of ‘peering over the shoulder of others’ working within the inflammatory, cancer and cardiovascular fields to identify molecules which selectively modulate these pathways, with the added constraint that these therapies must cross the blood-brain barrier. Finally, as a kinase known to function at the synapse, modulation of TNIK is also an attractive translational target [57].
Will modulation of the DISC1 pathway be relevant only to those carrying highly penetrant mutations? This is unlikely to be the situation, given the evidence that common variants impact on multiple current targets relevant for psychiatric drug development [26]. Taken together, and despite the ‘unfavourable’ start point of DISC1 itself, there are reasonable grounds for optimism that through the discovery of the DISC1 gene and the DISC1 interactome, we have a novel and promising set of opportunities for ‘conventional’ drug development in psychiatry. There are technical, safety and efficacy considerations to consider with all of these approaches, but the possibility of identifying mechanisms for treating these devastating disorders makes this an absolute scientific and clinical imperative.
A more challenging, if potentially more valid, approach would be to rework the question and take a genomically-informed, systems approach to stratified medicine. This means thinking of the DISC1 complex as the ‘target’ in both time and space [21]. It means developing physiological read outs and developing small molecules and/or biologics to restore and maintain homeostatic balance, first in model systems and then clinical studies. None of this will prove easy or come quickly, but if we are not to see this as an opportunity, where else should we look?
Caveats and limitations
By meeting the criteria for causality [20], DISC1 has provided a useful inroad into the biological understanding of other psychiatric risk genes and pathways, connecting ‘bottom up’ genetics and genomics to the ‘top down’ pharmacology, neurodevelopment, neurosignaling and neural circuitry – in short, helping to establish and promote an integrated systems neurobiology approach. But there are caveats and limitations, which are summarized in Box 1.
Box 1. Outstanding Questions.
What is the full spectrum of functional variation in the DISC1 gene and DISC1 interactors?
How do genetic variants in the DISC1 pathway impact on psychopathology, behaviour and cognition? Is the concept of DISCopathies valid and useful?
What are the biophysical properties of DISC1 and the DISC1 complex and how are these affected by common and rare genetic variants?
How is DISC1 transcription and isoform expression regulated throughout development and in adult neuronal cell lineages?
What is the nature of the DISC1 complex over the developmental time-course and at different sub-cellular locations?
How is DISC1 modified by phosphorylation and other post-translation mechanisms, and with what functional consequences?
What are the canonical and non-canonical functions of DISC1?
How do these functions affect normal and aberrant development and behavior?
Future challenges and directions
The recent demonstration of a phosphorylation mediated, developmental switch in DISC1 function [44] signals a new level of necessary enquiry. It raises the question of whether other forms of post-translational modification are critical in modulating the role of DISC1. Multiple mouse models are now available and have proved instructive [32], but a complete ‘null’ (either constitutive or regulatable) mutant and humanized models that recapitulate the t(1;11) as well as other disease associated mutations and variants will be valuable. For circuitry studies, rat models offer anatomical size advantages over the mouse and are now amenable to genetic [71] and optogenetic engineering [72]. For behavioral drug screening, the zebrafish has obvious appeal and is already proving promising for complex behavioural phenotypes of potential relevance to psychiatry [73, 74]. Arguably, the most valuable model to be developed will be patient-derived somatic cells carrying defined DISC1 mutations or variants reprogrammed to create specific cell types of defined neuronal lineage [75, 76]. Only once this full panoply of models is available can we realistically bridge the gap between basic and applied research. Box 2 summarises some of the outstanding questions for the DISC1 field over the next decade (to be reiterated in similar fashion for other robust genetic candidates in psychiatry).
Box 2. Future directions.
Modelling DISC1 clinical mutations and common variants in human cell culture by neuralising reprogrammed fibroblasts.
Humanising the laboratory mouse and other tractable animal species.
Combining cell-culture-based and in vivo models to refine our molecular and cellular understanding of DISC1 function during development, at the synapse, and in brain signaling and circuitry.
Testing for phenotypic rescue through genetic and chemical (small molecule) screens.
Modulating the biophysical properties of aberrant DISC1 and DISC1 complexes.
Translating these findings into improved strategies for diagnosis, prognosis and treatment by adapting treatments developed for other disciplines that target proteins known to interact with DISC1.
To conclude, the first 10 years of DISC1 research has unveiled through diverse experimental approaches rich seams of evidence for fundamental roles in psychiatry and neuroscience. The next 10 are set to be even more exciting, if challenging, as we seek to refine our knowledge and understanding, satisfy our curiosity, and contribute in a practical way to understanding and modifying brain development, human behavior and mental illness.
Glossary
- Autism
is a neurodevelopmental disorder with a strong genetic component, characterized by impaired social interaction and repetitive behaviours
- AKT/mTor
is an insulin and growth factor sensitive signaling pathway important in cell growth, survival and differentiation. mTOR is inhibited by rapamycin, a drug used to prevent transplant rejection
- Bipolar disorder
is a severe form of mental illness with a strong genetic component that affects approximately 1% of the population. It is characterized by swings of mood from extreme elevation to profound depression and was formerly referred to as manic depression
- CA1
is a sub-region of the hippocampus, densely packed with pyramidal neurons, adjacent to, but distinct from the dentate gyrus
- CaMKII
(calmodulin-dependent protein kinase II) is important for learning and memory and is expressed exclusively in the brain. The CaMKII promoter is commonly used to direct expression of transgenes in the brain
- Circuitry
is used in the context of the brain to describe the network of neuronal connections and synapses
- Construct validity
describes a well-founded concept as well as set of observations and measurements, in this case a mouse model, which corresponds accurately to the real situation, in this case, schizophrenia
- Cyclic AMP
(cAMP) is synthesized from ATP by adenylyl cyclase and catabolised to AMP by phosphodiesterases. cAMP is a second messenger widely involved in signal transduction
- Dendritic arborization
is the branching of dendrites preceding the formation of synapses
- Dentate Gyrus
is part of the hippocampal formation, important in memory formation, and a site of active neurogenesis in the adult brain
- Emetics
are substances that induce vomiting. As rodents lack a gag response, rolipram and similar modecules can be used in these experimental models, but not for patients in the clinic
- ENU
(ethynitrosourea) is a powerful chemical mutagen
- Epistasis
is a genetic term, indicating that the effects of one gene are modified by the effects of one or more other gene
- Event-related potential P300
is measured as a characteristic 300 millisceond delay between a stimulus and a response in the brain measured by electorencephalography (EEG). It is thought to measure cognitive function and decision making processes. In schizophrenia the P300 amplitude is typically reduced
- Excitatory Postsynaptic Currents
arise following temporary depolarization of postsynaptic membranes as a result of the opening of ligand-sensitive channels
- Forced Swim Test
sometimes referred to as the Porsolt test, is a measure of behavioral despair. Animals placed in a water-filled glass cylinder will swim and attempt to escape on first exposure. On second exposure, the time spent simply floating (a proxy for ‘despair’) is measured. Rodents swim longer if treated with antidepressants
- Genetics Paradox
in the case of DISC1, is the apparent contradiction that a single gene defect can have manifold effects across a spectrum of psychiatric disorders; the paradox is resolved on realization of the multiple neural proteins that bind to and are regulated by DISC1, thus one gene can have many functions and affect multiple cellular, pathological and clinical phenotypes
- Optogenetic Engineering
is a recently developed method, applicable in real time in vivo, in which a light activated channel is expressed under the control of a cell type specific promoter. Channel activity can thus be switched on (or off) is a selected neuronal type by exposure to light of a selective wavelength
- Pharmacological Models
refer to drug-evoked responses and behaviours that mimic a clinical condition
- Post-synaptic density
is a supra-molecular protein complex associated with the synapse comprising receptor, scaffold and signaling proteins
- Psychosis
is used to summarize the combination of hallucinations, delusions and thought disorder typically seen in schizophrenia and sometimes in bipolar disorder
- Prepulse Inhibition
is the well-described phenomenon in which a weak brain stimulus inhibits the response to a subsequent stronger stimulus. This is a measure of information filtering. Patients with schizophrenia tend to show a marked deficit in this ability to filter and inhibit response to stimuli
- Schizophrenia
is a severe form of mental illness with a strong genetic component that affects approximately 1% of the population and which is characterized by hallucinations, delusions, disorganized thoughts and cognitive deficit
- shRNA
stands for small or short hairpin RNA, molecules that can silence gene expression by binding to mRNA and directing RNA interference
- Synapse
is the junction between one neuron and another across which electrical and/or chemical signals pass
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Millar JK, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human Molecular Genetics. 2000;9(9):1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood DH, et al. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. American Journal of Human Genetics. 2001;69(2):428–33. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottesman, et al. Severe mental disorders in offspring with 2 psychiatrically ill parents. Archives of General Psychiatry. 2010;67(3):252–7. doi: 10.1001/archgenpsychiatry.2010.1. [DOI] [PubMed] [Google Scholar]
- 5.Chubb JE, et al. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13(1):36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 6.Johnstone M, et al. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophrenia Bulletin. 2011;37(1):14–20. doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2011 doi: 10.1016/j.neuropharm.2010.12.027. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares DC, et al. DISC1: structure, function and therapeutic potential for major mental illness. ACS Chemical Neuroscience. 2011 doi: 10.1021/cn200062k. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W, et al. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochemical and Biophysical Research Communications. 2008;367(3):700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- 10.Song W, et al. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neuroscience Letters. 2010;486(3):136–40. doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310(5751):1187–91. doi: 10.1126/science.1112915. [see comment] [DOI] [PubMed] [Google Scholar]
- 12.Menniti FS, et al. Phosphodiesterases in the CNS: targets for drug development. Nat Rev Drug Discov. 2006;5(8):660–70. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 13.Kamiya A, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nature Cell Biology. 2005;7(12):1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 14.Insel TR. Rethinking schizophrenia. Nature. 2010;468(7321):187–93. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 15.Leliveld SR, et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28(15):3839–45. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leliveld SR, et al. Oligomer Assembly of the C-Terminal DISC1 Domain (640–854) Is Controlled by Self-Association Motifs and Disease-Associated Polymorphism S704C. Biochemistry. 2009;48(32):7746–7755. doi: 10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- 17.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Molecular Psychiatry. 2007;12(1):74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 18.Boxall R, et al. DISC1 and Huntington’s disease--overlapping pathways of vulnerability to neurological disorder? PLoS ONE. 2011;6(1):e16263. doi: 10.1371/journal.pone.0016263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young-Pearse TL, et al. Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J Neurosci. 2010;30(31):10431–40. doi: 10.1523/JNEUROSCI.1445-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porteous D. Genetic causality in schizophrenia and bipolar disorder: out with the old and in with the new. Current Opinion in Genetics & Development. 2008;18(3):229–234. doi: 10.1016/j.gde.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Porteous D, Millar K. How DISC1 regulates postnatal brain development: girdin gets in on the AKT. Neuron. 2009;63(6):711–3. doi: 10.1016/j.neuron.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Hennah W, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Molecular Psychiatry. 2009;14(9):865–73. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- 23.Hennah W, et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16(5):453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 24.Burdick KE, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Human Molecular Genetics. 2008;17(16):2462–73. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicodemus KK, et al. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet. 2010;127(4):441–52. doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- 26.Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS ONE. 2009;4(3):e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakata K, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proceedings of the National Academy of Sciences. 2009;106(37):15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newburn EN, et al. Interactions of human truncated DISC1 proteins: implications for schizophrenia. Translational Psychiatry. 2011 doi: 10.1038/tp.2011.31. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipska BK, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Human Molecular Genetics. 2006;15(8):1245–58. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 30.Hyde TM, et al. Expression of GABA Signaling Molecules KCC2, NKCC1, and GAD1 in Cortical Development and Schizophrenia. J Neurosci. 2011;31(30):11088–95. doi: 10.1523/JNEUROSCI.1234-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaaro-Peled H, et al. Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32(9):485–95. doi: 10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly MP, Brandon NJ. Taking a bird’s eye view on a mouse model review: a comparison of findings from mouse models targeting DISC1 or DISC1-interacting proteins. Future Neurology. 2011;6(5):661–677. [Google Scholar]
- 33.Hikida T, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104(36):14501–6. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapcote SJ, et al. Behavioral Phenotypes of Disc1 Missense Mutations in Mice. Neuron. 2007;54(3):387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Brown SM, et al. Synaptic modulators Nrxn1 and Nrxn3 are disregulated in a Disc1 mouse model of schizophrenia. Mol Psychiatry. 2011;16(6):585–7. doi: 10.1038/mp.2010.134. [DOI] [PubMed] [Google Scholar]
- 36.Li W, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(46):18280–5. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abazyan B, et al. Prenatal interaction of mutant DISC1 and immune activation produces adult psychopathology. Biological Psychiatry. 2010;68(12):1172–81. doi: 10.1016/j.biopsych.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayhan Y, et al. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Molecular Psychiatry. 2011;16(3):293–306. doi: 10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niwa M, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65(4):480–9. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamiya A, et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Archives of General Psychiatry. 2008;65(9):996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda T, et al. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J Biol Chem. 2010;285(52):40554–61. doi: 10.1074/jbc.M110.179481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh KK, et al. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67(1):33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Y, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136(6):1017–31. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishizuka K, et al. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473(7345):92–6. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan X, et al. Disrupted-In-Schizophrenia 1 Regulates Integration of Newly Generated Neurons in the Adult Brain. Cell. 2007;130(6):1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JY, et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63(6):761–73. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enomoto A, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63(6):774–87. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 48.Meyer KD, Morris JA. Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009;18(17):3286–3297. doi: 10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomita K, et al. Disrupted-in-Schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Human Molecular Genetics. 2011 doi: 10.1093/hmg/ddr194. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 50.Murdoch H, et al. Isoform-Selective Susceptibility of DISC1/Phosphodiesterase-4 Complexes to Dissociation by Elevated Intracellular cAMP Levels. Journal of Neuroscience. 2007;27(35):9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlyle BC, et al. Co-ordinated action of DISC1, PDE4B and GSK3beta in modulation of cAMP signalling. Molecular Psychiatry. 2011 doi: 10.1038/mp.2011.17. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 52.Lipina TV, et al. Genetic and pharmacological evidence for schizophrenia-related Disc1 interaction with GSK-3. Synapse. 2011;65(3):234–48. doi: 10.1002/syn.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipina TV, et al. Synergistic interactions between PDE4B and GSK-3: DISC1 mutant mice. Neuropharmacology. 2011 doi: 10.1016/j.neuropharm.2011.02.020. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 54.Bradshaw NJ, et al. NDE1 and NDEL1: Multimerisation, alternate splicing and DISC1 interaction. Neurosci Lett. 2009;449(3):228–233. doi: 10.1016/j.neulet.2008.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradshaw NJ, et al. PKA Phosphorylation of NDE1 Is DISC1/PDE4 Dependent and Modulates Its Interaction with LIS1 and NDEL1. J Neurosci. 2011;31(24):9043–54. doi: 10.1523/JNEUROSCI.5410-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins DM, et al. Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through Protein Kinase A (PKA) Cell Signal. 2008;20(12):2356–69. doi: 10.1016/j.cellsig.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Molecular Psychiatry. 2010 doi: 10.1038/mp.2010.87. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi-Takagi A, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13(3):327–32. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee FH, et al. Disc1 point mutations in mice affect development of the cerebral cortex. J Neurosci. 2011;31(9):3197–206. doi: 10.1523/JNEUROSCI.4219-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahmoudi T, et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28(21):3329–40. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hussain NK, et al. MINK and TNIK differentially act on Rap2-mediated signal transduction to regulate neuronal structure and AMPA receptor function. J Neurosci. 2010;30(44):14786–94. doi: 10.1523/JNEUROSCI.4124-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maclaren EJ, et al. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Molecular & Cellular Neurosciences. 2011;47(2):93–9. doi: 10.1016/j.mcn.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olney JW, et al. NMDA receptor hypofunction model of schizophrenia. J Psychiatr Res. 1999;33(6):523–33. doi: 10.1016/s0022-3956(99)00029-1. [DOI] [PubMed] [Google Scholar]
- 64.Mohn AR, et al. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98(4):427–36. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 65.Ramsey AJ, et al. Impaired NMDA receptor transmission alters striatal synapses and DISC1 protein in an age-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(14):5795–800. doi: 10.1073/pnas.1012621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Namba T, et al. NMDA receptor regulates migration of newly generated neurons in the adult hippocampus via Disrupted-In-Schizophrenia 1 (DISC1) J Neurochem. 2011 doi: 10.1111/j.1471-4159.2011.07282.x. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Insel TR. Translating scientific opportunity into public health impact: a strategic plan for research on mental illness. Archives of General Psychiatry. 2009;66(2):128–33. doi: 10.1001/archgenpsychiatry.2008.540. [DOI] [PubMed] [Google Scholar]
- 68.Castagne V, et al. Preclinical behavioral models for predicting antipsychotic activity. Adv Pharmacol. 2009;57:381–418. doi: 10.1016/S1054-3589(08)57010-4. [DOI] [PubMed] [Google Scholar]
- 69.Kellendonk C, et al. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49(4):603–15. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 70.Dietsch GN, et al. Characterization of the inflammatory response to a highly selective PDE4 inhibitor in the rat and the identification of biomarkers that correlate with toxicity. Toxicol Pathol. 2006;34(1):39–51. doi: 10.1080/01926230500385549. [DOI] [PubMed] [Google Scholar]
- 71.Cui X, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29(1):64–7. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 72.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5(3):439–56. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rihel J, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327(5963):348–51. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiang CH, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Molecular Psychiatry. 2011;16(4):358–60. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–5. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]